Abstract
In multi-cellular organisms, gene expression is orchestrated by thousands of transcription factors (TF). Chromatin immunoprecipitation followed by sequencing (ChIP-seq) is a robust tool to investigate gene expression because this technique profiles in vivo protein-DNA interaction at a genome-wide scale. Eight years after the first ChIP-seq paper, there are limited reports of ChIP-seq experiments in plants, especially for sequence-specific DNA binding TFs This lag greatly prevents our understanding of transcriptional regulation in an entire kingdom. In order to bridge the technical gap, we describe a ChIP-seq procedure that we have successfully applied to dozens of sequence-specific DNA binding TFs. The basic protocol includes procedures to isolate nuclei, sonicate chromatin, immunoprecipitate TF-DNA complex, and recover ChIP-enriched DNA fragments. The support protocol also describes practices to optimize library preparation by a gel-free DNA size selection. Lastly, examples are given to optimize library amplification using real-time PCR.
Keywords: ChIP-seq, transcription factor, Arabidopsis
INTRODUCTION
ChIP-seq is a robust approach to investigate transcriptional regulation in vivo (Ren et al., 2000; Johnson et al., 2007). It is an improvement over ChIP-chip with better signal-to-noise ratio, unambiguous and genome-wide sequence information. ChIP-seq can be applied to any protein that is associated with chromatin, including histones, chromatin remodelers, RNA polymerases, mediators, general and sequence-specific DNA binding transcription factors. Sequence-specific DNA binding TFs are of particular interest because of their large number and functional diversity; yet they are also the most technically challenging to study because of their low protein abundance and rapid turnover in cells. The progress of ChIP-seq in plants has lagged far behind other eukaryotic organisms: a mere 1% of all ChIP-seq samples submitted to Gene Expression Omnibus (GEO) are from plants. Eight years after the publication of the initial ChIP-seq paper, fewer than 30 sequence-specific DNA binding TFs in Arabidopsis have been subjected to ChIP-seq (Heyndrickx et al., 2014; Fan et al., 2014; Pfeiffer et al., 2014). This unit aims to bridge this gap by providing a technical guidance of ChIP-seq experiments in plants.
BASIC PROTOCOL 1
Enrichment of transcription factor bound DNA fragments by chromatin immunoprecipitation
Introductory paragraph
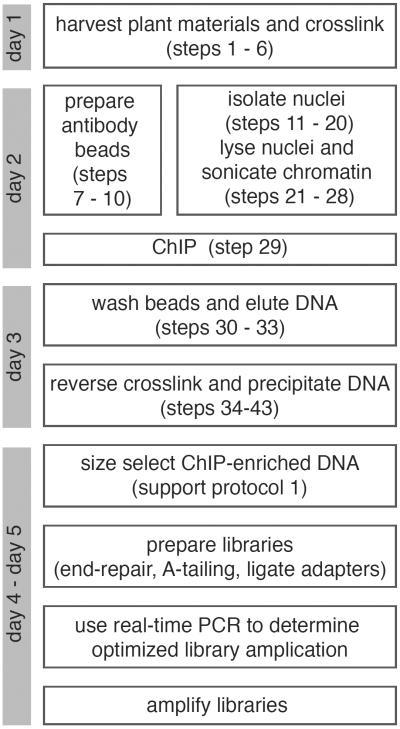
This protocol describes a step-by-step ChIP procedure. It contains experimental modules for cross-linking samples using formaldehyde, preparation of antibody beads, isolation of nuclei, sonication of chromatin, immunoprecipitation of the TF-DNA complex, and recovery of ChIP-enriched DNA fragments. Arabidopsis etiolated seedlings and floral buds are used as examples here. The protocol can also be applied to many other tissues and plant species. A general overview of ChIP-seq procedures is illustrated in Figure 1, and several key steps are shown in Figure 2.
Figure 1.
Flowchart of ChIP-seq procedures with a suggested timeline.
Figure 2.
Key steps to prepare chromatin for sonication.
(A) Plant samples were cross-linked by 1% formaldehyde under vacuum. (B) After homogenization by grinding in liquid nitrogen, etiolated (left) and light-grown (right) samples were resuspended in Extraction Buffer I and filtered by cheesecloth and Miracloth. (C) Pellets after centrifugation in Extraction Buffer I. (D) Pellets after centrifugation in Extraction Buffer II. (E) Setup of sucrose gradient using Extraction Buffer III. (F) Pellets after centrifugation in Extraction Buffer III. (G) Pellets after centrifugation in nuclei lysis buffer.
Materials
Sample material
Arabidopsis tissues.
Reagents and solutions
10 mM HEPES-NaOH, pH 7.4
formaldehyde (37% in H2O, Sigma Aldrich)
fixation buffer (see recipe)
2 M glycine
PBS-BSA (see recipe)
Dynabeads® protein G / Dynabeads protein A (Thermo Fisher Scientific)
ChIP-grade antibody (10 μg per ChIP)
Liquid nitrogen
Extraction buffer I (see recipe)
Extraction buffer II (see recipe)
Extraction buffer III (see recipe)
Nuclei lysis buffer (see recipe)
ChIP dilution buffer (see recipe)
Low salt wash buffer (see recipe)
High salt wash buffer (see recipe)
Final wash buffer (see recipe)
Elution buffer (see recipe)
Proteinase K (20 mg/ml, Thermo Fisher Scientific)
Phenol:chloroform:isoamyl alcohol (25:24:1, pH 8.0)
5 M NaCl
100% ethanol
glycogen (5 mg/ml)
Equipment
Vacuum pump (ME 4 NT diaphragm pump, VACUUBRAND)
Desiccator with a stopcock (Bel-Art)
DynaMag™-2 Magnet (Thermo Fisher Scientific)
Mortar
Pestle
Spatula
50 ml conical centrifuge tubes
Low binding DNase- and RNase-free microcentrifuge tubes and pipette tips
Cheesecloth
Miracloth (EMD Millipore)
Refrigerated centrifuge for 50 ml tubes
Refrigerated microcentrifuge
Bioruptor® (Diagenode)
Tube rotator
Thermomixer (Eppendorf)
Phase Lock Gel® heavy (5 PRIME) / MaXtract High Density (Qiagen)
Qubit® fluorometer (Thermo Fisher Scientific) / TapeStation (Agilent Technologies)
Note: use RNase- and DNase-free reagents, deionized distilled water, and low binding plasticware, and barrier pipette tips throughout the experiment.
Cross-link plant samples with 1% formaldehyde
- Harvest plant materials into fixation buffer. Use 30 ml fixation buffer per gram of plant material.Use fresh formaldehyde to achieve efficient and reproducible fixation. Avoid handling samples with amine-rich solutions such as Tris buffer before fixation. The amount of samples needed depends on the TF expression level and tissue type. In our hands, 3–4 grams of etiolated seedlings or 1–2 grams of flower buds are sufficient to ChIP many sequence-specific DNA binding TFs. More tissue can be used if the abundance of the TF is very low.
- Cross-link sample with fixation buffer under vacuum infiltration for 20 minutes. Break and re-apply vacuum after 5 and 15 minutes to facilitate penetration of formaldehyde into the sample (Figure 2A).Fixation time can be adjusted according to the thickness of plant tissue. In general, do not cross-link samples for more than 25 minutes for Arabidopsis tissues; otherwise over-fixation may decrease sonication efficiency.
Quench formaldehyde by adding 0.67 ml of 2 M fresh glycine to every 10 ml of fixation buffer. Apply vacuum for another 5 minutes.
Rinse sample with 10 mM HEPES-NaOH, pH7.4. Remove excess liquid from sample.
- (optional) Check cross-linked samples by western blot using antibody against TF-of-interest.Use uncross-linked sample as a control. A smeared band or bands of higher molecular weight is expected in cross-linked samples.
- Snap freeze sample in liquid nitrogen. Store at − 80 °C until use.Sample can be stored for at least a few weeks.
Preparation of antibody beads
Start this step 6+ hours before sonication.
-
7.For each ChIP, add 100 μl Dynabeads® protein G/A to a microcentrifuge tube.Choose Dynabeads® protein G or protein A according to their affinity for antibody species and subclasses of IgG.
-
8.
Wash beads with 1.5 ml pre-chilled PBS-BSA for three times. Collect beads by DynaMag™-2 Magnet and remove supernatant after each round of wash.
-
9.Mix 10 μg of ChIP-grade antibody and 250 μl of PBS-BSA with Dynabeads® protein G/A.The antibody is a key factor to a successful ChIP. Based on our experience, GFP tagged TFs ChIPped with a polyclonal GFP antibody (Thermo Fisher Scientific, cat. # A-11122) or FLAG tagged TFs ChIPped with a monoclonal FLAG antibody (Sigma Aldrich, cat. # F1804) usually give a strong signal and clean background. It is also crucial to include “mock IP” controls. If an antibody against a native protein is used in ChIP, mock IP can be carried out by using IgG from the same species in which the antibody was raised. If an antibody against an epitope tag is used, mock IP can either use the same antibody to ChIP wild-type samples, or even better use the same antibody to ChIP transgenic plants expressing the epitope tag alone.
-
10.
Bind antibody to Dynabeads® protein G/A on a tube rotator at 4 °C for 6+ hours.
Isolation of nuclei
-
11.Grind cross-linked samples from step 6 in liquid nitrogen to a fine powder.Carry out all procedures at 4 °C from step 11 to step 30. Many buffers contain detergent. Avoid foaming in all steps.
-
12.
Mix the powder with seven volumes of Extraction Buffer I in a 50 ml conical centrifuge tube.
-
13.
Cut cheesecloth and Miracloth to 4 inch squares. Layer three pieces of cheesecloth above one piece of Miracloth. Pre-wet cheesecloth and Miracloth with Extraction Buffer I.
-
14.
Filter plant homogenate from step 13 through cheesecloth and Miracloth into a fresh 50 ml conical centrifuge tube (Figure 2B).
-
15.
Repeat filtration through one layer of pre-wet Miracloth.
-
16.
Centrifuge filtrate at 2,880 g for 20 minutes (Figure 2C). Discard supernatant.
-
17.
Resuspend pellet gently and thoroughly with 1.2 ml of Extraction Buffer II. Transfer sample to a 1.5 ml microcentrifuge tube.
-
18.Centrifuge sample at 12,000 g for 10 minutes (Figure 2D). Discard supernatant.For light-grown samples, repeat steps 17 and 18 as needed if too many chloroplasts are present as evidenced by a deep green pellet.
-
19.
Resuspend pellet gently and thoroughly with 500 μl of Extraction Buffer III. Carefully layer the resuspended pellet on top of another 500 μl of Extraction Buffer III in a fresh microcentrifuge tube (Figure 2E).
-
20.
Centrifuge sample at 16,000 g for an hour (Figure 2F). Discard supernatant.
Nuclei lysis and chromatin sonication
-
21.
Resuspend pellet from step 20 gently and thoroughly with 300 μl of Nuclei Lysis Buffer.
-
22.Gently rotate sample for 20 minutes on the tube rotator.This is to increase sonication efficiency (F. Turck, personal communication).
-
23.Sonicate chromatin in Bioruptor® in cold room for 25 cycles of 30-second ON and 2-minute OFF at HIGH setting. Gently vortex and then centrifuge tubes after every 10 cycles of sonication.Sonication efficiency of Bioruptor® may vary greatly by model. Adjust settings accordingly for best performance. Carry out a western blot to ensure sonication setup does not destroy protein or protein-DNA complex. Wear ear protection as sonication generates high-frequency sound waves that may damage hearing.
-
24.
Centrifuge sonicated chromatin at maximum speed for 10 minutes (Figure 2G). Transfer nuclear lysate in supernatant to a fresh microcentrifuge tube.
-
25.
Repeat step 24.
-
26.
Dilute sonicated nuclear lysate with 4 volumes (ca. 1,200 μl) of ChIP dilution buffer.
-
27.
Collect antibody beads from step 10 on a magnet. Wash antibody beads with 1.5 ml PBS-BSA for three times. Collect beads on magnet after each round of wash. Discard supernatant.
-
28.
Freeze 30 μl diluted nuclear lysate as input sample. Mix the remaining lysate with antibody beads for ChIP.
ChIP and reverse crosslink
-
29.
Mix sonicated nuclear lysate with antibody beads overnight using tube rotator. Collect antibody beads on magnet. Discard supernatant **.
-
30.Wash beads for a total of six rounds with the following buffers. Collect beads on magnet and remove supernatant after each round of wash **.
- One round of quick wash, and one round of 5-minute wash on tube rotator with Low Salt Wash Buffer.
- One round of quick wash, and one round of 5-minute wash on tube rotator with High Salt Wash Buffer.
- One round of quick wash, and one round of 5-minute wash on tube rotator with Final Wash Buffer.
-
31.
Resuspend antibody beads with 100 μl of Elution Buffer. Incubate tubes at 65 °C for 15 minutes to elute TF-DNA complex. Collect beads on magnet. Transfer supernatant to a fresh tube.
-
32.
Repeat step 31. Combine eluates from steps 31 and 32 **.
-
33.
(** optional) Use a small aliquot of supernatant from IP, wash, and elution steps for western blot to ensure TF-of-interest is efficiently immunoprecipitated by the antibody.
-
34.
Take input sample from step 28 out of freezer. Add 170 μl of Elution Buffer.
-
35.
Reverse cross-linked samples from steps 32 and 34 in a Thermomixer at 65 °C for 6+ hours.
-
36.
Add 1 μl (20 μg) Proteinase K to each sample and incubate at 65 °C for 2 hours.
-
37.Transfer samples to 1.5 – 2 ml Phase Lock Gel® heavy tubes. Extract DNA with an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1, pH8.0).The manufacturer of 5 PRIME has a detailed protocol on how to handle Phase Lock Gel®. Handle phenol/chloroform/isoamy alcohol in chemical hood.
-
38.
Centrifuge Phase Lock Gel® heavy tubes at 12,000 g for 5 minutes. Recover the aqueous upper phase to a new microcentrifuge tube.
-
39.Add to samples 4 μl of 5 M NaCl, 2 μl of 5 μg/μl glycogen, and 3 volume (ca. 600 μl) of 100% ethanol. Invert tubes a few times to mix. Incubate samples at − 80 °C for 3+ hours.Use NaCl instead of NaOAc to prevent precipitation of SDS.
-
40.
Precipitate DNA by centrifuging at maximum speed at 4 °C for 30 minutes.
-
41.Wash pellet twice with 1 ml of 75% ethanol. Dry the pellet.Dried pellet may become transparent.
-
42.
Resuspend DNA pellet in 50 μl H2O.
-
43.Measure DNA concentration of input sample by Qubit or TapeStation to estimate the amount of DNA used in ChIP.Unlike input DNA, concentration of ChIPped DNA is often very low. ChIP of low abundance TFs from 3 grams of etiolated Arabidopsis seedlings may frequently enrich less than 5 ng of fragmented DNA.
SUPPORT PROTOCOL 1 (optional)
Optimization of ChIP-seq library preparation
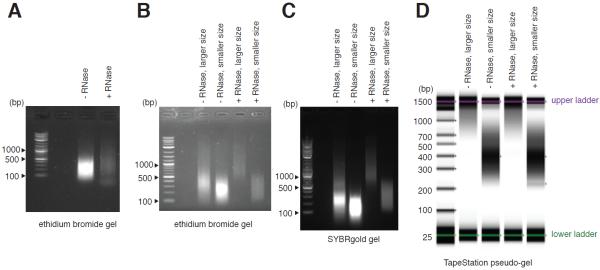
Many suppliers such as Illumina and New England BioLabs already have detailed protocols to construct ChIP-seq libraries. To avoid redundancy this protocol focuses on how to optimize the size distribution of ChIP-enriched DNA. A tight size distribution of ChIP-enriched DNA is desired. Excluding long fragments improves cluster formation on Illumina sequencer. Tight size distribution of ChIP-enriched DNA also allows more accurate prediction of fragment length and peak calling by analysis packages. A few examples are shown in Figure 3.
Figure 3.
Bead-based fractionation improves size distribution of sonicated DNA.
(A) Although RNA does not interfere with ChIP, RNase treatment allows an accurate estimate of size and concentration of sonicated DNA. (B–D) Bead-based size fractionation of DNA in (A) visualized by (B) 1.5% agarose-TAE gel containing ethidium bromide, (C) 1.5% agarose-TAE gel containing SYBRgold, and (D) TapeStation. Nucleic acid concentration appears to affect migration rate in the SYBRgold gel.
Materials
ChIP-enriched DNA, a few nanograms or more.
AMPure XP beads (Beckman Coulter)
80% (v/v) ethanol, freshly prepared.
Equipment
DynaMag™-2 Magnet (Thermo Fisher Scientific)
Thermomixer (Eppendorf)
Note: use RNase- and DNase-free reagents, deionized distilled water, and low binding plasticware, and barrier pipette tips throughout the experiment.
Double size selection of ChIP-enriched DNA
- Add 37.5 μl (0.75 volume) of AMPure XP beads to 50 μl of ChIP-enriched DNA in a 1.5 ml microcentrifuge tube.The volume of AMPure beads can be increased or decreased for a smaller or larger DNA size cutoff, respectively. We recommend empirically testing DNA size fractionation within the range from 0.65 volume to 0.85 volume of beads.
Vigorously vortex tube for 10 – 15 times. Incubate sample at room temperature for 10 minutes.
- Collect beads on magnet. Transfer supernatant to a fresh microcentrifuge tube.Supernatant contains DNA fragment smaller than 500 bp.
Add 62.5 μl AMPure beads to supernatant.
Vigorously vortex tube for 10 – 15 times. Incubate sample at room temperature for 10 minutes.
Collect beads on magnet. Discard supernatant.
Wash beads twice with freshly prepared 80% ethanol. Remove supernatant after each round of wash.
- Dry beads in a Thermomixer at 37 °C for 3-5 minutes.The purpose is to remove leftover ethanol from previous wash steps.
Gently resuspend dried beads with 50 μl H2O. Vigorously vortex tube for 10 – 15 times. Incubate sample at room temperature for 5 minutes.
Collect beads on magnet. Transfer supernatant containing the eluted DNA into a fresh microcentrifuge tube.
- Check the concentration and size of DNA on TapeStation (Figure 3D).If TapeStation is not available or if DNA concentration is too low, size-fractionate input sample from step 42 of the basic protocol and run a gel to infer the size of ChIP-enriched DNA. SYBRgold will interfere with DNA migration in the gel (Figure 3B vs Figure 3C). Therefore, we recommend running samples in ethidium bromide gel, or stain gel with SYBRgold after the run.
REAGENTS AND SOLUTIONS
Fixation buffer (1% (v/v) formaldehyde in 10 mM HEPES-NaOH).
PBS-BSA (1x PBS, 0.5 (w/v) bovine serum albumin), pre-chill before use.
- Extraction buffer I (10 mM Tris-HCl pH 7.4, 0.4 M sucrose, β-mercaptoethanol 0.035%*, 1 mM PMSF*, 50 uM MG132*, 1x Roche cOmplete protease inhibitor, EDTA-free*), pre-chill before use.* Add fresh before experiments. Same for extraction buffers II and III, nuclei lysis buffer and ChIP dilution buffer. Handle β-mercaptoethanol in chemical hood.
Extraction buffer II (10 mM Tris-HCl pH 7.4, 0.25 M sucrose, 1% Triton X-100, 10 mM MgCl2, 1mM PMSF*, 50 uM MG132*, 1x Roche cOmplete protease inhibitor, EDTA-free*), pre-chill before use.
Extraction buffer III (10 mM Tris-HCl pH 7.4, 1.7 M sucrose, 0.15% Triton X-100, 2 mM MgCl2, 1 mM PMSF*, 50 uM MG132*, 1x Roche cOmplete protease inhibitor, EDTA-free*), pre-chill before use.
Nuclei lysis buffer (50 mM Tris-HCl pH 7.4, 0.5% sarkosyl, 100 mM NaCl, 2 mM EDTA, 1 mM PMSF*, 50 μM MG132*, 1x Roche cOmplete protease inhibitor, EDTA-free*), pre-chill before use.
ChIP dilution buffer (50 mM Tris-HCl pH 7.4, 1.25% Triton X-100, 100 mM NaCl, 2 mM EDTA, 1 mM PMSF*, 50 μM MG132*, 1x Roche cOmplete protease inhibitor, EDTA-free*), pre-chill before use.
Low salt wash buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 2 mM EDTA, 0.5% Triton X-100), pre-chill before use.
High salt wash buffer (50 mM Tris-HCl pH 7.4, 500 mM NaCl, 2 mM EDTA, 0.5% Triton X-100), pre-chill before use.
Final wash buffer (50 mM Tris-HCl pH 7.4, 50 mM NaCl, 2 mM EDTA), pre-chill before use.
Elution buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA, 1% SDS)
COMMENTARY
Background Information
Chromatin immunoprecipitation has been used to study protein-DNA interactions for almost 40 years (Jackson, 1978). The technique has evolved multiple times by the advances of DNA detection methods. In the early days, ChIPped DNA were examined by low throughput approaches such as Southern blot or PCR (Solomon et al., 1988; Orlando and Paro, 1993; Hecht et al., 1996). Understanding protein-DNA interactions at a genome-wide scale only became possible after the invention of ChIP-chip by Ren et al. (Ren et al., 2000) and Iyer et al. (Iyer et al., 2001), via the development of high-density DNA microarrays (Schena et al., 1995). ChIP-chip has facilitated numerous breakthroughs in biology, including elucidation of transcriptional regulatory networks in Saccharomyces cerevisiae (Lee et al., 2002) and characterization of DNA regulatory sequences in human (Consortium et al., 2007). On the other hand, ChIP-chip suffers from all the limitations of microarray technology. For instance, the design of array depends on a priori knowledge of genome sequence; high background signal makes it difficult to detect weak protein-DNA interactions; cross-hybridization is problematic, especially for highly homologous or repetitive regions; to obtain micrograms of DNA for hybridization, ChIPped DNA needs to be amplified by several orders of magnitude and the process may introduce biases. In 2007, Johnson et al. addressed these issues by combining ChIP with high-throughput DNA sequencing technologies (Johnson et al., 2007). With the help of sophisticated statistical analysis (Landt et al., 2012), ChIP-seq has become the most prevalent method to confidently identify protein-DNA interaction in vivo. More recently, Rhee et al. developed an improved ChIP-seq protocol called ChIP-exo (Rhee and Pugh, 2011). ChIP-exo utilizes 5'-to-3' exonuclease to trim DNA to the precise protein binding location. Exonuclease also cleans up ChIP background by digesting naked DNA. However, the extra steps associated with exonuclease digestion often result in greater sample loss and subsequently low library complexity. As an answer to this problem, He et al.'s ChIP-nexus protocol improves the ligation efficiency during library preparation, and tracks DNA over-amplification by unique, randomized barcodes (He et al., 2015).
Several factors contribute to the difficulty of performing ChIP-seq in plants. First, unlike animal cells in which nuclei can be extracted by mild detergents, extraction of plant nuclei usually requires vigorous physical disruption because of cell walls. This is arguably the step that causes the greatest sample loss in a ChIP procedure and prevents parallel handling of samples. Secondly, plant tissues often contain high level of phenolic compounds and polysaccharides, which may be problematic for PCR amplification during library preparation. Thirdly, there is limited selection of ChIP-grade antibodies in plants, and as a consequence researchers have to spend months to generate transgenic lines to express epitope-tagged proteins before ChIP-seq experiments can be carried out. Besides this protocol, several other laboratories have also published detailed procedures of ChIP-chip or ChIP-seq for Arabidopsis (Kaufmann et al., 2010; Reimer and Turck, 2010). We recommend readers combine knowledge of all protocols to decide the best practice.
Critical Parameters and Troubleshooting
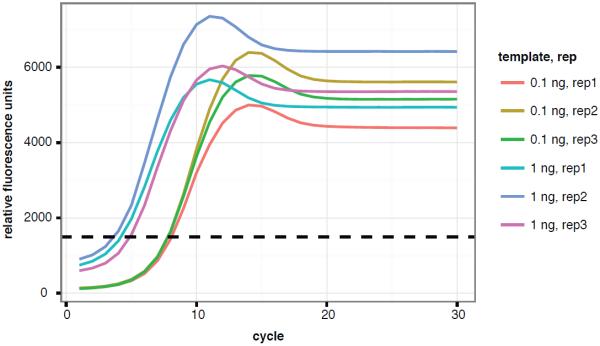
ChIP-seq is a long procedure. We include multiple quality control steps in the protocols to ensure its success. Formaldehyde fixation is the first key step in the experiment. Both under- and over-fixation will result in inefficient ChIP of transcription factors. Therefore, we suggest using fresh formaldehyde, applying accurate control of cross-link time, and measuring the level of cross-link by western blot (step 5, basic protocol). Ideally, sonication should shear DNA to a consistent, tightly distributed size smaller than 500 bp whereas still preserving the protein-DNA complexes. The size distribution of sheared DNA can be examined by TapeStation (step 43, basic protocol), and can be further tightened by a bead-based size selection (support protocol 1, Figure 3). Antibody quality is crucial for successful ChIP. We suggest monitoring the effectiveness of IP by examining the presence of TF-of-interest after IP, wash, and final elution (step 33, basic protocol). Finally, over-amplification may bias library composition. We suggest determining the number of PCR cycles by real-time PCR to avoid over-amplification of ChIP-seq libraries (Figure 4).
Figure 4.
Optimized library amplification determined by real-time PCR.
Adapter-ligated DNA of indicated concentration was used as template in real-time SYBRgreen PCR. Five to eight cycles and eight to eleven cycles of PCR will properly amplify 1 ng and 0.1 ng of templates, respectively.
Anticipated Results
Using the basic protocol, 3 grams of etiolated seedlings is expected to yield more than 10 μg of input DNA. Depending on the TF and antibody, the ChIP-enriched DNA can be 5 ng or less. Light-grown samples especially flowers will usually yield more DNA. In most cases, twelve or fewer cyclers of PCR is sufficient to amplify enough DNA for sequencing.
Time Considerations
Once the samples are harvested and cross-linked, it takes 4 – 6 days to complete ChIP. The most time consuming steps in the procedure are IP (usually overnight, can be shortened to a few hours if the immunoprecipitated protein is very abundant), reverse cross-linking (6 hours to overnight), ethanol precipitation of reverse cross-linked DNA (3 hours to overnight), and adaptor ligation (overnight).
ACKNOWLEDGEMENT
We thank F.Turck, K.N.Chang, S.C.Huang, H.Qiao, U. Padmale, N. Krogan, M. Zander, M. Lewsey, and M. Urich for providing useful discussions on ChIP and DNA size fractionation. L.S. was supported by Salk Pioneer postdoc fellowship. This work was supported by grants from NSF (MCB-1024999 to J.R.E.). J.R.E. is an investigator of the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation.
LITERATURE CITED
- Consortium EP, Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. Available at: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=17571346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M, Bai M-Y, Kim J-G, Wang T, Oh E, Chen L, Park CH, Son S-H, Kim S-K, Mudgett MB, et al. The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern-triggered immunity in Arabidopsis. The Plant cell. 2014;26:828–41. doi: 10.1105/tpc.113.121111. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3967043&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Johnston J, Zeitlinger J. ChIP-nexus enables improved detection of in vivo transcription factor binding footprints. Nat Biotechnol. 2015;33:395–401. doi: 10.1038/nbt.3121. Available at: http://dx.doi.org/10.1038/nbt.3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor sir3 from telomeric heterochromatin. Nature. 1996;383:92–6. doi: 10.1038/383092a0. Available at: http://www.ncbi.nlm.nih.gov/pubmed/8779721. [DOI] [PubMed] [Google Scholar]
- Heyndrickx KS, de Velde J, Van, Wang C, Weigel D, Vandepoele K. A Functional and Evolutionary Perspective on Transcription Factor. The Plant cell. 2014;26:3894–910. doi: 10.1105/tpc.114.130591. Available at: http://www.ncbi.nlm.nih.gov/pubmed/25361952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- Jackson V. Studies on histone organization in the nucleosome using formaldehyde as a reversible cross-linking agent. Cell. 1978;15:945–54. doi: 10.1016/0092-8674(78)90278-7. Available at: http://www.ncbi.nlm.nih.gov/pubmed/569554. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science (New York, N.Y.) 2007;316:1497–502. doi: 10.1126/science.1141319. Available at: http://www.ncbi.nlm.nih.gov/pubmed/17540862\n http://www.sciencemag.org/content/316/5830/1497. [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Pajoro A, Angenent GC. [Accessed June 12, 2011];Regulation of transcription in plants: mechanisms controlling developmental switches. Nature reviews. Genetics. 2010 11:830–42. doi: 10.1038/nrg2885. Available at: http://dx.doi.org/10.1038/nrg2885. [DOI] [PubMed] [Google Scholar]
- Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Research. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science (New York, N.Y.) 2002;298:799–804. doi: 10.1126/science.1075090. Available at: http://www.ncbi.nlm.nih.gov/pubmed/12399584. [DOI] [PubMed] [Google Scholar]
- Orlando V, Paro R. Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell. 1993;75:1187–1198. doi: 10.1016/0092-8674(93)90328-n. [DOI] [PubMed] [Google Scholar]
- Pfeiffer A, Shi H, Tepperman JM, Zhang Y, Quail PH. Combinatorial Complexity in a Transcriptionally-centered Signaling Hub in Arabidopsis. Molecular plant. 2014;7:1598–1618. doi: 10.1093/mp/ssu087. Available at: http://www.ncbi.nlm.nih.gov/pubmed/25122696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer JJ, Turck F. Genome-wide mapping of protein-DNA interaction by chromatin immunoprecipitation and DNA microarray hybridization (ChIP-chip). Part A: ChIP-chip molecular methods. Methods in molecular biology (Clifton, N.J.) 2010;631:139–60. doi: 10.1007/978-1-60761-646-7_12. Available at: http://www.ncbi.nlm.nih.gov/pubmed/20204874. [DOI] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-Wide Location and Function of DNA Binding Proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. Available at: http://www.sciencemag.org/content/290/5500/2306\n http://www.ncbi.nlm.nih.gov/pubmed/11125145\n http://www.sciencemag.org/content/290/5500/2306.abstract?ijkey=f2b39fc63d7b97e1ec55310ae121f014377c35b7&keytype2=tf_ipsecsha\n http://www.sciencemag.org/content/2. [DOI] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. Available at: http://dx.doi.org/10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative Monitoring of Gene Expression Patterns with a Complementary DNA Microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Solomon MJ, Larsen PL, Varshavsky A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988;53:937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]