Altering the expression of a vacuolar iron transporter doubles iron content in white wheat flour.
Abstract
Increasing the intrinsic nutritional quality of crops, known as biofortification, is viewed as a sustainable approach to alleviate micronutrient deficiencies. In particular, iron deficiency anemia is a major global health issue, but the iron content of staple crops such as wheat (Triticum aestivum) is difficult to change because of genetic complexity and homeostasis mechanisms. To identify target genes for the biofortification of wheat, we functionally characterized homologs of the VACUOLAR IRON TRANSPORTER (VIT). The wheat genome contains two VIT paralogs, TaVIT1 and TaVIT2, which have different expression patterns but are both low in the endosperm. TaVIT2, but not TaVIT1, was able to rescue the growth of a yeast (Saccharomyces cerevisiae) mutant defective in vacuolar iron transport. TaVIT2 also complemented a manganese transporter mutant but not a vacuolar zinc transporter mutant. By overexpressing TaVIT2 under the control of an endosperm-specific promoter, we achieved a greater than 2-fold increase in iron in white flour fractions, exceeding minimum legal fortification levels in countries such as the United Kingdom. The antinutrient phytate was not increased and the iron in the white flour fraction was bioavailable in vitro, suggesting that food products made from the biofortified flour could contribute to improved iron nutrition. The single-gene approach impacted minimally on plant growth and also was effective in barley (Hordeum vulgare). Our results show that by enhancing vacuolar iron transport in the endosperm, this essential micronutrient accumulated in this tissue, bypassing existing homeostatic mechanisms.
Iron is essential for plant growth and needed for a range of cellular processes involving electron transfer or redox-dependent catalysis (Kobayashi and Nishizawa, 2012). However, excess levels of iron are toxic to cells; therefore, organisms have evolved tight regulation and storage mechanisms. Plants store iron in ferritin or sequestered in vacuoles, with different species and tissues favoring one storage mechanism over another (Briat et al., 2010). Iron stored in seeds provides for essential iron enzymes during germination, before the seedling develops a root and is able to take up iron independently.
Iron is also an essential micronutrient for human nutrition, and over a billion people suffer from iron-deficiency anemia (WHO, 2008). Seeds such as rice (Oryza sativa), wheat (Triticum aestivum), and pulses are a major source of iron, especially in diets that are low in meat. To combat iron deficiency, more than 84 countries have legislation for the chemical fortification of flours milled from wheat, corn (Zea mays), and rice with iron salts or iron powder (www.ffinetwork.org/global_progress/index.php). A more sustainable approach is biofortification, or increasing the intrinsic micronutrient content of crops through traditional breeding or transgenic technology (Vasconcelos et al., 2017).
A key gene involved in iron loading in seeds, VACUOLAR IRON TRANSPORTER1 (VIT1), was first identified in Arabidopsis (Arabidopsis thaliana; Kim et al., 2006) as a homolog of yeast (Saccharomyces cerevisiae) Ca2+-SENSITIVE CROSS-COMPLEMENTER (CCC1), which transports iron into vacuoles (Li et al., 2001) and manganese into Golgi vesicles (Lapinskas et al., 1996). VIT1 is highly expressed in ripening Arabidopsis seeds and targets iron to the vacuoles of the endodermis and veins of the embryo (Kim et al., 2006; Roschzttardtz et al., 2009). Expression of Arabidopsis VIT1 also increases the manganese content of yeast cells (Kim et al., 2006), and it has a supporting role in manganese transport in Arabidopsis embryos (Eroglu et al., 2017). The VITs form a unique transporter family, found in plants, fungi, and protists such as the malarial parasite Plasmodium falciparum, but they are absent from metazoans (Slavic et al., 2016). VITs in plants share a high degree of sequence similarity and the capacity to transport iron, but their biological functions may differ. For example, TgVIT1 in tulip (Tulipa gesneriana) is involved in petal color determination (Momonoi et al., 2009). Due to their roles in iron storage, VITs are potentially good candidates for iron biofortification. Indeed, the expression of VIT1 from Arabidopsis controlled by a PATATIN promoter enhanced the iron content of cassava (Manihot esculenta) tubers 3- to 4-fold (Narayanan et al., 2015). Given the promise for biofortification, it is surprising that very few VITs from crop species have been characterized, particularly in cereals. Two VIT genes have been identified in rice, OsVIT1 and OsVIT2. These genes showed different expression patterns throughout the plant and in response to iron but were similar with respect to yeast complementation results. Knockout mutants accumulated more iron in the embryo, but this part of the grain is lost during processing to obtain white rice. The effect of overexpressing the OsVIT genes was not tested, and in fact, virtually nothing is known about the wider physiological effects of overexpressing VIT in plants (Ravet et al., 2009).
For the biofortification of cereal crops, simply increasing the iron content in grains is unlikely to increase their nutritional quality. Micronutrients are concentrated in the aleurone and seed coat, which are commonly removed in the production of polished rice or white wheat flour. The aleurone also is rich in phytate (myoinositol-1,2,3,4,5,6-hexakisphosphate), a phosphate storage molecule that is a major inhibitor of iron bioavailability in whole-grain products (Hurrell and Egli, 2010). On the other hand, phytate is low in the endosperm (O’Dell et al., 1972); therefore, this tissue should be targeted to increase bioavailable dietary iron in cereal food products. Previous biofortification strategies in wheat include the overexpression of ferritin, which increased iron levels 1.6- to 1.8-fold but with large variations per line (Singh et al., 2017). Because ferritin is localized in plastids, iron transport into plastids also needs to be up-regulated, and this may be a limiting factor in cereal grains. Elegant nanoscale secondary ion mass spectrometry studies showed that iron was concentrated in small vacuoles in the wheat aleurone, colocalizing with phosphorus, most likely in the form of phytate, but that some also localized in patches in the endosperm (Moore et al., 2012). Other biofortification strategies have focused on increasing the mobility of iron through the overexpression of nicotianamine synthase genes for the production of chelator molecules to translocate iron(II) and other divalent metals (Singh et al., 2017).
Here, we identified and functionally characterized TaVIT1 and TaVIT2, the two VIT paralogs found in the genome of bread wheat. The VIT genes differ in expression patterns and their ability to complement yeast metal transporter mutants. Based on these findings, we selected TaVIT2 for overexpression in the endosperm of wheat and barley (Hordeum vulgare), resulting in more than twice as much iron in white flour fractions but little impact on plant growth and grain number. Our results suggest that, by drawing iron into vacuoles in the endosperm, existing homeostasis mechanisms can be bypassed for a successful biofortification strategy.
RESULTS
Wheat Has Two Functionally Differentiated VIT Paralogs
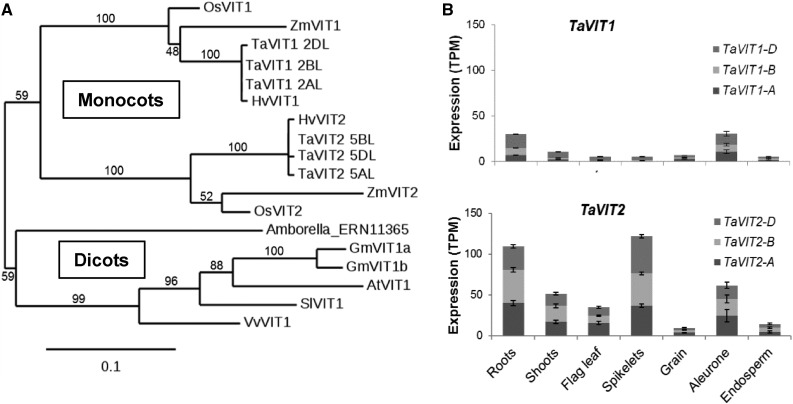
The newly sequenced and annotated wheat genome (Clavijo et al., 2017) offers the opportunity to make a complete inventory of putative metal transporters in wheat (Borrill et al., 2014). We found that wheat has two VIT genes (TaVIT1 and TaVIT2) on chromosome groups 2 and 5, respectively. As expected in hexaploid wheat, each TaVIT gene is represented by three copies (homoeologs) from the A, B, and D genomes that share 99% identity at the amino acid level (Supplemental Table S1; Supplemental Fig. S1). TaVIT1 and TaVIT2 have ∼87% amino acid identity with their closest rice homologs, OsVIT1 and OsVIT2, respectively. Phylogenetic analysis suggests an early evolutionary divergence of the two VIT genes, as there are two distinctly branching clades in the genomes of monocotyledonous species, in contrast to one clade in dicotyledons (Fig. 1A). The gene expression profiles of TaVIT1 and TaVIT2 were queried across 418 RNA sequencing (RNA-seq) samples (Supplemental Table S2). In general, all homoeologs of TaVIT2 were more highly expressed than TaVIT1 homoeologs (Fig. 1B). In the grains, TaVIT1 and TaVIT2 are both expressed in the aleurone, correlating with high levels of iron in this tissue, which is removed from white flours during the milling process. In contrast, the expression of TaVIT1 and TaVIT2 is very low in the starchy endosperm, the tissue from which white flour is extracted. Taken together, differences in phylogeny and expression pattern suggest that TaVIT1 and TaVIT2 may have distinct functions.
Figure 1.
The wheat genome encodes two VIT paralogs with different expression patterns. A, Phylogenetic tree of VIT genes from selected plant species: At, Arabidopsis thaliana; Gm, Glycine max (soybean); Hv, Hordeum vulgare (barley); Os, Oryza sativa (rice); Sl, Solanum lycopersicum (potato); Ta, Triticum aestivum (wheat); Vv, Vitis vinifera (grape); Zm, Zea mays (maize). Numbers above or below branches represent bootstrapping values for 100 replications. B, Gene expression profiles of TaVIT1 and TaVIT2 homoeologs using RNA-seq data from expVIP. Bars indicate mean transcripts per million (TPM) ± se. Full details and metadata are given in Supplemental Table S2.
TaVIT2 Facilitates the Transport of Iron and Manganese
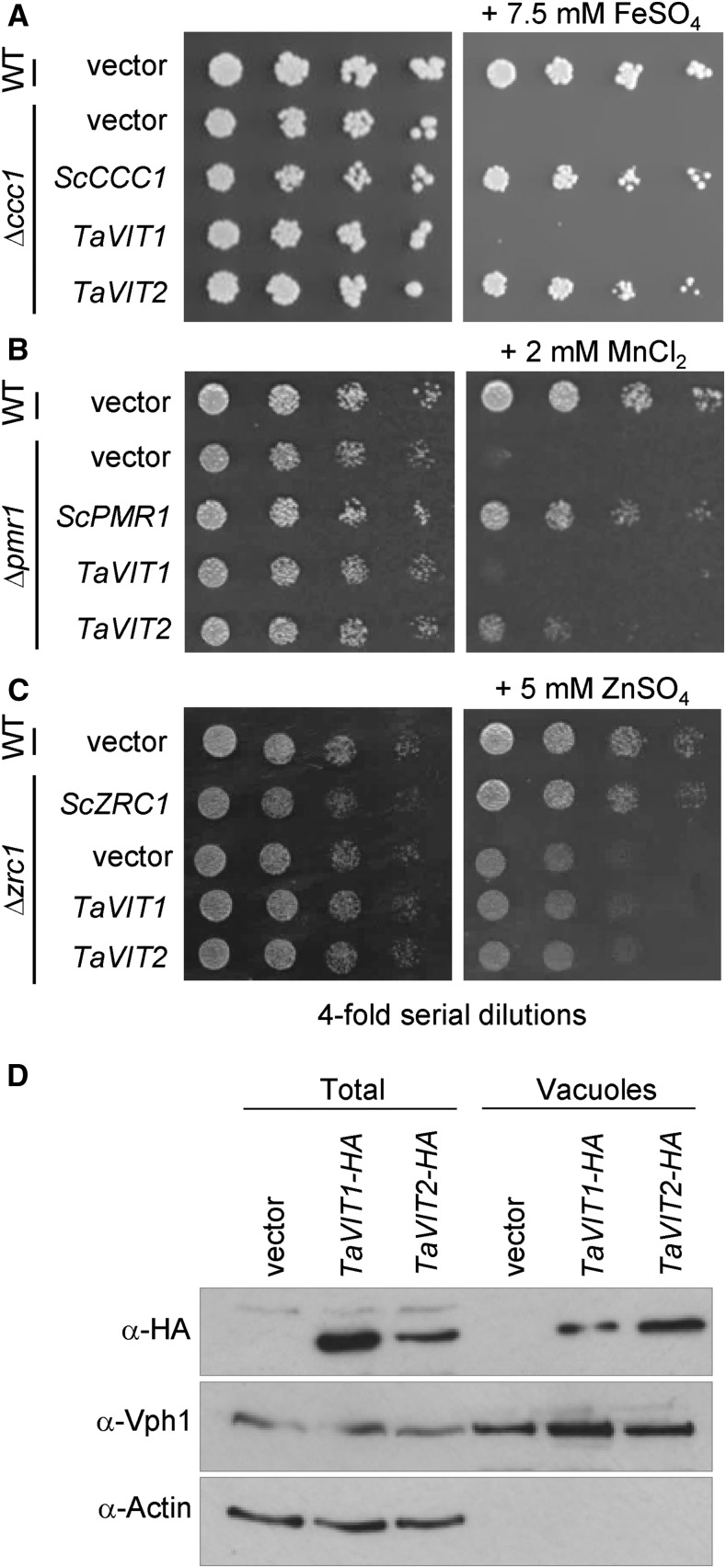
To test if the TaVIT proteins transport iron, the TaVIT1-B homoeolog and TaVIT2-D homoeolog, hereafter referred to as TaVIT1 and TaVIT2, respectively, were selected and expressed in yeast lacking the vacuolar iron transporter Ccc1. The Δccc1 yeast strain is sensitive to high concentrations of iron in the medium because of its inability to store iron in the vacuole. TaVIT2 fully rescued the growth of Δccc1 yeast exposed to a high concentration of FeSO4, but TaVIT1 was no different from the empty vector control (Fig. 2A). Yeast Ccc1 can transport both iron and manganese (Lapinskas et al., 1996). Therefore, we carried out yeast complementation using the Δpmr1 mutant, which is unable to transport manganese into Golgi vesicles and cannot grow in the presence of toxic levels of this metal (Lapinskas et al., 1995). We found that the expression of TaVIT2 in Δpmr1 yeast partially rescued the growth impairment on high concentrations of MnCl2, indicating that TaVIT2 can transport manganese (Fig. 2B). We also tested if TaVIT1 and TaVIT2 are able to rescue the growth of the yeast Δzrc1 strain, which is defective in vacuolar zinc transport, but neither TaVIT gene was able to rescue growth on high zinc concentrations (Fig. 2C).
Figure 2.
TaVIT2 facilitates iron and manganese transport. A to C, Yeast complementation assays of TaVIT1 and TaVIT2 in Δccc1 (A), Δpmr1 (B), and Δzrc1 (C) compared with yeast that is wild type (WT) for these three genes. The yeast (Sc) CCC1, PMR1, and ZRC1 genes were used as positive controls. Cells were spotted in a 4-fold dilution series and grown for 2 to 3 d on plates with or without 7.5 mm FeSO4 (Δccc1), 2 mm MnCl2 (Δpmr1), or 5 mm ZnSO4 (Δzrc1). D, Immunoblots of total and vacuolar protein fractions from yeast cells expressing hemagglutinin (HA)-tagged TaVIT1 or TaVIT2. The HA tag did not inhibit the function of TaVIT2, as it was able to complement Δccc1 yeast (data not shown). Vhp1 was used as a vacuolar marker, and the absence of actin shows the purity of the vacuolar fraction.
Western-blot analysis showed that both proteins were produced in yeast but that TaVIT1 and TaVIT2 might differ in their intracellular distribution (Fig. 2D). TaVIT2 was abundant in vacuolar membranes, cofractionating with the vacuolar marker protein Vph1. TaVIT1 also was found in the vacuolar membrane fraction, but based on higher abundance in the total fraction, it appeared that most of the TaVIT1 protein was targeted to other membranes. Closer inspection of the amino acid sequences revealed that TaVIT2 contains a universally conserved di-Leu motif for targeting to the vacuolar membrane (Bonifacino and Traub, 2003; Wang et al., 2014), which is absent from TaVIT1 (Supplemental Fig. S1B). Therefore, TaVIT1 may be able to transport iron but will not complement Δccc1 yeast. Instead, we tested if TaVIT1 was able to complement the Δfet3 yeast mutant, which is defective in high-affinity iron transport across the plasma membrane. Δfet3 mutants cannot grow on medium depleted of iron by the chelator bathophenanthrolinedisulfonic acid (BPS), but the expression of TaVIT1 rescued growth under these conditions (Supplemental Fig. S2). These data indicate that both TaVIT1 and TaVIT2 are able to transport iron but that their localization in the cell may differ.
Overexpression of TaVIT2 in the Endosperm of Wheat Specifically Increased the Iron Concentration in White Flour
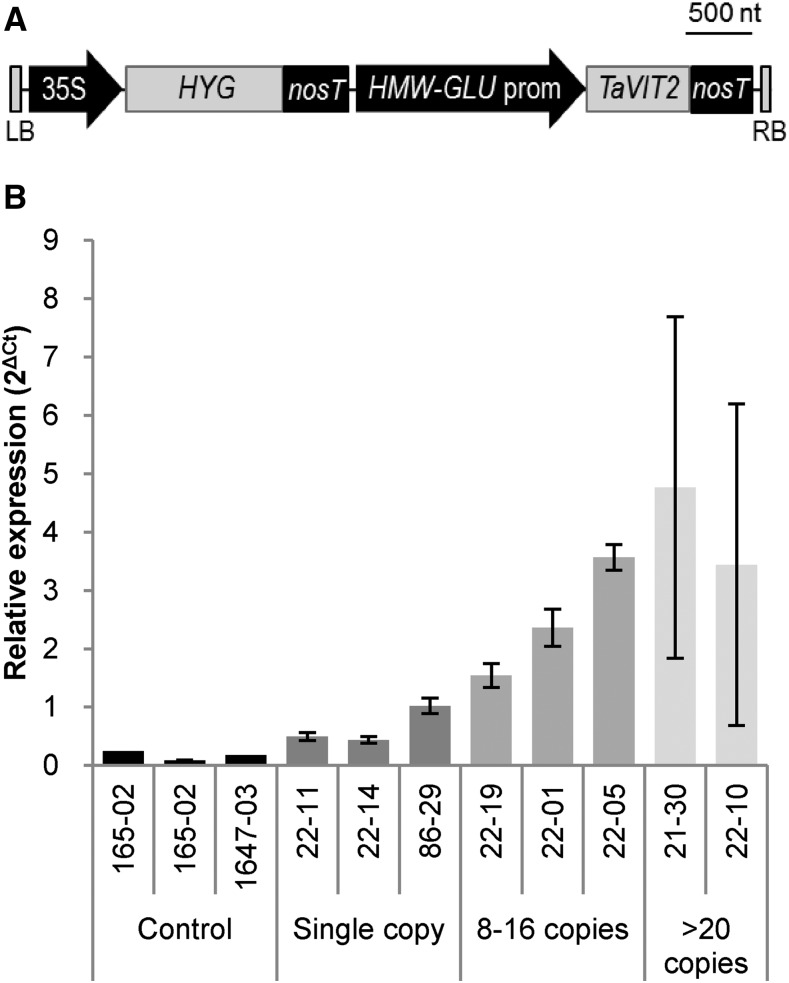
The functional characterization of TaVIT1 and TaVIT2 suggested that TaVIT2, as a bona fide iron transporter localized to vacuoles, is a good candidate for iron biofortification. We placed the TaVIT2 gene under the control of the wheat endosperm-specific promoter of the High Molecular Weight Glutenin-D1 (HMW) gene (Lamacchia et al., 2001) and transformed the construct together with a hygromycin resistance marker into the wheat cv Fielder (Fig. 3A). A total of 27 hygromycin-resistant plants were isolated, and the copy number of the transgene was determined by quantitative PCR. There were 10 lines with a single-copy insertion, and the highest number of insertions was 30. The transgene copy number correlated well with the expression of TaVIT2 in the developing grain (R2 = 0.6, P < 0.01; Fig. 3B; Supplemental Fig. S3). TaVIT2 expression was increased 3.8 ± 0.2-fold in single-copy lines and more than 20-fold in lines with multiple transgenes compared with nontransformed controls.
Figure 3.
Expression of TaVIT2 in cisgenic lines. A, Diagram of the transfer DNA construct: LB, left border; 35S, cauliflower mosaic virus 35S promoter; HYG, hygromycin resistance gene; nosT, nos terminator; HMW-GLU prom, high-molecular-weight glutenin-D1-1 promoter; TaVIT2, wheat VIT2-D gene; RB, right border; nt, nucleotides. B, Relative expression levels of TaVIT2 in developing grains at 10 d post-anthesis as determined by quantitative real-time PCR and normalized to housekeeping gene Traes_4AL_8CEA69D2F. Plant identification numbers and copy numbers of the HMW-TaVIT2 gene are given below the bars. Bars indicate means ± se of three independent biological replicates.
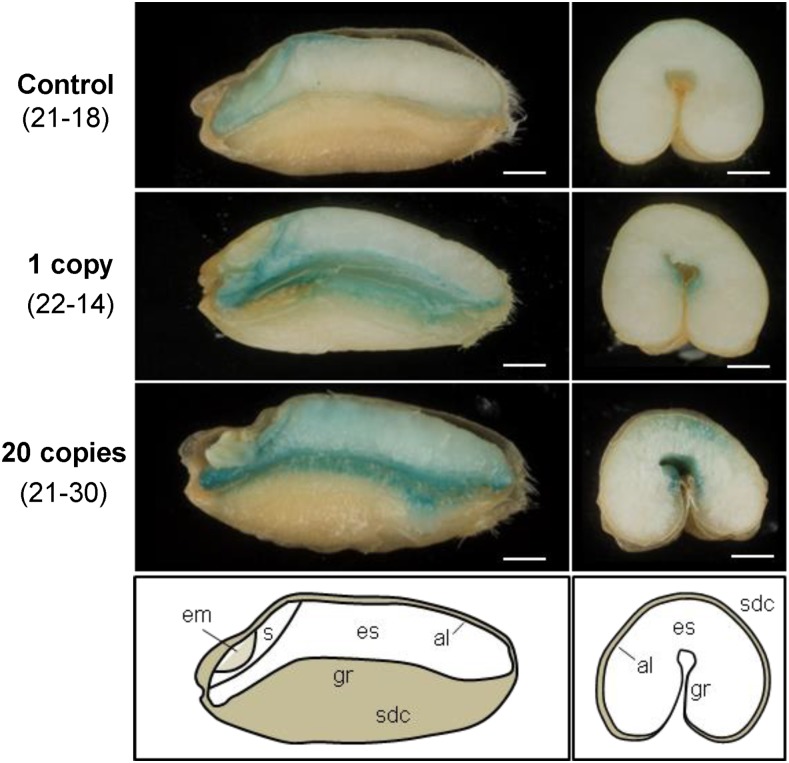
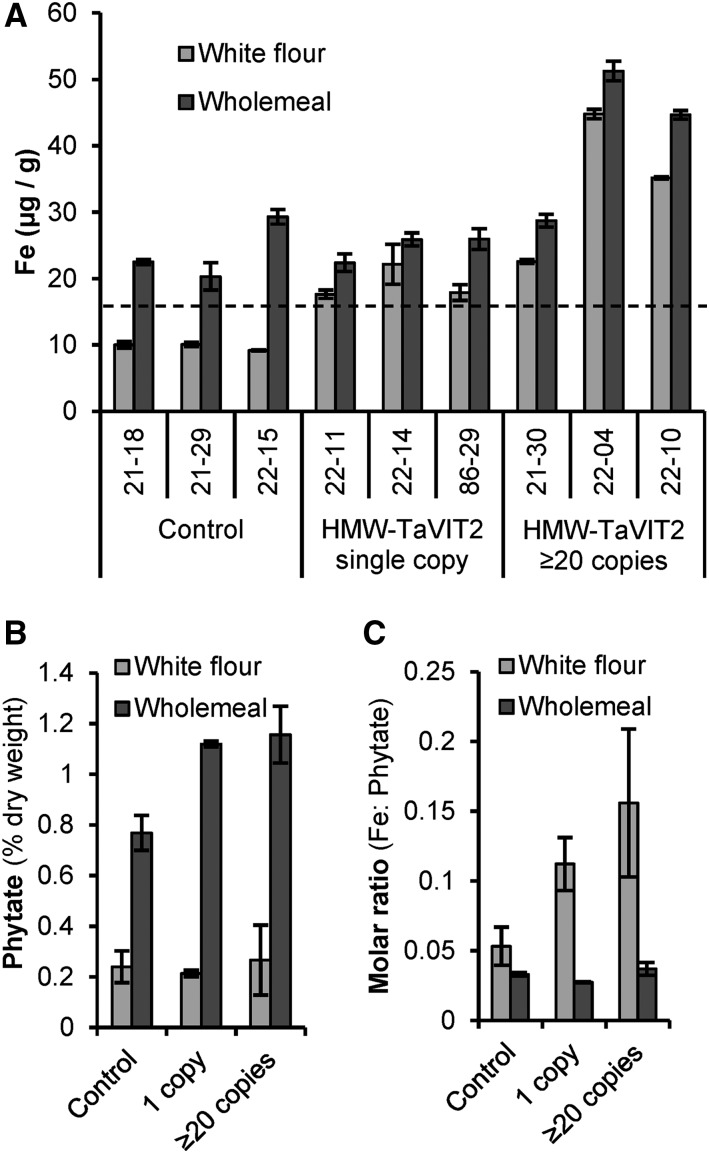
Mature wheat grains from transgenic lines and nontransformed controls were dissected with a platinum-coated blade and stained for iron using Perls’ Prussian Blue. In nontransformed controls, positive blue staining was visible in the embryo, scutellum, and aleurone layer, but the endosperm contained little iron (Fig. 4). In lines overexpressing TaVIT2, the Perls’ Prussian Blue staining was increased visibly, in particular around the groove and in patches of the endosperm. To quantify the amount of iron, grains from individual lines were milled to produce whole-meal flour, which was sieved to obtain a white flour fraction, followed by element analysis using inductively coupled plasma-optical emission spectroscopy (ICP-OES; Fig. 5A; Supplemental Table S3). Iron levels were consistently enhanced 2-fold in white flour, from 9.7 ± 0.3 µg g−1 in control lines to 21.7 ± 2.7 µg g−1 in lines with a single copy of HMW-TaVIT2 (P < 0.05). Additional transgene copies resulted in a similar 2-fold increase in iron, whereas lines with 20 or more copies contained 4-fold more iron than controls, to 41.5 ± 8.2 µg g−1 in white flour (P < 0.05). The iron content of whole-meal flour of single-insertion lines was similar to that of control lines but increased up to 2-fold in high-copy lines (P < 0.01). No statistically significant differences were found for other metals in single-copy HMW-TaVIT2 wheat grains, such as zinc, manganese, and magnesium (Supplemental Table S3), nor for the heavy metal contaminants cadmium and lead (Supplemental Table S4). In lines with 20 or more copies of HMW-TaVIT2, significant increases in all elements except manganese and lead were seen (P < 0.05), presumably as a secondary effect.
Figure 4.
Perls’ Prussian Blue staining for iron in grains transformed with HMW-TaVIT2. Grains from T0 wheat plants were dissected longitudinally (left) or transversely (right). al, Aleurone; em, embryo; es, endosperm; gr, groove; s, scutellum; sdc, seed coat. The transgene copy numbers and line numbers are indicated at left. Bars = 1 mm.
Figure 5.
Iron and phytate content of flour milled from HMW-TaVIT2 wheat lines. A, Iron concentrations in white and whole-meal flour from three control and six HMW-TaVIT2 lines. Bars represent means of two technical replicates ± sd. White flour from HMW-TaVIT2 lines has significantly more iron than that from control lines (n = 3–4, P < 0.001; for all data, see Supplemental Table S3). The dotted line at 16.5 μg g−1 iron indicates the minimum requirement for wheat flour sold in the United Kingdom. B, Phytate content of white and whole-meal flour of control and HMW-TaVIT2-expressing wheat. Bars represent means of two biological replicates ± sd. C, Molar ratio of iron to phytate in control and HMW-TaVIT2-expressing lines. Bars represent means of two biological replicates ± sd.
White Flour Has an Improved Iron-Phytate Ratio and the Iron Is Bioavailable
Because the high phytate content of cereal grains inhibits the bioavailability of minerals, we measured phytate levels in TaVIT2-overexpressing lines but found no significant increase in phytate in white flour (Fig. 5B), although there was a slight increase in phosphorus (Supplemental Table S3). There was also a small increase in phytate in whole-meal flour produced from those lines. Considering the 2-fold increase in iron, the iron-phytate molar ratio was improved 2-fold in white flour of HMW-TaVIT2 lines but unaffected in whole-meal flour (Fig. 5C).
To investigate the potential bioavailability of the iron, flour samples were subjected to simulated gastrointestinal digestion and the digests were applied to Caco-2 cells, a widely used cellular model of the small intestine (Glahn et al., 1998). For the purpose of this experiment, the availability of iron was maximized by treating the samples with phytase and by exposing the cells directly to the digestate after heat inactivation of the lytic enzymes. The increase in ferritin protein in Caco-2 cells after exposure to the digestate was used as a surrogate measure of iron availability. Iron from white flour was taken up by the Caco-2 cells, and more ferritin production was observed in cells exposed to samples from TaVIT2-overexpressing lines, although the values were variable between wheat lines (Supplemental Fig. S4). In contrast, the iron in whole-meal flour, although twice as high as in white flour, was not available for uptake, as noted previously (Eagling et al., 2014). Further analysis of breads baked from these flours is necessary to confirm that overexpression of TaVIT2 improves iron bioavailability. These data suggest that relocating iron into the endosperm may be more effective than increasing total iron in the grain as a biofortification strategy.
The High-Iron Phenotype Has Little Impact on Plant Growth and Is Maintained in T2 Grains
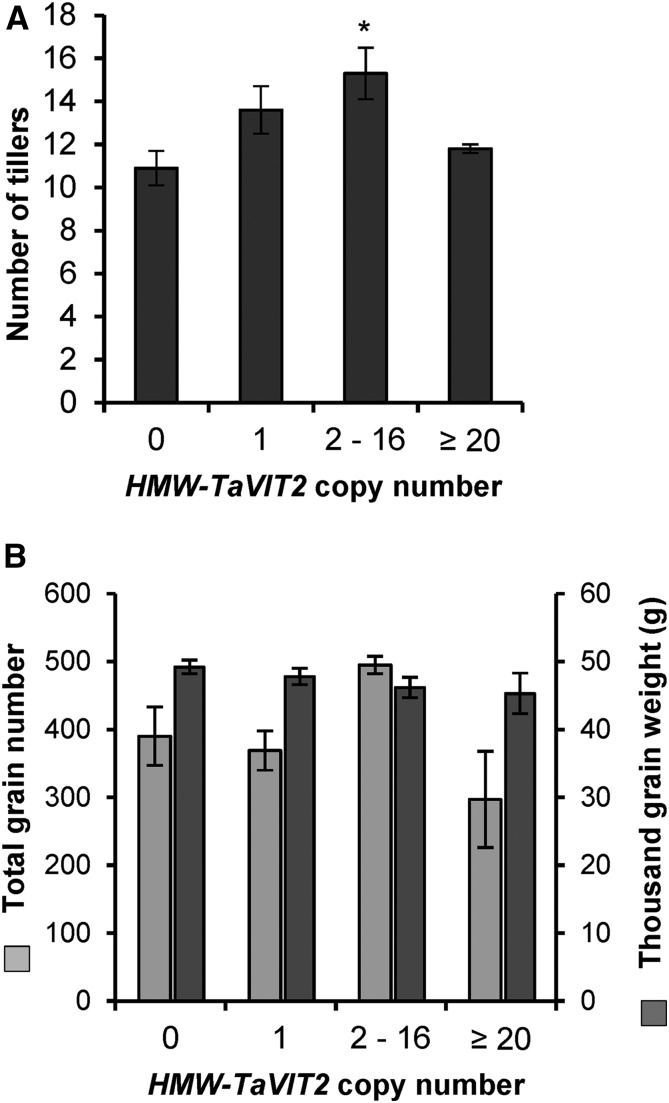
To investigate if TaVIT2 overexpression affected plant growth, we measured plant height, tiller number, grain size, number of grains per plant, and thousand-grain weight in TaVIT2-overexpressing lines and controls. None of these growth parameters was negatively affected by the HMW-TaVIT2 transgene in the T0 generation grown in controlled environment rooms (Fig. 6; Supplemental Table S5). Conversely, a statistically significant increase in tiller number was seen in plants containing two to 16 copies of the HMW-TaVIT2 transgene, to 15.3 ± 1.2 compared with the control of 10.9 ± 0.8 (P < 0.05, ANOVA; Supplemental Table S5). Analysis of further generations and field trials are required to confirm this effect and its potential impact on yield.
Figure 6.
Growth parameters of HMW-TaVIT2 wheat. The number of tillers (A) and seed output (B) of T0 wheat plants with indicated HMW-TaVIT2 copy numbers are shown. Bars indicate means ± se of the following numbers of biological replicates: zero gene copies, n = 9; one gene copy, n = 10; two to 16 gene copies, n = 9; 20 or more gene copies, n = 6. Further details are given in Supplemental Table S5. The asterisk indicates a significant difference from the negative control (one-way ANOVA with Tukey’s posthoc test: *, P < 0.05).
Seed from the first T0 transformant obtained (line 27-02, containing two copies of HMW-TaVIT2) was planted in a greenhouse to investigate the high-iron trait in the next generation (T1). The HMW-TaVIT2 transgene segregated in a 3:1 ratio (χ2 = 0.29). The growth of plants in the greenhouse was very different from that in controlled environment chambers, but there were no significant differences in growth and yield component traits for HMW-TaVIT2 plants compared with wild-type segregants or nontransformed controls (Supplemental Table S6). Though iron levels were overall higher in grain from greenhouse-grown plants, T2 grain still contained a 2-fold increase in iron in the white flour fraction compared to controls (P < 0.05; Supplemental Table S6). Taken together, endosperm-specific overexpression of TaVIT2 has no major growth defects, and the iron increase showed a similar trend in the next generation despite different growth conditions.
Expression of HMW-TaVIT2 in Barley Increases Grain Iron and Manganese Content
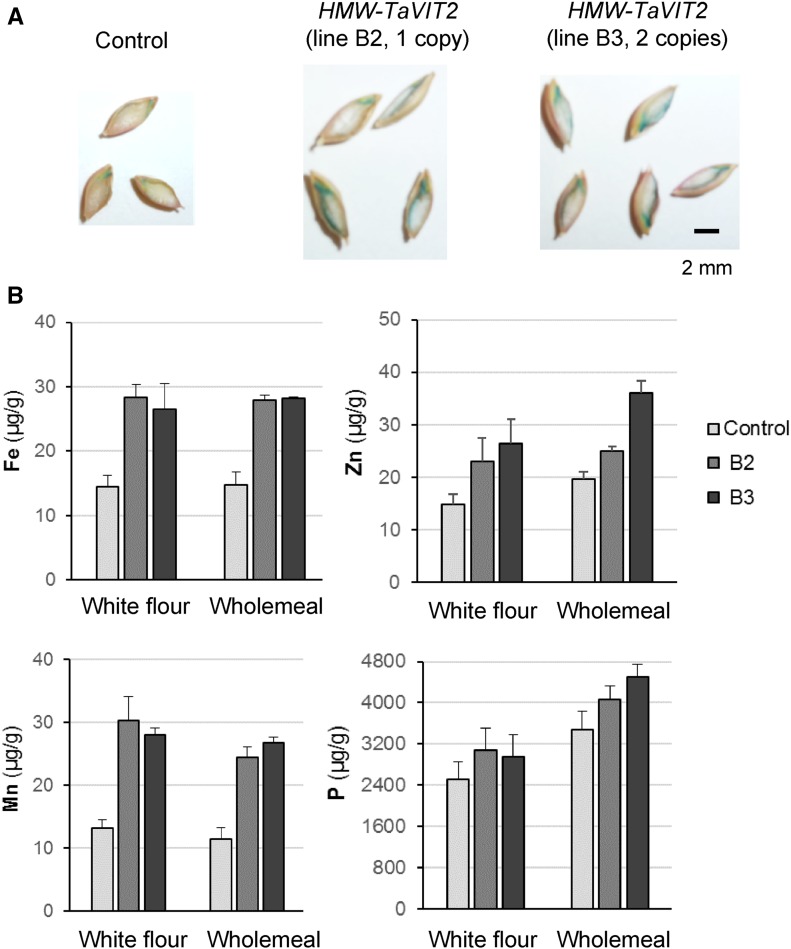
We also transformed barley cv Golden Promise with the HMW-TaVIT2 construct. The 12 transgenic plants had either one or two copies of the transgene and were indistinguishable from nontransformed controls with regard to vegetative growth and grain development. Staining with Perls’ Prussian Blue showed that, similar to wheat, there was more iron in transformed grains than in controls, and this tended to accumulate in the subaleurone region of the endosperm. To quantify the iron and other metals, lines B2 (one copy) and B3 (two copies) were selected for ICP-OES analysis and found to contain 2-fold more iron than the control in both white and whole-meal flour (Fig. 7). The white flour produced from barley contained relatively high levels of phosphorus, suggesting that there was some aleurone present, so the differences in minerals between white and whole-meal flours are not as pronounced as in wheat. Interestingly, in barley, there was also a 2-fold increase in manganese levels (Fig. 7). These results indicate that the ability of TaVIT2 to transport manganese, as observed in yeast (Fig. 2B), can be operational in plant tissue. Overall, our results indicate that endosperm-specific overexpression of TaVIT2 is a successful strategy for increasing the iron content in different cereal crop species.
Figure 7.
Endosperm-specific overexpression of TaVIT2 in barley. The TaVIT2-5DL gene from wheat under the control of the wheat HMW-GLU-1D-1 promoter (for full details, see Fig. 3A) was transformed into barley cv Golden Promise. Positive transformants were selected by hygromycin. A, Mature barley T1 grains of a control plant and two transgenic lines stained with Perls’ Prussian Blue for iron. B, Element analysis in white and whole-meal flours from a control and two HMW-TaVIT2-overexpressing barley plants. The values are means of two technical replicates ± sd.
DISCUSSION
The recently sequenced wheat genome greatly facilitates gene discovery in this economically important but genetically complex crop species. In a previous analysis (Borrill et al., 2014), we identified over 60 putative metal transporters and started with the functional characterization of VITs. We selected TaVIT1-B and TaVIT2-D for our studies. Each TaVIT gene has three homoeologs, but these share 99% amino acid identity, and those amino acids that differ are not conserved; therefore, we believe that our results are representative for all three homoeologs.
While TaVIT1 and TaVIT2 are ∼87% identical with OsVIT1 and OsVIT2, we found remarkable differences. Each rice VIT has the di-Leu motif involved in vacuolar targeting, and GFP fusion proteins showed vacuolar localization when transiently expressed in Arabidopsis protoplasts (Zhang et al., 2012). In wheat, only TaVIT2 has the di-Leu motif, and this correlated with the vacuolar localization of TaVIT2 in yeast. Another striking difference between rice and wheat VITs is the yeast complementation results. OsVIT1 and OsVIT2 partially complemented mutants in iron transport (Δccc1) and zinc transport (Δzrc1). In wheat, only TaVIT2 showed complementation of Δccc1, and we saw no evidence of zinc transport, similar to the metal specificity of the yeast ortholog. The growth defect of Δccc1 was completely rescued by TaVIT2, indicating efficient iron transport, in contrast to only weak complementation by the rice VIT genes. Unfortunately, the production of the rice VIT proteins in yeast was not verified by western-blot analysis (Zhang et al., 2012). Our initial experiments showed that wheat TaVIT1 was poorly expressed in yeast, so the sequence was codon optimized to remove codons that are rare in Saccharomyces cerevisiae (Supplemental Fig. S5). This greatly improved the expression of TaVIT1 to even higher levels than TaVIT2, but TaVIT1 still did not complement the yeast mutants in vacuolar Fe, Zn, or Mn transport. TaVIT1, however, did complement the Δfet3 yeast mutant (Supplemental Fig. S2). Yeast FET3 is part of a complex directing high-affinity Fe transport across the plasma membrane (Askwith et al., 1994). This suggests that TaVIT1 is indeed a functioning iron transporter but that it localizes mainly to a membrane other than the tonoplast. It will be interesting to identify the amino acid residues that determine metal specificity and/or localization in the VIT family. However, currently, there is no crystal structure of any of the VIT family members and no other good structural homology models. Recently, a first glimpse into the transport mechanism was provided, showing that P. falciparum VIT1 is an H+ antiporter with strong selectivity for Fe2+ (Slavic et al., 2016; Labarbuta et al., 2017).
Overexpression of the vacuolar iron transporter TaVIT2 in wheat endosperm was very effective in raising the iron concentration in this tissue. We hypothesize that increased sequestration of iron in the vacuoles creates a sink that then up-regulates the relocation of iron to that tissue. If the tissue normally stores iron in vacuoles rather than in ferritin, proteins and chelating molecules for iron mobilization into the vacuole already will be present. For a sink-driven strategy, timely expression of the gene in a specific tissue is essential: if the protein is produced constitutively, for example using the cauliflower mosaic virus 35S promoter, then it will draw iron into all tissues, not in one particular tissue. Interestingly, knockout mutants of VIT1 and VIT2 in rice accumulated more iron in the embryo (Zhang et al., 2012). A likely scenario is that iron distributed to the developing rice grain cannot enter the vacuoles in the aleurone (Kyriacou et al., 2014) and, thus, is diverted to the embryo. This finding further supports the idea that VITs play a key role in iron distribution in cereal grains. An additional advantage of endosperm-specific expression is that possible growth defects in vegetative tissues are likely to be avoided, as found in our studies.
Wheat and barley transformed with the same HMW-TaVIT2 construct showed surprising differences in the accumulation of iron and manganese. Wheat had a 2-fold increase in iron in the endosperm only, whereas barley contained 2-fold more iron in whole grains. Barley grains also contained 2-fold more manganese, but this element was not increased in wheat, even though TaVIT2 was found to transport both iron and manganese in yeast complementation assays. It is possible that the wheat HMW promoter has a different expression pattern in barley. If the promoter is activated in the aleurone cells in addition to the endosperm, this may lead to the observed higher iron concentrations in whole barley grains. The pattern of promoter activity can be further investigated with reporter constructs or by in situ hybridization specific for the transgene. It is also possible that wheat and barley differ in iron and manganese transport efficiency from roots to shoots, thus affecting the total amount of iron and manganese that is (re)mobilized to the grain.
In the Americas, Africa, and Asia, iron fortification of flours ranges from 30 to 44 µg g−1. In Europe, only the United Kingdom has a legal requirement for fortification: white and brown flours must contain at least 16.5 µg g−1 iron. We have now achieved this iron concentration in white flour produced from the single-copy HMW-TaVIT2 lines described here. More copies of TaVIT2 increased iron levels further but resulted in the accumulation of other metals. Moreover, with 20 or more transgene copies, there were fewer grains per plant. Combining endosperm-specific TaVIT2 overexpression with constitutive NAS overexpression may be one suitable approach to increase grain iron levels further. A combination strategy using overexpression of NAS2 and soybean (Glycine max) ferritin increased iron levels in polished rice more than 6-fold, from 2 to 15 μg g−1 in the field (Trijatmiko et al., 2016). However, combining NAS and FER overexpression in wheat did not show a synergistic effect: constitutive expression of the rice NAS2 gene resulted in 2.1-fold more iron in grains and 2.5-fold more iron in white flour, but coupled with endosperm-specific expression of FER, grain iron content was only 1.6- to 1.8-fold increased, similar to FER alone (Singh et al., 2017). As noted before, iron in wheat is mostly stored in vacuoles rather than ferritin, so increasing iron transport into vacuoles combined with increasing iron mobility is likely to be more effective. Nicotianamine also is reported to improve the bioavailability of iron (Zheng et al., 2010), which is a major determinant for the success of any biofortification strategy.
On a societal level, a major question is whether wheat biofortified using modern genetic techniques will be accepted by consumers. Our strategy used wheat genetic material (promoter and coding sequence) and, therefore, could be considered cisgenic. The HMW-TaVIT2 lines also contain DNA from species other than wheat, such as a hygromycin resistance gene of bacterial origin, but these regions can be removed using CRISPR technology, leaving only wheat DNA. In addition, the wheat lines described here are valuable tools to identify processes regulating the iron content of the grain. Identification of the transcription factors that control VIT expression would be helpful, but none has been identified so far in any plant species. Once more genetic components of the iron-loading mechanism into cereals have been identified, these can be targets of nontransgenic approaches such as TILLING (Krasileva et al., 2017).
MATERIALS AND METHODS
Identification of Wheat VIT Genes, Phylogenetic Analysis, and Analysis of RNA-Seq Data
The coding sequences of the wheat (Triticum aestivum) VIT genes were found by a BLAST search of the rice OsVIT1 (LOC_Os09g23300) and OsVIT2 (LOC_Os04g38940) sequences in Ensembl Plants (http://plants.ensembl.org). Full details of the wheat genes are given in Supplemental Table S1. Sequences of VIT genes from other species were found by a BLAST search of the Arabidopsis (Arabidopsis thaliana) AtVIT1 (AT2G01770) and rice (Oryza sativa) VIT sequences against the Ensembl Plants database. Amino acid alignments were performed using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). The tree was plotted with BioNJ with the Jones-Taylor-Thornton matrix and rendered using TreeDyn 198.3. RNA-seq data were obtained from the expVIP database (Borrill et al., 2016; http://www.wheat-expression.com). Full details of the data sets used are given in Supplemental Table S2.
Yeast Complementation
Coding DNA sequences for the wheat 2BL VIT1 homoeolog (TRIAE_CS42_2BL_TGACv1_129586_AA0389520) and the 5DL VIT2 homoeolog (TRIAE_CS42_5DL_TGACv1_433496_AA1414720) were synthesized and inserted into pUC57 vectors by Genscript. The wheat VIT genes were first synthesized with wheat codon usage, but TaVIT1 was poorly translated in yeast so was resynthesized with yeast codon usage including a 3× HA tag at the C-terminal end. Untagged codon-optimized TaVIT1 was amplified from this construct using primers TaVIT1co-XbaI-F and TaVIT1co-EcoRI-R (for primer sequences, see Supplemental Table S7). TaVIT2-HA was cloned by amplifying the codon sequence without the stop codon using primers TaVIT2-BamHI-F and TaVIT2(ns)-EcoRI-R and by amplifying the HA tag using primers HAT-EcoRI-F and HAT(Stop)-ClaI-R. The two DNA fragments were inserted into plasmid p416 behind the yeast (Saccharomyces cerevisiae) MET25 promoter (Mumberg et al., 1995).
Genes ScCCC1, ScFET3, ScPMR1, and ScZRC1 were cloned from yeast genomic DNA, using the primer pairs ScCCC1-BamHI-F and ScCCC1-EcoRI-R, ScFET3-XbaI-F and ScFET3-XhoI-R, ScPMR1-SpeI-F and ScPMR1-XhoI-R, and ScZRC1-XbaI-F andScZRC1-EcoRI-R, respectively. Following restriction digestion, the DNA fragments were ligated into vector p416-MET25 and confirmed by sequencing. All constructs were checked by DNA sequencing.
The Saccharomyces cerevisiae strain BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) was used in all yeast experiments. The wild type, Δccc1 (Li et al., 2001), Δzrc1 (MacDiarmid et al., 2003), Δpmr1 (Lapinskas et al., 1995), or Δfet3 (Askwith et al., 1994) was transformed with approximately 100 ng of DNA using the polyethylene glycol/lithium acetate method (Ito et al., 1983). Complementation analysis was performed via drop assays using overnight cultures of yeast grown in selective synthetic dextrose (SD) medium, diluted to approximately 1 × 106 cells mL−1, spotted in successive 4× dilutions onto SD plates containing appropriate supplements. Plates were incubated for 2 to 3 d at 30°C. Total yeast protein extraction was performed by alkaline lysis of overnight cultures (Kushnirov, 2000).
Preparation of Vacuoles from Yeast
Preparation of yeast vacuoles was performed using cell fractionation over a Suc gradient (Hwang et al., 2000; Nakanishi et al., 2001). Briefly, 1 L of yeast was grown in selective SD medium to an OD600 of 1.5 to 2 and then centrifigued at 4,000g for 10 min, washed in buffer 1 (0.1 m Tris-HCl, pH 9.4, 50 mm β-mercaptoethanol, and 0.1 m Glc), and resuspended in buffer 2 (0.9 m sorbitol, 0.1 m Glc, 50 mm Tris-MES, pH 7.6, 5 mm DTT, and 0.5× SD medium). Zymolyase 20T (Seikagaku) was added at a concentration of 0.05% (w/v), and cells were incubated for 2 h at 30°C with gentle shaking. After cell wall digestion, spheroplasts were centrifuged at 3,000g for 10 min and then washed in 1 m sorbitol before being resuspended in buffer 3 (40 mm Tris-MES, pH 7.6, 1.1 m glycerol, 1.5% [w/v] polyvinylpyrrolidone 40,000, 5 mm EGTA, 1 mm DTT, 0.2% [w/v] BSA, 1 mm phenylmethylsulfonyl fluoride [PMSF], and 1× protease inhibitor cocktail [Promega]) and homogenized on ice using a glass homogenizer. The homogenate was centrifuged at 2,000g for 10 min at 4°C, and the supernatant was transferred to fresh tubes, while the pellet was resuspended in fresh buffer 3 and centrifuged again. The supernatants were pooled and centrifuged at 150,000g for 45 min at 4°C to pellet microsomal membranes. For the preparation of vacuole-enriched vesicles, the pellet was resuspended in 15% (w/w) Suc in buffer 4 (10 mm Tris-MES, pH 7.6, 1 mm EGTA, 2 mm DTT, 25 mm KCl, 1.1 m glycerol, 0.2% [w/v] BSA, 1 mm PMSF, and 1× protease inhibitor cocktail), and this was layered onto an equal volume of 35% (w/w) Suc solution in buffer 4 before centrifugation at 150,000g for 2 h at 4°C. Vesicles were collected from the interface and diluted in buffer 5 (5 mm Tris-MES, pH 7.6, 0.3 m sorbitol, 1 mm DTT, 1 mm EGTA, 0.1 m KCl, 5 mm MgCl2, 1 mm PMSF, and 1× protease inhibitor cocktail). The membranes were centrifuged at 150,000g for 45 min at 4°C and resuspended in a minimal volume of buffer 6 (5 mm Tris-MES, pH 7.6, 0.3 m sorbitol, 1 mm DTT, 1 mm PMSF, and 1× protease inhibitor cocktail). Vesicles were snap frozen in liquid nitrogen and stored at −80°C.
Generation of Transgenic Plant Lines
The TaVIT2 gene was amplified using primers TaVIT2-NcoIF and TaVIT2-SpeIR and cloned into vector pRRes14_RR.301 containing the promoter sequence comprising nucleotides −1,187 to −3 with respect to the ATG start codon of the GLU-1D-1 gene, which encodes the high-molecular-weight glutenin subunit 1Dx5 (Lamacchia et al., 2001). The promoter-gene fragment was then cloned into vector pBract202 containing a hygromycin resistance gene and left border and right border elements for insertion into the plant genome (Smedley and Harwood, 2015). The construct was checked by DNA sequencing. Transformation into wheat (cv Fielder) and barley (Hordeum vulgare cv Golden Promise) was performed by the BRACT platform at the John Innes Centre using Agrobacterium tumefaciens-mediated techniques as described previously (Wu et al., 2003; Harwood et al., 2009). Transgene insertion and copy number in T0 plants were assessed by iDNA Genetics using quantitative PCR with a Taqman probe. For the T1 generation, the presence of the hygromycin resistance gene was analyzed by PCR with primers Hyg-F and Hyg-R.
Plant Growth and Quantitative Analysis
The first generation of transgenic plants (T0) was grown in a controlled environment room under 16 h of light (300 μmol m−2 s−1) at 18°C/8 h of dark at 15°C with 65% relative humidity. The next generation (T1) was grown in a glasshouse kept at approximately 20°C with 16 h of light. Wheat and barley plants were grown on a mix of 40% medium-grade peat, 40% sterilized soil, and 20% horticultural grit and fertilized with 1.3 kg m−3 PG Mix 14+16+18 (Yara UK) containing 0.09% Fe, 0.16% Mn, and 0.04% Zn. Ears from wheat and barley plants were threshed by hand, and grain morphometric characteristics, mass, and number were determined using a MARVIN universal grain analyzer (GTA Sensorik).
RNA Extraction and Quantitative Real-Time PCR
Samples of developing grain were taken at 10 d postanthesis and frozen in liquid nitrogen. RNA extraction was performed using phenol/chloroform extraction (Box et al., 2011). Developing grains were ground with a pestle and mortar under liquid nitrogen and mixed with RNA extraction buffer (0.1 m Tris-HCl, pH 8, 5 mm EDTA, 0.1 m NaCl, 0.5% [w/v] SDS, and 1% [v/v] 2-mercaptoethanol) and the Ambion Plant RNA Isolation Aid (ThermoFisher). Samples were centrifuged for 10 min at 15,000g, and the supernatant was added to 1:1 acidic phenol (pH 4.3):chloroform. After mixing and incubation at room temperature for 10 min, the upper phase was added to isopropanol containing 0.3 m sodium acetate. Samples were incubated at −80°C for 15 min and centrifuged for 30 min at 15,000g at 4°C. The supernatant was discarded, and the pellet was washed twice in 70% (v/v) ethanol and dried before being resuspended in RNase-free water. RNA was DNase treated using the TURBO DNase-free kit (ThermoFisher) according to the manufacturer’s instructions. DNase inactivation reagent was added, and the samples were centrifuged at 10,000g for 90 s. Supernatant containing RNA was retained. RNA was reverse transcribed using oligo(dT) primer and SuperScript II reverse transcriptase (ThermoFisher) according to the manufacturer’s instructions. Quantitative real-time PCR was used to analyze the expression of TaVIT2 and the housekeeping gene (HKG) Traes_4AL_8CEA69D2F, chosen because it was shown to have the most stable gene expression across grain development in over 400 RNA-seq samples (Borrill et al., 2016), using primer pairs qRT-TaVIT2-F/qRT-TaVIT2-R and qRT-HKG-F/qRT-HKG-R, respectively. Samples were run in a CFX96 Real-Time System (Bio-Rad) with the following conditions: 3 min at 95°C; 35 cycles of 5 s at 95°C, 10 s at 62°C, and 7 s at 72°C; and melt curve of 5 s at 65°C and 5 s at 95°C. TaVIT2 expression levels were normalized to the expression levels of the housekeeping gene and expressed as 2ΔCt.
Perls’ Prussian Blue Staining
Mature grains were dissected using a platinum-coated scalpel, stained for 45 min in Perls’ Prussian Blue staining solution (2% [w/v] potassium hexacyanoferate [II] and 2% [v/v] hydrochloric acid), and then washed twice in deionized water.
Flour Preparation, Element Analysis, and Phytate Determination
Barley grains were dehulled by hand, and all grains were coarsely milled using a coffee grinder and then ground into flour using a pestle and mortar. White flour fractions were obtained by passing the material through a 150-μm nylon mesh. Flour samples were dried overnight at 55°C and then digested for 1 h at 95°C in ultrapure nitric acid (55%, v/v) and hydrogen peroxide (6%, v/v). Samples were diluted 1:11 in ultrapure water and analyzed by ICP-OES (Vista-PRO CCD Simultaneous ICP-OES; Agilent) calibrated with standards: Zn, Fe, and Mg at 0.2, 0.4, 0.6, 0.8, and 1 mg L−1 and Mn and P at 1, 2, 3, 4, and 5 mg L−1. Soft winter wheat flour was used as a reference material (RM 8438; U.S. National Institute of Standards and Technology) and analyzed in parallel with all experimental samples. Phytate levels were determined using a phytic acid (total phosphorus) assay kit (Megazyme).
Bioavailability Assays in Caco-2 Cells
Caco-2 cells (HTB-37) were obtained from American Type Culture Collection and cultured as described previously (Rodriguez-Ramiro et al., 2017). Wheat flour samples were subjected to simulated gastrointestinal digestion as described (Glahn et al., 1998) with minor modifications. One gram of flour was added to 5 mL of pH 2 buffer saline solution (140 mmol L−1 NaCl and 5 mmol L−1 KCl) followed by the addition of pepsin (0.04 g mL−1). Ascorbic acid was added at a molar ratio of 1:20 to ensure the complete solubilization of released iron. Additionally, phytase (Megazyme) was added to fully degrade phytate (myoinositol hexakisphosphate). Samples were incubated at 37°C on a rolling table for 90 min. Next, the pH of the samples was gradually adjusted to pH 5.5, bile (0.007 g mL−1) and pancreatin (0.001 g mL−1) digestive enzymes were added, the pH was adjusted to 7, and the samples were incubated for an additional 1 h. At the end of the simulated digestion, samples were centrifuged at 3,000g for 10 min, the gastrointestinal enzymes were heated inactivated at 80°C for 10 min and centrifuged as before, and the resultant supernatant was used subsequently for iron uptake experiments similar to Bodnar et al. (2013) with little modification. A volume of 0.5 mL of wheat digestate was diluted in 0.5 mL of Eagle’s MEM and applied over Caco-2 cell monolayers grown on collagen-coated 12-well plates. Samples were incubated for 2 h at 37°C in a humidified incubator containing 5% CO2 and 95% air. After incubation, an additional 0.5 mL of MEM was added, and cells were incubated for a further 22 h prior to harvesting for ferritin analysis. To harvest the cells, the medium was removed by aspiration, and cells were rinsed with 18 Ω MilliQ water and subsequently lysed by scraping in 100 µL of Cellytic M (Sigma-Aldrich). Cell pellets were kept on ice for 15 min and stored at −80°C. For analysis, samples were thawed and centrifuged at 14,000g for 15 min. The supernatant containing the proteins was used for ferritin determination using the Spectro Ferritin ELISA (RAMCO) according to the manufacturer’s protocol. Ferritin concentrations were normalized to total cell protein using the Pierce Protein BCA protein assay (ThermoFisher).
All experiments were performed using the following controls: (1) a blank digestion without any wheat sample or added iron, and (2) a reference digestion of 50 µm ferrous sulfate heptahydrate (FeSO4∙7H2O) solubilized in 0.1 m HCl with 1,000 µm ascorbic acid.
Statistical Analysis
Statistical analyses (F test, ANOVA, Student’s t test, Kruskal-Wallis test, regression analysis, χ2) were performed using Microsoft Excel 2010 and Genstat 18th Edition. Unless stated otherwise in the text, P values were obtained from Kruskal-Wallis tests with Dunnett posthoc tests. When representative images are shown, the experiment was repeated at least three times with similar results.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: TaVIT1-A, TRIAE_CS42_2AL_TGACv1_094675_AA0301300; TaVIT1-B, TRIAE_CS42_2BL_TGACv1_129586_AA0389520; TaVIT1-D, TRIAE_CS42_2DL_TGACv1_159145_AA0533310; TaVIT2-A, TRIAE_CS42_5AL_TGACv1_374137_AA1191460; TaVIT2-B, TRIAE_CS42_5BL_TGACv1_406234_AA1342560; and TaVIT2-D, TRIAE_CS42_5DL_TGACv1_433496_AA1414720.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Gene models and protein sequences of wheat vacuolar iron transporters.
Supplemental Figure S2. TaVIT1 complements a yeast mutant defective in iron transport across the plasma membrane.
Supplemental Figure S3. Correlation between HMW-TaVIT2 transgene copy number and the expression of TaVIT2.
Supplemental Figure S4. Ferritin formation in Caco-2 cells incubated with phytase-treated flour digestates.
Supplemental Figure S5. Alignment of yeast codon-optimized TaVIT1 DNA sequence with the original sequence.
Supplemental Table S1. Wheat VIT genes identified in this study.
Supplemental Table S2. Expression analysis of TaVIT genes.
Supplemental Table S3. Element analysis of control and HMW-TaVIT2 wheat lines.
Supplemental Table S4. Heavy metals in control and HMW-TaVIT2 wheat lines.
Supplemental Table S5. Architectural and yield components of control and HMW-TaVIT2 T0 transformants.
Supplemental Table S6. Architectural and yield components of T1 plants segregating from a TaVIT2 overexpressor T0 plant.
Supplemental Table S7. List of primers.
Acknowledgments
We thank James Simmonds for technical assistance; Wendy Harwood for wheat transformation; Alison Huttly at Rothamsted Research for the HMW GLU-D1-1 promoter; Sylvaine Bruggraber at MRC-HNR and Graham Chilvers at the University of East Anglia for element analysis; and Tony Miller and Dale Sanders for helpful discussions.
Glossary
- RNA-seq
RNA sequencing
- ICP-OES
inductively coupled plasma-optical emission spectroscopy
- SD
synthetic dextrose
- PMSF
phenylmethylsulfonyl fluoride
Footnotes
This work was funded by HarvestPlus (J.M.C., J.B., and C.U.) and by the BBSRC Institute Strategic Programme Grant BB/J004561/1 (C.U. and J.B.) and DRINC2 grant BB/L025515/1 (I.R.-R., S.F.-T., and J.B.).
Articles can be viewed without a subscription.
References
- Askwith C, Eide D, Van Ho A, Bernard PS, Li L, Davis-Kaplan S, Sipe DM, Kaplan J (1994) The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell 76: 403–410 [DOI] [PubMed] [Google Scholar]
- Bodnar AL, Proulx AK, Scott MP, Beavers A, Reddy MB (2013) Iron bioavailability of maize hemoglobin in a Caco-2 cell culture model. J Agric Food Chem 61: 7349–7356 [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Traub LM (2003) Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem 72: 395–447 [DOI] [PubMed] [Google Scholar]
- Borrill P, Connorton JM, Balk J, Miller AJ, Sanders D, Uauy C (2014) Biofortification of wheat grain with iron and zinc: integrating novel genomic resources and knowledge from model crops. Front Plant Sci 5: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrill P, Ramirez-Gonzalez R, Uauy C (2016) expVIP: a customizable RNA-seq data analysis and visualization platform. Plant Physiol 170: 2172–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box MS, Coustham V, Dean C, Mylne JS (2011) Protocol: a simple phenol-based method for 96-well extraction of high quality RNA from Arabidopsis. Plant Methods 7: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briat JF, Duc C, Ravet K, Gaymard F (2010) Ferritins and iron storage in plants. Biochim Biophys Acta 1800: 806–814 [DOI] [PubMed] [Google Scholar]
- Clavijo BJ, Venturini L, Schudoma C, Accinelli GG, Kaithakottil G, Wright J, Borrill P, Kettleborough G, Heavens D, Chapman H, et al. (2017) An improved assembly and annotation of the allohexaploid wheat genome identifies complete families of agronomic genes and provides genomic evidence for chromosomal translocations. Genome Res 27: 885–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagling T, Wawer AA, Shewry PR, Zhao FJ, Fairweather-Tait SJ (2014) Iron bioavailability in two commercial cultivars of wheat: comparison between wholegrain and white flour and the effects of nicotianamine and 2′-deoxymugineic acid on iron uptake into Caco-2 cells. J Agric Food Chem 62: 10320–10325 [DOI] [PubMed] [Google Scholar]
- Eroglu S, Giehl RFH, Meier B, Takahashi M, Terada Y, Ignatyev K, Andresen E, Küpper H, Peiter E, von Wirén N (2017) Metal Tolerance Protein 8 mediates manganese homeostasis and iron reallocation during seed development and germination. Plant Physiol 174: 1633–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn RP, Lee OA, Yeung A, Goldman MI, Miller DD (1998) Caco-2 cell ferritin formation predicts nonradiolabeled food iron availability in an in vitro digestion/Caco-2 cell culture model. J Nutr 128: 1555–1561 [DOI] [PubMed] [Google Scholar]
- Harwood WA, Bartlett JG, Alves SC, Perry M, Smedley MA, Leyland N, Snape JW (2009) Barley transformation using Agrobacterium-mediated techniques. In Jones HD, Shewry PR, eds, Transgenic Wheat, Barley and Oats. Humana Press, Totowa, NJ, pp 137–147 [DOI] [PubMed] [Google Scholar]
- Hurrell R, Egli I (2010) Iron bioavailability and dietary reference values. Am J Clin Nutr 91: 1461S–1467S [DOI] [PubMed] [Google Scholar]
- Hwang I, Harper JF, Liang F, Sze H (2000) Calmodulin activation of an endoplasmic reticulum-located calcium pump involves an interaction with the N-terminal autoinhibitory domain. Plant Physiol 122: 157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A (1983) Transformation of intact yeast cells treated with alkali cations. J Bacteriol 153: 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SA, Punshon T, Lanzirotti A, Li L, Alonso JM, Ecker JR, Kaplan J, Guerinot ML (2006) Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science 314: 1295–1298 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Nishizawa NK (2012) Iron uptake, translocation, and regulation in higher plants. Annu Rev Plant Biol 63: 131–152 [DOI] [PubMed] [Google Scholar]
- Krasileva KV, Vasquez-Gross HA, Howell T, Bailey P, Paraiso F, Clissold L, Simmonds J, Ramirez-Gonzalez RH, Wang X, Borrill P, et al. (2017) Uncovering hidden variation in polyploid wheat. Proc Natl Acad Sci USA 114: E913–E921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov VV. (2000) Rapid and reliable protein extraction from yeast. Yeast 16: 857–860 [DOI] [PubMed] [Google Scholar]
- Kyriacou B, Moore KL, Paterson D, de Jonge MD, Howard DL, Stangoulis J, Tester M, Lombi E, Johnson AAT (2014) Localization of iron in rice grain using synchrotron x-ray fluorescence microscopy and high resolution secondary ion mass spectrometry. J Cereal Sci 59: 173–180 [Google Scholar]
- Labarbuta P, Duckett K, Botting CH, Chahrour O, Malone J, Dalton JP, Law CJ (2017) Recombinant vacuolar iron transporter family homologue PfVIT from human malaria-causing Plasmodium falciparum is a Fe2+/H+ exchanger. Sci Rep 7: 42850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamacchia C, Shewry PR, Di Fonzo N, Forsyth JL, Harris N, Lazzeri PA, Napier JA, Halford NG, Barcelo P (2001) Endosperm-specific activity of a storage protein gene promoter in transgenic wheat seed. J Exp Bot 52: 243–250 [PubMed] [Google Scholar]
- Lapinskas PJ, Cunningham KW, Liu XF, Fink GR, Culotta VC (1995) Mutations in PMR1 suppress oxidative damage in yeast cells lacking superoxide dismutase. Mol Cell Biol 15: 1382–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapinskas PJ, Lin SJ, Culotta VC (1996) The role of the Saccharomyces cerevisiae CCC1 gene in the homeostasis of manganese ions. Mol Microbiol 21: 519–528 [DOI] [PubMed] [Google Scholar]
- Li L, Chen OS, McVey Ward D, Kaplan J (2001) CCC1 is a transporter that mediates vacuolar iron storage in yeast. J Biol Chem 276: 29515–29519 [DOI] [PubMed] [Google Scholar]
- MacDiarmid CW, Milanick MA, Eide DJ (2003) Induction of the ZRC1 metal tolerance gene in zinc-limited yeast confers resistance to zinc shock. J Biol Chem 278: 15065–15072 [DOI] [PubMed] [Google Scholar]
- Momonoi K, Yoshida K, Mano S, Takahashi H, Nakamori C, Shoji K, Nitta A, Nishimura M (2009) A vacuolar iron transporter in tulip, TgVit1, is responsible for blue coloration in petal cells through iron accumulation. Plant J 59: 437–447 [DOI] [PubMed] [Google Scholar]
- Moore KL, Zhao FJ, Gritsch CS, Tosi P, Hawkesford MJ, McGrath SP, Shewry PR, Grovenor CRM (2012) Localisation of iron in wheat grain using high resolution secondary ion mass spectrometry. J Cereal Sci 55: 183–187 [Google Scholar]
- Mumberg D, Müller R, Funk M (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122 [DOI] [PubMed] [Google Scholar]
- Nakanishi Y, Saijo T, Wada Y, Maeshima M (2001) Mutagenic analysis of functional residues in putative substrate-binding site and acidic domains of vacuolar H+-pyrophosphatase. J Biol Chem 276: 7654–7660 [DOI] [PubMed] [Google Scholar]
- Narayanan N, Beyene G, Chauhan RD, Gaitán-Solis E, Grusak MA, Taylor N, Anderson P (2015) Overexpression of Arabidopsis VIT1 increases accumulation of iron in cassava roots and stems. Plant Sci 240: 170–181 [DOI] [PubMed] [Google Scholar]
- O’Dell BL, De Boland AR, Koirtyohann SR (1972) Distribution of phytate and nutritionally important elements among the morphological components of cereal grains. J Agric Food Chem 20: 718–723 [Google Scholar]
- Ravet K, Touraine B, Kim SA, Cellier F, Thomine S, Guerinot ML, Briat JF, Gaymard F (2009) Post-translational regulation of AtFER2 ferritin in response to intracellular iron trafficking during fruit development in Arabidopsis. Mol Plant 2: 1095–1106 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Ramiro I, Brearley CA, Bruggraber SFA, Perfecto A, Shewry P, Fairweather-Tait S (2017) Assessment of iron bioavailability from different bread making processes using an in vitro intestinal cell model. Food Chem 228: 91–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschzttardtz H, Conéjéro G, Curie C, Mari S (2009) Identification of the endodermal vacuole as the iron storage compartment in the Arabidopsis embryo. Plant Physiol 151: 1329–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SP, Keller B, Gruissem W, Bhullar NK (2017) Rice NICOTIANAMINE SYNTHASE 2 expression improves dietary iron and zinc levels in wheat. Theor Appl Genet 130: 283–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavic K, Krishna S, Lahree A, Bouyer G, Hanson KK, Vera I, Pittman JK, Staines HM, Mota MM (2016) A vacuolar iron-transporter homologue acts as a detoxifier in Plasmodium. Nat Commun 7: 10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley MA, Harwood WA (2015) Gateway-compatible plant transformation vectors. In Wang K, ed, Agrobacterium Protocols, Vol 1 Springer, New York, pp 1–15 [Google Scholar]
- Trijatmiko KR, Dueñas C, Tsakirpaloglou N, Torrizo L, Arines FM, Adeva C, Balindong J, Oliva N, Sapasap MV, Borrero J, et al. (2016) Biofortified indica rice attains iron and zinc nutrition dietary targets in the field. Sci Rep 6: 19792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasconcelos MW, Gruissem W, Bhullar NK (2017) Iron biofortification in the 21st century: setting realistic targets, overcoming obstacles, and new strategies for healthy nutrition. Curr Opin Biotechnol 44: 8–15 [DOI] [PubMed] [Google Scholar]
- Wang X, Cai Y, Wang H, Zeng Y, Zhuang X, Li B, Jiang L (2014) Trans-Golgi network-located AP1 gamma adaptins mediate dileucine motif-directed vacuolar targeting in Arabidopsis. Plant Cell 26: 4102–4118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2008) Global health risks, mortality and burden of disease attributable to selected major risks. http://www.who.int/healthinfo/global_burden_disease/GlobalHealthRisks_report_annex.pdf.
- Wu H, Sparks C, Amoah B, Jones HD (2003) Factors influencing successful Agrobacterium-mediated genetic transformation of wheat. Plant Cell Rep 21: 659–668 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xu YH, Yi HY, Gong JM (2012) Vacuolar membrane transporters OsVIT1 and OsVIT2 modulate iron translocation between flag leaves and seeds in rice. Plant J 72: 400–410 [DOI] [PubMed] [Google Scholar]
- Zheng L, Cheng Z, Ai C, Jiang X, Bei X, Zheng Y, Glahn RP, Welch RM, Miller DD, Lei XG, et al. (2010) Nicotianamine, a novel enhancer of rice iron bioavailability to humans. PLoS ONE 5: e10190. [DOI] [PMC free article] [PubMed] [Google Scholar]