The largest Brassicaceae genomes, forming the well-supported Hesperis clade, have descended from a common ancestor with seven chromosomes and exhibit considerable karyotype and chromosome number stasis.
Abstract
Clade E, or the Hesperis clade, is one of the major Brassicaceae (Crucifereae) clades, comprising some 48 genera and 351 species classified into seven tribes and is distributed predominantly across arid and montane regions of Asia. Several taxa have socioeconomic significance, being important ornamental but also weedy and invasive species. From the comparative genomic perspective, the clade is noteworthy as it harbors species with the largest crucifer genomes but low numbers of chromosomes (n = 5–7). By applying comparative cytogenetic analysis and whole-chloroplast phylogenetics, we constructed, to our knowledge, the first partial and complete cytogenetic maps for selected representatives of clade E tribes and investigated their relationships in a family-wide context. The Hesperis clade is a well-supported monophyletic lineage comprising seven tribes: Anchonieae, Buniadeae, Chorisporeae, Dontostemoneae, Euclidieae, Hesperideae, and Shehbazieae. The clade diverged from other Brassicaceae crown-group clades during the Oligocene, followed by subsequent Miocene tribal diversifications in central/southwestern Asia. The inferred ancestral karyotype of clade E (CEK; n = 7) originated from an older n = 8 genome, which also was the purported progenitor of tribe Arabideae (KAA genome). In most taxa of clade E, the seven linkage groups of CEK either remained conserved (Chorisporeae) or were reshuffled by chromosomal translocations (Euclidieae). In 50% of Anchonieae and Hesperideae species, the CEK genome has undergone descending dysploidy toward n = 6 (−5). These genomic data elucidate early genome evolution in Brassicaceae and pave the way for future whole-genome sequencing and assembly efforts in this as yet genomically neglected group of crucifer plants.
Already, the Romans prized the dame’s rocket (Hesperis matronalis) and stocks (Matthiola incana and Matthiola longipetala) for their delightful fragrances, which develop in the late afternoon and persist long through the evening and night. However, these plants and their close relatives, classified today as members of clade E, are not only attractive for their scent but also for their large, diversely colored flowers, decorating our gardens today (Matthiola spp.) as well as mainly Asian steppes, grasslands, rocky outcrops, and sparsely vegetated screes of high mountains (e.g. Chorispora, Clausia, Hesperis, Matthiola, Parrya, Solms-laubachia, and Tchihatchewia spp.; Fig. 1). On the less attractive side, several clade E species also are regarded as noxious weeds (Chorispora tenella and Strigosella africana) and invasive elements entering naturally occurring plant communities (Bunias orientalis and H. matronalis; Francis et al., 2009, CABI, 2012). According to the Global Naturalized Alien Flora database covering 843 regions worldwide (van Kleunen et al., 2015), the two most invasive clade E species are H. matronalis, reported to be naturalized in 97 regions, and B. orientalis in 53 regions, followed by M. incana (44 regions), S. africana (28 regions), and Euclidium syriacum (19 regions).
Figure 1.
Representatives of the seven tribes of clade E. A, A bouquet of Matthiola incana (ANCH). B, Bunias orientalis (BUNI). C, Euclidium syriacum (EUCL). D, Leiospora exscapa (EUCL). E, Tetracme quadricornis (EUCL). F, Strigosella africana (EUCL). G, Hesperis tristis (HESP). H, Tchihatchewia isatidea (HESP). I, Chorispora bungeana (CHOR). J, Parrya olgae (CHOR). K, Dontostemon elegans (DONT). L, Shehbazia tibetica (SHEH). Photographs are by T. Mandáková (A), K. Schneider (B), P.E. Yevseyenkov (C), I.E. Smelyansky (D), A.L. Ebel (E), S.V. Smirnov (F), L. Hoskovec (G), E. Rencová (H), P.A. Kosachev (I and K), N. Yu Beshko (J), and Q. Lin (L). All photographs are reproduced with the permission of their authors.
According to the most recent tribal treatment of Brassicaceae (Al-Shehbaz, 2012), lineage III (Beilstein et al., 2006) or clade E (Huang et al., 2016) includes seven tribes, namely Anastaticeae (ANAS; 13 genera/65 species), Anchonieae (ANCH; 10/75), Buniadeae (BUNI; one/two), Chorisporeae (CHOR; four/55), Dontostemoneae (DONT; two/17), Euclidieae (EUCL; 28/149), and Hesperideae (HESP; two/52), plus the recently described monotypic Shehbazieae (SHEH; one/one; German and Friesen, 2014). In congruence with some previous studies (for review, see German et al., 2011), this circumscription of lineage III was not fully supported by the multigene analysis of Huang et al. (2016), due to ANAS (Lobularia maritima) being positioned outside of the monophyletic clade E or Hesperis clade of six tribes (ANCH, BUNI, CHOR, DONT, EUCL, and HESP; SHEH was not studied but should be assigned here because it represents an ancient hybrid between CHOR and DONT). ANAS consistently clustered with representatives of Biscutelleae, Cochlearieae, and Iberideae, as a newly recognized clade C (Huang et al., 2016; Guo et al., 2017). Without ANAS, the seven tribes of the Hesperis clade include 48 genera and 351 species and represent 9% of the total species diversity of the family (BrassiBase; Kiefer et al., 2014).
The Hesperis clade has a special position among all Brassicaceae lineages and clades due to its unusual, more than 30-fold variation in genome size. Whereas most Brassicaceae species possess very small genomes with a mean size of 0.62 Gb (Lysak et al., 2009), the largest genomes have been found among clade E species (Lysak et al., 2009; Kiefer et al., 2014). Crucifer genomes larger than 2 Gb are represented by species of Bunias (BUNI), Clausia (DONT), Hesperis (HESP), and Matthiola (ANCH). The largest known genome of clade E and the whole family was estimated for H. matronalis (8 Gb; 2n = 24 and 28), whereas the smallest genomes in clade E (0.26 Gb) were reported for Diptychocarpus strictus (CHOR; 2n = 14) and E. syriacum (EUCL; 2n = 14). The two smallest genomes were chosen to be sequenced within the framework of the BMAP initiative (JGI Genome Portal; accessed January 31, 2017).
Despite its genomic, phylogenetic, and ecogeographical distinctiveness within the Brassicaceae, as well as its socioeconomic importance, virtually nothing is known about the origin and genome evolution of the Hesperis clade. Therefore, to our knowledge for the first time, we investigated genome evolution in tribes assigned to clade E by comparative chromosome painting, with the aim to reconstruct its ancestral genome and elucidate the genomic processes that have shaped the origin of this lineage. Our cytogenetic analyses, along with whole-chloroplast phylogeny, support the monophyly of the Hesperis clade, allowing us to construct, to our knowledge, the first cytogenomic maps and propose an ancestral genome for the lineage. This phylogenomic analysis is an important step toward achieving a better understanding of early genome evolution in the Brassicaceae.
RESULTS
Karyotypes of Clade E Species
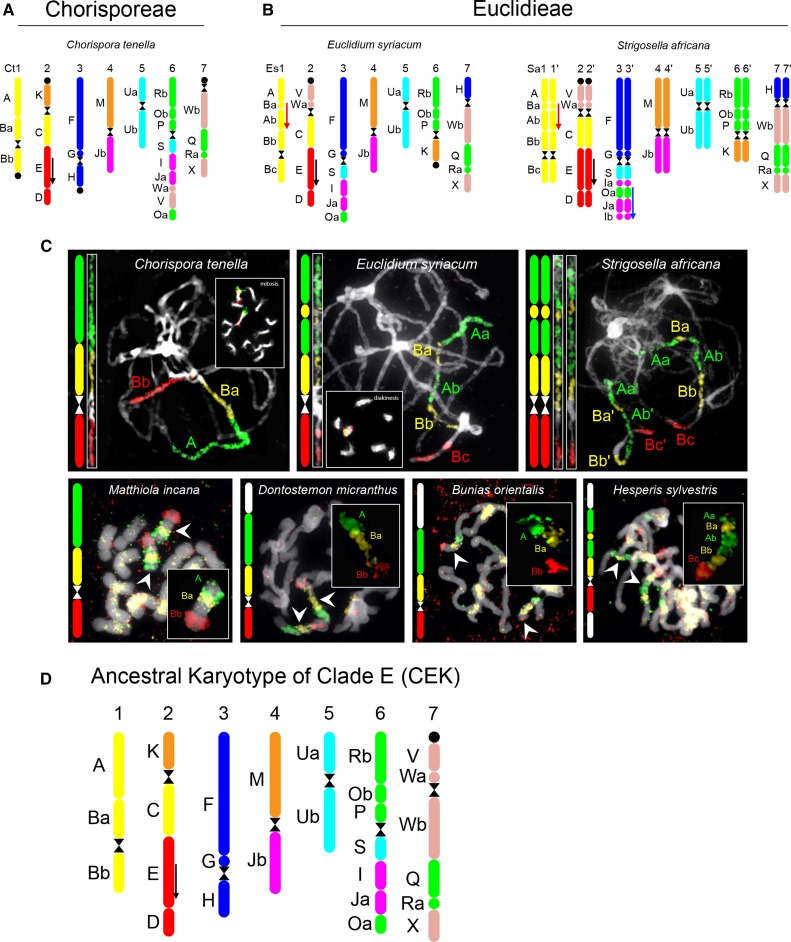
Comparative cytogenetic maps were constructed by chromosome painting for the following species: C. tenella (2n = 14; CHOR), E. syriacum (2n = 14; EUCL), and S. africana (2n = 28; EUCL; Fig. 2). The karyotypes were then compared with the reference ACK genome comprising eight chromosomes and 22 genomic blocks (GBs; Schranz et al., 2006; Lysak et al., 2016). In C. tenella, only chromosome Ct3 structurally resembled the ancestral chromosome AK3, whereas the remaining GB associations (except for D-E) were reshuffled by chromosomal rearrangements (Fig. 2A).
Figure 2.
Ideograms of the extant and ancestral genomes of clade E tribes and examples of cytogenetic analyses. A, Genome structure of C. tenella (CHOR). B, Genome structures of E. syriacum and the neotetraploid S. africana (both EUCL). Black arrows refer to the inverted collinearity of block E in relation to the ancestral crucifer karyotype (ACK); red arrows show a EUCL-specific paracentric inversion on chromosome 1; the blue arrow indicates a paracentric inversion differentiating chromosome 3 in E. syriacum and S. africana. C, Identification of genomic blocks A and B (linkage group 1) by comparative chromosome painting analysis on pachytene chromosomes (top three species) and mitotic chromosomes of seven clade E species. D, Ancestral karyotype of clade E (CEK). The color code and capital letters correspond to the eight chromosomes and 22 genomic blocks of ACK, respectively. The black circle marks the position of the 35S rDNA locus. Colors in C correspond to epifluorescence of biotin-, digoxigenin- and Cy3-labeled painting contigs. Chromosomes were counterstained by 4′,6-diamidino-2-phenylindole (DAPI).
In E. syriacum (Fig. 2B), none of its seven chromosomes retained the ACK structure; however, chromosomes Es4 and Es5 were structurally identical to Ct4 and Ct5 of C. tenella. Among the other five linkage groups, GBs on the upper arms of chromosomes Es3 and Es6 resembled the structures of Ct3 and Ct6 in C. tenella. Similarly, the upper arms of chromosomes Es2 and Es7 had the same GB composition as the bottom arms of Ct2 and Ct7 in C. tenella. Chromosome Es1 differed from its Ct1 homolog by a paracentric inversion on the upper arm (Fig. 2C).
The tetraploid genome of S. africana resembled that of Euclidium spp., with all but one homolog pair having the same structure. Chromosomes Sa3 and Sa3′ differed from the Es3 homolog by a paracentric inversion on the bottom arm (Fig. 2B).
As large-scale comparative chromosome painting (CCP) on pachytene chromosomes in ANCH, CHOR, DONT, and HESP genomes with a high repeat content was challenging (for details, see “Materials and Methods”), only the unique GB associations shared among karyotypes of Chorispora, Euclidium, and Strigosella (i.e. Ct1/Es1/Sa1, Ct4/Es4/Sa4, and Ct5/Es5/Sa5) were identified successfully on mitotic chromosomes of B. orientalis (2n = 14; BUNI), Dontostemon micranthus (2n = 14; DONT), Hesperis sylvestris (2n = 12; HESP), and M. incana (2n = 14; ANCH). CCP localization of linkage group 1 (GBs A and B) in the four species is shown in Figure 2C. Chromosome 1 in M. incana, D. micranthus, and B. orientalis resembled Ct1 in C. tenella, with the upper arm bearing GBs A and Ba and the bottom arm formed by Bb. In the two latter species, the terminal part of the upper arm (A-Ba) remained unlabeled after applying the painting probe for chromosome Ct1. In H. sylvestris, chromosome 1 was structurally similar to its homolog in EUCL species (Fig. 2B); however, its terminal parts were not painted by the probe corresponding to chromosome Es1. These findings suggest that chromosome 1 in BUNI, DONT, and HESP species participated in a taxon-specific translocation event(s). In H. sylvestris, the structure of chromosome 1 may indicate that this chromosome was formed via an insertion-like translocation event (nested chromosome insertion) responsible for the descending dysploidy from n = 7 to n = 6.
CCP with probes corresponding to homologs Ct4/Es4/Sa4 and Ct5/Es5/Sa5 did not uncover any specific chromosomal rearrangements in any of the ANCH, BUNI, DONT, and HESP species analyzed. Thus, these two chromosomes are shared by all the analyzed clade E species.
Ancestral Karyotype of Clade E
By comparing the karyotype structure of the analyzed species, we inferred a putative structure of the ancestral genome shared by all clade E tribes (Fig. 2D). The CEK genome had seven linkage groups and was structurally closest to the analyzed genome of C. tenella (Fig. 2A), whereby only a single reciprocal translocation differentiates the two genomes. Three reciprocal translocations transformed the CEK genome into the Euclidium/Strigosella karyotype (Fig. 2B; Supplemental Fig. S1).
ANAS Did Not Descend from CEK
As tribe ANAS was formerly treated as belonging to clade E, we attempted to identify CEK-specific GB associations on pachytene chromosomes of three ANAS species, namely Farsetia stylosa (2n = 20), Lobularia libyca (2n = 22), and Morettia canescens (2n = 22). However, we failed to identify three unique GB associations (i.e. A-B, M-Jb, and U) in the ANAS genomes analyzed. Instead, the three tested chromosomes of F. stylosa, L. libyca, and M. canescens exhibited ACK-derived associations of GBs (data not shown). As two genomic copies of each GB were consistently observed in haploid complements of ANAS species with the lowest known chromosome numbers for the tribe, these genomes probably originated by a whole-genome duplication event(s) not detected in clade E genomes.
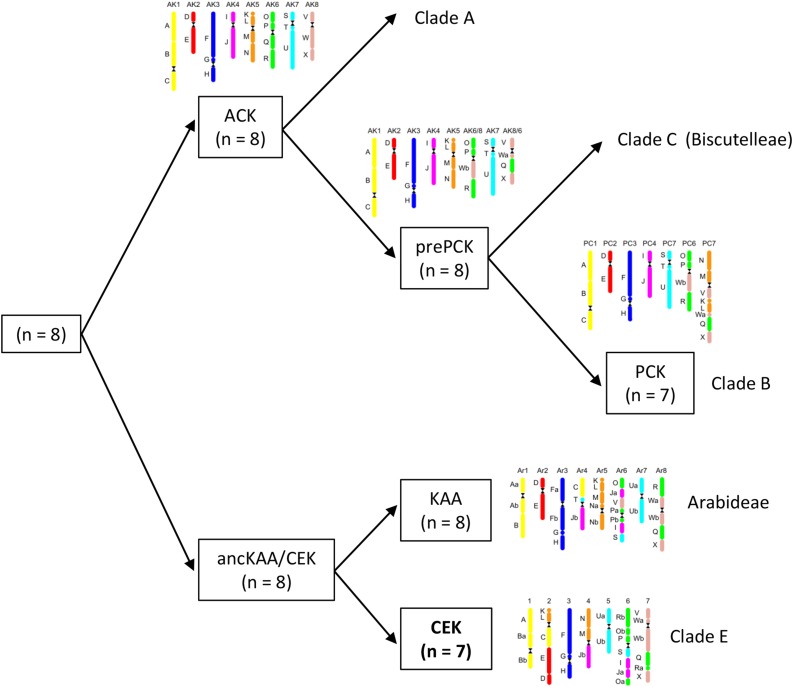
Comparison of CEK with Other Ancestral Genomes
After inferring CEK, we aimed to elucidate its closest relatives among the yet proposed crucifer ancestral genomes. The seven linkage groups of CEK hinted at descending dysploidy from an older n = 8 genome. We realized that chromosomes CEK_1 (GBs A-B), CEK_3 (F-G-H), and CEK_5 (Ua-Ub) resembled chromosomes KAA_1, KAA_3, and KAA_7 in the KAA genome of Arabis alpina (Willing et al., 2015). CEK and KAA share the structure of the bottom arm of chromosomes CEK_4 and KAA_4 (GB Jb), and the GB compositions of chromosomes CEK_7 and KAA_8 are notably similar. Chromosome CEK_3 has the same structure as its homologs in ACK, the proto-Calepineae karyotype (PCK; Lysak et al., 2016), and KAA (except for the different centromere position in KAA; Willing et al., 2015). GB association D-E can be identified as either an entire chromosome in ACK, PCK, and KAA or as a part of chromosome CEK_2. Altogether, extant as well as reconstructed chromosomal structures link the inferred CEK and KAA genomes of A. alpina (Willing et al., 2015). We propose that the two lineages (i.e. clade E and tribe Arabideae) descended from a genome with eight linkage groups (Fig. 3). This n = 8 ancestral genome presumably shared a common ancestor with ACK (n = 8), which was retained up to the current time in tribes of lineage I and reshuffled to form the pre-PCK genome (n = 8) of clade C (Geiser et al., 2016) and the PCK genome (n = 7) of clade B/lineage II (Mandáková and Lysak, 2008).
Figure 3.
Parsimonious evolutionary scenario of the origin of the CEK (n = 7) and KAA (n = 8) genomes from evolutionarily older n = 8 genome(s). The presumed relationships of these genomes to other inferred ancestral Brassicaceae genomes are outlined.
Clade E Is a Monophyletic Lineage with Miocene Tribal Diversification
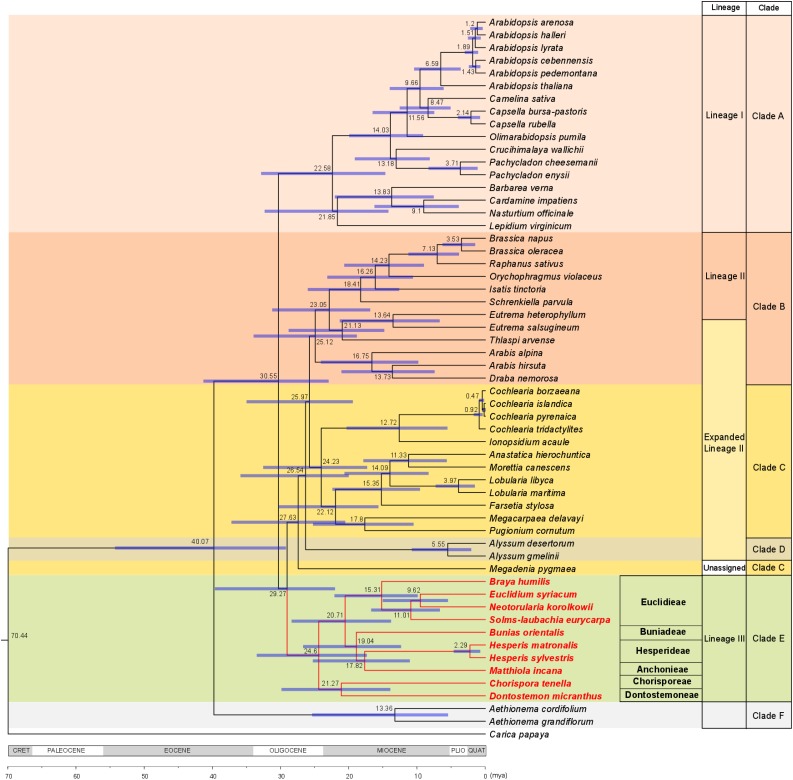
To corroborate the monophyly of clade E retrieved by cytogenetic analyses, we sequenced whole-chloroplast genomes of eight clade E species (representing six out of seven tribes) and four ANAS representatives. Our sequence data were analyzed together with all the whole-chloroplast data hitherto available for Brassicaceae species (Hohmann et al., 2015; Guo et al., 2017; GenBank accessions). In the phylogenetic tree (Fig. 4), the core Brassicaceae taxa were divided into two clades: clade A (lineage I) and all other crown-group clades. Within the latter group, clade E was retrieved as sister to the three remaining clades (clades B, C, and D) with high statistical support (Bayesian posterior probability of 100%). The ANAS genomes clustered together with other clade C genera outside of clade E.
Figure 4.
Brassicaceae family tree/chronogram showing the phylogenetic positions and divergence times of clade E tribes. A maximum clade credibility tree was produced by BEAST analysis based on whole-chloroplast sequence data of Brassicaceae taxa. Divergence times based on a relaxed clock log normal model are shown, with blue lines representing 95% high posterior density intervals. Classification to lineages and clades follows Franzke et al. (2011) and Huang et al. (2016), respectively.
Using four divergence time estimates (Magallón et al., 2015), we inferred the Aethionemeae-core Brassicaceae clade split to have occurred 40.07 million years ago (mya), with 95% high posterior density of 29.44 to 54.66 mya. The origin of clade E was dated to 29.27 mya (Oligocene), and the diversification of clade E tribes commenced at 24.60 mya in the Late Oligocene and continued throughout the Miocene (Fig. 4).
DISCUSSION
The Hesperis Clade Is a Well-Supported Monophyletic Lineage
Clade E, or the Hesperis clade, is an evolutionary unit defined by multiple parameters. (1) Morphologically, clade E species share multicellular glands (a unique character in the family), simple, nonauriculate leaves with blades usually gradually narrowing to a petiole, and often lobed stigmas with connivent lobes and/or filaments of median stamens united in pairs (Fig. 1). (2) The majority of species are native to Asia, with fewer taxa occurring in Europe and Africa and very few in North America. (3) The group includes the largest nuclear genomes in Brassicaceae, where increases in genome size usually are not associated with neopolyploidy. (4) The vast majority of clade E species has seven chromosome pairs (diploids) or chromosome complements based on x = 7 (polyploids). (5) The genome structures described here and the inferred ancestral genome (CEK) point to a monophyletic origin of the clade. (6) Phylogenetic analyses based on nuclear and chloroplast gene markers repeatedly retrieved the Hesperis clade as being a monophyletic lineage (Beilstein et al., 2006, 2008, 2010; German et al., 2009, 2011; Couvreur et al., 2010; Warwick et al., 2010; Huang et al., 2016; Guo et al., 2017; this study).
Our chloroplast tree, congruent with Beilstein et al. (2006, 2008, 2010), German et al. (2009), Couvreur et al. (2010), Huang et al. (2016), and Guo et al. (2017), showed that ANAS does not belong to clade E. The distinct phylogenetic history of ANAS also is supported by its base chromosome numbers equal to eight to 13 but not six or seven (BrassiBase; Kiefer et al., 2014) and by the absence of clade E-specific chromosomal rearrangements. Furthermore, this study and Mandáková et al. (2017) revealed that the extant diploid ANAS species represent diploidized mesotetraploid genomes. In contrast, no evidence for a mesopolyploid event in the ancestry of clade E was obtained.
Phylogenomic Evidence of Two Major Intraclade Branches
Within clade E, Huang et al. (2016) retrieved two subclades: the first one containing CHOR and DONT and the second one harboring ANCH, BUNI, EUCL, and HESP. SHEH, formed via an intertribal hybridization between CHOR and DONT (German and Friesen, 2014), should belong to the CHOR/DONT subclade. Species from the CHOR/DONT subclade have simple trichomes and often winged or margined seeds, whereas the larger subclade is characterized predominantly by branched trichomes and wingless seeds. The same dichotomy was retrieved in our chloroplast phylogeny, based however on only 10 chloroplast sequences. Although within clade E, topology differs considerably among authors, this split was often observed, as by Beilstein et al. (2006, 2008, 2010), Franzke et al. (2009), Couvreur et al. (2010), and partly by German et al. (2011). It could be assumed that the tribal dichotomy also is reflected by differences between reconstructed genome structures of CHOR and EUCL (Fig. 2, A and B), whereby the CHOR/DONT subclade would represent more ancestral, CEK-like genomes with a slower rate of karyotype evolution.
Oligocene Origin and Miocene Diversification of the Hesperis Clade
Our divergence time estimates based on chloroplast genes dated the origin of the Hesperis clade to the Oligocene, and its later diversification occurred throughout the Miocene. These time estimates are largely congruent with the purported Oligocene divergence of major Brassicaceae clades (Huang et al., 2016) as well as with other inferred emergence dates for clade E of 21.4 mya (Couvreur et al., 2010) and 21 mya (maximum stem age; Hohmann et al., 2015). Hohmann et al. (2015) estimated the emergence of clade E tribes at 17 mya, and the same estimate (17.2 mya) for the most recent common ancestor of clade E was reported by Huang et al. (2016). A Middle Miocene divergence (15 mya) also was proposed for the basal split within DONT (between Clausia and Dontostemon; Friesen et al., 2016). Because the vast majority of Aethionemeae, a sister clade to all other Brassicaceae clades, occurs in the Irano-Turanian region (predominantly in Turkey) and one of the main diversity hotspots of the family is located there as well, this region is often referred to as the cradle of the family (Hedge, 1976; Franzke et al., 2009, 2011; Couvreur et al., 2010). Extant taxa of Hesperis clade tribes also occur predominantly in the Ancient Mediterranean floristic subkingdom, especially in the western Asiatic subregion of the Irano-Turanian region (Takhtajan, 1986) or the Irano-Turanian region sensu (Hedge, 1976), which could mean that the emergence of the clade is close to that of the whole family. On the other hand, DONT and some EUCL genera demonstrate diversification in the eastern part of the Irano-Turanian floristic region (central Asiatic subregion sensu [Takhtajan, 1986] or outside the Irano-Turanian floristic region sensu [Hedge, 1976]) and even in mountainous areas of the eastern Asian region, assuming that the already early branching of clade E might be somewhat more eastern than the origin of the whole family. Generally, the increase in open habitats during the Late Oligocene/Early Miocene could have facilitated the diversification of clade E (Franzke et al., 2009). This might be particularly true for the evolution of DONT, apparently driven by the development of the Eurasian steppe belt (Friesen et al., 2016), enabling the genetic diversification of DONT, associated with a more eastward (mainly central Asian) distribution of the tribe. The diversification of several high-mountain and alpine genera of EUCL (e.g. Braya, Lepidostemon, Sisymbriopsis, and Solms-laubachia), also characterized by more eastern centers of present-day diversity, was probably triggered by uplifts of the Qinghai-Tibetan Plateau and mountains of the Hengduan-Himalayan region (Wen et al., 2014, and refs. therein). However, in all cases, current distribution patterns should be interpreted with caution. For example, even for the relatively young (Late Pliocene/Early Pleistocene) Solms-laubachia, an ancestral, more western distribution compared with its current center of diversity was detected by Yue et al. (2009).
Clade E and Early Genome Evolution in Brassicaceae
Unlike previous authors, Huang et al. (2016) claimed that clade E branched out early after the split of the Aethionemeae (clade F) from the crown group and that the clade is sister to all the remaining crown-group clades (ABCD). Within our chloroplast tree, interclade relationships are more ambiguous, with clade E being sister to clades B, C, and D and, again, this superclade being sister to clade A. This topology is congruent with the plastome phylogeny of Guo et al. (2017) and to a large extent is supported by the scenario proposed here of an ancient genome evolution (Fig. 3). In accord with Guo et al. (2017), our phylogenetic analysis retrieved clade C paraphyletic due to Megadenia being sister to clades B, C, and D.
All the inferred ancestral genomes in Brassicaceae have descended from a common post-At-α genome, which later diversified into an ancestral clade F genome and an ancestral genome (n = 8) shared by all crown-group clades (A–E). The evolution of the latter genome is still rather elusive due to the lack of genomic data on clades C (except for Biscutelleae; Geiser et al., 2016) and D. Comparisons of structurally characterized modern genomes of clades A, B, C, and E plus Arabideae suggest that the ancestral crown-group genome further evolved into ACK (n = 8; Schranz et al., 2006) and another n = 8 genome shared by Arabideae and clade E. ACK either remained conserved in clade A (Lysak et al., 2006, 2016; Mandáková et al., 2013), was altered by a reciprocal translocation in clade C (pre-PCK of Biscutelleae; Geiser et al., 2016), or underwent descending dysploidy toward the PCK genome of clade B (n = 7; Mandáková and Lysak, 2008; Cheng et al., 2013; Mandáková et al., 2015). Although CEK of clade E and KAA of Arabideae (Willing et al., 2015) share some unique genomic features (Fig. 3), this genomic affinity is not corroborated by plastome phylogeny (Guo et al., 2017; Fig. 4), and more work is needed to settle this discrepancy.
Trends of Genome Evolution in Clade E
Based on chromosome counts collated by BrassiBase (Kiefer et al., 2014), 96% to 100% of BUNI, CHOR, DONT, and EUCL species possessed genomes based on x = 7 (usually 2n = 14 or 28). In HESP, 57% of available counts corresponded to x = 7, 36% to x = 6, and 1% to x = 5. A very similar pattern was observed in ANCH, with x = 7 in 50% of chromosome counts, 44% corresponding to x = 6, and 4% corresponding to x = 5 (note that 5.3% and 2.5% of 2n = 16 counts in HESP and ANCH, respectively, are most likely miscounts of 2n = 14). The prevalence of x = 7 across all tribes further justifies CEK as an ancestral genome of the Hesperis clade and points to its apparent stasis. This is demonstrated by almost identical genomes of Euclidium and Strigosella, also suggesting that intratribal diversification was not associated with major chromosomal reshuffling. It remains to be seen whether the strong tendency for descending dysploidy from n = 7 to n = 6 (−5) in Hesperis and Matthiola could be associated with speciation events in these genera. A comparable karyotype and chromosome number stasis was reported previously for clade B (expanded lineage II; Mandáková and Lysak, 2008), despite containing some 25 tribes (Al-Shehbaz, 2012). Such genomes represent well-tuned genetic systems that have not been affected by major genomic alterations for the last 20 million years. In both clades, the lack of genome repatterning and extensive descending dysploidies also can be attributed to the absence of mesopolyploid whole-genome duplications. Independent polyploidizations frequently triggered major genomic rearrangements and descending dysploidies across the Brassicaceae (Mandáková et al., 2017).
MATERIALS AND METHODS
Plant Material
Plants used for cytogenetic and/or phylogenetic analyses were grown from seeds or collected in the field (for origins, see Supplemental Table S1).
Chromosome Preparation
Inflorescences containing young flower buds were collected into fixative (3:1, 96% ethanol:glacial acetic acid) and kept at −20°C until needed. Mitotic and meiotic chromosome preparations were prepared from anthers as described by Mandáková and Lysak (2016a). Preparations were staged using a phase-contrast microscope, and suitable slides containing tapetal mitoses and/or meiosis I chromosomes were postfixed in 4% formaldehyde in distilled water for 10 min and air dried. Chromosome preparations were treated with 100 μg mL−1 RNase in 2× SSC (20× SSC = 3 m sodium chloride and 300 mm trisodium citrate, pH 7) for 60 min and with 0.1 mg mL−1 pepsin in 0.01 m HCl at 37°C for 3 to 15 min, then postfixed in 4% formaldehyde in 2× SSC for 10 min, washed in 2× SSC twice for 5 min, and dehydrated in an ethanol series (70%, 90%, and 100%, 2 min each).
CCP
For CCP in CHOR and EUCL species, chromosome-specific BAC clones of Arabidopsis (Arabidopsis thaliana) were grouped into contigs according to 22 GBs of ACK (Lysak et al., 2016). To determine and characterize paracentric inversions (see chromosomes Ct2, Es1, Es2, Sa1, Sa1′, Sa2, Sa2′, Sa3, and Sa3′ in Fig. 2, A and B) and splits of block I (see chromosomes Sa3 and Sa3′ in Fig. 2B; Supplemental Fig. S1B), BAC contigs corresponding to GBs A, B, E, and I were subdivided after initial CCP experiments into smaller, differentially labeled contigs. We were not able to detect block T, probably due to its close proximity to (peri)centromeric heterochromatin. Arabidopsis BAC clone T15P10 (AF167571), bearing 35S rRNA gene repeats, was used for in situ localization of nucleolar organizer regions and Arabidopsis clone pCT 4.2 (M65137). All DNA probes were labeled with biotin-, digoxigenin-, or Cy3-dUTP by nick translation as described by Mandáková and Lysak (2016b). A total of 100 ng of labeled DNA of each selected BAC clone was pooled together, ethanol precipitated, dissolved in 20 µL of hybridization mixture containing 50% formamide and 10% dextran sulfate in 2× SSC, and pipetted onto each chromosome preparation. The slides were heated at 80°C for 2 min and incubated at 37°C overnight. Hybridized probes were visualized either as direct fluorescence of Cy3-dUTP (yellow) or through fluorescently labeled antibodies against biotin-dUTP (red) and digoxigenin-dUTP (green), as described by Mandáková and Lysak (2016b). Chromosomes were counterstained with DAPI (2 µg mL−1) in Vectashield antifade. Fluorescent signals were analyzed and photographed using a Zeiss AxioImager epifluorescence microscope equipped with a CoolCube camera (MetaSystems). Individual images were merged and processed using Photoshop CS software (Adobe Systems). Painted chromosomes in Figure 2C were straightened using the plugin Straighten Curved Objects in ImageJ (Kocsis et al., 1991).
CCP on pachytene chromosomes in ANCH, CHOR, DONT, and HESP species with large genomes resulted in nonspecific hybridization signals covering multiple chromosomes. This was probably caused by a high repeat content of these genomes and/or high levels of chromosome heterochromatinization. However, a modified CCP protocol enabled us to identify common GB associations of CHOR and EUCL on mitotic chromosomes of Bunias orientalis, Dontostemon micranthus, Hesperis sylvestris, and Matthiola incana. The combinations of BAC contigs building up chromosomes Ct1/Es1/Sa1/Sa1′, Ct4/Es4/Sa4/Sa4′, and Ct5/Es5/Sa5/Sa5′ in Chorispora, Euclidium, and Strigosella, respectively, were used as painting probes. The following modifications were applied: (1) the concentration of each selected labeled BAC clone was increased 5 times (500 ng per slide); (2) denaturation time was increased (4 min); (3) hybridization times at 37°C were prolonged (68–72 h); and (4) stringent posthybridization washing was prolonged (three washes in 20% formamide in 2× SSC, 10 min each time). After CCP, chromosomes were counterstained with half-concentrated DAPI (1 µg mL−1) in Vectashield antifade.
Chloroplast Genome de Novo Assembly
Leaf material of 12 species, presumably belonging to clade E, and that of Alyssum gmelinii (Supplemental Table S1) was harvested and dried using silica gel. For the de novo assembly of chloroplast genomes, reads from low-coverage whole-genome sequencing (Illumina; 2× 100 bp, 2× 350 bp) were used. Chloroplast reads make up to 6% of all reads. Quality filtering (Phred score > 20 and cutoff value of 80%), adaptor trimming, and converting fastq to fasta were performed using the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/).
For each species, chloroplast genome reads were identified by BLAST software (Altschul et al., 1990). All raw reads were aligned (using BLASTn) on the reference genome of Arabidopsis (AP000423) and Lobularia maritima (NC_009274), and only reads with positive hits were used for de novo assembly. Before de novo assembly, chloroplast reads were down sampled to 100× coverage (i.e. 150,000 100-bp paired-end reads and 45,000 350-bp paired-end reads). De novo assembly was performed by Ray assembler (Boisvert et al., 2010), and sequence gaps in scaffolds were attempted to be filled by Gapfiller (Boetzer and Pirovano, 2012). All contigs were mapped to the reference chloroplast genome of Arabidopsis by Geneious 8.1.7 (Kearse et al., 2012), and all chloroplast reads were then mapped back to the consensus, with sequences being checked manually to remove misalignments and mismatches between the newly assembled and reference genomes.
Genome Annotation
Annotations of all 13 chloroplast genomes were performed on the Dual Organellar GenoMe Annotator (DOGMA; Wyman et al., 2004). Each DOGMA annotation was manually corrected for the start and stop codons or intron/exon junctions by comparison with known homologous chloroplast genes, and tRNA genes were checked by ARAGORN (Laslett and Canback, 2004).
Phylogenetic Analysis
For phylogenetic analysis, we used the alignment published by Hohmann et al. (2015), whole-chloroplast sequences of 14 Brassicaceae species from GenBank, and our 13 newly assembled chloroplast genomes. From assembled and annotated genomes, genes used by Hohmann et al. (2015) were extracted, aligned to the Arabidopsis reference genome, and start/stop codons, introns, and insertions/deletions were removed. Extracted genes were then ordered and added to the nexus of 72 species of Hohmann et al. (2015). Sequence alignments have been deposited in the Dryad Digital Repository (http://dx.doi.org/10.5061/dryad.54df2). Divergence time estimation was conducted in BEAST version 2.4.4 (Bouckaert et al., 2014) using independent site and clock models. Vitis vinifera was defined as an outgroup. We used four fossil constraints, as used by Magallón et al. (2015) and Hohmann et al. (2015), and a normal distribution was used for these. We ran two independent MCMC runs with 300,000,000 generations each, sampled every 30,000 generations. LogCombiner version 1.8.3 was used to combine trees from the two runs, and 10% of trees were discarded as burn in. TreeAnnotator version 1.8.3 was used to generate a maximum clade credibility tree. All phylogenetic analyses were computed through the Cyberinfrastructure for Phylogenetic Research portal (http://www.phylo.org/; Miller et al., 2010). Visualization was performed in FigTree version 1.4.2 (Rambaut, 2014).
Accession Numbers
Newly assembled chloroplast sequences from this article can be found in GenBank under accession numbers KY912021 to KY912032 and MF169880 (A. gmelinii).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Parsimoniously reconstructed origins of chromosomes in clade E species.
Supplemental Table S1. Collection data for the species used in this study.
Acknowledgments
We thank the core Global Naturalized Alien Flora project members for providing data on naturalized species of clade E and Kateřina Beková for help with analyzing sequence data. Several of our colleagues (see Fig. 1 legend) are acknowledged for giving us permission to reproduce their photographs of clade E species.
Glossary
- ANAS
Anastaticeae
- ANCH
Anchonieae
- BUNI
Buniadeae
- CHOR
Chorisporeae
- DONT
Dontostemoneae
- EUCL
Euclidieae
- HESP
Hesperideae
- SHEH
Shehbazieae
- GB
genomic block
- CCP
comparative chromosome painting
- mya
million years ago
- DAPI
4′,6-diamidino-2-phenylindole
Footnotes
This work was supported by a research grant from the Czech Science Foundation (grant no. P501/12/G090) and the CEITEC 2020 (grant no. LQ1601) project. D.A.G. was supported by a research grant from the DFG (grant no. KO2302-13/1,2).
References
- Al-Shehbaz IA. (2012) A generic and tribal synopsis of the Brassicaceae (Cruciferae). Taxon 61: 931–954 [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Beilstein MA, Al-Shehbaz IA, Kellogg EA (2006) Brassicaceae phylogeny and trichome evolution. Am J Bot 93: 607–619 [DOI] [PubMed] [Google Scholar]
- Beilstein MA, Al-Shehbaz IA, Mathews S, Kellogg EA (2008) Brassicaceae phylogeny inferred from phytochrome A and ndhF sequence data: tribes and trichomes revisited. Am J Bot 95: 1307–1327 [DOI] [PubMed] [Google Scholar]
- Beilstein MA, Nagalingum NS, Clements MD, Manchester SR, Mathews S (2010) Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 18724–18728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetzer M, Pirovano W (2012) Toward almost closed genomes with GapFiller. Genome Biol 13: R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert S, Laviolette F, Corbeil J (2010) Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol 17: 1519–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ (2014) BEAST 2: a software platform for Bayesian evolutionary analysis. PLOS Comput Biol 10: e1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CABI (2012) Bunias orientalis (Turkish warty-cabbage). Datasheet Invasive Species Compendium. http://www.cabi.org/isc/datasheet/109130
- Cheng F, Mandáková T, Wu J, Xie Q, Lysak MA, Wang X (2013) Deciphering the diploid ancestral genome of the mesohexaploid Brassica rapa. Plant Cell 25: 1541–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvreur TLP, Franzke A, Al-Shehbaz IA, Bakker FT, Koch MA, Mummenhoff K (2010) Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae). Mol Biol Evol 27: 55–71 [DOI] [PubMed] [Google Scholar]
- Francis A, Cavers PB, Warwick SI (2009) The biology of Canadian weeds, 140: Hesperis matronalis L. Can J Plant Sci 89: 189–204 [Google Scholar]
- Franzke A, German D, Al-Shehbaz IA, Mummenhoff K (2009) Arabidopsis family ties: molecular phylogeny and age estimates in Brassicaceae. Taxon 58: 425–437 [Google Scholar]
- Franzke A, Lysak MA, Al-Shehbaz IA, Koch MA, Mummenhoff K (2011) Cabbage family affairs: the evolutionary history of Brassicaceae. Trends Plant Sci 16: 108–116 [DOI] [PubMed] [Google Scholar]
- Friesen N, German DA, Hurka H, Herden T, Oyuntsetseg B, Neuffer B (2016) Dated phylogenies and historical biogeography of Dontostemon and Clausia (Brassicaceae) mirror the palaeogeographic history of the Eurasian steppe. J Biogeogr 43: 738–749 [Google Scholar]
- Geiser C, Mandáková T, Arrigo N, Lysak MA, Parisod C (2016) Repeated whole-genome duplication, karyotype reshuffling, and biased retention of stress-responding genes in Buckler mustards. Plant Cell 28: 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- German DA, Friesen N, Neuffer B, Al-Shehbaz IA, Hurka H (2009) Contribution to ITS phylogeny of the Brassicaceae, with a special reference to some Asian taxa. Plant Syst Evol 283: 33–56 [Google Scholar]
- German DA, Friesen NW (2014) Shehbazia (Shehbazieae, Cruciferae), a new monotypic genus and tribe of hybrid origin from Tibet. Turczaninowia 17: 17–23 [Google Scholar]
- German DA, Grant JR, Lysak MA, Al-Shehbaz IA (2011) Molecular phylogeny and systematics of the tribe Chorisporeae (Brassicaceae). Plant Syst Evol 294: 65–86 [Google Scholar]
- Guo X, Liu J, Hao G, Zhang L, Mao K, Wang X, Zhang D, Ma T, Hu Q, Al-Shehbaz IA, et al. (2017) Plastome phylogeny and early diversification of Brassicaceae. BMC Genomics 18: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedge IC. (1976) A systematic and geographical survey of the Old World Cruciferae. In Vaughan JG, Macleod AJ, Jones BMG, eds, The Biology and Chemistry of the Cruciferae. Academic Press, London, pp 1–45 [Google Scholar]
- Hohmann N, Wolf EM, Lysak MA, Koch MA (2015) A time-calibrated road map of Brassicaceae species radiation and evolutionary history. Plant Cell 27: 2770–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CH, Sun R, Hu Y, Zeng L, Zhang N, Cai L, Zhang Q, Koch MA, Al-Shehbaz I, Edger PP, et al. (2016) Resolution of Brassicaceae phylogeny using nuclear genes uncovers nested radiations and supports convergent morphological evolution. Mol Biol Evol 33: 394–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer M, Schmickl R, German DA, Mandáková T, Lysak MA, Al-Shehbaz IA, Franzke A, Mummenhoff K, Stamatakis A, Koch MA (2014) BrassiBase: introduction to a novel knowledge database on Brassicaceae evolution. Plant Cell Physiol 55: e3. [DOI] [PubMed] [Google Scholar]
- Kocsis E, Trus BL, Steer CJ, Bisher ME, Steven AC (1991) Image averaging of flexible fibrous macromolecules: the clathrin triskelion has an elastic proximal segment. J Struct Biol 107: 6–14 [DOI] [PubMed] [Google Scholar]
- Laslett D, Canback B (2004) ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32: 11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak MA, Berr A, Pecinka A, Schmidt R, McBreen K, Schubert I (2006) Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc Natl Acad Sci USA 103: 5224–5229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak MA, Koch MA, Beaulieu JM, Meister A, Leitch IJ (2009) The dynamic ups and downs of genome size evolution in Brassicaceae. Mol Biol Evol 26: 85–98 [DOI] [PubMed] [Google Scholar]
- Lysak MA, Mandáková T, Schranz ME (2016) Comparative paleogenomics of crucifers: ancestral genomic blocks revisited. Curr Opin Plant Biol 30: 108–115 [DOI] [PubMed] [Google Scholar]
- Magallón S, Gómez-Acevedo S, Sánchez-Reyes LL, Hernández-Hernández T (2015) A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytol 207: 437–453 [DOI] [PubMed] [Google Scholar]
- Mandáková T, Kovarík A, Zozomová-Lihová J, Shimizu-Inatsugi R, Shimizu KK, Mummenhoff K, Marhold K, Lysak MA (2013) The more the merrier: recent hybridization and polyploidy in Cardamine. Plant Cell 25: 3280–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T, Li Z, Barker MS, Lysak MA (2017) Diverse genome organization following 13 independent mesopolyploid events in Brassicaceae contrasts with convergent patterns of gene retention. Plant J 91: 3–21 [DOI] [PubMed] [Google Scholar]
- Mandáková T, Lysak MA (2008) Chromosomal phylogeny and karyotype evolution in x=7 crucifer species (Brassicaceae). Plant Cell 20: 2559–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T, Lysak MA (2016a) Chromosome preparation for cytogenetic analyses in Arabidopsis. Curr Protoc Plant Biol 1: 1–9 [DOI] [PubMed] [Google Scholar]
- Mandáková T, Lysak MA (2016b) Painting of Arabidopsis chromosomes with chromosome-specific BAC clones. Curr Protoc Plant Biol 1: 359–371 [DOI] [PubMed] [Google Scholar]
- Mandáková T, Singh V, Krämer U, Lysak MA (2015) Genome structure of the heavy metal hyperaccumulator Noccaea caerulescens and its stability on metalliferous and nonmetalliferous soils. Plant Physiol 169: 674–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, November 14, 2010, 1–8 [Google Scholar]
- Rambaut A. (2014) FigTree version 1.4.2. http://tree.bio.ed.ac.uk/software/figtree/
- Schranz ME, Lysak MA, Mitchell-Olds T (2006) The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci 11: 535–542 [DOI] [PubMed] [Google Scholar]
- Takhtajan AL. (1986) Floristic Regions of the World. University of California Press, Berkeley [Google Scholar]
- van Kleunen M, Dawson W, Essl F, Pergl J, Winter M, Weber E, Kreft H, Weigelt P, Kartesz J, Nishino M, et al. (2015) Global exchange and accumulation of non-native plants. Nature 525: 100–103 [DOI] [PubMed] [Google Scholar]
- Warwick SI, Mummenhoff K, Sauder CA, Koch MA, Al-Shehbaz IA (2010) Closing the gaps: phylogenetic relationships in the Brassicaceae based on DNA sequence data of nuclear ribosomal ITS region. Plant Syst Evol 285: 209–232 [Google Scholar]
- Wen J, Zhang JQ, Nie ZL, Zhong Y, Sun H (2014) Evolutionary diversification of plants on the Qinghai-Tibetan Plateau. Front Genet 5: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing E-M, Rawat V, Mandáková T, James GV, Nordström KJ, Maumus F, Becker C, Warthmann N, Chica C, Szarzynska B, et al. (2015) Genome expansion of Arabis alpina linked with retrotransposition and reduced symmetric DNA methylation. Nat Plants 1: 14023. [DOI] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL (2004) Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20: 3252–3255 [DOI] [PubMed] [Google Scholar]
- Yue JP, Sun H, Baum DA, Li JH, Al-Shehbaz IA, Ree R (2009) Molecular phylogeny of Solms-laubachia (Brassicaceae) s.l., based on multiple nuclear and plastid DNA sequences, and its biogeographic implications. J Syst Evol 47: 402–415 [Google Scholar]