Abstract
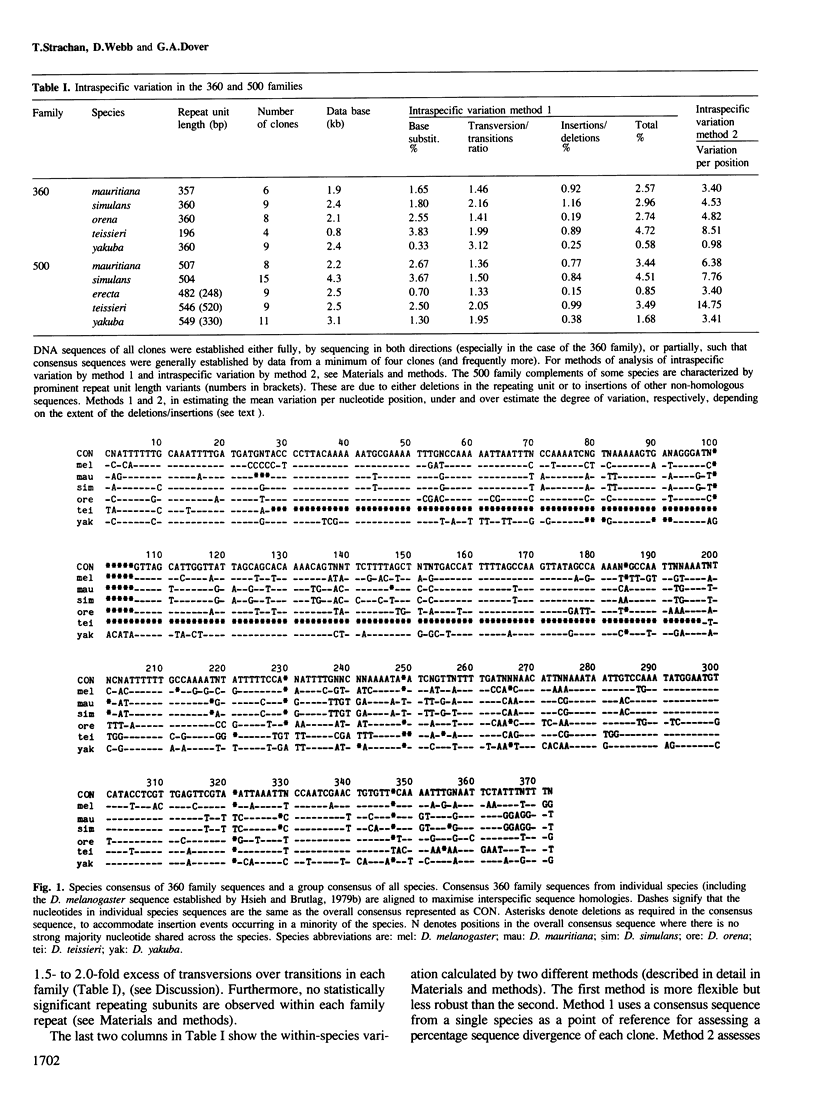
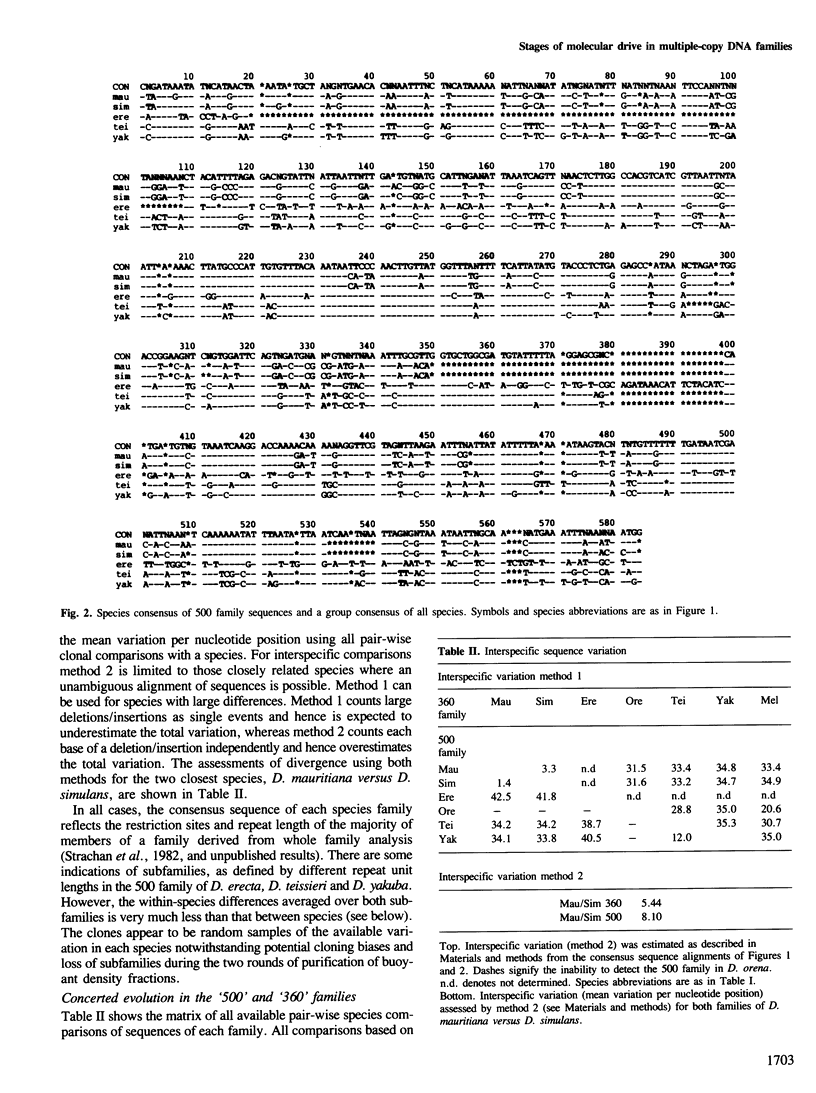
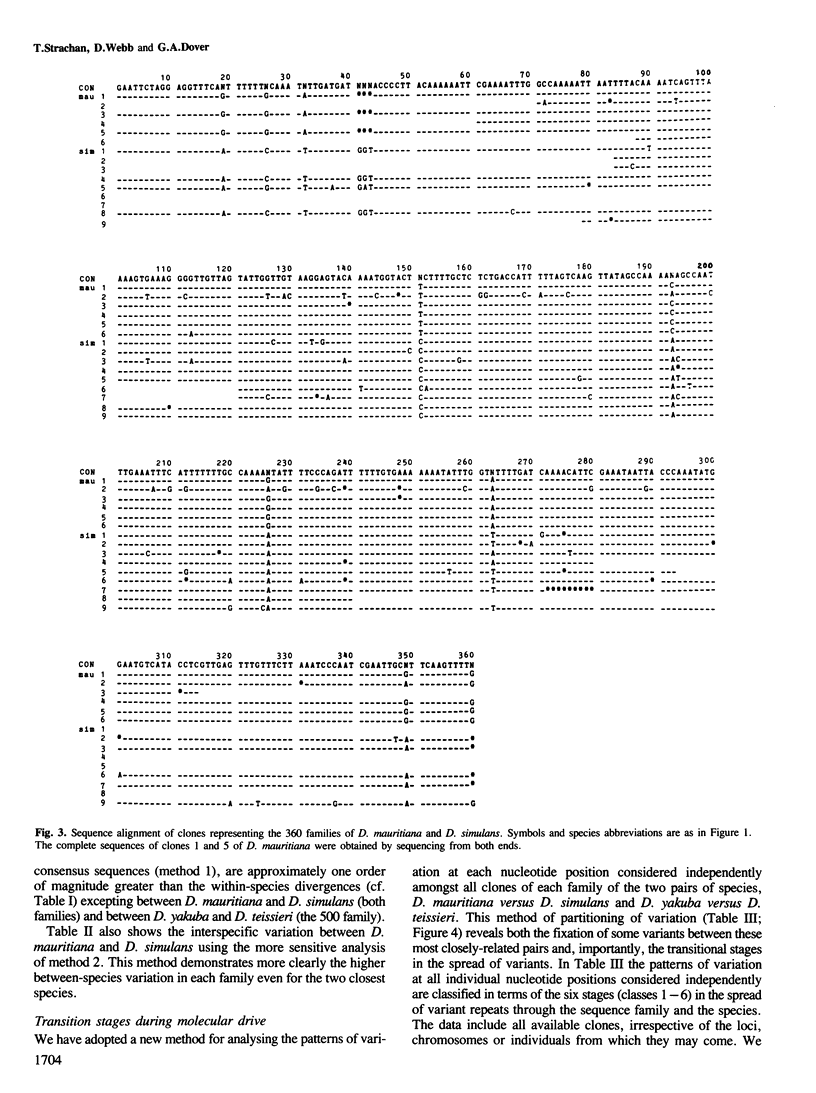
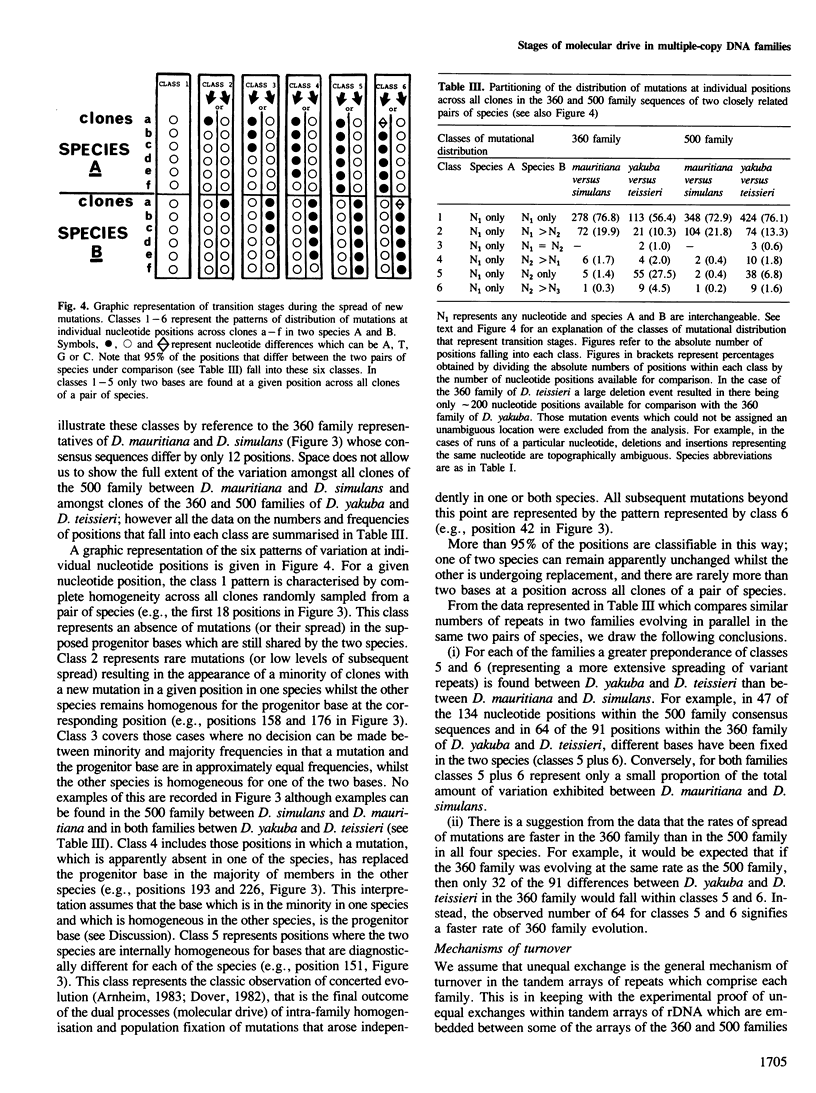
Multigene and non-genic DNA families are in a state of turnover and hence are continually being replaced throughout a population by new variant repeats. To quantify such molecular processes, in the absence of selection, it is necessary to find and compare stages of transistion during the homogenization of at least two non-genic families evolving in parallel in a closely related group of species. Detailed sequence analysis of patterns of variation, at each nucleotide position considered independently, amongst repeats of two tandem DNA families from seven related Drosophila species, reveals all stages of transition during the spread of randomly produced variant repeats. Variant repeats are found at different stages of homogenization and fixation in a population, irrespective of the loci, chromosomes or individuals from which they were cloned. Differences between the families in the relatively small number of variants at each transition stage and the greater number of fully homogenized and fixed variants between species of greater divergence indicate that the process of spread (molecular drive) is rapid relative to the mutation rate and occurs at seemingly different constant rates for each family. Occasional gene conversions, in addition to unequal exchanges, have contributed to family turnover. The significance of these results to the evolution of functional multigene families and divergence and conservation of sequences is discussed.
Keywords: multigene families, molecular drive, concerted evolution, evolution rates, Drosophila
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. M., Scheller R. H., Posakony J. W., McAllister L. B., Trabert S. G., Beall C., Britten R. J., Davidson E. H. Repetitive sequences of the sea urchin genome. Distribution of members of specific repetitive families. J Mol Biol. 1981 Jan 5;145(1):5–28. doi: 10.1016/0022-2836(81)90332-6. [DOI] [PubMed] [Google Scholar]
- Barnes S. R., Webb D. A., Dover G. The distribution of satellite and main-band DNA components in the melanogaster species subgroup of Drosophila. I. Fractionation of DNA in actinomycin D and distamycin A density gradients. Chromosoma. 1978 Aug 14;67(4):341–363. doi: 10.1007/BF00285965. [DOI] [PubMed] [Google Scholar]
- Brown S. D., Dover G. A. Conservation of sequences in related genomes of Apodemus: constraints on the maintenance of satellite DNA sequences. Nucleic Acids Res. 1979 Jun 11;6(7):2423–2434. doi: 10.1093/nar/6.7.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. D., Dover G. Organization and evolutionary progress of a dispersed repetitive family of sequences in widely separated rodent genomes. J Mol Biol. 1981 Aug 25;150(4):441–466. doi: 10.1016/0022-2836(81)90374-0. [DOI] [PubMed] [Google Scholar]
- Brutlag D. L. Molecular arrangement and evolution of heterochromatic DNA. Annu Rev Genet. 1980;14:121–144. doi: 10.1146/annurev.ge.14.120180.001005. [DOI] [PubMed] [Google Scholar]
- Coen E. S., Dover G. A. Multiple Pol I initiation sequences in rDNA spacers of Drosophila melanogaster. Nucleic Acids Res. 1982 Nov 11;10(21):7017–7026. doi: 10.1093/nar/10.21.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen E. S., Dover G. A. Unequal exchanges and the coevolution of X and Y rDNA arrays in Drosophila melanogaster. Cell. 1983 Jul;33(3):849–855. doi: 10.1016/0092-8674(83)90027-2. [DOI] [PubMed] [Google Scholar]
- Coen E. S., Thoday J. M., Dover G. Rate of turnover of structural variants in the rDNA gene family of Drosophila melanogaster. Nature. 1982 Feb 18;295(5850):564–568. doi: 10.1038/295564a0. [DOI] [PubMed] [Google Scholar]
- Coen E., Strachan T., Dover G. Dynamics of concerted evolution of ribosomal DNA and histone gene families in the melanogaster species subgroup of Drosophila. J Mol Biol. 1982 Jun 15;158(1):17–35. doi: 10.1016/0022-2836(82)90448-x. [DOI] [PubMed] [Google Scholar]
- Dover G. A., Flavell R. B. Molecular coevolution: DNA divergence and the maintenance of function. Cell. 1984 Oct;38(3):622–623. doi: 10.1016/0092-8674(84)90255-1. [DOI] [PubMed] [Google Scholar]
- Dover G. Molecular drive: a cohesive mode of species evolution. Nature. 1982 Sep 9;299(5879):111–117. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- Fedoroff N. V. On spacers. Cell. 1979 Apr;16(4):697–710. doi: 10.1016/0092-8674(79)90086-2. [DOI] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M., Clarke L., Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982 May;29(1):235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- Goad W. B., Kanehisa M. I. Pattern recognition in nucleic acid sequences. I. A general method for finding local homologies and symmetries. Nucleic Acids Res. 1982 Jan 11;10(1):247–263. doi: 10.1093/nar/10.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood L., Campbell J. H., Elgin S. C. The organization, expression, and evolution of antibody genes and other multigene families. Annu Rev Genet. 1975;9:305–353. doi: 10.1146/annurev.ge.09.120175.001513. [DOI] [PubMed] [Google Scholar]
- Hsieh T., Brutlag D. L. A protein that preferentially binds Drosophila satellite DNA. Proc Natl Acad Sci U S A. 1979 Feb;76(2):726–730. doi: 10.1073/pnas.76.2.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T., Brutlag D. Sequence and sequence variation within the 1.688 g/cm3 satellite DNA of Drosophila melanogaster. J Mol Biol. 1979 Dec 5;135(2):465–481. doi: 10.1016/0022-2836(79)90447-9. [DOI] [PubMed] [Google Scholar]
- Jones C. W., Kafatos F. C. Accepted mutations in a gene family: evolutionary diversification of duplicated DNA. J Mol Evol. 1982;19(1):87–103. doi: 10.1007/BF02100227. [DOI] [PubMed] [Google Scholar]
- Kedes L. H. Histone genes and histone messengers. Annu Rev Biochem. 1979;48:837–870. doi: 10.1146/annurev.bi.48.070179.004201. [DOI] [PubMed] [Google Scholar]
- Long E. O., Dawid I. B. Repeated genes in eukaryotes. Annu Rev Biochem. 1980;49:727–764. doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- McDonell M. W., Simon M. N., Studier F. W. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J Mol Biol. 1977 Feb 15;110(1):119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- Miklos G. L., Gill A. C. Nucleotide sequences of highly repeated DNAs; compilation and comments. Genet Res. 1982 Feb;39(1):1–30. doi: 10.1017/s0016672300020711. [DOI] [PubMed] [Google Scholar]
- Ohta T., Dover G. A. Population genetics of multigene families that are dispersed into two or more chromosomes. Proc Natl Acad Sci U S A. 1983 Jul;80(13):4079–4083. doi: 10.1073/pnas.80.13.4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T., Dover G. A. The cohesive population genetics of molecular drive. Genetics. 1984 Oct;108(2):501–521. doi: 10.1093/genetics/108.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M. F. Highly repeated sequences in mammalian genomes. Int Rev Cytol. 1982;76:67–112. doi: 10.1016/s0074-7696(08)61789-1. [DOI] [PubMed] [Google Scholar]
- Smith A. J. DNA sequence analysis by primed synthesis. Methods Enzymol. 1980;65(1):560–580. doi: 10.1016/s0076-6879(80)65060-5. [DOI] [PubMed] [Google Scholar]
- Spradling A. C., Rubin G. M. Drosophila genome organization: conserved and dynamic aspects. Annu Rev Genet. 1981;15:219–264. doi: 10.1146/annurev.ge.15.120181.001251. [DOI] [PubMed] [Google Scholar]
- Strachan T., Coen E., Webb D., Dover G. Modes and rates of change of complex DNA families of Drosophila. J Mol Biol. 1982 Jun 15;158(1):37–54. doi: 10.1016/0022-2836(82)90449-1. [DOI] [PubMed] [Google Scholar]
- Strachan T., Sodoyer R., Damotte M., Jordan B. R. Complete nucleotide sequence of a functional class I HLA gene, HLA-A3: implications for the evolution of HLA genes. EMBO J. 1984 Apr;3(4):887–894. doi: 10.1002/j.1460-2075.1984.tb01901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartof K. D. Unequal mitotic sister chromatid exchange and disproportionate replication as mechanisms regulating ribosomal RNA gene redundancy. Cold Spring Harb Symp Quant Biol. 1974;38:491–500. doi: 10.1101/sqb.1974.038.01.053. [DOI] [PubMed] [Google Scholar]
- Trick M., Dover G. A. Unexpectedly slow homogenisation within a repetitive DNA family shared between two subspecies of tsetse fly. J Mol Evol. 1984;20(3-4):322–329. doi: 10.1007/BF02104738. [DOI] [PubMed] [Google Scholar]


