Abstract
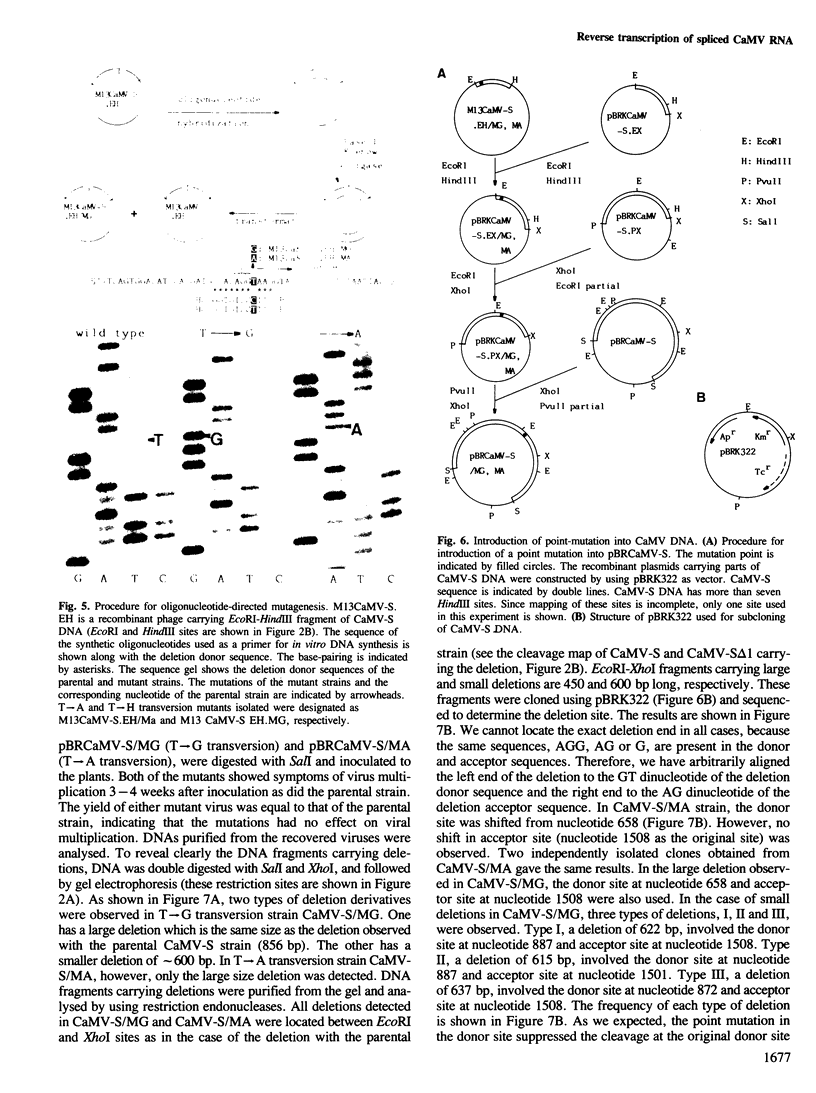
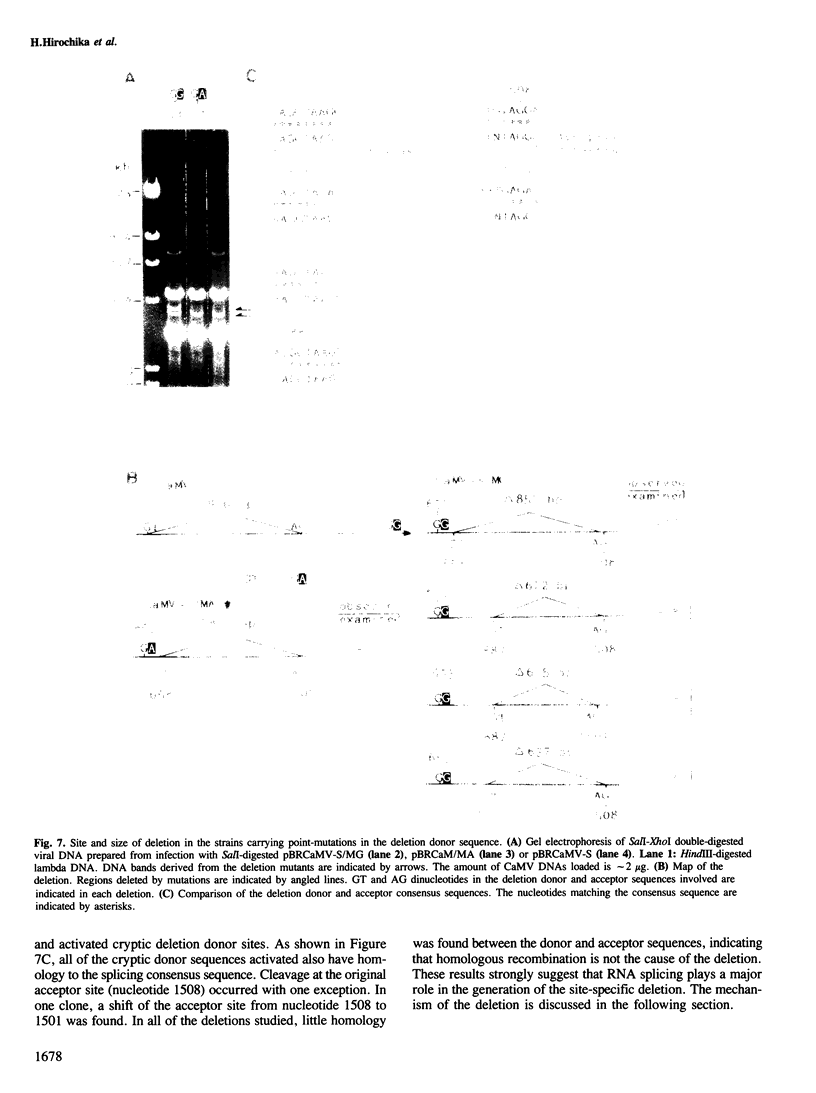
A frequent site-specific deletion was observed in the life cycle of cauliflower mosaic virus (S strain). Analysis of the sequence around the deletion site and the parental sequence implied that the deletion was promoted at sequences similar to the donor and acceptor consensus sequences of RNA splicing, designated as the deletion donor and acceptor sequences, respectively. To elucidate the mechanism of this site-specific deletion, point mutations were introduced into the deletion donor sequence (GT to GG or GA transversion). Deletion at the original deletion donor site did not occur in these mutants, instead, new (cryptic) donor sites were activated. All of these activated cryptic sites had sequences similar to the splicing consensus sequence. In all cases except one, the original deletion acceptor site was used. These results can be most readily explained by postulating that the site-specific deletion occurs by reverse transcription of spliced viral RNA. This frequent site-specific deletion was not observed in other strains. For a virus which replicates by reverse transcription, a mechanism to regulate the rate of splicing is required to ensure the intactness of the viral genome. We discuss the possibility that the S strain has a mutation in this regulatory mechanism.
Keywords: cauliflower mosaic virus, RNA splicing, reverse transcription, deletion mutation, mutagenesis
Full text
PDF
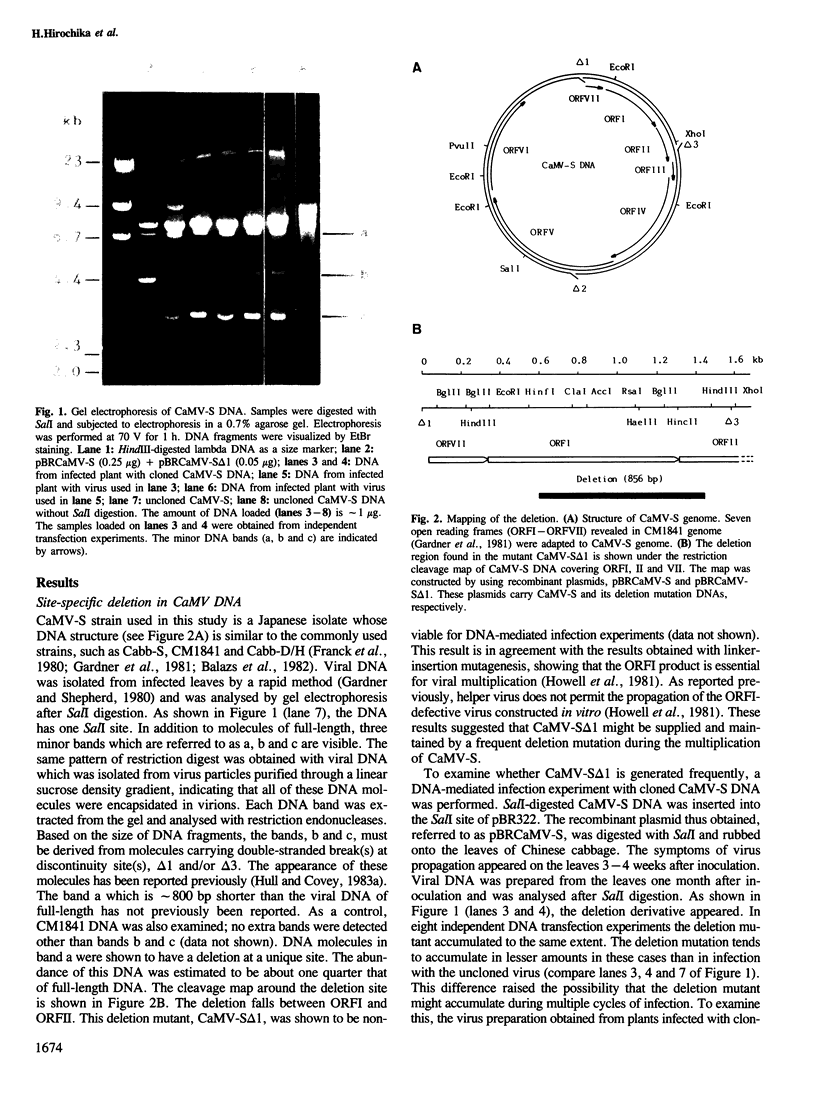

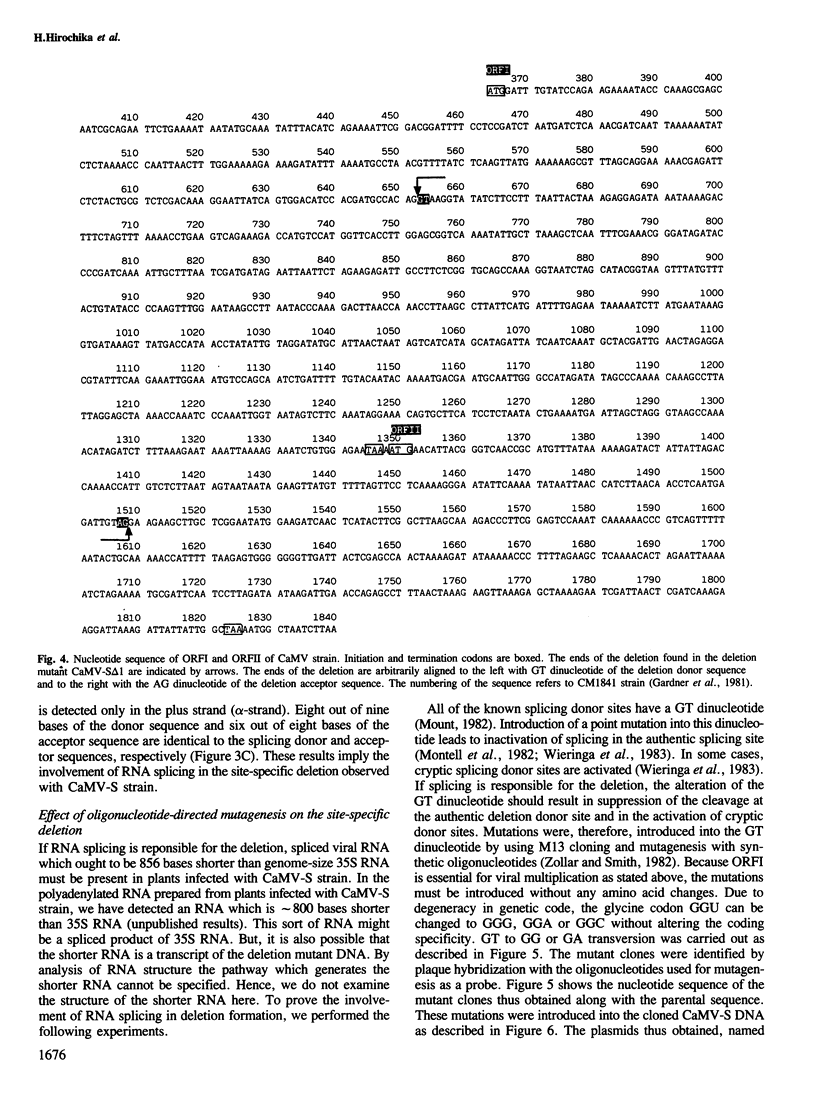


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balàzs E., Guilley H., Jonard G., Richards K. Nucleotide sequence of DNA from an altered-virulence isolate D/H of the cauliflower mosaic virus. Gene. 1982 Oct;19(3):239–249. doi: 10.1016/0378-1119(82)90013-0. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condit C., Hagen T. J., McKnight T. D., Meagher R. B. Characterization and preliminary mapping of cauliflower mosaic virus transcripts. Gene. 1983 Nov;25(1):101–108. doi: 10.1016/0378-1119(83)90172-5. [DOI] [PubMed] [Google Scholar]
- Covey S. N., Lomonossoff G. P., Hull R. Characterisation of cauliflower mosaic virus DNA sequences which encode major polyadenylated transcripts. Nucleic Acids Res. 1981 Dec 21;9(24):6735–6747. doi: 10.1093/nar/9.24.6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck A., Guilley H., Jonard G., Richards K., Hirth L. Nucleotide sequence of cauliflower mosaic virus DNA. Cell. 1980 Aug;21(1):285–294. doi: 10.1016/0092-8674(80)90136-1. [DOI] [PubMed] [Google Scholar]
- Gardner R. C., Howarth A. J., Hahn P., Brown-Luedi M., Shepherd R. J., Messing J. The complete nucleotide sequence of an infectious clone of cauliflower mosaic virus by M13mp7 shotgun sequencing. Nucleic Acids Res. 1981 Jun 25;9(12):2871–2888. doi: 10.1093/nar/9.12.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilley H., Dudley R. K., Jonard G., Balàzs E., Richards K. E. Transcription of Cauliflower mosaic virus DNA: detection of promoter sequences, and characterization of transcripts. Cell. 1982 Oct;30(3):763–773. doi: 10.1016/0092-8674(82)90281-1. [DOI] [PubMed] [Google Scholar]
- Guilley H., Richards K. E., Jonard G. Observations concerning the discontinuous DNAs of cauliflower mosaic virus. EMBO J. 1983;2(2):277–282. doi: 10.1002/j.1460-2075.1983.tb01417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn T., Richards K., Geneviève-Lebeurier Cauliflower mosaic virus on its way to becoming a useful plant vector. Curr Top Microbiol Immunol. 1982;96:194–236. [PubMed] [Google Scholar]
- Hull R., Covey S. N. Characterisation of cauliflower mosaic virus DNA forms isolated from infected turnip leaves. Nucleic Acids Res. 1983 Mar 25;11(6):1881–1895. doi: 10.1093/nar/11.6.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. D., Barkan A., Mertz J. E. Nucleotide sequence analysis of the recombinant joints in 16 naturally arising deletion mutants of simian virus 40. Virology. 1982 Dec;123(2):464–469. doi: 10.1016/0042-6822(82)90281-1. [DOI] [PubMed] [Google Scholar]
- Leis J. P., Scheible P., Smith R. E. Correlation of RNA binding affinity of avian oncornavirus p19 proteins with the extent of processing of virus genome RNA in cells. J Virol. 1980 Sep;35(3):722–731. doi: 10.1128/jvi.35.3.722-731.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner M. R., Boyle J. A., Mount S. M., Wolin S. L., Steitz J. A. Are snRNPs involved in splicing? Nature. 1980 Jan 10;283(5743):220–224. doi: 10.1038/283220a0. [DOI] [PubMed] [Google Scholar]
- Majors J. E., Varmus H. E. Nucleotide sequencing of an apparent proviral copy of env mRNA defines determinants of expression of the mouse mammary tumor virus env gene. J Virol. 1983 Sep;47(3):495–504. doi: 10.1128/jvi.47.3.495-504.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Montell C., Fisher E. F., Caruthers M. H., Berk A. J. Resolving the functions of overlapping viral genes by site-specific mutagenesis at a mRNA splice site. Nature. 1982 Feb 4;295(5848):380–384. doi: 10.1038/295380a0. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S., Steitz J. Lessons from mutant globins. Nature. 1983 Jun 2;303(5916):380–381. doi: 10.1038/303380a0. [DOI] [PubMed] [Google Scholar]
- Olszewski N. E., Guilfoyle T. J. Nuclei purified from cauliflower mosaic virus-infected turnip leaves contain subgenomic, covalently closed circular cauliflower mosaic virus DNAs. Nucleic Acids Res. 1983 Dec 20;11(24):8901–8914. doi: 10.1093/nar/11.24.8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer P., Hohn T. Involvement of reverse transcription in the replication of cauliflower mosaic virus: a detailed model and test of some aspects. Cell. 1983 Jul;33(3):781–789. doi: 10.1016/0092-8674(83)90020-x. [DOI] [PubMed] [Google Scholar]
- Richards K. E., Guilley H., Jonard G. Further characterization of the discontinuities in cauliflower mosaic virus DNA. FEBS Lett. 1981 Nov 2;134(1):67–70. doi: 10.1016/0014-5793(81)80552-2. [DOI] [PubMed] [Google Scholar]
- Rogers J., Wall R. A mechanism for RNA splicing. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1877–1879. doi: 10.1073/pnas.77.4.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A. Conversion of RNA to DNA in mammals: Alu-like elements and pseudogenes. Nature. 1983 Feb 10;301(5900):471–472. doi: 10.1038/301471a0. [DOI] [PubMed] [Google Scholar]
- Shimotohno K., Temin H. M. Loss of intervening sequences in genomic mouse alpha-globin DNA inserted in an infectious retrovirus vector. Nature. 1982 Sep 16;299(5880):265–268. doi: 10.1038/299265a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tanaka H., Muraki M., Honda K., Nakazato S. Some improvements in the separation technique for the chemical syntheses of oligodeoxyribonucleotides. Nucleic Acids Symp Ser. 1982;(11):73–76. [PubMed] [Google Scholar]
- Toh H., Hayashida H., Miyata T. Sequence homology between retroviral reverse transcriptase and putative polymerases of hepatitis B virus and cauliflower mosaic virus. 1983 Oct 27-Nov 2Nature. 305(5937):827–829. doi: 10.1038/305827a0. [DOI] [PubMed] [Google Scholar]
- Varmus H. E. Form and function of retroviral proviruses. Science. 1982 May 21;216(4548):812–820. doi: 10.1126/science.6177038. [DOI] [PubMed] [Google Scholar]
- Wieringa B., Meyer F., Reiser J., Weissmann C. Unusual splice sites revealed by mutagenic inactivation of an authentic splice site of the rabbit beta-globin gene. Nature. 1983 Jan 6;301(5895):38–43. doi: 10.1038/301038a0. [DOI] [PubMed] [Google Scholar]
- Yang V. W., Lerner M. R., Steitz J. A., Flint S. J. A small nuclear ribonucleoprotein is required for splicing of adenoviral early RNA sequences. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1371–1375. doi: 10.1073/pnas.78.3.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis using M13-derived vectors: an efficient and general procedure for the production of point mutations in any fragment of DNA. Nucleic Acids Res. 1982 Oct 25;10(20):6487–6500. doi: 10.1093/nar/10.20.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]