M. domestica polyphenol oxidase 1 (MdPPO1) was recombinantly expressed in its latent form (Lys1–Ser504) and successfully mutated at four different positions around the active centre which have been proposed to be decisive for the activity of the enzyme. The wild-type MdPPO1 and two of the mutants were successfully crystallized. In crystallo activity tests demonstrated the importance of these amino acids for the activity of the enzyme.
Keywords: polyphenol oxidase, plant tyrosinase, latent pro-enzyme, type 3 copper enzyme, activity controllers
Abstract
Tyrosinases are type 3 copper enzymes that belong to the polyphenol oxidase (PPO) family and are able to catalyze both the ortho-hydroxylation of monophenols and their subsequent oxidation to o-quinones, which are precursors for the biosynthesis of colouring substances such as melanin. The first plant pro-tyrosinase from Malus domestica (MdPPO1) was recombinantly expressed in its latent form (56.4 kDa) and mutated at four positions around the catalytic pocket which are believed to influence the activity of the enzyme. Mutating the amino acids, which are known as activity controllers, yielded the mutants MdPPO1-Ala239Thr and MdPPO1-Leu243Arg, whereas mutation of the so-called water-keeper and gatekeeper residues resulted in the mutants MdPPO1-Glu234Ala and MdPPO1-Phe259Ala, respectively. The wild-type enzyme and two of the mutants, MdPPO1-Ala239Thr and MdPPO1-Phe259Ala, were successfully crystallized, leading to single crystals that diffracted to 1.35, 1.55 and 1.70 Å resolution, respectively. All crystals belonged to space group P212121, exhibiting similar unit-cell parameters: a = 50.70, b = 80.15, c = 115.96 Å for the wild type, a = 50.58, b = 79.90, c = 115.76 Å for MdPPO1-Ala239Thr and a = 50.53, b = 79.76, c = 116.07 Å for MdPPO1-Phe259Ala. In crystallo activity tests with the crystals of the wild type and the two mutants were performed by adding the monophenolic substrate tyramine and the diphenolic substrate dopamine to crystal-containing drops. The effects of the mutation on the activity of the enzyme were observed by colour changes of the crystals owing to the conversion of the substrates to dark chromophore products.
1. Introduction
Tyrosinases (TYRs) are members of the type 3 copper enzyme family, which also includes catechol oxidases (COs) and aurone synthase (AUS). TYRs, COs and AUS are summarized under the umbrella term polyphenol oxidases (PPOs). These enzymes are widely distributed in bacteria, fungi, plants and animals (Mayer, 2006 ▸; Tran et al., 2012 ▸; Kaintz, Mauracher et al., 2014 ▸; Pretzler et al., 2015 ▸). Tyrosinases are able to catalyze the ortho-hydroxylation of monophenols to o-diphenols (monophenolase activity; EC 1.14.18.1) coupled with the subsequent two-electron oxidation of o-diphenols to the corresponding o-quinones (diphenolase activity; EC 1.10.3.1) (Mason, 1955 ▸; Mayer et al., 1966 ▸). The o-diphenols formed in the hydroxylation step remain in the active centre and are oxidized to the quinonic state (Ramsden & Riley, 2014 ▸). During the TYR-mediated hydroxylation and oxidation of one molecule of monophenol, one molecule of dioxygen is reduced to water. COs lack the monophenolase activity and are thus only capable of oxidizing o-diphenols. The resulting o-quinones, which are the products of all PPOs, are highly reactive and represent the starting material for the biosynthesis of colouring substances such as melanin (Mason, 1948 ▸; Rodríguez-López et al., 1992 ▸). Thus, both TYRs and COs are involved in the browning reaction of fruits and vegetables, which represents a major concern in the food industry. The third type 3 copper enzyme AUS from the petals of Coreopsis grandiflora (cgAUS1; Molitor, Mauracher, Pargan et al., 2015 ▸; Kaintz, Molitor et al., 2014 ▸; Kaintz et al., 2015 ▸) shows monophenolase activity towards chalcone substrates (for example isoliquiritigenin) but not towards classical tyrosinase substrates such as tyrosine and tyramine, and has therefore been classified as a CO. Moreover, cgAUS1 has been crystallized and structurally analyzed (Molitor, Mauracher & Rompel, 2015 ▸; Molitor, Mauracher & Rompel, 2016 ▸; Molitor, Bijelic & Rompel, 2016 ▸).
Plant PPOs are known to be expressed as latent pro-enzymes in vivo exhibiting a molecular weight of ∼64–68 kDa and consist of three domains: a signal peptide (in a minority of plant PPOs) or a transit peptide containing a thylakoid transfer domain (in the majority of plant PPOs) (∼4–9 kDa), a catalytically active domain (∼40 kDa) and a shielding C-terminal domain (∼15–19 kDa) (Tran et al., 2012 ▸; Fig. 1 ▸ a). The enzyme is activated in vivo by a hitherto unknown proteolytic cleavage process (Fig. 1 ▸ a) in which the C-terminus is separated from the active domain, leading to a substrate-accessible catalytic pocket (Flurkey & Inlow, 2008 ▸). The enzyme can also be activated in vitro using proteases (Gandía-Herrero et al., 2005b ▸; Pretzler et al., 2017 ▸; Kampatsikas et al., 2017 ▸), an acidic pH (Valero & García-Carmona, 1992 ▸; Kampatsikas et al., 2017 ▸), fatty acids (Sugumaran & Nellaiappan, 1991 ▸) or detergents such as sodium dodecyl sulfate (SDS; Gandía-Herrero et al., 2005a ▸; Kampatsikas et al., 2017 ▸). The maturation process activating the enzyme in vivo and the function of the C-terminal domain are still a matter of debate.
Figure 1.
(a) Schematic representation of the primary structure of recombinant latent MdPPO1 (56.4 kDa). The active domain of the enzyme is coloured red and the conserved regions (histidines) of the copper-coordinating motifs are presented in purple. The C-terminal domain is coloured green and the putative position of proteolytic activation between the active domain and the C-terminus is displayed in blue. (b) Structural representation of the active centre of the wild-type MdPPO1 from homology modelling shows the conserved histidines coordinating CuA (His86, His107 and His116) and CuB (His238, His242 and His272) and the positions of the mutated residues, namely the two activity controllers alanine (Ala239) and leucine (Leu243), both in purple, the water keeper glutamic acid (Glu234) in light green and the gatekeeper phenylalanine (Phe259) in red. The homology model of MdPPO1 was obtained in the same way as described in Kampatsikas et al. (2017 ▸).
Another ongoing discussion is the structural basis for the lack of monophenolase activity in COs, as the crystal structures of diverse COs (Klabunde et al., 1998 ▸; Virador et al., 2010 ▸; Molitor, Mauracher & Rompel, 2015 ▸, 2016 ▸) and TYRs (Matoba et al., 2006 ▸; Sendovski et al., 2011 ▸; Ismaya, Rozeboom, Schurink et al., 2011 ▸; Ismaya, Rozeboom, Weijn et al., 2011 ▸; Mauracher et al., 2014a ▸,b ▸; Zekiri, Bijelic et al., 2014 ▸; Bijelic et al., 2015a, ▸ b ▸) revealed an astoundingly high similarity of their active sites. In recent decades some theories based on the position and/or presence of certain amino acid residues within the active site have been postulated, but none of these were able to identify structural features which could explain the lack of monophenolase activity in COs (Kanteev et al., 2015 ▸; Pretzler & Rompel, 2017 ▸). One of these amino acids is the so-called gatekeeper residue, which is located directly above the first copper in the active site (CuA) and acts as a stabilizer for incoming substrates via hydrophobic T-shaped π–π interactions (Bijelic et al., 2015a, ▸ b ▸; Molitor, Mauracher & Rompel, 2016 ▸). Another residue is the water keeper, which is assumed to stabilize a conserved water molecule that is responsible for the putatively monophenolase-decisive deprotonation of incoming monophenolic substrates (Goldfeder et al., 2014 ▸).
Recently, cDNA encoding a PPO pro-enzyme has been cloned from apple leaves (Malus domestica; MdPPO1; ENA LT718522) and was heterologously expressed in Escherichia coli (Kampatsikas et al., 2017 ▸). MdPPO1 was subsequently purified and characterized, revealing monophenolase activity towards tyramine and tyrosine and thus classifying MdPPO1 as a TYR (Kampatsikas et al., 2017 ▸). The study also revealed the presence of further amino acids that are relevant for the enzyme’s activity, which are located next to the first and second histidines of copper B (CuB) and are therefore termed activity controllers. Together with the two abovementioned residues, they can control the activity of the enzyme (monophenolase or diphenolase activity).
In this study, four mutants of MdPPO1 were produced recombinantly and purified in order to study their effect on the activity of the enzyme, namely MdPPO1-Ala239Thr, MdPPO1-Leu243Arg, MdPPO1-Glu234Ala and MdPPO1-Phe259Ala. The first two mutations, MdPPO1-Ala239Thr and MdPPO1-Leu243Arg, affect the nonconserved activity-controller residues Ala239 and Leu243 (Kampatsikas et al., 2017 ▸). MdPPO1-Glu234Ala is the result of mutating the water keeper Glu234, and mutation of the gatekeeper Phe259 yielded the mutant MdPPO1-Phe259Ala (see Fig. 1 ▸ b). During the subsequent crystallization trials, the wild-type MdPPO1 and two of the four mutants, MdPPO1-Ala239Thr and MdPPO1-Phe259Ala, were successfully crystallized, representing the first crystals of recombinantly expressed latent plant TYR. Crystals of wild-type MdPPO1, MdPPO1-Ala239Thr and MdPPO1-Phe259Ala diffracted to 1.35, 1.55 and 1.70 Å resolution, respectively. Furthermore, the crystals were soaked with a monophenolic (tyramine) and a diphenolic (dopamine) substrate using sodium dodecyl sulfate (SDS) as an activator in order to perform in crystallo activity tests. This is possible as the products of the PPO reaction, quinones, further react to dark chromophores, leading to the browning of the crystals. This procedure was well tolerated by the crystals without damage. The in crystallo activity tests revealed the importance of the mutated positions, as both mutants significantly affect the activity of the enzyme, providing valuable insights into the catalytic mechanism of plant PPOs.
2. Materials and methods
2.1. Preparation of MdPPO1
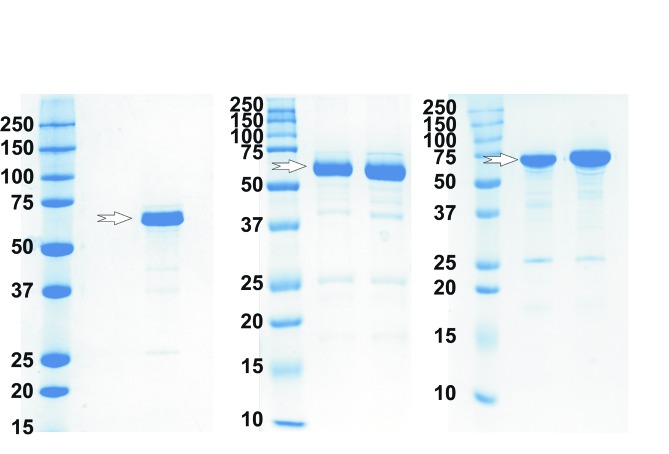
Production of the target enzyme MdPPO1 was performed as described previously (Kampatsikas et al., 2017 ▸). Briefly, the gene encoding latent MdPPO1 (without the signal peptide; residues Lys1–Ser504) was inserted into the expression vector pGEX-6P-1 (GE Healthcare) and N-terminally fused with the glutathione S-transferase (GST) tag of the vector. A protease (HRV3C) recognition sequence (LEVLFQ|GP) was present between the GST tag and the target enzyme for subsequent tag removal. Competent E. coli BL21 cells were transformed with the vector and cultured at 310 K until an OD600 of 0.6–0.8 was reached, at which point the temperature was reduced to 293 K and protein expression was induced by the addition of 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and 0.5 mM copper(II) sulfate. Expression was continued for additional 30–40 h at 293 K until the cell density reached an OD600 of 7–10. The cells were collected by centrifugation and lysed by the freeze–thaw technique. The enzyme was purified using a GSTrap FF column (GE Healthcare) followed by GST-tag cleavage with HRV3C protease. Further purification was performed using a second GSTrap FF column. The protein was concentrated to about 5–10 mg ml−1 in a buffer consisting of 50 mM Tris–HCl, 200 mM NaCl pH 7.5. Target-enzyme production is summarized in Table 1 ▸. For the production of the four mutants, primers were designed which are summarized in Table 1 ▸. The clone of the wild type in the pGEX-6P-1 vector was used as a template for the construction of the mutants and the Q5 Site-Directed Mutagenesis Kit (NEB) was used to prepare the mutants. Expression and purification of the mutants was performed as described for the wild-type MdPPO1; the purity level of the produced mutants was controlled with SDS–PAGE (Fig. 2 ▸).
Table 1. Production information for wild-type MdPPO1 and four mutants.
The amino acids in red indicate the positions of the respective mutations.
| Protein | Wild-type MdPPO1 | MdPPO1-Ala239Thr | MdPPO1-Leu243Arg | MdPPO1-Glu234Ala | MdPPO1-Phe259Ala |
|---|---|---|---|---|---|
| Source organism | M. domestica cv. Golden Delicious | M. domestica cv. Golden Delicious | M. domestica cv. Golden Delicious | M.domestica cv. Golden Delicious | M. domestica cv. Golden Delicious |
| DNA source | cDNA | Plasmid MdPPO1 in pGEX-6P-1 | Plasmid MdPPO1 in pGEX-6P-1 | Plasmid MdPPO1 in pGEX-6P-1 | Plasmid MdPPO1 in pGEX-6P-1 |
| Forward primer | 5′-AGCCTATAGCCCCACCAGACG-3′ | 5′-GACACCACACACGCCGGTTCATTTATG-3′ | 5′-GCCGGTTCATAGATGGACCGGTG-3′ | 5′-TCCATCGCGGGGACACCA-3′ | 5′-GGGAATGCTTACTCCGCTGGT-3′ |
| Reverse primer | 5′-CTAAGAAGCAAATTCAATCTTGATACCACCAA-3′ | 5′-CCCTCGATGGAGCCGC-3′ | 5′-GCGTGTGGTGTCCCCTCG-3′ | 5′-GCCGCCACCAGGGTC-3′ | 5′-CATGTCCTCAAAGTTGGGCTG-3′ |
| Cloning vector | pGEX-6P-1 | pGEX-6P-1 | pGEX-6P-1 | pGEX-6P-1 | pGEX-6P-1 |
| Expression vector | pGEX-6P-1 | pGEX-6P-1 | pGEX-6P-1 | pGEX-6P-1 | pGEX-6P-1 |
| Expression host | E. coli BL21 | E. coli BL21 | E. coli BL21 | E. coli BL21 | E. coli BL21 |
| Complete amino acid sequence of the construct produced | MSPILGYWKIKGLVQPTRLLLEYLEEKYEEHLYERDEGDKWRNKKFELGLEFPNLPYYIDGDVKLTQSMAIIRYIADKHNMLGGCPKERAEISMLEGAVLDIRYGVSRIAYSKDFETLKVDFLSKLPEMLKMFEDRLCHKTYLNGDHVTHPDFMLYDALDVVLYMDPMCLDAFPKLVCFKKRIEAIPQIDKYLKSSKYIAWPLQGWQATFGGGDHPPKSDLEVLFQGPKPIAPPDVSKCGPADLPQGAVPTNCCPPPSTKIIDFKLPAPAKLRIRPPAHAVDQAYRDKYYKAMELMKALPDDDPRSFKQQAAVHCAYCDGAYDQVGFPELELQIHNSWLFFPFHRYYLYFFEKILGKLINDPTFALPFWNWDSPAGMPLPAIYADPKSPLYDKLRSANHQPPTLVDLDYNGTEDNVSKETTINANLKIMYRQMVSNSKNAKLFFGNPYRAGDEPDPGGGSIEGTPHAPVHLWTGDNTQPNFEDMGNFYSAGRDPIFFAHHSNVDRMWSIWKTLGGKRTDLTDSDWLDSGFLFYNENAELVRVKVRDCLETKNLGYVYQDVDIPWLSSKPTPRRAKVALSKVAKKLGVAHAAVASSSKVVAGTEFPISLGSKISTVVKRPKQKKRSKKAKEDEEEILVIEGIEFDRDVAVKFDVYVNDVDDLPSGPDKTEFAGSFVSVPHSHKHKKKMNTILRLGLTDLLEEIEAEDDDSVVVTLVPKFGAVKIGGIKIEFAS | MSPILGYWKIKGLVQPTRLLLEYLEEKYEEHLYERDEGDKWRNKKFELGLEFPNLPYYIDGDVKLTQSMAIIRYIADKHNMLGGCPKERAEISMLEGAVLDIRYGVSRIAYSKDFETLKVDFLSKLPEMLKMFEDRLCHKTYLNGDHVTHPDFMLYDALDVVLYMDPMCLDAFPKLVCFKKRIEAIPQIDKYLKSSKYIAWPLQGWQATFGGGDHPPKSDLEVLFQGPKPIAPPDVSKCGPADLPQGAVPTNCCPPPSTKIIDFKLPAPAKLRIRPPAHAVDQAYRDKYYKAMELMKALPDDDPRSFKQQAAVHCAYCDGAYDQVGFPELELQIHNSWLFFPFHRYYLYFFEKILGKLINDPTFALPFWNWDSPAGMPLPAIYADPKSPLYDKLRSANHQPPTLVDLDYNGTEDNVSKETTINANLKIMYRQMVSNSKNAKLFFGNPYRAGDEPDPGGGSIEGTPHTPVHLWTGDNTQPNFEDMGNFYSAGRDPIFFAHHSNVDRMWSIWKTLGGKRTDLTDSDWLDSGFLFYNENAELVRVKVRDCLETKNLGYVYQDVDIPWLSSKPTPRRAKVALSKVAKKLGVAHAAVASSSKVVAGTEFPISLGSKISTVVKRPKQKKRSKKAKEDEEEILVIEGIEFDRDVAVKFDVYVNDVDDLPSGPDKTEFAGSFVSVPHSHKHKKKMNTILRLGLTDLLEEIEAEDDDSVVVTLVPKFGAVKIGGIKIEFAS | MSPILGYWKIKGLVQPTRLLLEYLEEKYEEHLYERDEGDKWRNKKFELGLEFPNLPYYIDGDVKLTQSMAIIRYIADKHNMLGGCPKERAEISMLEGAVLDIRYGVSRIAYSKDFETLKVDFLSKLPEMLKMFEDRLCHKTYLNGDHVTHPDFMLYDALDVVLYMDPMCLDAFPKLVCFKKRIEAIPQIDKYLKSSKYIAWPLQGWQATFGGGDHPPKSDLEVLFQGPKPIAPPDVSKCGPADLPQGAVPTNCCPPPSTKIIDFKLPAPAKLRIRPPAHAVDQAYRDKYYKAMELMKALPDDDPRSFKQQAAVHCAYCDGAYDQVGFPELELQIHNSWLFFPFHRYYLYFFEKILGKLINDPTFALPFWNWDSPAGMPLPAIYADPKSPLYDKLRSANHQPPTLVDLDYNGTEDNVSKETTINANLKIMYRQMVSNSKNAKLFFGNPYRAGDEPDPGGGSIEGTPHAPVHRWTGDNTQPNFEDMGNFYSAGRDPIFFAHHSNVDRMWSIWKTLGGKRTDLTDSDWLDSGFLFYNENAELVRVKVRDCLETKNLGYVYQDVDIPWLSSKPTPRRAKVALSKVAKKLGVAHAAVASSSKVVAGTEFPISLGSKISTVVKRPKQKKRSKKAKEDEEEILVIEGIEFDRDVAVKFDVYVNDVDDLPSGPDKTEFAGSFVSVPHSHKHKKKMNTILRLGLTDLLEEIEAEDDDSVVVTLVPKFGAVKIGGIKIEFAS | MSPILGYWKIKGLVQPTRLLLEYLEEKYEEHLYERDEGDKWRNKKFELGLEFPNLPYYIDGDVKLTQSMAIIRYIADKHNMLGGCPKERAEISMLEGAVLDIRYGVSRIAYSKDFETLKVDFLSKLPEMLKMFEDRLCHKTYLNGDHVTHPDFMLYDALDVVLYMDPMCLDAFPKLVCFKKRIEAIPQIDKYLKSSKYIAWPLQGWQATFGGGDHPPKSDLEVLFQGPKPIAPPDVSKCGPADLPQGAVPTNCCPPPSTKIIDFKLPAPAKLRIRPPAHAVDQAYRDKYYKAMELMKALPDDDPRSFKQQAAVHCAYCDGAYDQVGFPELELQIHNSWLFFPFHRYYLYFFEKILGKLINDPTFALPFWNWDSPAGMPLPAIYADPKSPLYDKLRSANHQPPTLVDLDYNGTEDNVSKETTINANLKIMYRQMVSNSKNAKLFFGNPYRAGDEPDPGGGSIAGTPHAPVHLWTGDNTQPNFEDMGNFYSAGRDPIFFAHHSNVDRMWSIWKTLGGKRTDLTDSDWLDSGFLFYNENAELVRVKVRDCLETKNLGYVYQDVDIPWLSSKPTPRRAKVALSKVAKKLGVAHAAVASSSKVVAGTEFPISLGSKISTVVKRPKQKKRSKKAKEDEEEILVIEGIEFDRDVAVKFDVYVNDVDDLPSGPDKTEFAGSFVSVPHSHKHKKKMNTILRLGLTDLLEEIEAEDDDSVVVTLVPKFGAVKIGGIKIEFAS | MSPILGYWKIKGLVQPTRLLLEYLEEKYEEHLYERDEGDKWRNKKFELGLEFPNLPYYIDGDVKLTQSMAIIRYIADKHNMLGGCPKERAEISMLEGAVLDIRYGVSRIAYSKDFETLKVDFLSKLPEMLKMFEDRLCHKTYLNGDHVTHPDFMLYDALDVVLYMDPMCLDAFPKLVCFKKRIEAIPQIDKYLKSSKYIAWPLQGWQATFGGGDHPPKSDLEVLFQGPKPIAPPDVSKCGPADLPQGAVPTNCCPPPSTKIIDFKLPAPAKLRIRPPAHAVDQAYRDKYYKAMELMKALPDDDPRSFKQQAAVHCAYCDGAYDQVGFPELELQIHNSWLFFPFHRYYLYFFEKILGKLINDPTFALPFWNWDSPAGMPLPAIYADPKSPLYDKLRSANHQPPTLVDLDYNGTEDNVSKETTINANLKIMYRQMVSNSKNAKLFFGNPYRAGDEPDPGGGSIEGTPHAPVHLWTGDNTQPNFEDMGNAYSAGRDPIFFAHHSNVDRMWSIWKTLGGKRTDLTDSDWLDSGFLFYNENAELVRVKVRDCLETKNLGYVYQDVDIPWLSSKPTPRRAKVALSKVAKKLGVAHAAVASSSKVVAGTEFPISLGSKISTVVKRPKQKKRSKKAKEDEEEILVIEGIEFDRDVAVKFDVYVNDVDDLPSGPDKTEFAGSFVSVPHSHKHKKKMNTILRLGLTDLLEEIEAEDDDSVVVTLVPKFGAVKIGGIKIEFAS |
Figure 2.
SDS–PAGE with 7 µg of enzyme under reducing conditions. (a) Wild-type MdPPO1, (b) MdPPO1-Ala239Thr and MdPPO1-Leu243Arg, and (c) MdPPO1-Glu234Ala and MdPPO1-Phe259Ala. The arrows indicate the position of the enzyme. Lanes M contain molecular-mass markers (labelled in kDa).
2.2. Protein crystallization
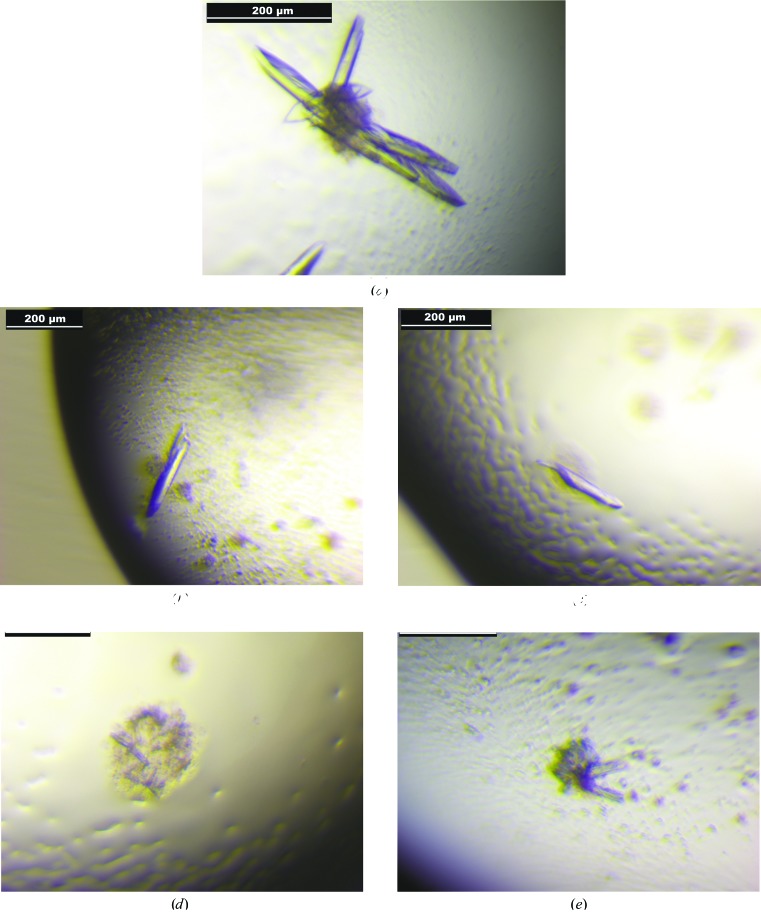
Initial screening for suitable crystallization conditions for wild-type MdPPO1 was performed by the sitting-drop vapour-diffusion method using a nanodispenser robot (Gryphon, Art Robbins) and 96-well Crystal Quick plates (Greiner Bio-One). A wide range of crystallization conditions were screened by using a variety of commercially available screening kits (JBScreen Classic 1–10, JBScreen Membrane 1–3 and Pi-PEG Screen HTS from Jena Bioscience). The screening procedure yielded some promising hits, which were further optimized manually by applying the hanging-drop vapour-diffusion technique using 15-well EasyXtal plates (Qiagen). Single crystals of wild-type MdPPO1 and of the mutants MdPPO1-Ala239Thr and MdPPO1-Phe259Ala (Fig. 3 ▸) were grown at 20°C by mixing 1 µl protein solution (5–10 mg ml−1) with 2 µl reservoir solution (50 mM Tris–HCl pH 7.0, 19–21% PEG 3350). Crystals usually appeared after 10–15 d. Crystallization information is summarized in Table 2 ▸. Crystallization trials of the mutants MdPPO1-Leu243Arg and MdPPO1-Glu234Ala applying the above-described methods yielded only low-quality crystals which were not suitable for adequate data collection (Fig. 3 ▸).
Figure 3.
Crystals of (a) wild-type MdPPO1, (b) MdPPO1-Ala239Thr, (c) MdPPO1-Phe259Ala, (d) MdPPO1-Glu234Ala and (e) MdPPO1-Leu243Ala.
Table 2. Crystallization conditions for latent wild-type MdPPO1 and the mutants MdPPO1-Ala239Thr, MdPPO1-Leu243Ala, MdPPO1-Glu234Ala and MdPPO1-Phe259Ala.
| Method | Vapour diffusion (hanging drop) |
| Plate type | 15-well EasyXtal plates (Qiagen) |
| Temperature (K) | 293 |
| Protein concentration (mg ml−1) | 5–10 |
| Buffer composition of protein solution | 50 mM Tris–HCl pH 7.5, 200 mM NaCl |
| Composition of reservoir solution | 50 mM Tris–HCl pH 7.0, 19–21% PEG 3350 |
| Volume and ratio of drop | 1 µl protein solution, 2 µl reservoir solution |
| Volume of reservoir (µl) | 500 |
2.3. Data collection and processing
Crystals of latent wild-type MdPPO1, MdPPO1-Ala239Thr and MdPPO1-Phe259Ala were mounted in nylon loops and flash-cooled in liquid nitrogen after quick soaking in cryoprotectant solution composed of 50 mM Tris–HCl pH 7.5, 200 mM NaCl, 20% PEG 3350, 20–25% PEG 1500. Data collection for wild-type MdPPO1 was carried out at 100 K on beamline ID23 at the ESRF, Grenoble, France (Table 3 ▸), whereas data for the mutants MdPPO1-Ala239Thr and MdPPO1-Phe259Ala were collected at 100 K on beamline ID30A-3/MASSIF-3 at the ESRF, Grenoble, France (Table 3 ▸). The data sets were processed (indexing, integration and scaling) with XDS (Kabsch, 2010 ▸). All crystals belonged to space group P212121, which was determined using POINTLESS (Evans, 2011 ▸) from the CCP4 suite (Winn et al., 2011 ▸).
Table 3. Data-collection and processing statistics for wild-type MdPPO1, MdPPO1-Ala239Thr and MdPPO1-Phe259Ala.
Values in parentheses are for the outer shell.
| Protein | Wild-type MdPPO1 | MdPPO1-Ala239Thr | MdPPO1-Phe259Ala |
|---|---|---|---|
| Diffraction source | ID23, ESRF | ID30A-3, ESRF | ID30A-3, ESRF |
| Wavelength (Å) | 0.97242 | 0.96770 | 0.96770 |
| Temperature (K) | 100 | 100 | 100 |
| Detector | PILATUS 6M | EIGER 4M | EIGER 4M |
| Crystal-to-detector distance (mm) | 189.76 | 118.16 | 118.16 |
| Rotation range per image (°) | 0.1 | 0.1 | 0.1 |
| Total rotation range (°) | 320 | 300 | 300 |
| Exposure time per image (s) | 0.125 | 0.05 | 0.05 |
| Space group | P212121 | P212121 | P212121 |
| a, b, c (Å) | 50.70, 80.15, 115.96 | 50.58, 79.90, 115.76 | 50.53, 79.76, 116.07 |
| Mosaicity (°) | 0.173 | 0.073 | 0.199 |
| Resolution range (Å) | 46.45–1.346 | 49.95–1.550 | 34.81–1.698 |
| Total No. of reflections | 1214526 (117878) | 725427 (73222) | 566074 (53670) |
| No. of unique reflections | 104709 (10123) | 67568 (6580) | 51889 (5071) |
| Completeness (%) | 99.51 (97.62) | 98.2 (96.8) | 98.6 (97.70) |
| Multiplicity | 11.6 (11.6) | 10.7 (11.1) | 10.9 (10.6) |
| 〈I/σ(I)〉 | 11.72 (2.02) | 11.90 (1.50) | 6.67 (1.06) |
| R r.i.m. | 0.120 (1.174) | 0.119 (1.687) | 0.257 (1.698) |
| R p.i.m † | 0.034 (0.337) | 0.036 (0.495) | 0.076 (0.505) |
| CC1/2 ‡ | 0.996 (0.708) | 0.999 (0.564) | 0.992 (0.478) |
| Overall B factor from Wilson plot (Å2) | 14.00 | 20.27 | 18.89 |
R
p.i.m. = 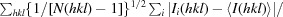
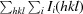 , where Ii(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations of reflection hkl.
, where Ii(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations of reflection hkl.
CC1/2 is defined as the correlation coefficient between two random half data sets, as described by Karplus & Diederichs (2012 ▸).
2.4. In crystallo activity tests
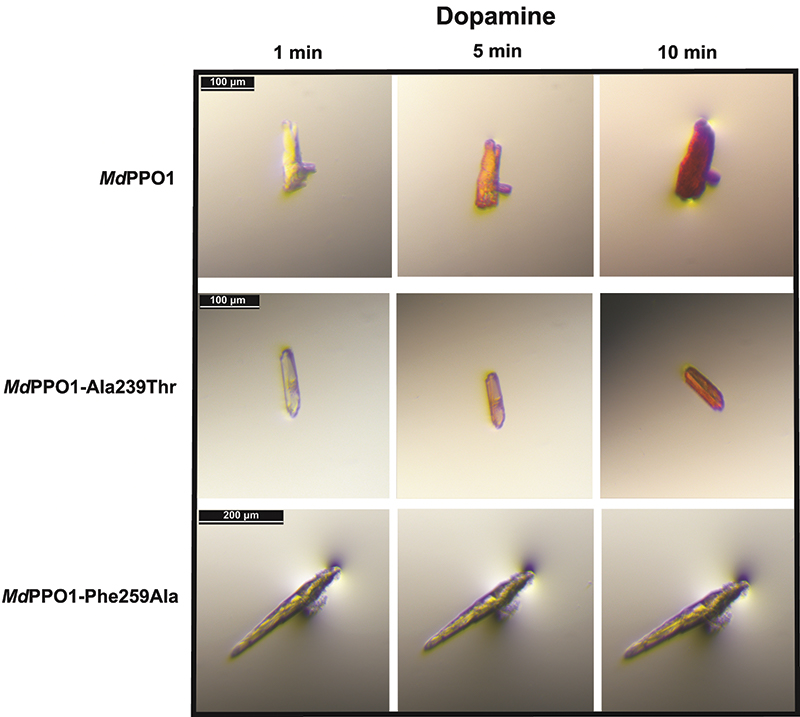
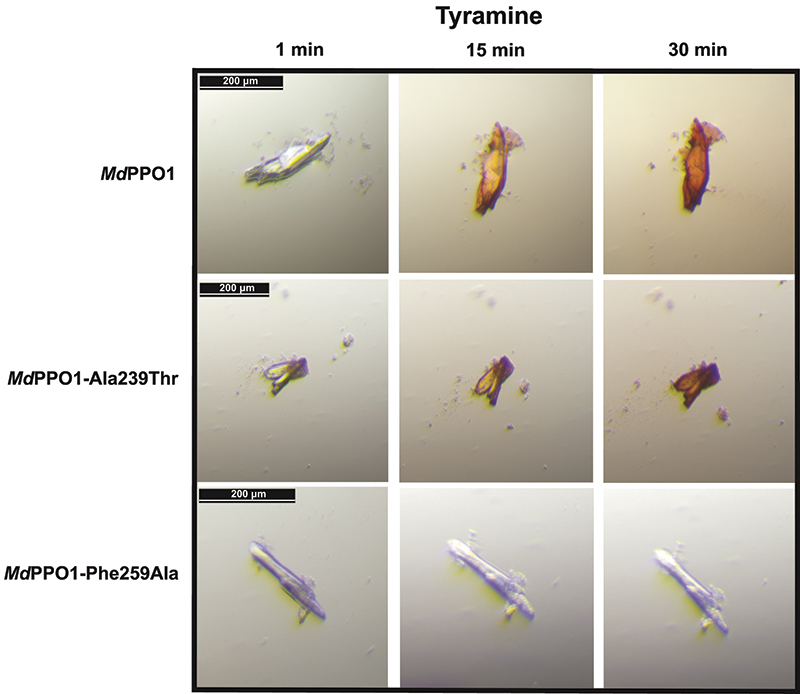
PPO reactions produce unstable quinones which further polymerize to chromophoric substances, and therefore it was possible to detect differences in activity between wild-type MdPPO1 and the mutants in crystallo. For this reason, crystals of the wild type and its mutants were transferred to drops consisting of the mother liquor with an additional 3 mM SDS, which is needed to activate the latent enzyme, and 10 mM substrate, namely tyramine or dopamine, to detect monophenolase and diphenolase activity, respectively. After the transfer of a crystal to a substrate-containing drop, pictures were taken during the reaction with dopamine (Fig. 4 ▸) or tyramine (Fig. 5 ▸). In this way, the conversion of the crystals was followed and differences were observed between the wild type and the mutants in the intensity and speed of the colouration.
Figure 4.
In crystallo activity tests: photographs of crystals of wild-type MdPPO1 and the mutants MdPPO1-Ala239Thr and MdPPO1-Phe259Ala with 10 mM dopamine and 3 mM SDS as an activator. Crystal photographs were taken after 1, 5 and 10 min, respectively.
Figure 5.
In crystallo activity tests: photographs of crystals of wild-type MdPPO1 and the mutants MdPPO1-Ala239Thr and MdPPO1-Phe259Ala with 10 mM tyramine and 3 mM SDS as an activator. Crystal photographs were taken after 1, 15 and 30 min, respectively.
3. Results and discussion
The method described by Kampatsikas et al. (2017 ▸) was used to produce sufficient amounts (225 mg per litre of culture) of wild-type MdPPO1 in its latent form as well as the mutants MdPPO1-Ala239Thr, MdPPO1-Leu243Arg, MdPPO1-Glu234Ala and MdPPO1-Phe259Ala. For crystallization reasons, wild-type MdPPO1 was screened with a total of 408 crystallization conditions, which led to four promising hits: (i) 4.3%(w/v) PEG 4000 and 25%(w/v) PEG 1500 in 50 mM sodium acetate buffer pH 4.8; (ii) 5.7%(v/v) PEG 350 monomethyl ether (MME) and 21.4%(w/v) PEG 3000 in 50 mM MES buffer pH 6.0; (iii) 2.5%(w/v) PEG 1000 and 25.7%(w/v) PEG 2000 MME in 50 mM sodium acetate buffer pH 4.8; and (iv) 2.5%(w/v) PEG 1500 and 25.7%(w/v) PEG 3000 in 50 mM sodium acetate buffer pH 5.2. The initial crystals were further optimized, resulting in large but twinned crystals. However, it was possible to separate single crystals from the twinned crystal clusters, thus obtaining single crystals that were suitable for data collection (Fig. 3 ▸ a). Similar conditions were tested for the crystallization of the four mutants. Crystals of MdPPO1-Ala239Thr and MdPPO1-Phe259Ala were obtained under the same conditions as used for the wild type (Table 2 ▸, Figs. 3 ▸ b and 3 ▸ c), while MdPPO1-Glu234Ala and MdPPO1-Leu243Ala only formed microcrystals that hardly diffracted and failed to produce evaluable data during the X-ray diffraction experiment (Figs. 3 ▸ d and 3 ▸ e). The crystals were obtained from solutions containing PEGs of different molecular masses as the precipitation agent. The utilization of PEG as a precipitation agent is the only constant among all published crystallization conditions of fungal tyrosinases (Ismaya, Rozeboom, Schurink et al., 2011 ▸; Ismaya, Rozeboom, Weijn et al., 2011 ▸; Mauracher et al., 2014a ▸,b ▸) and bacterial tyrosinases (Matoba et al., 2006 ▸; Sendovski et al., 2011 ▸) as well as of plant catechol oxidases (Klabunde et al., 1998 ▸; Virador et al., 2010 ▸; Molitor, Mauracher & Rompel, 2015 ▸) and plant tyrosinases (Zekiri, Molitor et al., 2014 ▸; Zekiri, Bijelic et al., 2014 ▸; Bijelic et al., 2015a, ▸ b ▸). The obtained crystals of wild-type MdPPO1 and the mutants MdPPO1-Ala239Thr and MdPPO1-Phe259Ala diffracted X-rays to 1.35, 1.55 and 1.70 Å resolution, respectively. Data-collection and processing statistics are summarized in Table 3 ▸, revealing that the data sets are of good quality. Phasing applying the molecular-replacement approach and subsequent structure determination is currently under way. Several promising models for the molecular-replacement step are available according to the results of a BLAST search (Altschul et al., 1990 ▸): tyrosinase from Juglans regia (sequence identity 66.6%; UniProt C0LU17; PBD entry 5ce9; Zekiri, Bijelic et al., 2014 ▸; Bijelic et al., 2015a, ▸ b ▸), catechol oxidase from Vitis vinifera (sequence identity 59.2%; UniProt P43311; PDB entry 2p3x; Virador et al., 2010 ▸), catechol oxidase from Ipomoea batatas (sequence identity 53.0%; UniProt Q9ZP19; PDB entry 1bt3; Klabunde et al., 1998 ▸) and aurone synthase from C. grandiflora (sequence identity 43.0%; UniProt A0A075DN54; PDB entry 4z11; Molitor, Mauracher & Rompel, 2015 ▸, 2016 ▸). The latter has previously been used for homology modelling of MdPPO1 in Kampatsikas et al. (2017 ▸).
The in crystallo activity test showed that the wild type is active towards both substrates, as expected and described in Kampatsikas et al. (2017 ▸). This was obvious owing to the rapid and intense colouration of the wild-type crystal with both dopamine (Fig. 4 ▸) and tyramine (Fig. 5 ▸). MdPPO1-Ala239Thr represents a mutation that affects one of the previously described activity-controller residues, namely Ala239, which was mutated from a small hydrophobic alanine to a polar threonine residue in order to influence the activity, as it was suggested that the polarity of the environment of the active site might be an important factor in the activity specificity of plant PPOs (Fig. 1 ▸ b). According to the in crystallo activity test, this mutation has only a minor impact on the diphenolase activity as the speed and intensity of crystal browning is comparable to that of the wild type (Fig. 4 ▸). However, this mutation led to significantly slower (1–2 min) and less intense browning with the monophenol tyramine, indicating a decrease in monophenolase activity. The crystal of the mutant MdPPO1-Ala239Thr maintains the monophenolase activity, indicating that this mutation is not sufficient to adequately diminish the reaction with tyramine. The mutation of the bulky gatekeeper Phe259 to a small alanine (Phe259Ala) led to a dramatic change in both diphenolase (Fig. 4 ▸) and monophenolase (Fig. 5 ▸) activity. In both cases no browning of the crystals was observed, indicating that this mutation leads to loss of both activities over the measured time. This observation is in accordance with the theory that an aromatic system at the gatekeeper position of plant PPOs is important for the stabilization and the orientation of the substrate. The importance of this conserved phenylalanine for the activity of plant PPOs is in contrast to PPOs from other kingdoms (fungi and bacteria), where this position is not occupied by a phenylalanine.
The crystal structure of MdPPO1 will be the first of a plant tyrosinase in its latent form. The C-terminal structure will be of great interest as it could provide highly valuable information about the further functions of the C-terminal domain (besides its active-site shielding role) and the putative maturation site of the enzyme. This could enhance knowledge about the entire maturation process of plant tyrosinases. Furthermore, structure determination, comprehensive enzyme kinetic assays and the preparation of enzyme–inhibitor complex crystals for the wild type and the mutants are currently in progress and will hopefully provide some new structural insights into the monophenolase and diphenolase discrepancy between TYRs and COs and thus contribute to this longstanding scientific issue. The structures might reveal significant differences between the wild type and the mutants which could be used to deduce the reason for their differences in activity, which will be determined exactly using Michaelis–Menten kinetics. The differences in both the structures and the enzymatic activities upon mutation will improve knowledge of the catalytic event in plant PPOs; also, the crystal structures of inhibitor complexes of the enzyme may provide information to resolve many of the queries about substrate binding in the active centre of PPOs.
Acknowledgments
We thank the staff at the ESRF and EMBL Grenoble, especially Dr Gianluca Santoni for assistance and support during the beamtime at beamline ID23 and Dr Ulrich Zander for support at beamline ID30A-3/MASSIF-3. Moreover, Dr Christian Molitor is greatly acknowledged for his support during data collection and for invaluable discussions.
References
- Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990). J. Mol. Biol. 215, 403–410. [DOI] [PubMed]
- Bijelic, A., Pretzler, M., Molitor, C., Zekiri, F. & Rompel, A. (2015). Angew. Chem. 127, 14889–14893. [DOI] [PMC free article] [PubMed]
- Bijelic, A., Pretzler, M., Molitor, C., Zekiri, F. & Rompel, A. (2015). Angew. Chem. Int. Ed. 54, 14677–14680. [DOI] [PMC free article] [PubMed]
- Evans, P. R. (2011). Acta Cryst. D67, 282–292. [DOI] [PMC free article] [PubMed]
- Flurkey, W. H. & Inlow, J. K. (2008). J. Inorg. Biochem. 102, 2160–2170. [DOI] [PubMed]
- Gandía-Herrero, F., Jiménez-Atiénzar, M., Cabanes, J., García-Carmona, F. & Escribano, J. (2005a). J. Agric. Food Chem. 53, 6825–6830. [DOI] [PubMed]
- Gandía-Herrero, F., Jiménez-Atiénzar, M., Cabanes, J., García-Carmona, F. & Escribano, J. (2005b). Biol. Chem. 386, 601–607. [DOI] [PubMed]
- Goldfeder, M., Kanteev, M., Isaschar-Ovdat, S., Adir, N. & Fishman, A. (2014). Nature Commun. 5, 4505. [DOI] [PubMed]
- Ismaya, W. T., Rozeboom, H. J., Schurink, M., Boeriu, C. G., Wichers, H. & Dijkstra, B. W. (2011). Acta Cryst. F67, 575–578. [DOI] [PMC free article] [PubMed]
- Ismaya, W. T., Rozeboom, H. J., Weijn, A., Mes, J. J., Fusetti, F., Wichers, H. J. & Dijkstra, B. W. (2011). Biochemistry, 50, 5477–5486. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kaintz, C., Mauracher, S. G. & Rompel, A. (2014). Adv. Protein Chem. Struct. Biol. 97, 1–35. [DOI] [PubMed]
- Kaintz, C., Mayer, R. L., Jirsa, F., Halbwirth, H. & Rompel, A. (2015). FEBS Lett. 589, 789–797. [DOI] [PMC free article] [PubMed]
- Kaintz, C., Molitor, C., Thill, J., Kampatsikas, I., Michael, C., Halbwirth, H. & Rompel, A. (2014). FEBS Lett. 588, 3417–3426. [DOI] [PMC free article] [PubMed]
- Kampatsikas, I., Bijelic, A., Pretzler, M. & Rompel, A. (2017). Sci. Rep. https://doi.org/10.1038/s41598-017-08097-5.
- Kanteev, M., Goldfeder, M. & Fishman, A. (2015). Protein Sci. 24, 1360–1369. [DOI] [PMC free article] [PubMed]
- Karplus, P. A. & Diederichs, K. (2012). Science, 336, 1030–1033. [DOI] [PMC free article] [PubMed]
- Klabunde, T., Eicken, C., Sacchettini, J. C. & Krebs, B. (1998). Nature Struct. Mol. Biol. 5, 1084–1090. [DOI] [PubMed]
- Mason, H. S. (1948). J. Biol. Chem. 172, 83–99. [PubMed]
- Mason, H. S. (1955). Adv. Enzymol. Relat. Areas Mol. Biol. 16, 105–184.
- Matoba, Y., Kumagai, T., Yamamoto, A., Yoshitsu, H. & Sugiyama, M. (2006). J. Biol. Chem. 281, 8981–8990. [DOI] [PubMed]
- Mauracher, S. G., Molitor, C., Al-Oweini, R., Kortz, U. & Rompel, A. (2014a). Acta Cryst. F70, 263–266. [DOI] [PMC free article] [PubMed]
- Mauracher, S. G., Molitor, C., Al-Oweini, R., Kortz, U. & Rompel, A. (2014b). Acta Cryst. D70, 2301–2315. [DOI] [PMC free article] [PubMed]
- Mayer, A. M. (2006). Phytochemistry, 67, 2318–2331. [DOI] [PubMed]
- Mayer, A. M., Harel, E. & Ben-Shaul, R. (1966). Phytochemistry, 5, 783–789.
- Molitor, C., Bijelic, A. & Rompel, A. (2016). Chem. Commun. 52, 12286–12289. [DOI] [PMC free article] [PubMed]
- Molitor, C., Mauracher, S. G., Pargan, S., Mayer, R. L., Halbwirth, H. & Rompel, A. (2015). Planta, 242, 519–537. [DOI] [PMC free article] [PubMed]
- Molitor, C., Mauracher, S. G. & Rompel, A. (2015). Acta Cryst. F71, 746–751. [DOI] [PMC free article] [PubMed]
- Molitor, C., Mauracher, S. G. & Rompel, A. (2016). Proc. Natl Acad. Sci. USA, 113, E1806–E1815. [DOI] [PMC free article] [PubMed]
- Pretzler, M., Bijelic, A. & Rompel, A. (2015). Reference Module in Chemistry, Molecular Sciences and Chemical Engineering. https://doi.org/10.1016/B978-0-12-409547-2.11521-5.
- Pretzler, M., Bijelic, A. & Rompel, A. (2017). Sci. Rep. 7, 1810. [DOI] [PMC free article] [PubMed]
- Pretzler, M. & Rompel, A. (2017). Inorg. Chim. Acta. https://doi.org/10.1016/j.ica.2017.04.041.
- Ramsden, C. A. & Riley, P. A. (2014). Bioorg. Med. Chem. 22, 2388–2395. [DOI] [PubMed]
- Rodríguez-López, J. N., Tudela, J., Varón, R., García-Carmona, F. & García-Cánovas, F. (1992). J. Biol. Chem. 267, 3801–3810. [PubMed]
- Sendovski, M., Kanteev, M., Ben-Yosef, V. S., Adir, N. & Fishman, A. (2011). J. Mol. Biol. 405, 227–237. [DOI] [PubMed]
- Sugumaran, M. & Nellaiappan, K. (1991). Biochem. Biophys. Res. Commun. 176, 1371–1376. [DOI] [PubMed]
- Tran, L. T., Taylor, J. S. & Constabel, C. P. (2012). BMC Genomics, 13, 395. [DOI] [PMC free article] [PubMed]
- Valero, E. & García-Carmona, F. (1992). Biochem. J. 286, 623–626. [DOI] [PMC free article] [PubMed]
- Virador, V. M., Reyes Grajeda, J. P., Blanco-Labra, A., Mendiola-Olaya, E., Smith, G. M., Moreno, A. & Whitaker, J. R. (2010). J. Agric. Food Chem. 58, 1189–1201. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Zekiri, F., Bijelic, A., Molitor, C. & Rompel, A. (2014). Acta Cryst. F70, 832–834. [DOI] [PMC free article] [PubMed]
- Zekiri, F., Molitor, C., Mauracher, S. G., Michael, C., Mayer, R. L., Gerner, C. & Rompel, A. (2014). Phytochemistry, 101, 5–15. [DOI] [PMC free article] [PubMed]