The multiple alternative oxidase isoforms from Arabidopsis are differentially activated by various metabolites, suggesting isoform-specific functions.
Abstract
Mitochondrial alternative oxidase (AOX) in plants is a non-proton-motive ubiquinol oxidase that is activated by redox mechanisms and 2-oxo acids. A comparative analysis of the AOX isoenzymes AOX1A, AOX1C, and AOX1D from Arabidopsis (Arabidopsis thaliana) revealed that cysteine residues, CysI and CysII, are both involved in 2-oxo acid activation, with AOX1A activity being more increased by 2-oxo acids than that of AOX1C and AOX1D. Substitution of cysteine in AOX1A by glutamate mimicked its activation by pyruvate or glyoxylate, but not in AOX1C and AOX1D. CysIII, only present in AOX1A, is not involved in activation by reduction or metabolites, but substitutions at this position affected activity. AOX1A carrying a serine residue at position CysI was activated by succinate, while correspondingly substituted variants of AOX1C and AOX1D were insensitive. Activation by glutamate at CysI and CysII is consistent with the formation of the thiohemiacetal, while succinate activation after changing CysI to serine suggests hemiacetal formation. Surprisingly, in AOX1A, replacement of CysI by alanine, which cannot form a (thio)hemiacetal, led to even higher activities, pointing to an alternative mechanism of activation. Taken together, our results demonstrate that AOX isoforms are differentially activated and that activation at CysI and CysII is additive.
As a consequence of their sessile nature, plants are exposed to changing environmental conditions on a daily and a seasonal basis, requiring adjustments of cellular metabolism to maintain the balance between energy generation and consumption. Various abiotic and biotic stress conditions, such as high light, drought, salinity, chilling, and pathogen attack, lead to the enhanced production and accumulation of reactive oxygen species, due to metabolic imbalances in plant cells (Suzuki et al., 2012). The avoidance of oxidative stress is achieved by several mechanisms (Aro et al., 1993; Shikanai, 2007; Hanke and Hase, 2008). Here, we will focus on nonplastidic mechanisms to balance the ATP/NAD(P)H ratio and to maintain redox homeostasis, with the alternative respiratory pathway in plant mitochondria playing an important role. This non-energy-conserving pathway of electron transfer consists of five enzymes: four NAD(P)H dehydrogenases for the oxidation of excess NAD(P)H and the alternative oxidase (AOX; Møller and Rasmusson, 1998; Møller, 2001; Rasmusson et al., 2004). AOX represents an alternative terminal oxidase found in all plants, many fungi, some protists, mollusks, nematodes, and chordates (Stenmark and Nordlund, 2003; McDonald and Vanlerberghe, 2006; Moore and Albury, 2008) and mediates cyanide-resistant respiration. AOX catalyzes the oxidation of ubiquinol and reduces oxygen to water without concomitant proton translocation by bypassing complex III, cytochrome c, and complex IV of the cytochrome c respiratory pathway (Finnegan et al., 2004). As a consequence, electron flow to AOX significantly reduces ATP generation via respiration, dissipating most energy as heat (Sluse and Jarmuszkiewicz, 1998; Affourtit et al., 2002). Homodimeric AOX is localized at the inner mitochondrial membrane with its catalytic centers oriented toward the matrix (Juszczuk and Rychter, 2003).
Genomes of many higher plants encode several AOX isoforms. The gene expression of these isoforms is tissue specific, depends on the developmental stage of the plant, changes in cellular metabolism as well as various abiotic and biotic stress conditions, and is highly responsive to dysfunctions in the mitochondrial respiratory metabolism (Juszczuk and Rychter, 2003; Millenaar and Lambers, 2003; Zarkovic et al., 2005; Clifton et al., 2006; Van Aken et al., 2009; Feng et al., 2013; Vanlerberghe, 2013). Arabidopsis (Arabidopsis thaliana) possesses five genes encoding AOX proteins that can be divided into two subfamilies: AOX1A to AOX1D and AOX2 (Polidoros et al., 2009; Pu et al., 2015). Transcripts of the main isoform AOX1A are abundant throughout all tissues and developmental stages of the plant. In parallel to AOX1A, AOX1C also is expressed ubiquitously in all developmental stages throughout plant development, albeit at a very low level. While AOX1B transcripts are abundant mainly in inflorescences, AOX1D transcripts are found predominantly during the early vegetative stage, flowering, and in senescent leaves. In contrast, AOX2 expression seems to be limited to developmental stages associated with the presence of seeds: for instance, in late stages of silique maturation and early stages of seed germination (Guo et al., 2004; Clifton et al., 2006). AOX2 from Arabidopsis also can be targeted to chloroplasts by its native targeting signal and can functionally substitute for the plastid terminal oxidase in a suppression screen (Fu et al., 2012).
Studies in Arabidopsis and tobacco (Nicotiana tabacum) clearly revealed a role for AOX in the dissipation of excessive reducing power while maintaining photosynthesis under high-light stress (Florez-Sarasa et al., 2011, 2016; Dahal et al., 2014, 2015; Vanlerberghe et al., 2016). In Arabidopsis, aox1a knockout plants cannot be complemented by the expression of other isogenes (Strodtkötter et al., 2009; Kühn et al., 2015). For instance, although the expression of AOX1D is induced, it cannot functionally replace the missing AOX1A. In contrast to the wild type, Arabidopsis aox1a knockout plants do not survive treatment by antimycin A, an inhibitor of electron transport at the site of cytochrome bc1 in complex III (Alexandre and Lehninger, 1984; Campo et al., 1992; Maguire et al., 1992; Xia et al., 1997; Pham et al., 2000; Strodtkötter et al., 2009; Kühn et al., 2015). Moreover, when electron transport through the cytochrome c respiratory chain is limited using a surrogate mutant approach, an additional inactivation of aox1a leads to a more severe growth phenotype, even though AOX1D is highly expressed at the transcript and protein levels (Kühn et al., 2015).
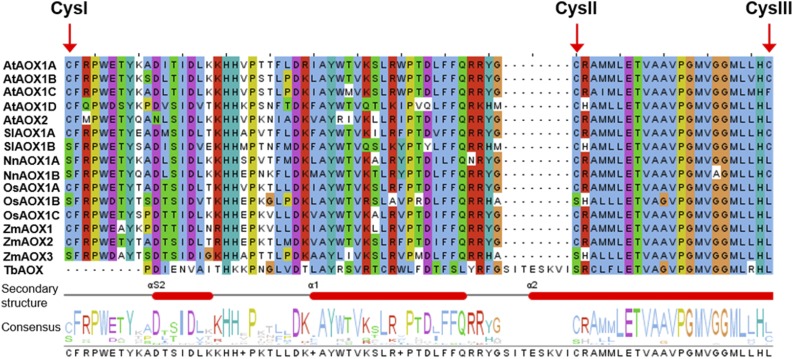
Besides transcriptional regulation, AOX activity has been shown to be posttranslationally regulated (Millar et al., 1993, 1996; Umbach and Siedow, 1993, 1996; Day and Wiskich, 1995; Day et al., 1995; Rhoads et al., 1998; Siedow and Umbach, 2000; Umbach et al., 2006; Selinski et al., 2016). Most AOX isoforms possess two highly conserved Cys residues (CysI and CysII) in the N-terminal domain of the protein. While AOX1A and AOX1B from Arabidopsis and AOX isoforms from some other plant species, like tomato (Solanum lycopersicum) and lotus (Nelumbo nucifera), possess a third Cys residue (CysIII) near the catalytic di-iron center, most AOX isoforms contain a Leu residue at this position, except AOX1C from Arabidopsis, in which a Phe residue is present (Fig. 1; Supplemental Fig. S1). While the formation of an intermolecular disulfide bond between CysI of each monomer causes the inactivation of AOX1A under oxidizing conditions, reducing conditions lead to a noncovalently linked dimer that is the active AOX1A form. The reduced and active forms of AOX1A can be further activated by 2-oxo acids, most notably pyruvate, through the formation of a thiohemiacetal with CysI (Millar et al., 1993; Umbach and Siedow, 1993, 1996; Rhoads et al., 1998; Umbach et al., 2006; Selinski et al., 2016). The balance between oxidized and reduced AOX1A protein depends on the redox state of the NAD(P)H pool, which itself is linked to mitochondrial metabolism (Vanlerberghe et al., 1995; Juszczuk and Rychter, 2003).
Figure 1.
Section of a multiple sequence alignment of AOX isoforms. Amino acid sequences of AOX isoforms from different plant species were aligned using the bioinformatic tools Clustal Omega and Jalview (Waterhouse et al., 2009; Sievers et al., 2011). The region including all Cys residues of plant AOX isoproteins is shown (for total alignment, see Supplemental Fig. S1). The top five sequences show AOX isoforms from Arabidopsis (AtAOX). The following lines illustrate sequences of tomato (SlAOX), lotus (NnAOX), rice (OsAOX), and maize (ZmAOX). The last line represents AOX from Trypanosoma brucei (TbAOX). Note that SlAOX1B, NnAOX1A, NnAOX1B, OsAOX1B, and ZmAOX3 possess a Ser residue at the position of CysI of AOX isoforms from Arabidopsis.
However, not all plant AOX isoforms contain a Cys residue at position I, like, for instance, AOX1B from tomato, AOX1A and AOX1B from lotus, AOX1B from rice (Oryza sativa), and AOX3 from maize (Zea mays; Fig. 1; Supplemental Fig. S1). These isoforms possess instead a Ser residue at the position of CysI. In consequence, AOX isoproteins from tomato and lotus were shown to be insensitive to 2-oxo acids but can be stimulated by the dicarboxylic acid succinate (Djajanegara et al., 1999; Ito et al., 1997; Karpova et al., 2002; Holtzapffel et al., 2003; Grant et al., 2009). Comparable to AOX isoforms that naturally possess a Ser residue at CysI, Arabidopsis AOX1A shows activation by succinate and insensitivity toward pyruvate or glyoxylate after substitution of CysI by Ser (Djajanegara et al., 1999).
Studies on the posttranslational activation were only carried out for the main isoform AOX1A from Arabidopsis (Millar et al., 1993; Umbach and Siedow, 1993, 1996; Rhoads et al., 1998; Umbach et al., 2006; Selinski et al., 2016). In this work, a detailed comparison of the role of the different Cys residues in posttranslational activation of the three Arabidopsis isoforms AOX1A, AOX1C, and AOX1D was performed using a sensitive experimental setup with prolonged linear time intervals based on Escherichia coli membranes enriched in individual AOX isoforms after heterologous expression (Selinski et al., 2016). Differences in activity of these isoforms and their corresponding mutant derivatives were determined using metabolites present in mitochondria known to stimulate AOX activity.
RESULTS
Substitution of Conserved Cys Residues Affects Basal Activities of AOX1A, AOX1C, and AOX1D Strongly
Due to the identical in planta localization of AOX isoforms, it is not possible to measure specific activities of each isoform in isolated plant mitochondria. Therefore, each AOX isoform was recombinantly expressed in E. coli strain BHH8, and membrane vesicles enriched in individual AOX isoenzymes were isolated (Selinski et al., 2016). The E. coli strain used is a cytochrome bo and bd-I oxidase double mutant (lacking two of three endogenous terminal oxidases), which enables isoform-specific oxygen consumption measurements by combining NADH dehydrogenases and the ubiquinol pool of the E. coli respiratory chain with the heterologously expressed AOX isoform, using a Clark-type oxygen electrode. The experimental setup described by Selinski et al. (2016) allows for the determination of constant activity rates with prolonged linear reaction time in BHH8 membrane vesicles enriched in AOX isoproteins. In addition to the basal activities of individual AOX isoforms and their derivatives, activity increases upon the addition of various effectors not affecting the respiration of E. coli itself (Selinski et al., 2016). Specific AOX activities were obtained by relating oxygen consumption to the signal intensities of AOX isoproteins determined by immunoblot analyses, as described in detail by Selinski et al. (2016).
In order to enable a comparative characterization of the posttranslational activation of AOX isoforms from Arabidopsis, various single, double, and triple substitutions of CysI, CysII, and/or CysIII (PheIII for AOX1C and LeuIII for AOX1D) were generated for AOX1A, AOX1C, and AOX1D. To simplify the nomenclature of constructs used in this study, the one-letter code for amino acids was used to describe the composition at sites I, II, and III occurring in native and mutant forms combined with a three-figure letter code, resulting in CCC for AOX1A wild type (WT), CCF for AOX1C-WT, and CCL for AOX1D-WT, respectively (Fig. 1; Supplemental Figs. S2–S4). The following substitutions were carried out. (1) CysI, CysII, and/or CysIII (PheIII for AOX1C and LeuIII for AOX1D) were substituted by Ser or Ala, maintaining a comparable size, while no disulfide bridge or thiohemiacetal can be formed. In addition, in the presence of Ala, hydrophilic interactions in close vicinity may be reduced. (2) CysI, CysII, and/or CysIII (PheIII for AOX1C and LeuIII for AOX1D) were substituted by Glu to introduce a negative charge, which potentially mimics a similarly sized thiohemiacetal formed in the presence of pyruvate at the corresponding position. (3) Substitution of Cys residues by the positively charged Lys was used as a control for the oppositely charged Glu. Although CysIII is not present in AOX1C and AOX1D, substitutions at this position were carried out in both isoforms for comparison with the respective substitutions in AOX1A.
Substitution of Single Cys Residues at Various Positions Leads to Increased Basal Activities in AOX1A and AOX1C But Not in AOX1D
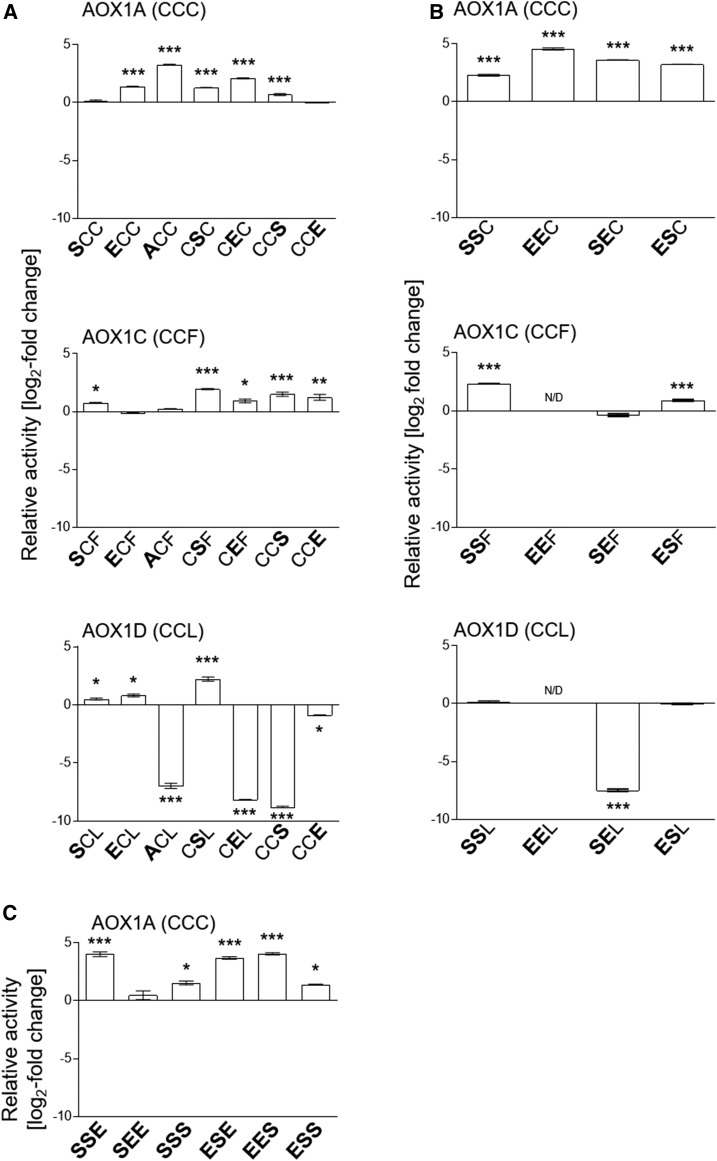
To test the influence of substitutions of Cys residues on AOX basal activities (in the absence of effectors) on DTT-reduced AOX1A, AOX1C, and AOX1D, relative activities represented as log2-fold change values were calculated on the basis of the activity of the corresponding AOX wild-type protein (Fig. 2; Supplemental Figs. S2–S4). Substitution of CysI for Ser resulted in no change in the basal activity for AOX1A (i.e. AOX1A-SCC was unaffected), while AOX1C and AOX1D showed increased activities after substitution of CysI by Ser (AOX1C-SCF and AOX1D-SCL; Fig. 2A). In AOX1A-ACC with Ala in place of CysI, the activity was increased significantly, whereas AOX1C-ACF activity remained at the same level as in the AOX1C wild-type protein, and in AOX1D-ACL, it was decreased significantly (100-fold; Fig. 2A). In addition, substitutions of CysI by Glu led to increased basal activities of AOX1A-ECC and AOX1D-ECL, while the activity of AOX1C-ECF remained unchanged (Fig. 2A).
Figure 2.
Effects of amino acid substitutions on AOX activities. AOX isoenzymes with different single (A), double (B), or triple (C) substitutions of CysI, CysII, and/or CysIII (PheIII or LeuIII) were heterologously expressed in E. coli strain BHH8 and analyzed for oxygen consumption in isolated membrane vesicles under reducing conditions. As a basis for specific AOX activity, respiration rates were normalized for each sample using densitometry on immunoblots (Selinski et al., 2016). Graphed values (log2 fold change; mutant/wild type) represent averages from two to five independent protein induction experiments with the appropriate se. Asterisks indicate that the differences (*, P < 0.05; **, P < 0.01; and ***, P < 0.001) between the basal activities of wild-type protein and the activities of substituted AOX derivatives are statistically significant as determined by one-way ANOVA with posthoc Tukey’s honestly significant difference (HSD) test. Wild types are as follows: AOX1A, CCC; AOX1C, CCF; and AOX1D, CCL. Substitutions are presented in the one-letter code for amino acids in enlarged boldface letters.
The substitution of CysII with Ser led to increased basal activities of all three AOX isoforms, AOX1A-CSC, AOX1C-CSF, and AOX1D-CSL. Glu at CysII in AOX1A-CEC and AOX1C-CEF resulted in increased basal activities, while in AOX1D-CEL, activity was decreased significantly (200-fold; Fig. 2A).
Substitution at position III of AOX1A and AOX1C by Ser (AOX1A-CCS and AOX1C-CCS) led to activation. In contrast, the basal activity of AOX1D-CCS was again decreased significantly (400-fold) compared with AOX1D-CCL (Fig. 2A). Furthermore, substitution of CysIII (PheIII for AOX1C and LeuIII for AOX1D) by Glu did not influence the basal activity of AOX1A, whereas it was increased in AOX1C-CCE compared with AOX1C-WT and was less active in AOX1D-CCE than in AOX1D-WT (Fig. 2A).
In summary, these data indicate that AOX1A and AOX1C and, to a lesser extent, AOX1D can be activated by substitutions mimicking a thiohemiacetal at CysI and CysII. In addition, CysIII in AOX1A, which is not present in AOX1C and AOX1D, represents another site of potential activation. However, the type of amino acids introduced by substitution appears to be important at all three positions.
Double Substitutions of CysI and CysII Have an Additive Effect on Basal Activity Compared with Corresponding Wild-Type Proteins Most Pronounced in AOX1A
Besides the effects of single substitutions on basal activities, we observed high impact of double substitutions of Cys residues in AOX1A, AOX1C, and AOX1D, which were much more pronounced than the single substitutions alone (Fig. 2B; Supplemental Fig. S2–S4). In AOX1A, substitutions of CysI and CysII by Ser and/or Glu led to an additive increase in the protein’s basal activity. For instance, the basal activities of AOX1A-SSC, -EEC, -SEC, and -ESC displayed a greater fold increase in activity compared with the single mutants (Fig. 2, A and B; e.g. the activity of AOX1A-SCC was unchanged, while AOX1A-CSC activity was increased 2-fold compared with the wild type but 5-fold in AOX1A-SSC). A similar pattern was seen with AOX1A-EEC compared with AOX1A-ECC or AOX1A-CEC, with AOX1A-EEC exhibiting the highest basal activity of all tested Cys substitutions, with a 24-fold increase in activity compared with the wild type (Fig. 2B). Based on the fact that CysIII is present only in AOX1A, proteins with triple substitutions were analyzed in more detail for this isoform. In comparison with proteins containing two exchanges, the substitution of CysIII in AOX1A did not significantly change the basal activity, except for AOX1A-SSE, which was increased, and AOX1A-SEE, which was decreased 10-fold compared with AOX1A-SEC (Fig. 2, B and C).
In AOX1C, double substitutions of CysI and CysII predominantly led to moderately increased basal activities compared with the single-substituted variants. For instance, AOX1C-SCF and AOX1C-CSF activities were increased 2- and 4-fold, respectively, while AOX1C-SSF exhibited the highest activity, with a 5-fold increase (Fig. 2, A and B; Supplemental Fig. S3). However, AOX1C-SEF was less active compared with the corresponding single-substituted proteins, with a 2-fold decreased activity compared with AOX1C-SCF and AOX1C-CEF (Fig. 2, A and B). In comparison with AOX1A, triple substitutions of AOX1C showed increased activities compared with AOX1C-CCF, while substitution of Phe at position III did not significantly change basal activities compared with double-substituted AOX1C (Supplemental Fig. S3). Nevertheless, AOX1C-SSS displayed a 3-fold increase compared with the corresponding double-substituted AOX1C-SSF (Supplemental Fig. S3). In contrast to AOX1A and AOX1C, double substitutions of AOX1D exhibited decreased activities compared with single substitutions (e.g. AOX1D-SSL activity was unchanged compared with AOX1D-SCL but decreased 5-fold compared with AOX1D-CSL; Fig. 2, A and B). Triple substitutions mainly yielded a less active protein compared with single-substituted AOX1D, but there was no difference in activity between double- and triple-substituted AOX1D (Supplemental Fig. S4).
Taken together, substitutions of CysI and CysII have an additive effect on basal activities when compared with the corresponding wild-type proteins. Therefore, as described above for AOX isoforms carrying single substitutions, the double-substituted proteins additionally support the view that CysI and CysII are both involved in the activation of AOX1A, AOX1C, and AOX1D. Notably, substitutions of highly conserved CysI and/or CysII did not destabilize AOX1A and AOX1C but yielded increased basal activities.
Activation by 2-Oxo Acids Occurs at CysI and CysII in AOX1A, AOX1C, and AOX1D
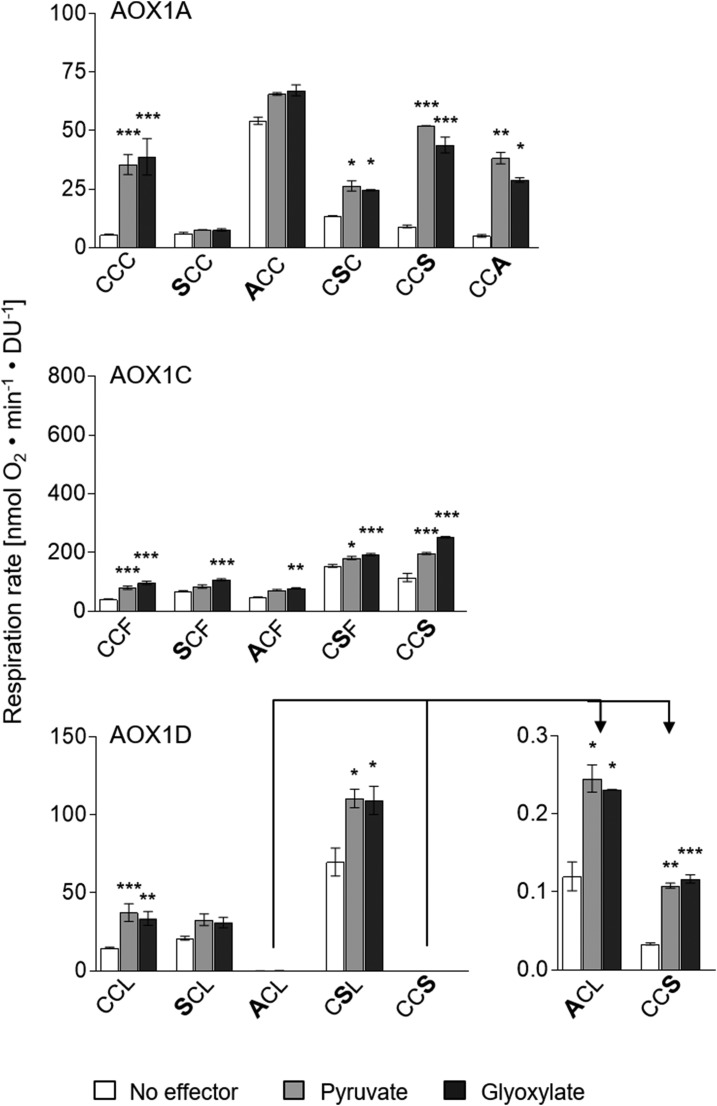
In previous studies, AOX1A from Arabidopsis was shown to be activatable by 2-oxo acids, which is consistent with the formation of a thiohemiacetal with CysI (Umbach and Siedow, 1993, 1996; Rhoads et al., 1998; Umbach et al., 2006), whereas comparable analyses for other AOX isoforms determining their sensitivity toward pyruvate and glyoxylate are still lacking. Therefore, the influence of 2-oxo acids on the activity of AOX1A, AOX1C, and AOX1D was investigated and compared with each other. While the AOX1A-CCC wild-type protein exhibited a 6- to 7-fold increase in activity after treatment with pyruvate or glyoxylate compared with its basal activity (no effector), AOX1C-CCF and AOX1D-CCL wild-type activities were increased only by a factor of 2- to 3-fold (Fig. 3). This indicates that the AOX1A-CCC wild-type protein is more prone to be activated by 2-oxo acids than AOX1C-CCF and AOX1D-CCL wild-type proteins.
Figure 3.
Identification of Cys resi-dues involved in 2-oxo acid stimula-tion of AOX1A, AOX1C, and AOX1D isoproteins. Oxygen consumption measurements and calculations of specific respiration rates were performed as described in the legend for Figure 2. Measurements were performed as technical duplicates or triplicates. Asterisks indicate that the differences (*, P < 0.05; **, P < 0.01; and ***, P < 0.001) between the basal activity (no effector) and activities in the presence of the effectors pyruvate or glyoxylate are statistically significant as determined by two-way ANOVA with posthoc Tukey’s HSD test. Wild types are as follows: AOX1A, CCC; AOX1C, CCF; and AOX1D, CCL. Substitutions are presented in the one-letter code for amino acids in enlarged boldface letters. DU, Density units.
Due to the fact that the activity of each AOX isoform can be stimulated by pyruvate and glyoxylate, single Cys substitutions and the influence of 2-oxo acids on their activities were analyzed to enable the identification of the Cys residue(s) involved in the formation of the thiohemiacetal. AOX1A was insensitive to pyruvate and glyoxylate after substitution of CysI by Ser or Glu (AOX1A-SCC or -ECC; Figs. 3 and 4; Supplemental Fig. S2), indicating that the proposed formation of a thiohemiacetal occurs at CysI in AOX1A. However, CysII could be identified as an additional site of 2-oxo acid activation of AOX1A. Although AOX1A-CSC was still activatable by pyruvate or glyoxylate, this variant only exhibited a 2- to 3-fold increase in activity after addition of the effectors, while the activity of the wild-type protein was increased 6- to 7-fold (Fig. 3). This suggests that AOX1A is less sensitive toward 2-oxo acid activation when CysII is lacking and, therefore, supports the notion that pyruvate and glyoxylate can bind to both CysI and CysII, with CysI being the primary site of metabolite activation. CysIII is unlikely to be an additional activation site for pyruvate or glyoxylate, because an AOX1A mutant protein containing an Ala or Ser residue at this position (AOX1A-CCS or AOX1A-CCA) exhibited a 6- to 7-fold increase in activity after treatment with these effectors, as is the case for the AOX1A wild-type protein (AOX1A-CCC; Fig. 3). In contrast, AOX1A-CCE and -CCK were insensitive to pyruvate and glyoxylate (Fig. 4; Supplemental Fig. S2). However, this is possibly due to the larger size as well as the charge of these amino acid residues, which seem to negatively affect the accessibility of CysI and/or CysII of AOX1A, since a Ser or Ala residue at this position does not influence AOX1A activation by pyruvate or glyoxylate.
Figure 4.
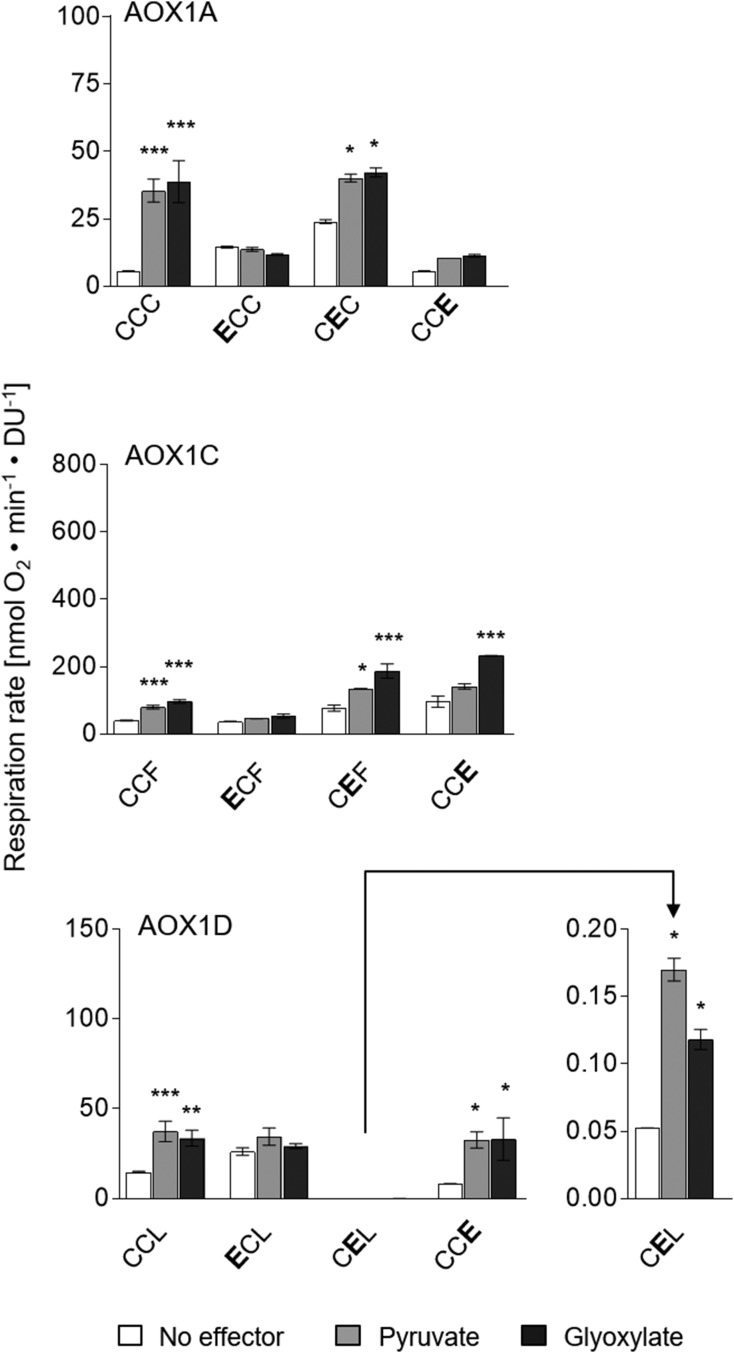
Simulation of the thiohemiacetal in AOX1A, AOX1C, and AOX1D. The influence of amino acid substitutions by Glu on AOX activity and its sensitivity to 2-oxo acids was analyzed as described in the legend for Figure 2. Measurements were performed as technical duplicates or triplicates. Asterisks indicate that the differences (*, P < 0.05; **, P < 0.01; and ***, P < 0.001) between the basal activity (no effector) and activities in the presence of effectors are statistically significant as determined by two-way ANOVA with posthoc Tukey’s HSD test. Wild types are as follows: AOX1A, CCC; AOX1C, CCF; and AOX1D, CCL. Substitutions are presented in the one-letter code for amino acids in enlarged boldface letters. DU, Density units.
Activation of AOX1C by 2-oxo acids, especially by pyruvate, was less pronounced after substitution of CysI by Ser (AOX1C-SCF) or Ala (AOX1C-ACF), indicating that CysI is involved in activation by pyruvate and glyoxylate. However, these mutant proteins could still be activated significantly by glyoxylate, but to a lesser extent (20%–50% increase of basal activity) compared with AOX1C wild-type protein (Fig. 3). The same activation pattern emerged for AOX1C-CSF. After substitution of CysII by Ser, the activation of AOX1C-CSF by pyruvate or glyoxylate was less pronounced than in the wild-type protein, but it was still present (20% increase of basal activity). In this context, it is important to mention that both proteins already showed increased basal activities compared with the wild-type protein (2-fold increase for AOX1C-SCF and 4-fold increase for AOX1C-CSF), which could be explained by the loss of redox regulatory Cys residues (CysI or CysII), yielding enzymes that are not inactivated by oxidation any longer (Fig. 2). Nevertheless, the AOX1C-CCS protein exhibited a 2- to 3-fold increase in activity after treatment with 2-oxo acids, as is the case for wild-type AOX1C-CCF. Therefore, it can be concluded that, comparable to AOX1A, the proposed formation of a thiohemiacetal at CysI and CysII in AOX1C leads to an activation of the catalytic activity.
AOX1D was less sensitive to pyruvate or glyoxylate after substitution of CysI by Ser (AOX1D-SCL; 50%–60% increase of basal activity; Fig. 3), whereas AOX1D-ACL and -ECC showed the same activation pattern as the wild-type protein AOX1D-CCL, although both exhibited a 100-fold lower basal activity (Fig. 3; Supplemental Fig. S4). This result is consistent with the formation of a thiohemiacetal at CysI of AOX1D. However, AOX1D-CSL also showed a reduced activation by 2-oxo acids (60% increase) compared with the AOX1D-CCL wild-type protein (Fig. 3), leading to the conclusion that, also in the case of AOX1D, both Cys residues, CysI and CysII, are involved in the proposed formation of a thiohemiacetal. The activation pattern of AOX1D-CCS and -CCE is consistent with this hypothesis. Both mutant proteins are activated by pyruvate or glyoxylate to the same extent as the wild-type protein (2- to 3-fold increase), although the basal activity of AOX1D-CCS is much lower than that of wild-type AOX1D-CCL (Figs. 3 and 4; Supplemental Fig. S4). Taken together, these results demonstrate that both CysI and CysII contribute to the activation of AOX1A, AOX1C, and AOX1D by pyruvate and glyoxylate, with CysI appearing to be strongly involved in metabolite activation while CysII represents a secondary, less effective activation site. Furthermore, AOX1D-ACL, -CCS, and -CEL can be activated in the same manner by pyruvate or glyoxylate as the wild-type protein, despite its drastically decreased basal activities. This indicates that the active site in the catalytic di-iron center as well as the two Cys residues, CysI and CysII, function as distinct units that are influenced individually.
The Substitution of CysI or CysII by Glu Mimics the Formation of the Thiohemiacetal in AOX1A But Only Partially in AOX1C and AOX1D
A thiohemiacetal and a Glu residue are similar: both are negatively charged and of nearly the same size, especially when the formation of a thiohemiacetal occurs between pyruvate and a Cys residue. Various AOX1A, AOX1C, and AOX1D derivatives containing a Glu residue at the position of CysI, CysII, or CysIII (PheIII for AOX1C and LeuIII for AOX1D) were generated (Fig. 4; Supplemental Figs. S2–S4). These AOX variants were used to mimic the presence of a thiohemiacetal within the protein, which is expected to result in constitutively active AOX proteins and would further support the presence of a thiohemiacetal at CysI and CysII in AOX1A, AOX1C, and AOX1D.
After substitution of CysI by Glu (AOX1A-ECC), AOX1A is not further stimulated by pyruvate and glyoxylate but exhibited a 2- to 3-fold increase in basal activity compared with the corresponding wild-type protein AOX1A-CCC (Figs. 2 and 4). However, the substitution of CysI by Glu only partially mimics the formation of a thiohemiacetal in AOX1A, resulting in a 2- to 3-fold increase in activity, while the activity of AOX1A-CCC wild-type protein was increased 6- to 7-fold by these 2-oxo acids (Fig. 4). By comparison, substitution of CysII by Glu in AOX1A (AOX1A-CEC) enabled a 4-fold increase in basal activity, which could be further activated by pyruvate or glyoxylate, leading to activities comparable to those of the wild-type protein activated with 2-oxo acids (Figs. 2 and 4). In this case, the effectors may bind to CysI, leading to further activation of AOX1A-CEC. All data demonstrate that AOX1A is activated by 2-oxo acids at CysI and CysII, as proposed above after analyzing single substitutions, in which Cys residues have been exchanged with Ser (Fig. 3). Furthermore, the double-substituted AOX1A (AOX1A-EEC) possessing a Glu residue at both positions, CysI and CysII, showed a drastically increased basal activity compared with the AOX1A wild-type protein (20- to 25-fold increase) as well as the corresponding single-substituted mutant proteins (5- to 10-fold increase; Supplemental Fig. S2), again supporting the view that AOX1A is activated at both CysI and CysII.
Comparable to AOX1A-ECC, AOX1D-ECL could not be further stimulated by pyruvate and glyoxylate. Its activity was increased 2- to 3-fold compared with AOX1D-CCL (Figs. 2 and 4). This increase in basal activity after substitution of CysI by Glu was similar to the effect of pyruvate and glyoxylate on the activity of the AOX1D wild-type protein (Fig. 4). In contrast, AOX1D-CEL and AOX1D-CCE were less active than the wild-type protein (200- and 2-fold decreased basal activity, respectively), but the activation by pyruvate or glyoxylate of both mutant proteins remained similar to the activation pattern of AOX1D-CCL (Fig. 4). Although it appears that the substitution of CysI by Glu can mimic the formation of the thiohemiacetal in AOX1D, this does not seem to be the case for CysII. Nonetheless, substituting Ser for CysII in AOX1D suggests that both Cys residues are sensitive to activation with pyruvate and glyoxylate (Fig. 3). In this context, it is important to mention that substitution of CysII by Ser or Glu (AOX1D-CSL or -CEL) has a major impact on the basal activity (Ser activating by a factor of 5 and Glu reducing by a factor of 200), suggesting that a substitution of CysII changes the conformation of the catalytic di-iron center, while the influence of CysI on the catalytic center and, therefore, the activation of the catalytic activity by 2-oxo acids remains unchanged.
In contrast to AOX1A and AOX1D, the substitution of CysI by Glu in AOX1C cannot mimic the presence of the thiohemiacetal. The basal activity of AOX1C-ECF remained at the same level as the wild-type protein. In addition, AOX1C-ECF could not be activated by pyruvate and glyoxylate (Figs. 2 and 4), whereas AOX1C-CEF and -CCE variants showed a similar increase in activity to the wild-type protein after the addition of 2-oxo acids (Fig. 4). However, the basal activity of both mutant proteins was increased by a factor of 2 after substitution of CysII or PheIII by Glu (Figs. 2 and 4), which suggests that Glu can possibly mimic the formation of a thiohemiacetal at the position of CysII or PheIII in AOX1C. However, since AOX1C and AOX1D wild-type proteins possess Phe and Leu, respectively, at position III (Fig. 1; Supplemental Fig. S1), the formation of a thiohemiacetal at this position is not possible in the wild-type proteins.
AOX1A, But Not AOX1C and AOX1D, Is Specifically Activated by Succinate after Substitution of CysI by Ser
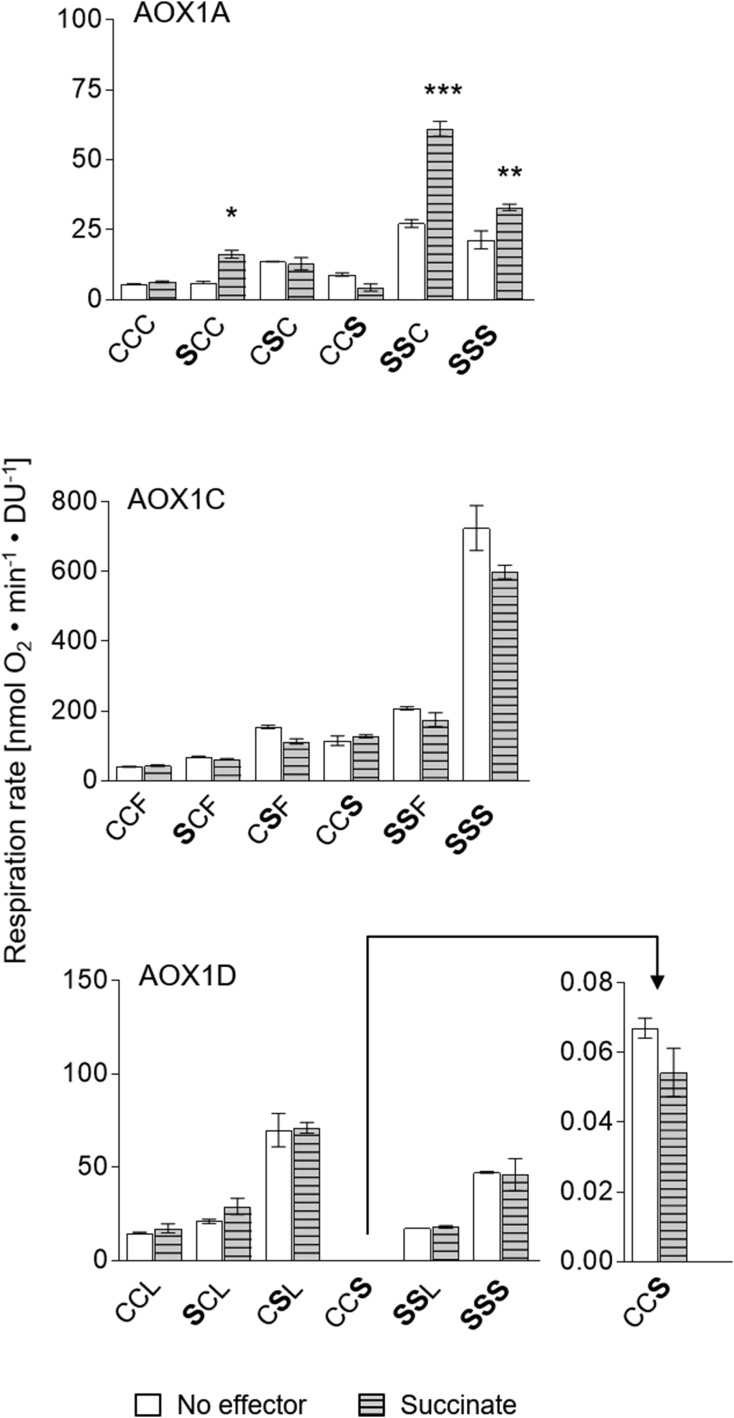
Djajanegara et al. (1999) and Umbach et al. (2002) showed that the dicarboxylic acid succinate activates AOX1A from Arabidopsis after substitution of CysI by Ser or Ala, while the wild-type protein is insensitive to succinate. Other plant species, such as tomato, lotus, rice, and maize, naturally possess AOX isoforms containing a Ser residue at the position of CysI from Arabidopsis (Fig. 1; Supplemental Fig. S1). These AOX isoforms were shown to be insensitive to 2-oxo acids but stimulated by succinate (Ito et al., 1997; Karpova et al., 2002; Holtzapffel et al., 2003; Grant et al., 2009). Consequently, various single-, double-, and triple-substituted derivatives of AOX1A, AOX1C, and AOX1D were systematically analyzed for their sensitivity toward succinate (Fig. 5).
Figure 5.
Influence of the dicarboxylic acid succinate on AOX activities. The impact of the dicarboxylic acid succinate on AOX1A, AOX1C, and AOX1D wild-type proteins and their corresponding Cys substitutions was analyzed. Assays of AOX activity were performed as described in the legend for Figure 2. Measurements were carried out as technical duplicates or triplicates. Asterisks indicate that the differences (*, P < 0.05; **, P < 0.01; and ***, P < 0.001) between the basal activity (no effector) and activities in the presence of the effector succinate are statistically significant as determined by two-way ANOVA with posthoc Tukey’s HSD test. Wild types are as follows: AOX1A, CCC; AOX1C, CCF; and AOX1D, CCL. Substitutions are presented in the one-letter code for amino acids in enlarged boldface letters. DU, Density units.
Various single-, double-, or triple-substituted AOX1A proteins possessing a Ser residue at the position of CysI (AOX1A-SCC, -SSC, -SSS, -SSA, and -SSK) were activated by succinate, while these mutant proteins could not be stimulated by 2-oxo acids (Fig. 5; Supplemental Fig. S2). In contrast, the AOX1A-CCC wild-type protein as well as AOX1A-CSC and -CCS derivatives were insensitive to succinate but activated by pyruvate or glyoxylate (Figs. 3 and 5). In addition, the presence of a Glu residue at position II and/or position III prevented succinate activation (Supplemental Fig. S2), probably due to repulsion by the additional negative charges generated by the presence of a Glu residue and the simultaneous addition of the dicarboxylic acid succinate. Although the activity of AOX1A-SCC, -SSC, -SSS, -SSA, and -SSK was increased significantly by succinate (2-fold), it has to be mentioned that the activity of these substituted proteins was not as sensitive toward succinate as the AOX1A wild-type protein (CCC) to 2-oxo acids (6- to 7-fold increase in activity; compare Figs. 2 and 5). However, the AOX1A-ACC protein also was activatable by succinate (Supplemental Fig. S2), whereas some AOX1A mutants (AOX1A-SSE, -SEE, and -SEC) remained insensitive to succinate even though a Ser residue was present at position I (Supplemental Fig. S2). Succinate activation, therefore, seems to be dependent on the protein structure and/or the amino acid environment in close proximity to CysI. Nevertheless, these results again strongly support the view that CysI is the primary site of metabolite activation.
In contrast to AOX1A, the isoforms AOX1C and AOX1D were insensitive to succinate after substitution of CysI and/or CysII (AOX1C-SCF, -CSF, and -SSF and AOX1D-SCL, -CSL, and -SSL; Fig. 5; Supplemental Figs. S3 and S4) as well as after substitution of the amino acid at position III (-CCS and -SSS) by Ser (Fig. 5). These data demonstrate that the amino acid environment and/or the protein structure in close vicinity to CysI differs between AOX isoforms, thus enabling the specific activation of AOX1A-SCC, -SSS, -SSA, -SSK, and -ACC by succinate, while AOX1C and AOX1D as well as the correspondingly substituted derivatives are insensitive to this effector.
DISCUSSION AND CONCLUSION
The posttranslational regulation of AOX was first reported in 1993 (Millar et al., 1993; Umbach and Siedow, 1993), and specific studies on AOX1A from Arabidopsis showed that this enzyme is posttranslationally activated by a disulfide/thiol switch and by the addition of 2-oxo acids such as the metabolic intermediates pyruvate and glyoxylate. It was postulated that this activation occurs via the formation of a thiohemiacetal between CysI and 2-oxo acids (Rhoads et al., 1998; Umbach et al., 2002, 2006). Prior to this study, little was known about the posttranslational activation of other AOX isoforms of Arabidopsis. AOX1C transcripts are present (but at lower abundance) in the same tissues and developmental stages as AOX1A. AOX1D expression is increased in aox1a knockout mutants from Arabidopsis (especially after restriction of the cytochrome c respiratory pathway). Yet, as shown by Strodtkötter et al. (2009) and Kühn et al. (2015), AOX1D only partially compensates for the lack of AOX1A (Clifton et al., 2006; Strodtkötter et al., 2009; Kühn et al., 2015). This suggests that differences in posttranslational activation of AOX isoforms are more likely to occur than differences in transcriptional regulation. Therefore, we focused on the analysis of AOX1A regulation as well as the activation of AOX1C and AOX1D and their corresponding Cys substitution derivatives with the aim to define the activation mechanisms of the different AOX isoforms.
AOX activity depends on the mitochondrial redox state (Vanlerberghe et al., 1995), which determines the level of reduced and active AOX protein. The activity of reduced AOX, in turn, depends on carbon metabolism and the concomitant level of AOX effectors such as pyruvate and glyoxylate (Millar et al., 1993, 1996; Umbach and Siedow, 1993, 1996; Day and Wiskich, 1995; Day et al., 1995; Rhoads et al., 1998; Siedow and Umbach, 2000; Umbach et al., 2006; Selinski et al., 2016). In vivo, the activity of AOX does not seem to be related directly to protein abundance. The basal activity of AOX1A and AOX1C wild-type proteins seems to be kept inherently low, because various substitutions lead to significant increases in basal activities (Fig. 2). This indicates that substitutions in AOX1A and AOX1C do not disturb the stability of the enzyme per se. In contrast, the same substitutions lead to a decrease in the basal activity of AOX1D (Fig. 2), possibly due to a poor accessibility of the substrate to the catalytic center produced by conformational changes or due to an instability in conformation (at least in close proximity to the catalytic di-iron center). Although AOX1D activity is decreased significantly in most cases after introducing substitutions at the positions of CysI, CysII, and/or LeuIII, these altered proteins still can be stimulated by pyruvate and glyoxylate, indicating that these substituted derivatives are functional, albeit at a much lower level (Supplemental Fig. S4). Compared with the cytochrome c-mediated respiratory pathway, electron flow through AOX reduces ATP generation via respiration dissipating most energy as heat (Sluse and Jarmuszkiewicz, 1998; Affourtit et al., 2002). Therefore, it is important to ensure that electron flow through this wasteful energy pathway is optimized under normal growth conditions, while its activity can be fine-tuned by redox mechanisms as well as metabolites, depending on the environmental requirements (Rasmusson et al., 2009).
Substitution of CysI and/or CysII by Ser or Ala affects not only the organic acid activation of AOX but also the redox-dependent inactivation of AOX (Djajanegara et al., 1999), because the formation of an intermolecular disulfide bond between CysI of each monomer can no longer take place. In addition, the exchange of CysI and/or CysII leads to a more active protein with altered basal activity in AOX1A, AOX1C, and AOX1D (Figs. 2 and 3). In contrast to AOX1C-SCF and AOX1D-SCL, the substitution of CysI by Ser does not significantly influence the basal activity of AOX1A-SCC, although AOX1A was shown to be activated at CysI (Millar et al., 1993; Umbach and Siedow, 1993, 1996; Rhoads et al., 1998; Umbach et al., 2006; Selinski et al., 2016). This observation may lead to the assumption that an ester bond is formed between Ser at position I and succinate. However, we assume that this mechanism does not occur, because the AOX1A-ACC derivative also is sensitive to succinate (Supplemental Fig. S2), although Ala does not provide a functional group that is needed for the formation of a hemiacetal. Therefore, the detailed mechanism of succinate activation remains elusive. However, due to the fact that various AOX1C and AOX1D mutants remain insensitive to succinate after the substitution of CysI by Ser or Ala, we postulate that the amino acid environment around CysI, which differs between AOX isoforms, determines the mode of activation by various effectors. This was also suggested by Holtzapffel et al. (2003), who pointed out that the hydroxyl group of the Ser residue is not sufficiently reactive to form a strong bond with the succinate molecule. Therefore, to stabilize its interaction with the enzyme, other amino acids in close vicinity must play a role. These other residues also may be important in relation to interactions with other organic acids. It should be noted that both groups, Holtzapffel et al. (2003) and Grant et al. (2009), have shown that the activating effect of succinate on oxygen consumption is not due to succinate serving as an electron donor for the electron transport chain via complex II, because AOX activation by succinate occurs even in the presence of malonate, an inhibitor of succinate dehydrogenase.
Umbach et al. (2002, 2006) first reported that AOX1A is activatable by 2-oxo acids at CysI and CysII. However, CysI appeared to be the primary site of activation. Our results confirm this. AOX1A and AOX1C, carrying double and triple substitutions at the positions of the conserved Cys residues, exhibit the highest basal activities of all corresponding Cys variants tested (Fig. 2; Supplemental Figs. S2–S4), whereas that is not the case for AOX1D derivatives. For instance, AOX1A-EEC is the most active derivative compared with wild-type, single-, and triple-substituted proteins of AOX1A. This suggests that the introduced negative charges (in this case Glu) present at both positions, CysI and CysII, can mimic the formation of a thiohemiacetal with pyruvate and/or glyoxylate, leading to a more active protein. Furthermore, it could be expected that both effectors bind simultaneously to both CysI and CysII, a mechanism that enables a further increase of AOX1A activity. When both effectors were added sequentially (either pyruvate followed by glyoxylate or vice versa) to the AOX1A wild-type protein (AOX1A-CCC), AOX1A-SCC, or AOX1A-ACC, the activity was maintained at the same level as after the addition of pyruvate or glyoxylate alone (data not shown). This was also the case for AOX1C-CCF, -SCF, and -ACF as well as for AOX1D-CCL, -SCL, and -ACL (data not shown). An additive effect of the two effectors on AOX activity could not be detected. Apparently, both activation sites for pyruvate and glyoxylate, at CysI and CysII, respectively, were already occupied by the initially added effector. Only in the case of differential affinities of CysI and CysII for pyruvate or glyoxylate could an additive effect of both effectors be expected. Umbach et al. (2006) hypothesized that glyoxylate interacts with both CysI and CysII, while pyruvate activation is limited to CysI in AOX1A. However, we could not observe those differences in pyruvate or glyoxylate activation for AOX1A, AOX1C, and AOX1D (Fig. 3; Supplemental Figs. S2–S4).
The different substitutions suggest differences in the structure between AOX isoforms. In AOX1A, CysI is known to be involved in inactivation of the enzyme complex under oxidizing conditions by the formation of a disulfide-linked homodimer (Rhoads et al., 1998; Umbach et al., 2002, 2006). It is likely that the N-terminal region plays an important role in the mode of activation by various effectors, leading to differential fine-regulation by metabolites, as shown for AOX1A-SCC compared with AOX1C-SCF and AOX1D-SCL (Fig. 5). Furthermore, CysI seems to be the primary site of metabolite activation in all three AOX isoforms, because pyruvate and glyoxylate activation is largely reduced after its substitution, while CysII substitutions have a smaller influence on the activation by these effectors (Fig. 3; Supplemental Figs. S2–S4). CysII seems to form the center of a second independent region of activation, which plays a subordinate role in 2-oxo acid activation compared with CysI. This conclusion is supported by the results obtained with the various AOX derivatives (Figs. 2–4) and by other studies of constitutively active AOX1A proteins highlighting these regions (Rhoads et al., 1998; Umbach et al., 1998, 2002, 2006). AOX1A-ECC and -CEC mutants possess increased basal activities, indicating that a negative charge (in this case Glu) present at both positions can mimic the formation of a thiohemiacetal, leading to a more active protein and, furthermore, showing additive effects in AOX1A-EEC (Fig. 2).
Our study supports the notion that the amino acid composition around CysIII of AOX1A (PheIII for AOX1C and LeuIII for AOX1D), which was not included in former studies, also plays a role in enzyme activation, although AOX activation by metabolites seems to be independent of the specific amino acid present at this position (Figs. 1 and 3). A positive or negative charge introduced at CysIII (AOX1A-CCE or -CCK) did not affect AOX1A activity (Fig. 2). In contrast, AOX1C mutant proteins containing a Glu residue at position II or III (AOX1C-CEF or -CCE) showed increased basal activities. Negative charges introduced at position II or III might, therefore, lead to conformational changes of the second α-helix of AOX1C (Fig. 1; Supplemental Fig. S1), resulting in a higher accessibility of the catalytic di-iron center of the protein or in a decrease in interaction between both AOX1C monomers, yielding, in both cases, a more active protein. On the other hand, substitution at position II (CysII) or III (LeuIII) in AOX1D had the reverse effect (Fig. 2), leading to fewer interactions between both monomers or to a less accessible active center.
Overall, our analyses point to an AOX homodimer that has a low basal activity but can be activated by substitutions and/or effectors, especially at CysI and CysII, to increase enzyme activity. The detailed mechanisms remain elusive, but all results support the view that allosteric effects within the neighboring regions act independently and are not exactly limited to CysI, CysII, and CysIII (PheIII for AOX1C and LeuIII for AOX1D) and, therefore, are additive when combined, to achieve enhanced AOX activity under a variety of different conditions that require an increased flux through the alternative pathway.
However, a simple model for activation cannot be deduced. While a substitution of CysI by Glu activates AOX, as previously proposed to occur via the formation of a thiohemiacetal, Ala substitution at CysI results in a greater activation, although Ala cannot form a thiohemiacetal mimicking the carboxylate charge as does the substitution with Glu. This also has been observed in tobacco for AOX1A, where substitution of Cys-126 (equivalent to CysI) with Ala resulted in a highly active AOX in suspension cells but not in isolated mitochondria (Vanlerberghe et al., 1998). Furthermore, glyoxylate activates as efficiently as pyruvate, and a thiohemiacetal formed with glyoxylate would produce an alcoholic carbon at CysI and would be sterically similar to a Ser or Thr residue at that position. A Ser residue at CysI does not lead to activation by pyruvate or glyoxylate but instead by succinate. Thiohemiacetals are most commonly observed at the active/catalytic site in enzymes such as plant betaine aldehyde dehydrogenase (Zárate-Romero et al., 2016) or human UDP-Glc 6-dehydrogenase (Egger et al., 2012) and are transient in the catalytic cycle, as in the case of glyceraldehyde 3-phosphate dehydrogenase. To date, we cannot detect the formation of a thiohemiacetal by mass spectrometry in AOX, although it was detected in crystal structures of betaine aldehyde dehydrogenase (Zárate-Romero et al., 2016) or human UDP-Glc 6-dehydrogenase (Egger et al., 2012), but at present, crystal structures are not available for plant AOX proteins. Based on the results presented in this study, it is proposed that the activation of AOX can occur through any alteration that leads to an improved accessibility of the prosthetic group for its substrate oxygen within the catalytic site. This can involve the formation of a thiohemiacetal, as proposed previously (Rhoads et al., 1998; Umbach et al., 1998, 2002, 2006), but other alterations also can induce changes in the tertiary and quaternary structures of AOX to achieve the same, or an increased, effect on the activity of AOX proteins. Importantly, the degree of activation at CysI or CysII via thiohemiacetal formation (if it occurs) is dependent on the nature of other amino acids in the close vicinity of the Cys residues. An important consequence of this model for activation is that the three AOX1 isoforms can be activated differentially in vivo, influenced by the type of effector and the amino acids differing between the three isoforms. This molecular analysis of in vitro activities is consistent with in vivo studies in five different plant species, concluding that species-specific differences in alternative oxidase activity are due to differential posttranslational regulation of AOX (Florez-Sarasa et al., 2016). Reduced Cys residues in AOX appear to act as a restraint, and its substitutions simulate activation by organic acids. After modification/substitution, an activated enzyme is stably established and cannot easily be reoxidized under the stressful conditions when its activity is required.
MATERIALS AND METHODS
RNA Extraction and Reverse Transcription
Arabidopsis (Arabidopsis thaliana) ecotype Columbia plants were cultivated for 6 weeks in a growth chamber in soil under short-day conditions with 7.5 h of light and a light intensity of 150 µmol m−2 s−1 at 20°C. Total RNA was isolated from 100 mg of frozen leaf material using TRI-Reagent (RNA-DNA-protein isolation reagent; Applied Biosystems) as described in the protocol of the supplier. According to the manufacturer’s instructions (Macherey-Nagel), a DNase digest was performed to remove genomic DNA after RNA isolation. From 5 µg of total RNA, cDNA was synthesized using oligo(dT) as primers provided with the kit, according to the instructions of the supplier (RevertAid First Strand cDNA Synthesis Kit; Fermentas).
Construction of Plasmids and Site-Directed Mutagenesis of AOX Isoforms
Construction of plasmids p537 and p583 was carried out according to Selinski et al. (2016). For cloning of the mature wild-type AOX1C (At3g27620) and AOX1D (At1g32350), transit peptides were identified using MITOPROT (Claros and Vincens, 1996). Transit peptides were determined to comprise 44 codons and 40 codons in the 5′ region of the coding sequence for AOX1C and AOX1D, respectively. For the amplification of mature AOX1C and AOX1D, the primers AOX1C for or AOX1D for (including an NdeI site) and AOX1C rev or AOX1D rev (including an EcoRI site; Supplemental Table S1) were used, which amplify AOX1C starting at codon 45 and AOX1D at codon 41. After PCR amplification, fragments were ligated into blunt-restricted vector pJET1.2, according to the instructions of the supplier (CloneJET PCR Cloning Kit; Thermo Fisher Scientific). After NdeI/EcoRI restriction, fragments (AOX1C, 865 bp; AOX1D, 844 bp) were introduced into correspondingly restricted vector pET-22b (+) (Novagen), leading to plasmids p583 (AOX1C in pET-22b) and p537 (AOX1D in pET-22b). Constructs were verified by sequencing through the ligation sites.
The substitutions of Cys residues at positions 64 (CysI), 114 (CysII), and/or 136 (CysIII) of mature AOX1A (formerly Cys-78, Cys-128, and Cys-150; Rhoads et al., 1998; Umbach et al., 1998, 2002), residues at positions 47 (CysI), 97 (CysII), and/or 119 (Phe) of mature AOX1C, and residues at positions 41 (CysI), 91 (CysII), and/or 113 (Leu) of mature AOX1D were carried out via site-directed mutagenesis. The new numbering of Cys residues in AOX1A is due to differences in length of the transit peptide (14 amino acids [Kumar and Söll, 1992] versus 62 amino acids [The Arabidopsis Information Resource; Selinski et al., 2016]) and to a differential specification of the starting point, counting the amino acid residues in front of the first Cys residue (Cys-78 includes the transit peptide [Kumar and Söll, 1992] and Cys-64 excludes the transit peptide of AOX1A). However, both genes start at the same codon encoding an Ala residue (Kumar and Söll, 1992; Selinski et al., 2016). For mutagenesis, plasmids p536 (AOX1A in pET-22b; Selinski et al., 2016), p537, and p583 were amplified via PCR using PfuUltra II Fusion HS DNA-Polymerase (Agilent Technologies) and specific mutagenesis primers for amplification (Supplemental Table S1). Following PCR, products were treated for 1 h at 37°C with DpnI to eliminate the maternal DNA template. Constructs were verified by sequencing.
Activity Measurements of Recombinantly Expressed Alternative Oxidases
Cell growth, protein expression, Escherichia coli membrane vesicle isolation, and alternative oxidase activity measurements with concomitant immunoblot analysis and calculations were carried out according to Selinski et al. (2016) with minor modifications. Briefly, electrocompetent cells of E. coli strain BHH8 lacking internal cytochrome bo and bd-I oxidases (Selinski et al., 2016) were transformed with plasmids p536 (AOX1A in pET-22b; At3g22370; Selinski et al., 2016), p537, p583, or their corresponding derivatives and, in each case, with pT7Pol26 (Mertens et al., 1995) as a second plasmid. Cells were aerobically grown in Luria-Bertani medium with ampicillin (100 μg mL−1) and kanamycin (50 μg mL−1) at 37°C to an optical density of 0.4 to 0.6. After induction of AOX expression for 1 h with 0.3 mm isopropyl-β-d-1-thiogalactopyranoside, cells were harvested, frozen in liquid nitrogen, and stored at −80°C. Inverted membrane vesicles were prepared at 4°C according to Brandt et al. (2013). Isolated membranes were resuspended in 300 μL of TMG buffer (50 mm Tris-HCl [pH 7.5], 10 mm MgCl2, and 10% [v/v] glycerol), and the concentration of the membrane protein was determined using a small-scale approach of the bicinchoninic acid assay according to the protocol of the supplier (Pierce).
Freshly prepared inverted membrane vesicles were used directly (without freezing) for AOX activity measurements, which were performed in NKM buffer (150 mm NaCl, 50 mm potassium phosphate buffer, pH 7, 10 mm KCl, and 5 mm MgCl2). Oxygen consumption was measured at a constant temperature of 25°C and a stirrer speed of 65 rpm using a Clark-type oxygen electrode (Clark, 1956). After reaching a linear rate, an incubation step of 5 min started after adding 10 µL of inverted membrane vesicles and 1 mm potassium cyanide. Within the first 30 s of incubation, 5 mm DTT and 5 mm of the appropriate effector (pyruvate, glyoxylate, or succinate) were added. After this incubation step, 5 mm NADH was added to start respiratory oxygen consumption. After 2 min within the linear range, 0.75 mm salicylhydroxamic acid was supplied into the reaction chamber to inhibit respiration by AOX. To specifically determine the content of AOX protein present in each sample of inverted membrane vesicles, immunoblot analyses were carried out using polyclonal antibodies raised against the conserved C-terminal consensus motif from plant AOX isoforms (AOX 1/2; Agrisera; host, rabbit) and a secondary antibody [goat anti-rabbit IgG, Fc (clone: pAb)-Cy3; Dianova] for detection. Calculations of specific AOX activities were carried out as described in detail by Selinski et al. (2016). The specific activities of AOX isoforms were defined as nanomoles of oxygen consumed per minute and AOX density units instead of milligrams of membrane vesicle protein to compensate for small changes in the amount of AOX protein present at the E. coli membrane, mostly due to variations in the induced expression of the differently mutated aox genes.
Statistical Analysis
Statistical evaluations were conducted by means of the two-way ANOVA with posthoc Tukey’s HSD test integrated in GraphPad Prism 7 (GraphPad Software). Differences with P < 0.05, P < 0.01, and P < 0.001 were considered as significant and indicated as *, **, and ***, respectively.
Accession Numbers
Sequence data for the genes from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following numbers: AT3G22370 (AOX1A), AT3G27620 (AOX1C), and AT1G32350 (AOX1D).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Alignment of AOX isoforms.
Supplemental Figure S2. Overview of AOX1A activities, various Cys mutants, and the effect of 2-oxo acids.
Supplemental Figure S3. Overview of AOX1C activities, various Cys mutants, and the effect of 2-oxo acids.
Supplemental Figure S4. Overview of AOX1D activities, various Cys mutants, and the effect of 2-oxo acids.
Supplemental Table S1. Primers used in this study.
Acknowledgments
We thank Brigitte Herkenhoff-Hesselmann for expert technical assistance, Saskia Höfler and Marco Krüger for experimental support, and David A. Day for critical reading of the article.
Glossary
- HSD
honestly significant difference
Footnotes
This work was supported by the Collaborative Research Center 944 ‘Physiology and Dynamics of Cellular Microcompartments’ (to R.S. and G.D.-H.), the government of Lower Saxonia (Lichtenberg fellowship to J.S.), the Frauenförderpool of Osnabrück University (to J.S.), and the Australian Research Council Centre of Excellence Program in Plant Energy Biology (grant no. CE140100008 to J.W.).
Articles can be viewed without a subscription.
References
- Affourtit C, Albury MS, Crichton PG, Moore AL (2002) Exploring the molecular nature of alternative oxidase regulation and catalysis. FEBS Lett 510: 121–126 [DOI] [PubMed] [Google Scholar]
- Alexandre A, Lehninger AL (1984) Bypasses of the antimycin a block of mitochondrial electron transport in relation to ubisemiquinone function. Biochim Biophys Acta 767: 120–129 [DOI] [PubMed] [Google Scholar]
- Aro EM, Virgin I, Andersson B (1993) Photoinhibition of photosystem II: inactivation, protein damage and turnover. Biochim Biophys Acta 1143: 113–134 [DOI] [PubMed] [Google Scholar]
- Brandt K, Maiwald S, Herkenhoff-Hesselmann B, Gnirß K, Greie JC, Dunn SD, Deckers-Hebestreit G (2013) Individual interactions of the b subunits within the stator of the Escherichia coli ATP synthase. J Biol Chem 288: 24465–24479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo ML, Kinnally KW, Tedeschi H (1992) The effect of antimycin A on mouse liver inner mitochondrial membrane channel activity. J Biol Chem 267: 8123–8127 [PubMed] [Google Scholar]
- Clark LC. (1956) Electrochemical device for chemical analysis. US Patent Application No. 2913386
- Claros MG, Vincens P (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 241: 779–786 [DOI] [PubMed] [Google Scholar]
- Clifton R, Millar AH, Whelan J (2006) Alternative oxidases in Arabidopsis: a comparative analysis of differential expression in the gene family provides new insights into function of non-phosphorylating bypasses. Biochim Biophys Acta 1757: 730–741 [DOI] [PubMed] [Google Scholar]
- Dahal K, Martyn GD, Vanlerberghe GC (2015) Improved photosynthetic performance during severe drought in Nicotiana tabacum overexpressing a nonenergy conserving respiratory electron sink. New Phytol 208: 382–395 [DOI] [PubMed] [Google Scholar]
- Dahal K, Wang J, Martyn GD, Rahimy F, Vanlerberghe GC (2014) Mitochondrial alternative oxidase maintains respiration and preserves photosynthetic capacity during moderate drought in Nicotiana tabacum. Plant Physiol 166: 1560–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day DA, Whelan J, Millar AH, Siedow JN, Wiskich JT (1995) Regulation of the alternative oxidase in plants and fungi. Aust J Plant Physiol 22: 497–509 [Google Scholar]
- Day DA, Wiskich JT (1995) Regulation of alternative oxidase activity in higher plants. J Bioenerg Biomembr 27: 379–385 [DOI] [PubMed] [Google Scholar]
- Djajanegara I, Holtzapffel R, Finnegan PM, Hoefnagel MHN, Berthold DA, Wiskich JT, Day DA (1999) A single amino acid change in the plant alternative oxidase alters the specificity of organic acid activation. FEBS Lett 454: 220–224 [DOI] [PubMed] [Google Scholar]
- Egger S, Chaikuad A, Klimacek M, Kavanagh KL, Oppermann U, Nidetzky B (2012) Structural and kinetic evidence that catalytic reaction of human UDP-glucose 6-dehydrogenase involves covalent thiohemiacetal and thioester enzyme intermediates. J Biol Chem 287: 2119–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Guan D, Sun K, Wang Y, Zhang T, Wang R (2013) Expression and signal regulation of the alternative oxidase genes under abiotic stresses. Acta Biochim Biophys Sin (Shanghai) 45: 985–994 [DOI] [PubMed] [Google Scholar]
- Finnegan PM, Soole KL, Umbach AL (2004) Alternative mitochondrial electron transport proteins in higher plants. In DA Day, AH Millar, J Whelan, eds, Plant Mitochondria: From Genome to Function. Springer, Dordrecht, The Netherlands, pp 163–230 [Google Scholar]
- Florez-Sarasa I, Flexas J, Rasmusson AG, Umbach AL, Siedow JN, Ribas-Carbo M (2011) In vivo cytochrome and alternative pathway respiration in leaves of Arabidopsis thaliana plants with altered alternative oxidase under different light conditions. Plant Cell Environ 34: 1373–1383 [DOI] [PubMed] [Google Scholar]
- Florez-Sarasa I, Ribas-Carbo M, Del-Saz NF, Schwahn K, Nikoloski Z, Fernie AR, Flexas J (2016) Unravelling the in vivo regulation and metabolic role of the alternative oxidase pathway in C3 species under photoinhibitory conditions. New Phytol 212: 66–79 [DOI] [PubMed] [Google Scholar]
- Fu A, Liu H, Yu F, Kambakam S, Luan S, Rodermel S (2012) Alternative oxidases (AOX1a and AOX2) can functionally substitute for plastid terminal oxidase in Arabidopsis chloroplasts. Plant Cell 24: 1579–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant N, Onda Y, Kakizaki Y, Ito K, Watling J, Robinson S (2009) Two Cys or not two Cys? That is the question. Alternative oxidase in the thermogenic plant sacred lotus. Plant Physiol 150: 987–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Cai Z, Gan S (2004) Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ 27: 521–549 [Google Scholar]
- Hanke GT, Hase T (2008) Variable photosynthetic roles of two leaf-type ferredoxins in Arabidopsis, as revealed by RNA interference. Photochem Photobiol 84: 1302–1309 [DOI] [PubMed] [Google Scholar]
- Holtzapffel RC, Castelli J, Finnegan PM, Millar AH, Whelan J, Day DA (2003) A tomato alternative oxidase protein with altered regulatory properties. Biochim Biophys Acta 1606: 153–162 [DOI] [PubMed] [Google Scholar]
- Ito Y, Saisho D, Nakazono M, Tsutsumi N, Hirai A (1997) Transcript levels of tandem-arranged alternative oxidase genes in rice are increased by low temperature. Gene 203: 121–129 [DOI] [PubMed] [Google Scholar]
- Juszczuk IM, Rychter AM (2003) Alternative oxidase in higher plants. Acta Biochim Pol 50: 1257–1271 [PubMed] [Google Scholar]
- Karpova OV, Kuzmin EV, Elthon TE, Newton KJ (2002) Differential expression of alternative oxidase genes in maize mitochondrial mutants. Plant Cell 14: 3271–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn K, Yin G, Duncan O, Law SR, Kubiszewski-Jakubiak S, Kaur P, Meyer E, Wang Y, Small CC, Giraud E, et al. (2015) Decreasing electron flux through the cytochrome and/or alternative respiratory pathways triggers common and distinct cellular responses dependent on growth conditions. Plant Physiol 167: 228–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar AM, Söll D (1992) Arabidopsis alternative oxidase sustains Escherichia coli respiration. Proc Natl Acad Sci USA 89: 10842–10846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire JJ, Kagan VE, Packer L (1992) Electron transport between cytochrome c and alpha tocopherol. Biochem Biophys Res Commun 188: 190–197 [DOI] [PubMed] [Google Scholar]
- McDonald AE, Vanlerberghe GC (2006) Origins, evolutionary history, and taxonomic distribution of alternative oxidase and plastoquinol terminal oxidase. Comp Biochem Physiol Part D Genomics Proteomics 1: 357–364 [DOI] [PubMed] [Google Scholar]
- Mertens N, Remaut E, Fiers W (1995) Tight transcriptional control mechanism ensures stable high-level expression from T7 promoter-based expression plasmids. Biotechnology (N Y) 13: 175–179 [DOI] [PubMed] [Google Scholar]
- Millar AH, Hoefnagel M, Day DA, Wiskich JT (1996) Specificity of the organic acid activation of alternative oxidase in plant mitochondria. Plant Physiol 111: 613–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Wiskich JT, Whelan J, Day DA (1993) Organic acid activation of the alternative oxidase of plant mitochondria. FEBS Lett 329: 259–262 [DOI] [PubMed] [Google Scholar]
- Millenaar FF, Lambers H (2003) The alternative oxidase: in vivo regulation and function. Plant Biol 5: 2–15 [Google Scholar]
- Møller IM. (2001) Plant mitochondria and oxidative stress: electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu Rev Plant Physiol Plant Mol Biol 52: 561–591 [DOI] [PubMed] [Google Scholar]
- Møller IM, Rasmusson AG (1998) The role of NADP in the mitochondrial matrix. Trends Plant Sci 3: 21–27 [Google Scholar]
- Moore AL, Albury MS (2008) Further insights into the structure of the alternative oxidase: from plants to parasites. Biochem Soc Trans 36: 1022–1026 [DOI] [PubMed] [Google Scholar]
- Pham NA, Robinson BH, Hedley DW (2000) Simultaneous detection of mitochondrial respiratory chain activity and reactive oxygen in digitonin-permeabilized cells using flow cytometry. Cytometry 41: 245–251 [DOI] [PubMed] [Google Scholar]
- Polidoros AN, Mylona PV, Arnholdt-Schmitt B (2009) Aox gene structure, transcript variation and expression in plants. Physiol Plant 137: 342–353 [DOI] [PubMed] [Google Scholar]
- Pu XJ, Lv X, Lin HH (2015) Unraveling the evolution and regulation of the alternative oxidase gene family in plants. Dev Genes Evol 225: 331–339 [DOI] [PubMed] [Google Scholar]
- Rasmusson AG, Fernie AR, van Dongen JT (2009) Alternative oxidase: a defence against metabolic fluctuations? Physiol Plant 137: 371–382 [DOI] [PubMed] [Google Scholar]
- Rasmusson AG, Soole KL, Elthon TE (2004) Alternative NAD(P)H dehydrogenases of plant mitochondria. Annu Rev Plant Biol 55: 23–39 [DOI] [PubMed] [Google Scholar]
- Rhoads DM, Umbach AL, Sweet CR, Lennon AM, Rauch GS, Siedow JN (1998) Regulation of the cyanide-resistant alternative oxidase of plant mitochondria: identification of the cysteine residue involved in alpha-keto acid stimulation and intersubunit disulfide bond formation. J Biol Chem 273: 30750–30756 [DOI] [PubMed] [Google Scholar]
- Selinski J, Hartmann A, Höfler S, Deckers-Hebestreit G, Scheibe R (2016) Refined method to study the posttranslational regulation of alternative oxidases from Arabidopsis thaliana in vitro. Physiol Plant 157: 264–279 [DOI] [PubMed] [Google Scholar]
- Shikanai T. (2007) Cyclic electron transport around photosystem I: genetic approaches. Annu Rev Plant Biol 58: 199–217 [DOI] [PubMed] [Google Scholar]
- Siedow JN, Umbach AL (2000) The mitochondrial cyanide-resistant oxidase: structural conservation amid regulatory diversity. Biochim Biophys Acta 1459: 432–439 [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluse FE, Jarmuszkiewicz W (1998) Alternative oxidase in the branched mitochondrial respiratory network: an overview on structure, function, regulation, and role. Braz J Med Biol Res 31: 733–747 [DOI] [PubMed] [Google Scholar]
- Stenmark P, Nordlund P (2003) A prokaryotic alternative oxidase present in the bacterium Novosphingobium aromaticivorans. FEBS Lett 552: 189–192 [DOI] [PubMed] [Google Scholar]
- Strodtkötter I, Padmasree K, Dinakar C, Speth B, Niazi PS, Wojtera J, Voss I, Do PT, Nunes-Nesi A, Fernie AR, et al. (2009) Induction of the AOX1D isoform of alternative oxidase in A. thaliana T-DNA insertion lines lacking isoform AOX1A is insufficient to optimize photosynthesis when treated with antimycin A. Mol Plant 2: 284–297 [DOI] [PubMed] [Google Scholar]
- Suzuki N, Koussevitzky S, Mittler R, Miller G (2012) ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ 35: 259–270 [DOI] [PubMed] [Google Scholar]
- Umbach AL, Gonzàlez-Meler MA, Sweet CR, Siedow JN (2002) Activation of the plant mitochondrial alternative oxidase: insights from site-directed mutagenesis. Biochim Biophys Acta 1554: 118–128 [DOI] [PubMed] [Google Scholar]
- Umbach AL, Ng VS, Siedow JN (2006) Regulation of plant alternative oxidase activity: a tale of two cysteines. Biochim Biophys Acta 1757: 135–142 [DOI] [PubMed] [Google Scholar]
- Umbach AL, Rhoads DM, Sweet CR, Siedow JN (1998) Investigation of regulation of alternative oxidase activity using site-directed mutagenesis. In Møller IM, Gardeström P, Glimelius K, Glaser E, eds, Plant Mitochondria: From Gene to Function. Backhuys Publishers, Leiden, The Netherlands, pp 531–535 [Google Scholar]
- Umbach AL, Siedow JN (1993) Covalent and noncovalent dimers of the cyanide-resistant alternative oxidase protein in higher plant mitochondria and their relationship to enzyme activity. Plant Physiol 103: 845–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach AL, Siedow JN (1996) The reaction of the soybean cotyledon mitochondrial cyanide-resistant oxidase with sulfhydryl reagents suggests that alpha-keto acid activation involves the formation of a thiohemiacetal. J Biol Chem 271: 25019–25026 [DOI] [PubMed] [Google Scholar]
- Van Aken O, Giraud E, Clifton R, Whelan J (2009) Alternative oxidase: a target and regulator of stress responses. Physiol Plant 137: 354–361 [DOI] [PubMed] [Google Scholar]
- Vanlerberghe GC. (2013) Alternative oxidase: a mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. Int J Mol Sci 14: 6805–6847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC, Day DA, Wiskich JT, Vanlerberghe AE, McIntosh L (1995) Alternative oxidase activity in tobacco leaf mitochondria (dependence on tricarboxylic acid cycle-mediated redox regulation and pyruvate activation). Plant Physiol 109: 353–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC, Martyn GD, Dahal K (2016) Alternative oxidase: a respiratory electron transport chain pathway essential for maintaining photosynthetic performance during drought stress. Physiol Plant 157: 322–337 [DOI] [PubMed] [Google Scholar]
- Vanlerberghe GC, McIntosh L, Yip JYH (1998) Molecular localization of a redox-modulated process regulating plant mitochondrial electron transport. Plant Cell 10: 1551–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ (2009) Jalview version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia D, Yu CA, Kim H, Xia JZ, Kachurin AM, Zhang L, Yu L, Deisenhofer J (1997) Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science 277: 60–66 [DOI] [PubMed] [Google Scholar]
- Zárate-Romero A, Murillo-Melo DS, Mújica-Jiménez C, Montiel C, Muñoz-Clares RA (2016) Reversible, partial inactivation of plant betaine aldehyde dehydrogenase by betaine aldehyde: mechanism and possible physiological implications. Biochem J 473: 873–885 [DOI] [PubMed] [Google Scholar]
- Zarkovic J, Anderson SL, Rhoads DM (2005) A reporter gene system used to study developmental expression of alternative oxidase and isolate mitochondrial retrograde regulation mutants in Arabidopsis. Plant Mol Biol 57: 871–888 [DOI] [PubMed] [Google Scholar]