Abstract
Molecular tethers span the nuclear envelope to mechanically connect the cytoskeleton and nucleoskeleton. These bridge-like tethers, termed LInkers of Nucleoskeleton and Cytoskeleton (LINC) complexes, consist of SUN proteins at the inner nuclear membrane and KASH proteins at the outer nuclear membrane. LINC complexes are central to a variety of cell activities including nuclear positioning and mechanotransduction, and LINC-related abnormalities are associated with a spectrum of tissue-specific diseases, termed laminopathies or envelopathies. Protocols used to study the biochemical and structural characteristics of core elements of SUN-KASH complexes are described here to facilitate further studies in this new field of cell biology.
1. Introduction
The nuclear envelope (NE) physically separates the nucleus from the cytoplasm, generating two distinct compartments. Molecular exchange between the nucleoplasm and cytoplasm is mediated by nuclear pore complexes, which act as selective permeability barriers. Mechanical communication between the nucleus and cytoplasm involves specific tethers, termed LInkers of Nucleoskeleton and Cytoskeleton (LINC) complexes, that span the NE. LINC complexes are formed by a family of KASH (Klarsicht, ANC-1, and Syne Homology) proteins embedded in the outer nuclear membrane (ONM) that interact within the NE lumen with SUN (Sad1 and UNC-84) proteins, which span the inner nuclear membrane (INM). SUN and KASH proteins each project from the NE and directly bind to components of the nucleoskeleton and the cytoskeleton, respectively. These mechanical connections are critically important in a wide range of activities such as nuclear migration and anchorage, meiotic chromosome movements, the centrosome-nucleus connection, signal transduction and DNA repair (Burke & Roux, 2009; Chang, Worman, & Gundersen, 2015; Luxton & Starr, 2014; Rothballer & Kutay, 2013; Starr & Fridolfsson, 2010).
KASH proteins are tail-anchored, single-span transmembrane proteins, mostly found at the ONM. The C-terminal “KASH motif” comprises the transmembrane helix and the adjacent luminal segment, which consists of 8–30 residues, depending on the specific nesprin gene and species (Starr & Han, 2002). Vertebrate KASH proteins are often called nesprins (NE spectrin repeat proteins), since their cytoplasmic portions typically contain numerous spectrin repeats. The cytoplasmic extensions vary greatly in size due to alternative splicing and transcription initiation of multiple nesprin genes (Zhang et al., 2001). The two longest (‘giant’; 0.8–1.0 MDa) nesprin isoforms, Nesprin-1G and Nesprin-2G, each bind to actin filaments via calponin homology (CH) domains at their N terminus. Coupling of actin filaments to LINC complexes has been best visualized in migrating fibroblasts, where SUN2-Nesprin-2G complexes assemble into linear arrays at the NE and form so-called TAN (transmembrane actin-associated nuclear) lines. Formation of TAN lines is instrumental in moving the nucleus rearward by coupling to retrogradely-moving actin cables (Luxton, Gomes, Folker, Vintinner, & Gundersen, 2010; Luxton, Gomes, Folker, Worman, & Gundersen, 2011). The much shorter protein Nesprin-3α binds plectin, which in turn binds to cytoplasmic intermediate filaments and/or actin. Since Nesprin-3α also binds to the CH domains of Nesprin-1G and Nesprin-2G, a nesprin scaffold is proposed to form around the nucleus that might play a role in regulating nuclear size (Lu et al., 2012). Nesprin-4 interacts with microtubules through kinesin-1, and is proposed to function specifically in ear development and hearing (Horn, Brownstein, et al., 2013a). Yet another tissue-specific KASH protein, Nesprin-5, binds to microtubules through dynein and functions during meiotic chromosome pairing in germ cells (Horn, Kim, et al., 2013b). Finally, a recently recognized sixth KASH protein in zebrafish, lymphoid restricted membrane protein (LRMP), is involved in pronuclear congression during fertilization (Lindeman & Pelegri, 2012).
Similarly most organisms also encode several SUN homologs. Of the five known mammalian SUN proteins, SUN1 and SUN2 are widely expressed (Crisp et al., 2006; Padmakumar et al., 2005), whereas SUN3, SUN4, and SUN5 are expressed during spermatogenesis in testis (Göb, Schmitt, Benavente, & Alsheimer, 2010). SUN proteins have at least one transmembrane helix, which typically anchors them in the INM. The C-terminal ~20 kDa SUN domain, preceded by a predicted coiled-coil segment of variable length, are both located in the NE lumenal space. The N terminus of SUN proteins extends into the nucleoplasm, and binds lamins (nuclear intermediate filament proteins). Mammalian SUN proteins are known to bind to A-type lamins, while interaction with B-type lamins is relatively weak (Crisp et al., 2006; Haque et al., 2006). Although lamina attachment restricts diffusion of the SUNs (Ostlund et al., 2009), lamins do not seem to be the only factors anchoring LINC complexes. Indeed, both SUN1 and SUN2 are properly localized in the absence of A- and B-type lamins (Crisp et al., 2006; Haque et al., 2006; Padmakumar et al., 2005). Lamin associated proteins such as emerin and SAMP1 likely help anchor the LINC complexes (Borrego-Pinto et al., 2012; Chang, Folker, Worman, & Gundersen, 2013), and the intricate interplay among these proteins is not yet understood. Mutations in A-type lamins, emerin, SUNs and nesprins can each disrupt nucleocytoskeletal coupling and are also genetically linked to laminopathies such as skeletal and/or cardiac muscular dystrophies, lipodystrophy, dysplasia or segmental progeroid (‘accelerated aging’) disorders (Worman, 2012). The striated muscle disease EDMD (Emery-Dreifuss muscular dystrophy), and the premature aging syndrome HGPS (Hutchinson-Gilford progeria syndrome) are examples where perturbed functioning of LINC complexes and associated factors contributes to pathology (Bione et al., 1994; Bonne et al., 1999; Zhang et al., 2007) (Puckelwartz et al., 2009) (Haque et al., 2010) (Chen et al., 2012). Further structural and biochemical characterization of LINC complexes is needed to understand the molecular basis of these disorders.
Recent crystallographic studies established the structure of the core element of the LINC complex, namely the SUN-KASH interaction (Wang et al, 2012; Zhou et al, 2012). Human SUN2 proteins form a triple-stranded coiled-coil stalk to generate a trimeric structure that positions the adjacent, C-terminal β-sandwich-shaped SUN domains to form a globular trefoil. The helical stalk assumes an unusual, right-handed supercoil that positions the SUN domains in a KASH-binding competent state. Three KASH peptides are bound at the three interfaces between adjacent SUN protomers in the trefoil, immediately explaining why monomeric SUN does not bind a KASH peptide. Altogether, the core of the LINC complex is a heterohexameric SUN3-KASH3 complex. The carboxyl group of the terminal residue in the KASH peptide is specifically recognized by the SUN domain, thereby explaining why KASH peptides are always located at the C terminus of a protein. A cysteine residue at the N terminus of the KASH peptide can form a disulfide bridge with a conserved cysteine on the SUN domain, presumably enhancing the mechanical strength of the complex (Sosa, Rothballer, Kutay, & Schwartz, 2012).
Although these SUN2-KASH1/2 structures revealed crucial information about LINC complex assembly, much more work is needed to fully understand the SUN-KASH interactome. Specific methods for the purification, biochemical analysis, and structure determination of apo-SUN2, and SUN2-KASH complexes, are detailed below. In addition to SUN2-KASH1/2 complexes, which have been crystallized, we also describe strategies for purifying SUN2 complexes with KASH3, KASH4, KASH5 or KASH6.
2. Purification of SUN proteins and SUN-KASH complexes
2.1 Construct design
We use the pETDuet-1 bacterial expression system (EMD Biosciences) to produce SUN and KASH domain proteins from two different multiple cloning sites (MCS). The cDNA encoding each SUN domain protein is cloned into the first MCS (MCS1), and the cDNA encoding the KASH domain protein is cloned into the second MCS (MCS2), either as single open reading frames for isolation of the apo proteins or in tandem for isolation of SUN-KASH complexes. When cloned together, the dual cassette expression enables SUN-KASH interaction already in the bacterial cell, facilitating isolation of stoichiometric complexes.
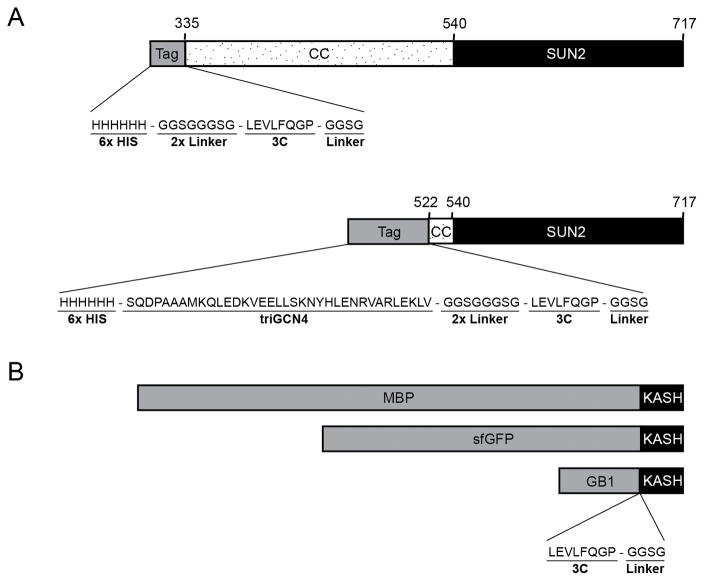
We generated N-terminally 6xHis-tagged SUN2 fragments for expression from MCS1. These fragments contain lumenal portions of SUN2. Since SUN2 fragments that lack significant portions of the predicted coiled-coil regions might become monomeric in solution, prohibiting KASH binding, short SUN2 fragments are fused to a coiled-coil fragment of engineered tri-GCN4 (Ciani et al., 2010) at their N termini to restore KASH-binding competence (Fig. 1A). The crystallized part of SUN2 (residues 522–717) is such an example (Sosa et al., 2012). This engineering is not required for longer SUN2 constructs with a native extended coiled-coil stalk, which form stable trimers in solution. A cleavage site for 3C protease is inserted near the N terminus of each SUN2 fragment to facilitate removal of the fusion tag after purification.
Figure 1.
Schematic drawing of the expression constructs for human SUN2 (A), and KASH (B). Each SUN2 fragment includes the SUN domain (residues 540-717) and preceding lumenal segments of different length, predicted to form coiled-coils (CC). The SUN2 (residue 335-717) fragment is N-terminally fused to a 3C-cleavable 6xHis tag, whereas a 6xHis:triGCN4 tag is used for a shorter SUN2 fragment (residues 522-717). KASH motifs are C-terminally attached to 3C-cleavable MBP, superfolder GFP (sfGFP) or GB1 tags. Short flexible linkers are incorporated around the 3C cleavage sites to enhance removal of the fusion tags.
For purification of SUN2-KASH complexes, we clone lumenal portions of KASH motifs into the MCS2 site of the pETDuet-1 vector (Fig. 1B). KASH1/2/3/4 peptides can be attached to 3C-cleavable maltose binding protein (MBP) tags (di Guan, Li, Riggs, & Inouye, 1988) at their N termini. MBP-tagging helps in various ways. First, it typically yields super-stoichiometric expression of the MBP-KASH fusion protein compared to SUN2, which enables the isolation of stoichiometric SUN2-KASH complexes in large amounts. Second, the MBP moiety provides an orthogonal affinity-tag after Ni2+-pulldown to isolate stoichiometric complexes in high purity (see Section 2.3).
We faced problems removing the fusion tags during the purification of SUN2-KASH complexes. To increase the efficiency of tag removal, we introduced flexible Gly/Ser-rich linkers on either side of the 3C-cleavage sites (Fig. 1). This linker strategy proved useful for cutting off the 6xHis-triGCN4 tags, but did not improve the removal of MBP tags. As explained in Section 2.3, this problem was solved in some cases by cleaving the MBP tag in multiple steps, followed by chromatography. For example, the SUN2-KASH1/2 crystals were obtained from MBP-tagged complexes (Sosa et al., 2012). However, isolation of the other SUN-KASH complexes (SUN2-KASH3/4/5/6) was more problematic. For these complexes we tested different tags including superfolder (sf) GFP (Pédelacq, Cabantous, Tran, Terwilliger, & Waldo, 2006), thioredoxin (LaVallie, Lu, Diblasio-Smith, Collins-Racie, & McCoy, 2000) and GB1 (Huth et al., 1997). The GB1 tag significantly enhanced cleavage efficiency, and was used to purify SUN2-KASH5/6 complexes (Fig. 1B). Advantages of using different tags during purification will be explained in detail in Section 2.3.
2.2 Purification of human apo-SUN2
We purified human apo-SUN2 (residues 335–717) and apo-SUN2 (residues 522–717) in E. coli strain LOBSTR-BL21(DE3)-RIL (Kerafast, Inc, Boston MA) (Andersen, Leksa, & Schwartz, 2013) for increased purity, using the following protocol.
Inoculate 3–6 ml Lysogeny Broth (LB) with a single LOBSTR-BL21(DE3)-RIL colony that was heat-shock transformed with the SUN2-expressing plasmid. Include ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml) to select for the pETDuet-1 derived SUN2-expressing plasmid and the RIL plasmid (Agilent Technologies), respectively. Grow this starter culture overnight at 30°C.
The next morning, inoculate 1 L of LB medium containing 0.4% (w/v) glucose, ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml) with the overnight culture. Grow these bacteria in a 2-L baffled Erlenmeyer flask at 37°C in a shaker to an OD600 of 0.6–0.8, then transfer to 18°C and incubate 20 minutes longer. Then add IPTG (0.2 mM final) to induce protein expression, and shake overnight at 18°C.
The next morning, record the OD600 (usually between 6–8) and then harvest. Pellet cells by centrifugation at 6,000 rpm for 6 min (e.g., Sorvall SLA-3000 rotor). Resuspend the bacterial pellet (20 ml lysis buffer per 1000 OD600) in ice-cold lysis buffer (50 mM potassium phosphate pH 8.0, 400 mM NaCl, and 40 mM imidazole). Note that lysis and all subsequent steps should be done at 4°C with pre-chilled solutions.
Resuspend the bacteria homogenously to obtain a clump-free cell suspension, then process using a cell homogenizer (Constant Systems) at 25 kpsi. Mix the collected lysate immediately with 0.1 M PMSF (50 μl per 10 ml lysate) and add 250 units of TurboNuclease (Eton Bioscience).
Centrifuge the lysate at 9,500 rpm for 25 min (e.g., Sorvall SLA-600TC rotor), and recover the supernatant. Mix the supernatant with Ni2+ Sepharose 6 Fast Flow (GE Healthcare) slurry equilibrated with lysis buffer. Use approximately 1 ml Ni2+ resin per 1000 OD600 of cells.
Gently stir the mixture for 30 min, collect the Ni2+-resin in a 50 ml conical tube via several spins using a tabletop centrifuge, and then batch-wash the Ni2+-resin three times with 40 ml lysis buffer. Pour the Ni2+-Sepharose slurry into a disposable Pierce column (Thermo Scientific), and wash with 6x resin bed volumes of lysis buffer via gravity flow.
Once the column is drained, elute proteins using a 6x resin bed volume of elution buffer (10 mM Tris/HCl pH 8.0, 150 mM NaCl and 250 mM imidazole).
Concentrate that eluted protein to a final volume of ~10 ml using a centrifugal concentrator, and then purify by size exclusion chromatography on a HiLoad 26/60 Superdex S200 column (GE Healthcare) in a buffer containing 10 mM Tris/HCl pH 8.0 and 150 mM NaCl. Pool the peak corresponding to His6-tagged SUN2 is pooled, and mix with 3C protease at an enzyme:protein ratio of 1:50 (w/w). Overnight incubation with 3C protease is generally sufficient to remove the fusion tags, but this should be verified by SDS-PAGE analysis of a small aliquot.
Concentrate the cleaved SUN2 protein and purify again by size exclusion chromatography on a HiLoad 26/60 Superdex S200 column in 10 mM Tris/HCl pH 8.0, and 150 mM NaCl. Note that apo-SUN2 (residues 335–717) and apo-SUN2 (residues 522–717) elute differently without their fusion tags. Apo-SUN2 (residues 335–717) includes the entire predicted coiled-coil region and elutes as a homotrimer, as confirmed by analytical ultracentrifugation (Sosa et al., 2012). By contrast, apo-SUN2 (residues 522–717) behaves as a monomer after the His6-tri-GCN4 tag is removed, although this behavior is buffer dependent (see Section 3.2). These observations support the notion that the coiled-coil region (residues 335–540) adjacent to the SUN domain (residues 540–717) helps stabilize the SUN homotrimer and, hence, the KASH-binding-competent oligomeric state.
2.3 Purification of human SUN2-KASH complexes
Protocols for purifying SUN2 (residues 522–717)-KASH1-6 complexes are described here and shown schematically (Fig. 2).
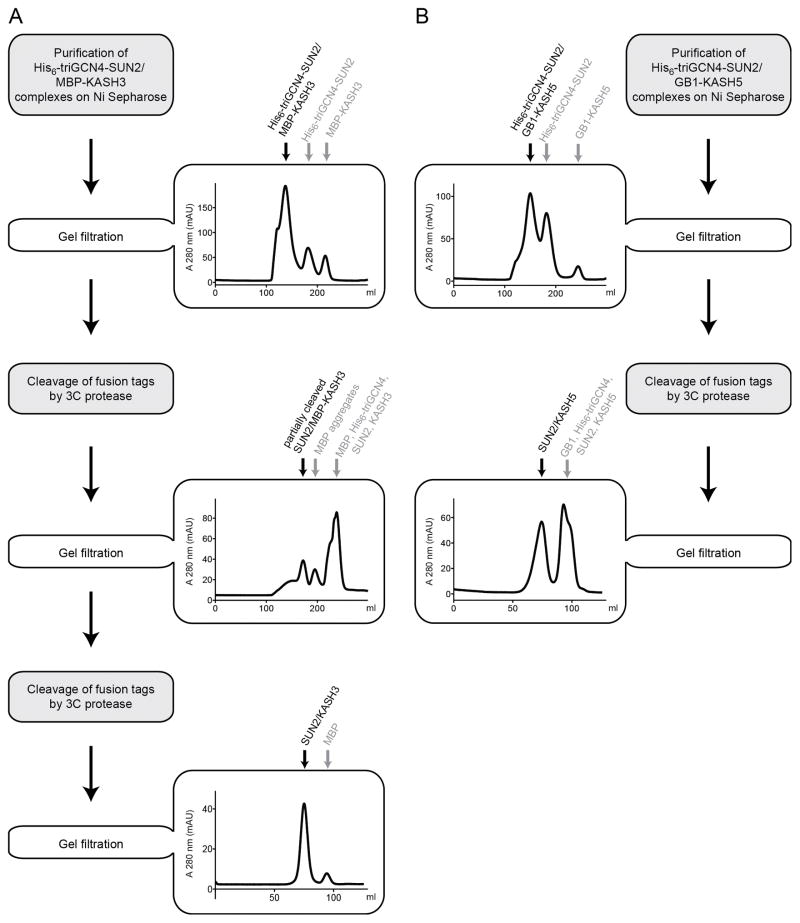
Figure 2.
Purification schemes for SUN2-KASH complexes. (A) Purification of the SUN2-KASH3 complex is illustrated to exemplify the strategy used for SUN2 / MBP-KASH constructs. A representative gel filtration elution profile at each step is shown, and the proteins eluted under each peak are indicated. (B) Purification of the SUN2-KASH5 complex is illustrated to demonstrate the strategy used for SUN2 / GB1-KASH constructs.
Steps 1–7 described in section 2.2 remain essentially the same, with one important difference: we now include 1 mM KCl in the Ni2+ elution buffer (and all subsequent buffers) since a K+ ion is likely to be coordinated in the cation loop of SUN2-KASH complexes (Sosa et al., 2012).
Similar to apo-SUN2, purify SUN2-KASH Ni2+eluates by size exclusion chromatography using a Superdex S200 column equilibrated with 10 mM Tris/HCl pH 8.0, 150 mM NaCl, and 1 mM KCl. This step helps remove aggregates, unbound SUN2, and KASH. Alternatively, the Ni2+ eluate can be subjected to a second purification on amylose resin if KASH is MBP-tagged. In this scenario, imidazole is removed by dialyzing the Ni2+ eluate against 10 mM Tris/HCl pH 8.0, 150 mM NaCl and 1 mM KCl prior to binding to amylose, and the SUN2-KASH complexes are then eluted in the presence of 10 mM maltose.
Mix the resulting SUN2-KASH complexes with 3C protease at an enzyme:substrate ratio of 1:25–1:50 (w/w). Retain an aliquot of uncut sample for SDS-PAGE analysis. After incubating 16 hours, resolve a small aliquot of SUN-KASH complexes by 15% SDS-PAGE to estimate cleavage efficiency. If necessary, add fresh 3C protease and incubate for at least 8 hours.
(a) The above procedure yields MBP-tagged SUN-KASH complexes that are often incompletely cleaved. Cleaved MBP can also form soluble aggregates of ill-defined size that partially co-elute with SUN-KASH complexes during gel filtration. To overcome both problems, and improve SUN-KASH complex homogeneity, we cleave the MBP tag in two steps (Fig. 2A). First, load the partially cleaved SUN-KASH complex from step 2.3.3 on a Superdex S200 column (in standard gel filtration buffer, e.g., 10 mM Tris/HCl pH 8.0, 150 mM NaCl, and 1 mM KCl) and purify. To avoid protein precipitation before column loading, the protein concentration should not exceed 1–2 mg/ml at this step. The partially cleaved SUN-KASH complex elutes in a fairly non-homogeneous peak during gel filtration, and is collected with contaminants. Second, pool the SUN-KASH complex-containing fractions, and cleave again with 3C protease at an enzyme:substrate ratio of approximately 1:50 (w/w). This second cleavage step should completely remove the fusion tags from SUN2-KASH1/2/3/4 complexes (verify by analytical SDS-PAGE). Re-concentrate these purified complexes, and then load onto a HiLoad 16/60 Superdex S200 column and purify using standard gel filtration buffer. SUN-KASH complexes elute in a single peak, and are now suitable for crystallization studies or biochemical assays. (b) The above strategies failed to yield pure and homogeneous complexes in the case of SUN2-KASH5/6. We tested alternatives to MBP-tags that could sustain super-stoichiometric expression of KASH5/6 peptides, relative to SUN, and could be removed more efficiently by 3C protease in step 2.3.3. Two candidates, sfGFP and thioredoxin, both reduced KASH-fusion protein expression to sub-stoichiometric levels compared to SUN (data not shown). However, the GB1 tag met both criteria, and was completely removed by proteolysis as observed by SDS-PAGE analysis. In a final step, the resulting complexes are concentrated and purified via gel filtration (Fig. 2B). We have not tried the GB1 tag with other SUN-KASH complexes, but suspect it may have consistent advantages over the MBP tag.
3. Structural analysis of human SUN2 and SUN2-KASH1/2 complexes
3.1 Crystallization and structure determination
We deposited three crystal structures in the Protein Data Bank representing human apo-SUN2 (PDB ID: 4DXT), and SUN2-KASH1/2 complexes (PDB ID: 4DXR/ 4DXS) (Sosa et al., 2012). Independently, apo-SUN2 (PDB ID: 3UNP) and SUN2-KASH2 (PDB ID: 4FI9) structures were solved by the Wang and Zhou labs (Wang et al, 2012; Zhou et al, 2012). Crystallization in all labs was performed by the hanging-drop vapor diffusion method in distinct experiment drop compositions. Apo-SUN2 crystals have grown in 16% (w/v) polyethylene glycol (PEG) 3350 and 200 mM potassium thiocyanate at 18°C in our lab, whereas the Wang group crystallized it in 100 mM imidazole, 1 M sodium acetate pH 6.5 and 10 mM YCl3 at 4°C. The resulting structures are overall very similar, except for a varying conformation of the unstructured KASH ‘lid’ (SUN2 residues 567-587) in the apo-form.
The KASH lid of the SUN domain becomes ordered, and adopts a β-hairpin form, only upon binding to KASH. Reflecting the high similarity between KASH1 and KASH2 in length and amino acid composition, the lid adopts an identical conformation in SUN2-KASH1/2 complexes. Although KASH1/2 peptides are not involved in crystal-packing contacts, SUN2-KASH1 and SUN2-KASH2 complexes crystallize in different conditions. In our lab, the SUN2-KASH1 complex was crystallized in 100 mM HEPES pH 7.4, 7% (w/v) PEG 4000, 10% 1,6-hexanediol and 0.25% n-decyl-b-D-maltoside (DM), whereas SUN2-KASH2 complex crystals were grown in 100 mM HEPES pH 7.5, 200 mM ammonium acetate, 25% 2-propanol and 0.3% DM. The Zhou lab, on the other hand, crystallized the SUN2-KASH2 complex in 50 mM MgCl2, 100 mM HEPES pH 7.5, 6% (w/v) polyethylene glycol monomethyl ether 5000, and 19.5 mM methyl-6-O-(N-heptylcarbamoyl)-a-D-glucopyranoside (HECAMEG).
Interestingly, apo-SUN2 as well as SUN2-KASH complexes, pack in rhombohedral crystals such that a trefoil-to-trefoil assembly occurs between neighboring SUN2 homotrimers, with the coiled-coil stalks pointing away in opposite directions. The major difference between these crystals is that the distance between neighboring apo-SUN2 trimers is much smaller than that of the SUN2-KASH1/2 complexes, due to the conformational change in the KASH-lid upon binding to KASH. Therefore, apo-SUN2 crystals have a lower solvent content, which might explain why they tend to diffract to higher resolution. Because the KASH peptide is not directly involved in crystal packing, there is a good chance that other SUN2-KASH complexes can be structurally characterized under similar crystallization conditions.
3.2 In vitro binding experiments
Interactions between SUN and KASH proteins have been studied using in vitro binding protocols established for apo-SUN2 and KASH2. For these studies apo-SUN2 (residues 522-717) and KASH2 (residues 6863-6885) are each purified separately. Apo-SUN2 is purified as described in section 2.2. We purify KASH2 as a fusion to the C-terminus of 6xHis-sfGFP; this 6xHis-sfGFP-KASH2 polypeptide (‘sfGFP-KASH2’) is first purified by Ni2+-affinity as described for apo-SUN constructs. The Ni2+-eluate is further purified by gel filtration using a Superdex S75 column equilibrated into 10 mM Tris/HCl pH 8.0 and 150 mM NaCl. sfGFP-KASH2 purified in this manner yields two approximately equal-intensity bands on SDS-PAGE, with a small size difference. Only the larger species binds SUN, suggesting the truncation is C-terminal and eliminates (part of) the KASH-peptide. Since these two forms of sfGFP-KASH2 are present at a ratio of about 1:1 (by SDS-PAGE), for qualitative binding assays we use a molar stoichiometry of 2:1 (sfGFP-KASH2 to apo-SUN2) to ensure a roughly 1:1 ratio of apo-SUN2 and binding-competent sfGFP-KASH2.
Binding is measured by analytical gel filtration on a Superdex S200 HR10/300 column (Fig. 3). Prior to injecting the sample, apo-SUN2, and apo-SUN2 incubated with sfGFP-KASH2, are dialyzed into the gel filtration buffer overnight. For each gel filtration run, a total volume of 500 μl is loaded onto the column; each sample contains 50 μM SUN2 and 100 μM sfGFP-KASH2 (to achieve a 1:1 ratio of SUN2 to binding-competent undegraded sfGFP-KASH). In the chromatogram, the sfGFP-KASH2 fusion protein is monitored by absorbance at 488 nm, and total protein absorption is measured at 280 nm.
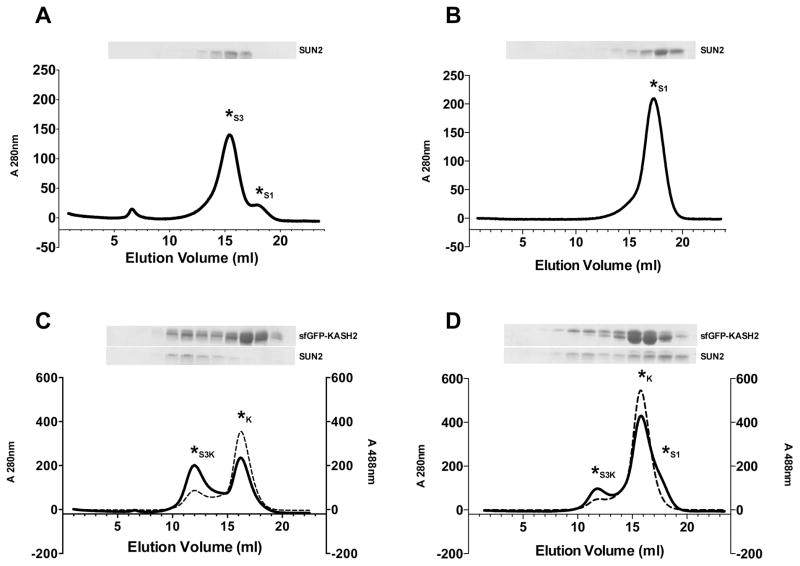
Figure 3.
In vitro SUN-KASH binding experiments. SUN-KASH binding was analyzed by gel filtration on a Superdex S200 HR10/300 column. Each panel shows a representative gel filtration profile and corresponding SDS-PAGE as follows: (A) Apo-SUN2 in buffer 1. (B) Apo-SUN2 in buffer 2. (C) Apo-SUN2 and sfGFP-KASH2 in buffer 1. (D) Apo-SUN2 and sfGFP-KASH2 in buffer 2. Asterisks are used to mark each peak, denoting the SUN2 monomer peak as S1, the SUN2 trimer as S3, the SUN2 trimer bound to sfGFP-KASH2 as S3K, and unbound sfGFP-KASH2 as K. Solid lines on each chromatogram represent the 280 nm trace; dashed lines represent the 488 nm trace. Buffer 1 contains 20 mM HEPES pH 8.0 and 100 mM KCl. Buffer 2 contains 10 mM Tris/HCl pH 8.0, 150 mM NaCl and 1 mM KCl. Each Coomassie-stained SDS-PAGE gel shows 1 ml fractions covering the 7–18 ml elution segment. SUN oligomerization is buffer-dependent. Buffers that enable SUN trimerization are required for SUN-KASH binding experiments.
The results of such binding experiments are shown in Figure 3. When apo-SUN2 (residues 522-717) is run in buffer 1 (20 mM HEPES/NaOH pH 8.0, 100 mM KCl), it elutes primarily as a trimer (‘S3’; Fig. 3A); a small fraction elutes as a monomer (‘S1’; Fig. 3A). Buffer 1 was previously used for in vitro SUN-KASH binding assays, however the effect of monomer-trimer equilibrium on KASH binding was not discussed (Zhou et al, 2012). In contrast, apo-SUN2 elutes as a monomer when using buffer 2 (10 mM Tris/HCl pH 8.0, 150 mM NaCl, 1 mM KCl) (Fig. 3B). SUN trimerization is a prerequisite for KASH binding, as shown by gel filtration analysis of apo-SUN2 / sfGFP-KASH2 mixtures. Using buffer 1, we obtain a stoichiometric SUN2 / sfGFP-KASH2 complex, and excess sfGFP-KASH2 elutes as a separate peak (‘K’; Fig. 3C). Using buffer 2, only a fraction of SUN2 elutes as an assembled complex with sfGFP-KASH2, whereas the majority remains unbound (Fig. 3D). Thus, buffer conditions play an important role in SUN-KASH binding experiments. This buffer dependence is particularly critical for SUN constructs that lack portions of the coiled-coil element.
4. Conclusions and pitfalls
Our biochemical understanding of SUN-KASH assemblies is still incomplete, and many interesting questions remain open. The buffer-dependence of SUN proteins in vitro is not yet understood; indeed the mechanisms of LINC complex assembly and disassembly may be strongly influenced by the unique microenvironment of the NE/ER lumen, which current in vitro conditions do not reproduce. The specificity and strength of SUN-KASH interactions are relatively unexamined, yet further analysis of SUN-KASH proteins from different species can potentially help resolve this problem. Finally, apart from the SUN-KASH core, structural knowledge about most other regions of LINC complexes is limited— an open area of research with fascinating implications for understanding the nuclear envelope and mechanisms of human laminopathy diseases.
Acknowledgments
We thank Brian A. Sosa for developing the initial protocols for purification and crystallization of human SUN2-KASH1/2 complexes. This work was supported by a grant from the NIH (1RO1-AR065484).
References
- Andersen KR, Leksa NC, Schwartz TU. Optimized E. coli expression strain LOBSTR eliminates common contaminants from His-tag purification. Proteins. 2013;81:1857–1861. doi: 10.1002/prot.24364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bione S, Maestrini E, Rivella S, Mancini M, Regis S, Romeo G, Toniolo D. Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat Genet. 1994;8:323–327. doi: 10.1038/ng1294-323. [DOI] [PubMed] [Google Scholar]
- Bonne G, Di Barletta MR, Varnous S, Becane HM, Hammouda EH, Merlini L, et al. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat Genet. 1999;21:285–288. doi: 10.1038/6799. [DOI] [PubMed] [Google Scholar]
- Borrego-Pinto J, Jegou T, Osorio DS, Auradé F, Gorjánácz M, Koch B, et al. Samp1 is a component of TAN lines and is required for nuclear movement. J Cell Sci. 2012;125:1099–1105. doi: 10.1242/jcs.087049. [DOI] [PubMed] [Google Scholar]
- Burke B, Roux KJ. Nuclei take a position: managing nuclear location. Dev Cell. 2009;17:587–597. doi: 10.1016/j.devcel.2009.10.018. [DOI] [PubMed] [Google Scholar]
- Chang W, Folker ES, Worman HJ, Gundersen GG. Emerin organizes actin flow for nuclear movement and centrosome orientation in migrating fibroblasts. Mol Biol Cell. 2013;24:3869–3880. doi: 10.1091/mbc.E13-06-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Worman HJ, Gundersen GG. Accessorizing and anchoring the LINC complex for multifunctionality. J Cell Biol. 2015;208:11–22. doi: 10.1083/jcb.201409047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Chi YH, Mutalif RA, Starost MF, Myers TG, Anderson SA, et al. Accumulation of the inner nuclear envelope protein Sun1 is pathogenic in progeric and dystrophic laminopathies. Cell. 2012;149:565–577. doi: 10.1016/j.cell.2012.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, et al. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Guan C, Li P, Riggs PD, Inouye H. Vectors that facilitate the expression and purification of foreign peptides in Escherichia coli by fusion to maltose-binding protein. Gene. 1988;67:21–30. doi: 10.1016/0378-1119(88)90004-2. [DOI] [PubMed] [Google Scholar]
- Göb E, Schmitt J, Benavente R, Alsheimer M. Mammalian sperm head formation involves different polarization of two novel LINC complexes. PloS One. 2010;5:e12072. doi: 10.1371/journal.pone.0012072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque F, Lloyd DJ, Smallwood DT, Dent CL, Shanahan CM, Fry AM, et al. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol Cell Biol. 2006;26:3738–3751. doi: 10.1128/MCB.26.10.3738-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque F, Mazzeo D, Patel JT, Smallwood DT, Ellis JA, Shanahan CM, Shackleton S. Mammalian SUN protein interaction networks at the inner nuclear membrane and their role in laminopathy disease processes. J Biol Chem. 2010;285:3487–3498. doi: 10.1074/jbc.M109.071910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn HF, Brownstein Z, Lenz DR, Shivatzki S, Dror AA, Dagan-Rosenfeld O, et al. The LINC complex is essential for hearing. J Clinical Investigation. 2013a;123:740–750. doi: 10.1172/JCI66911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn HF, Kim DI, Wright GD, Wong ESM, Stewart CL, Burke B, Roux KJ. A mammalian KASH domain protein coupling meiotic chromosomes to the cytoskeleton. J Cell Biol. 2013b;202:1023–1039. doi: 10.1083/jcb.201304004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huth JR, Bewley CA, Jackson BM, Hinnebusch AG, Clore GM, Gronenborn AM. Design of an expression system for detecting folded protein domains and mapping macromolecular interactions by NMR. Protein Science : a Publication of the Protein Society. 1997;6:2359–2364. doi: 10.1002/pro.5560061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVallie ER, Lu Z, Diblasio-Smith EA, Collins-Racie LA, McCoy JM. Thioredoxin as a fusion partner for production of soluble recombinant proteins in Escherichia coli. Methods in Enzymology. 2000;326:322–340. doi: 10.1016/s0076-6879(00)26063-1. [DOI] [PubMed] [Google Scholar]
- Lindeman RE, Pelegri F. Localized products of futile cycle/lrmp promote centrosome-nucleus attachment in the zebrafish zygote. Current Biology. 2012;22:843–851. doi: 10.1016/j.cub.2012.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Schneider M, Neumann S, Jaeger VM, Taranum S, Munck M, et al. Nesprin interchain associations control nuclear size. Cell Molec Life Sciences. 2012;69:3493–3509. doi: 10.1007/s00018-012-1034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton GWG, Starr DA. KASHing up with the nucleus: novel functional roles of KASH proteins at the cytoplasmic surface of the nucleus. Curr Opin Cell Biol. 2014;28:69–75. doi: 10.1016/j.ceb.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton GWG, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 2010;329:956–959. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton GWG, Gomes ER, Folker ES, Worman HJ, Gundersen GG. TAN lines: a novel nuclear envelope structure involved in nuclear positioning. Nucleus (Austin, Tex) 2011;2:173–181. doi: 10.4161/nucl.2.3.16243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostlund C, Folker ES, Choi JC, Gomes ER, Gundersen GG, Worman HJ. Dynamics and molecular interactions of linker of nucleoskeleton and cytoskeleton (LINC) complex proteins. J Cell Sci. 2009;122:4099–4108. doi: 10.1242/jcs.057075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmakumar VC, Libotte T, Lu W, Zaim H, Abraham S, Noegel AA, et al. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J Cell Sci. 2005;118:3419–3430. doi: 10.1242/jcs.02471. [DOI] [PubMed] [Google Scholar]
- Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nature Biotechnology. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- Puckelwartz MJ, Kessler E, Zhang Y, Hodzic D, Randles KN, Morris G, et al. Disruption of nesprin-1 produces an Emery Dreifuss muscular dystrophy-like phenotype in mice. Hum Mol Genet. 2009;18:607–620. doi: 10.1093/hmg/ddn386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothballer A, Kutay U. The diverse functional LINCs of the nuclear envelope to the cytoskeleton and chromatin. Chromosoma. 2013;122:415–429. doi: 10.1007/s00412-013-0417-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa BA, Rothballer A, Kutay U, Schwartz TU. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell. 2012;149:1035–1047. doi: 10.1016/j.cell.2012.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Fridolfsson HN. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol. 2010;26:421–444. doi: 10.1146/annurev-cellbio-100109-104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Han M. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science. 2002;298:406–409. doi: 10.1126/science.1075119. [DOI] [PubMed] [Google Scholar]
- Wang W, Shi Z, Jiao S, Chen C, Wang H, Liu G, Wang Q, Zhao Y, Greene MI, Zhou Z. Structural insights into SUN-KASH complexes across the nuclear envelope. Cell Res. 2012;22:1440–1452. doi: 10.1038/cr.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ. Nuclear lamins and laminopathies. J Pathology. 2012;226:316–325. doi: 10.1002/path.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Bethmann C, Worth NF, Davies JD, Wasner C, Feuer A, et al. Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum Mol Genet. 2007;16:2816–2833. doi: 10.1093/hmg/ddm238. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Skepper JN, Yang F, Davies JD, Hegyi L, Roberts RG, et al. Nesprins: a novel family of spectrin-repeat-containing proteins that localize to the nuclear membrane in multiple tissues. J Cell Sci. 2001;114:4485–4498. doi: 10.1242/jcs.114.24.4485. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Du X, Cai Z, Song X, Zhang H, Mizuno T, Suzuki E, Yee MR, Berezov A, Murali R, Wu SL, Karger BL, Greene MI, Wang Q. Structure of Sad1-UNC84 homology (SUN) domain defines features of molecular bridge in nuclear envelope. J Biol Chem. 2012;287:5317–5326. doi: 10.1074/jbc.M111.304543. [DOI] [PMC free article] [PubMed] [Google Scholar]