Abstract
Hemorrhagic shock is a leading cause of morbidity and mortality worldwide. Significant blood loss may lead to decreased blood pressure and inadequate tissue perfusion with resultant organ failure and death, even after replacement of lost blood volume. One reason for this high acuity is that the fundamental mechanisms of shock are poorly understood. Proteomic and metabolomic approaches have been used to investigate the molecular events occurring in hemorrhagic shock but, to our knowledge, a systematic analysis of the transcriptomic profile is missing. Therefore, a pilot analysis using paired-end RNA sequencing was used to identify changes that occur in the blood transcriptome of rats subjected to hemorrhagic shock after blood reinfusion. Hemorrhagic shock was induced using a Wigger’s shock model. The transcriptome of whole blood from shocked animals shows modulation of genes related to inflammation and immune response (Tlr13, Il1b, Ccl6, Lgals3), antioxidant functions (Mt2A, Mt1), tissue injury and repair pathways (Gpnmb, Trim72) and lipid mediators (Alox5ap, Ltb4r, Ptger2) compared with control animals. These findings are congruent with results obtained in hemorrhagic shock analysis by other authors using metabolomics and proteomics. The analysis of blood transcriptome may be a valuable tool to understand the biological changes occurring in hemorrhagic shock and a promising approach for the identification of novel biomarkers and therapeutic targets.
Impact statement
This study provides the first pilot analysis of the changes occurring in transcriptome expression of whole blood in hemorrhagic shock (HS) rats. We showed that the analysis of blood transcriptome is a useful approach to investigate pathways and functional alterations in this disease condition. This pilot study encourages the possible application of transcriptome analysis in the clinical setting, for the molecular profiling of whole blood in HS patients.
Keywords: Rats, hemorrhagic shock, RNAseq, transcriptome, reperfusion, whole blood
Introduction
Hemorrhagic shock (HS) results in significant morbidity and mortality worldwide.1,2 It is a leading cause of death with a global financial burden of $518 billion/year. Characterized by inadequate tissue perfusion and oxygen delivery, despite decades of research, there exists no consensus as to the fundamental mechanisms of shock, nor has an optimal treatment strategy been achieved.1 Clinical intervention is aimed at restoring patient hemodynamics by re-establishing adequate systemic arterial pressure, cardiac function and organ perfusion. However, reperfusion after circulatory shock is frequently followed by a systemic severe inflammatory response syndrome that may lead to multi-organ failure (MOF)3 and death.
The trigger mechanisms that induce tissue damage in MOF at the molecular level are unclear. The microvasculature is a target of shock, being very sensitive to the deleterious effects of hypoxia induced by ischemia and reoxygenation following reperfusion.4 Molecular, cellular and biochemical changes have been reported in endothelial cells lining blood vessels exposed to I/R.5 These include endothelial glycocalyx shedding,6 excess reactive oxygen species (ROS) production,7 and leukocyte activation.4
In shock, cardiovascular function is impaired and the circulating components of this system continuously interact with all cells and tissues of the body. Circulating blood may reflect mechanisms occurring in tissues and organs directly involved in the disease process as well as modulate these mechanisms. The potential of blood to be used as a “fingerprint” for different disease states has been demonstrated in several conditions.8 In HS, modified expression of genes and pathways in whole blood may have the potential to highlight both specific functions activated by circulating cells and general mechanisms of cellular adaptation to altered physiological conditions. Whole blood samples, that include white and red blood cells and platelets, have the advantage of being easily accessible and thus are a potentially valuable source for biomarker identification in the clinical setting. The use of whole blood allows a global overview of the transcriptome, when there is no specific interest in the individual contributions of each cell type.
In this paper, we present the results of a pilot study aimed at profiling the transcriptome of peripheral whole blood of HS rats compared to healthy controls by means of RNAseq technology.
Materials and methods
Experimental protocol
The experimental protocol was conducted as described in detail in Aletti et al.9 Briefly, six male Wistar rats (300–450 g, Harlan Laboratories, Inc., Indianapolis, IN) were randomly assigned to either a control (CTRL, n = 3) or a HS (n = 3) group. All rats were anesthetized (xylazine, 4 mg/kg; ketamine 75 mg/kg i.m.), and the right femoral vein and artery were cannulated for blood withdrawal and intravenous supplemental anesthesia, and for continuous monitoring of arterial pressure, respectively. Body temperature was maintained at 37℃ via water-heated support and heat blanket. Animals were allowed 5 min for hemodynamic stabilization after induction of anesthesia and vascular line placement, and then were heparinized (1 unit heparin/cc estimated total blood volume, estimated at 6% body weight) to ensure patency of vascular access lines. The HS model chosen for this experimental study is the Wiggers’ model. Hypovolemia was achieved by withdrawal of blood from the femoral vein (0.5 cc/min) to a target mean arterial pressure (MAP) of 35 mmHg, and maintained for 2 h. The shed blood was then reinfused (0.5 cc/min), and the animals were monitored for an additional 2 h, before euthanasia (Beuthanasia-D, 120 mg/kg, Merck Animal Health). The shed blood was maintained at room temperature during the hypovolemic period (∼22℃) and gently warmed to 37℃ before reinfusion to minimize the temperature gradient.
CTRL animals were treated identically except for actual hemorrhage. Death was confirmed by verification of cardiac arrest after thoracotomy.
The animal protocol was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at the University of California, San Diego and conforms to the Guide for the Care and Use of Laboratory Animals, 8th edition, by the National Institutes of Health (2011).
Blood collection and RNA extraction
At the conclusion of the 120-min observation period which followed blood reperfusion (before euthanasia), venous blood was collected from each animal in EDTA-coated tubes and maintained on ice. Four hundred microliters of whole blood was immediately added to 400 µl of denaturing solution (Ambion, USA), in duplicate. Total RNA was extracted from 800 µl of treated blood with MirVana Paris Kit (Ambion, USA). RNA quality control was performed on all RNA samples with an electrophoretic run on Agilent Bioanalyzer instrument using the RNA 6000 Nano Kit (Agilent, Santa Clara, CA). RNA Integrity Number (RIN) was determined for every sample, and all samples were considered suitable for processing based on the RNA integrity (RIN > 8). RNA concentration was estimated through spectrophotometric measurement using a Nanoquant Infinite M200 instrument (Tecan, Austria).
As regards healthy controls, a blood sample was withdrawn at the end of a period of observation under anesthesia of the same duration as the shock experiment.
Library preparation for RNA sequencing
Sequencing libraries were prepared using the TruSeq Stranded Total RNA with Ribo-Zero Globin Kit (Illumina, San Diego, CA) using 900 ng of total RNA as input. The kit uses oligo-attached magnetic beads to remove rRNA and globin mRNA from total RNA. The remaining RNAs were purified, fragmented at 94℃ for 8 min and primed with random hexamers for cDNA synthesis. Multiple indexing adapters were ligated to the ends of ds cDNAs that were then amplified with 10 PCR cycles. Final libraries were validated and quantified with the DNA1000 kit on Agilent Bioanalyzer Instrument. Pooled libraries were sequenced on the HiSeq2500 Instrument producing 50 × 2 bp paired end reads.
Analysis of sequencing data
Raw reads were first checked for quality using FASTQC (v0.11.2) (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Input reads were then aligned to the rat reference genome (Rnov 6.0, ensembl release 82) using STAR aligner (v2.4.2).10 We tuned indexing and alignment parameters in order to deal with 50 × 2 bp paired end reads and obtain only uniquely mapped fragments. We used the rattus norvegicus ensembl v.82 gene transfer file (GTF) as reference annotation for alignment and calculation of sequenced fragments overlapping features. In particular, we performed strand-specific fragments counting (-s 2 and –minOverlap 2) using featureCounts (v.1.5.0)11 available in the Subread package. The gene counts matrix was then processed in R environment by DESeq2 12 (v1.10.1) and EdgeR13 (v3.2.1) packages. Gene count matrix pre-processing and differentially expressed genes (DEGs) identification were performed in parallel with DESeq2 and EdgeR adopting their default normalization methods (median of ratios and trimmed mean of M values (TMM), respectively). In the analysis performed with EdgeR, we required at least three samples having normalized read counts of 3 cpm (three counts per million aligned fragments), in order to include a gene in the differential expression analysis. We selected DEGs with p.adj value (FDR) < 0.05. Final DEGs list includes genes common to both pipelines. All DEGs were checked by visual inspection using Integrative Genome Viewer. Genes with inconsistency between reads mapping coverage and reference gene model were discarded from downstream data analyses.
Downstream bioinformatics analysis
We used ClueGO v2.2.5 software14 to identify enriched biological processes, starting from the list of identified DEGs. We used as input the list of DEGs gene symbols. Parameters were set up in order to decrease redundancy by fusing-related terms that share similar gene sets. We applied the default embedded statistical test in ClueGO that is an overrepresentation analysis (ORA) based on a two-sided hypergeometric test that uses Bonferroni as multiple testing correction. Enriched terms were grouped through a Kappa Statistics (kappa score threshold 0.4) in order to reduce the complexity of the results. Only enriched clusters with a p-value < 0.05 were considered statistically significant.
Molecular functions of DEGs were investigated with a manual approach, and DEGs were systematically searched for citations in PubMed database, using “ischemia and reperfusion” and “hemorrhagic shock” as keywords.
Results
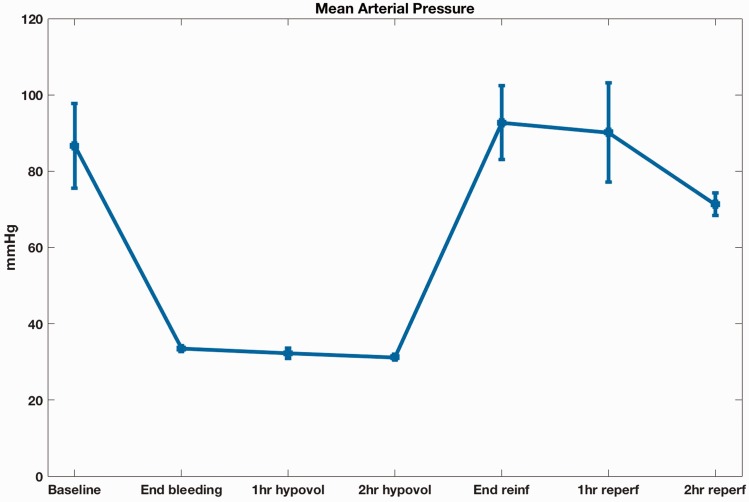
To induce HS, blood was withdrawn to a target MAP of 35 mmHg, and maintained for 2 h, at which time the shed blood was reinfused. The animals were observed for 2 h following reinfusion and blood samples for RNAseq were taken at the end of this period. During this time, despite the restoration of euvolemia and the initial recovery of basal BP levels, BP progressively decreased (Figure 1).
Figure 1.
Mean arterial pressure tracing during the acute hemorrhagic shock experiment (A color version of this figure is available in the online journal.)
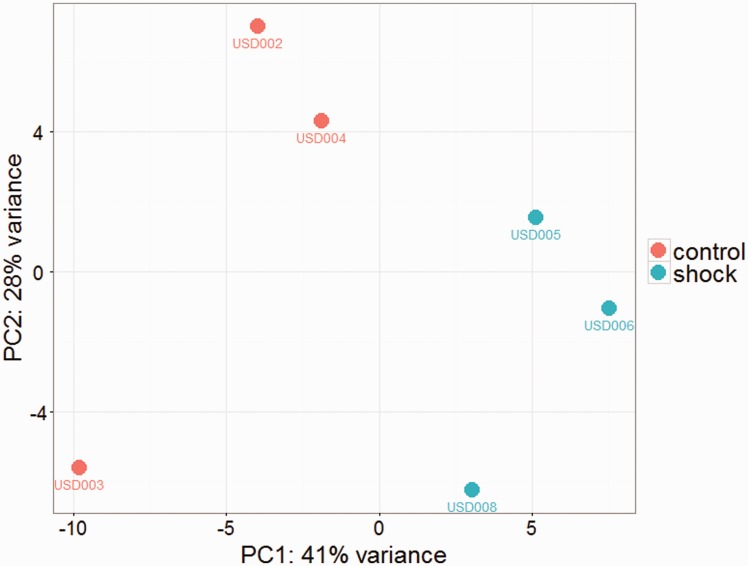
In the RNAseq experiment, on average, 66.91 ± 8.83 M reads per sample were obtained (91.93% ± 2.73 mapping to Rnov6) and 11.14 ± 3.43 M fragment counts per sample mapping uniquely and unambiguously to Rnov6 ensembl v.82 gtf annotation file. According to the threshold used to consider a gene expressed (cpm > 3 in at least three samples), we found an average of 8364 ± 119 expressed genes in the six animals. We performed a principal component analysis (PCA) on the top 500 most variable genes in HS and CTRL rats. The PCA plot (Figure 2) shows the two groups of rats clearly separated on the first PC.
Figure 2.
Principal component analysis (PCA) on the top 500 most variable genes in HS and CTRL rats (A color version of this figure is available in the online journal.)
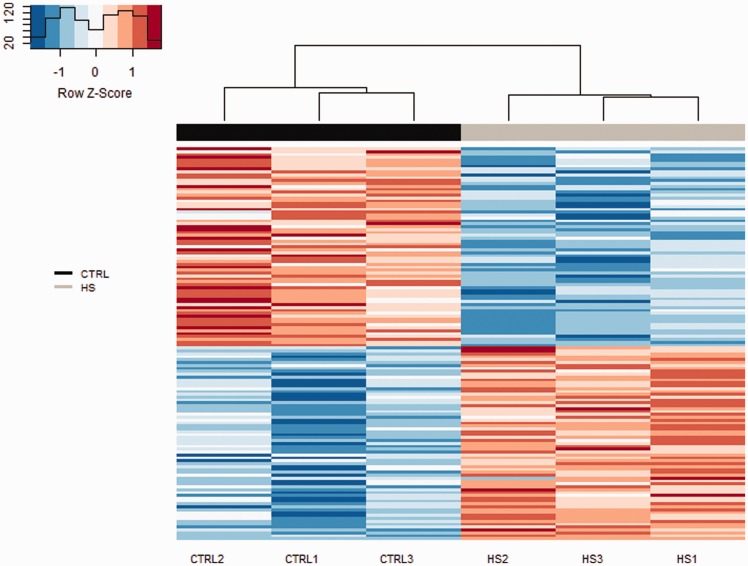
The gene expression profile of HS and CTRL samples were compared to identify DEGs. We identified 136 DEGs (FDR < 0.05) (Figure 3, Supplementary Table 1), with 67 upregulated and 69 downregulated genes in HS compared with CTRL rats.
Figure 3.
Heatmap of gene expression for 136 genes differentially expressed between HS and CTRL rats. Each column represents an animal (CTRL and HS), with animals clustered by similarity using a hierarchical clustering. The gene expression level is represented by a color scale gradient ranging from blue (lower expression) to red (higher expression) (A color version of this figure is available in the online journal.)
Functional analysis using ClueGO detected the enrichment of seven gene ontology clusters: leukocyte chemotaxis, positive regulation of I-κB kinase in the NF-κB signaling pathway, negative regulation of cellular response to insulin stimulus, response to zinc ion, cholesterol metabolic processes, tricarboxylic acid cycle and regulation of extrinsic apoptotic signaling pathway (Table 1).
Table 1.
Results of gene ontology overrepresentation analysis (ORA)
| GOID | GOTerm | Cluster | Adj.pval | Gene symbols |
|---|---|---|---|---|
| GO:0006099 | Tricarboxylic acid cycle | 1 | 1.90E-03 | Dlst, Idh2, Pdha1 |
| GO:0032675 | Regulation of interleukin-6 production | 2 | 9.60E-05 | Bank1, Hyal2, Il1b, Tlr7, Tnfaip3 |
| GO:0032722 | Positive regulation of chemokine production | 2 | 9.60E-05 | Ffar2, Il1b, Tlr7 |
| GO:0032755 | Positive regulation of interleukin-6 production | 2 | 9.60E-05 | Hyal2, Il1b, Tlr7 |
| GO:0032757 | Positive regulation of interleukin-8 production | 2 | 9.60E-05 | Ffar2, Hyal2, Il1b, Tlr7 |
| GO:0050729 | Positive regulation of inflammatory response | 2 | 9.60E-05 | Alox5ap, CcI6, Ffar2, Hyal2, Il1b |
| GO:0002675 | Positive regulation of acute inflammatory response | 2 | 9.60E-05 | Alox5ap, Ffar2, Il1b |
| GO:0043123 | Positive regulation of I-kappaB kinase/NF-kappaB signaling | 2 | 9.60E-05 | Il18r1, Il1b, Ltbr, Nfkbie, Prkcb, Tlr7 |
| GO:0072604 | Interleukin-6 secretion | 2 | 9.60E-05 | Bank1, Hyal2, Il1b |
| GO:0042307 | Positive regulation of protein import into nucleus | 2 | 9.60E-05 | Hyal2, Il18r1, Il1b, Tlr7 |
| GO:0042346 | Positive regulation of NF-kappaB import into nucleus | 2 | 9.60E-05 | Il18r1, Il1b, Tlr7 |
| GO:0030595 | Leukocyte chemotaxis | 3 | 5.50E-04 | C5ar1, CcI6, Ffar2, Il1b, Jaml, Lgals3, Swap70 |
| GO:0097529 | Myeloid leukocyte migration | 3 | 5.50E-04 | C5ar1, CcI6, Il1b, Jaml, Lgals3, Swap70 |
| GO:0030593 | Neutrophil chemotaxis | 3 | 5.50E-04 | C5ar1, CcI6, Il1b, Jaml, Lgals3 |
| GO:1900077 | Negative regulation of cellular response to insulin stimulus | 4 | 5.50E-04 | Il1b, Mzb1, Prkcb, Trim72 |
| GO:2001236 | Regulation of extrinsic apoptotic signaling pathway | 5 | 2.50E-05 | Hyal2, Il1b, Inhba, Itga6, Lgals3, Ltbr, Tnfaip3, Traf1 |
| GO:2001237 | Negative regulation of extrinsic apoptotic signaling pathway | 5 | 2.50E-05 | Il1b, Itga6, Lgals3, Tnfaip3 |
| GO:2001238 | Positive regulation of extrinsic apoptotic signaling pathway | 5 | 2.50E-05 | Hyal2, Inhba, Ltbr |
| GO:0008203 | Cholesterol metabolic process | 6 | 4.70E-03 | Cebpa, Fdft1, Hmgcr, Ldlr |
| GO:0010043 | Response to zinc ion | 7 | 2.10E-03 | Ass1, Hvcn1, Mt1, Mt2A |
Note: Summary table of ClueGO analysis showing overrepresented gene ontology groups. For each enriched GO term, information is provided about: identification of gene ontologies terms (GOID, GO term); the cluster that include the GO term (Cluster); multiple testing adjusted p value obtained with Bonferroni stepwise correction (adj.pval); the genes symbols from the uploaded cluster that were associated with the term (gene symbols).
ClueGO analysis, complemented with a literature-based search, highlighted 39 DEGs, whose functions are related to inflammation, immune system, lipid and amino acid metabolism, ROS scavenging, tissue repair and inhibition of protease activity (Table 2). Some of the identified DEGs have been previously linked to HS and/or ischemia-reperfusion. In detail, we identified the involvement of genes encoding Pattern Recognition Receptors (Tlr13, Clec4b2, Tlr7, Ly86), and inflammation-related genes (Il1b, Ccl6, Il18r1, Lgals3, Hyal2, Hpk1, Jam2, Amica1, C5ar, Cfd, Tnfaip3, Nfkbie). In HS rats, functions of the adaptive immune system were downregulated (Cd79b, Ighm, RT1-Ba, RT1-Da, RT1-Db1, RT1-N3 and Ms4a1). A change was found in the expression level of genes related to the lipid pathway (Alox5ap, Ltb4r, Ptger2, Ffar2, Ldlr) and amino acid metabolism (Ass1, Bcat1, Ggt1, Prodh). We also observed in HS rats the upregulation of genes with protective functions, including ROS scavenging (Nqo1, Mt2A, Mt1), inhibition of neutrophil protease activity (Olfm4, Cst7) and tissue repair (Gpnmb, Trim72).
Table 2.
Differentially expressed genes and functional pathways
| DEGs | Functions |
|---|---|
| Tlr13, Clec4b2, Tlr7, Ly86 | Pattern recognition receptors |
| Il1b, Ccl6, Il18r1, Lgals3, Hyal2, Hpk1, Jam2, Amica1, C5ar, Cfd, Tnfaip3, Nfkbie | Acute inflammation |
| Cd79b, Ighm, RT1-Ba, RT1-Da, RT1-Db1, RT1-N3 and Ms4a1 | Adaptive immune functions |
| Alox5ap, Ltb4r, Ptger2, Ffar2, Ldlr | Lipid pathway |
| Ass1, Bcat1, Ggt1, Prodh | Amino acid metabolism |
| Nqo1, Mt2A, Mt1 | ROS scavenging |
| Gpnmb, Trim72 | Tissue and cell membrane repair |
| Olfm4, Cst7 | Proteases activity inhibition |
Note: Selection of differentially expressed genes involved in relevant functions in the pathophysiology of hemorrhagic shock.
Discussion
The aim of the present pilot study was to describe the effects of blood reperfusion following prolonged hypovolemia on the blood transcriptome in an acute experimental model of HS. We used a well-studied model of HS that has blood pressure as primary end point.15
Blood samples for transcriptome profiling were collected at the end of the two post reinfusion hours, to assess the acute effect of HS.
The 136 genes that significantly changed their expression between CTRL and HS rats are predominantly related to acute inflammation, pattern recognition receptors, immune response, ROS scavenging, tissue injury and repair. Shock induces a systemic inflammatory response after the primary injury. Injured tissues release damage-associated molecular patterns (DAMPs), the alarmins that promote the inflammatory response, the production of pro-inflammatory cytokines and immune cell chemotaxis.16 In our study, Tlr13, an innate pattern-recognition orphan receptor expressed by innate immune cells in rodents, was upregulated in HS rats. Our data confirm the observation of Mira et al.,17 who detected the upregulation of Tlr13 in a murine model of HS/polytrauma, suggesting a role of TLR13 in binding DAMPs released by injured tissues after trauma. Similarly, the C-type lectin Clec4b2, a member of the C-type lectin receptors that functions as a pattern recognition receptor, was upregulated in HS rats. Conversely, downregulation was observed for toll-like receptor 7 (Tlr7) that binds single stranded viral RNA18 and for lymphocyte antigen 86 (Ly86 alias Md-1), an accessory molecule involved in inhibition of TLR signaling.19 Along with modifications of the expression of pattern recognition receptors, we observed in HS the downregulation of Il1b (interleukin1 beta) and the upregulation of Ccl6 (a CC chemokine) and Il18r1 (interleukin 18 receptor), all which play a key role in acute inflammation. Il1b was downregulated in HS rats compared with controls, although it is a well-known proinflammatory cytokine, suggesting that the inflammatory response induced by HS in blood does not necessarily require the specific action of this signaling molecule. Ccl6 is an important mediator in the recruitment of macrophages, responsible for initiating and maintaining macrophage infiltration during the inflammatory process,20 whereas Il18r1 is the receptor of the powerful proinflammatory cytokine Il18. Lgals3, which is upregulated in HS rats, encodes Galectin-3, a member of the galectin family of carbohydrate-binding proteins secreted by immune cells. Galectin-3 acts as an alarmin and is involved in neutrophil activation and adhesion, chemoattraction of monocytes, opsonization of apoptotic neutrophils and activation of mast cells.21 Galectin-3 has been proposed as an independent predictor of mortality and rehospitalization in acute heart failure patients.22 Ischemia/reperfusion (I/R) that occurs in HS with resuscitation is also characterized by the modification of the interaction between leukocytes and endothelial cells that leads to migration of neutrophils in tissues.4 Among the molecules involved in neutrophil adhesion functions, the hematopoietic progenitor kinase 1 gene (Hpk1 alias Map4k1)23 was upregulated in HS rats as well as Jam2, a junctional adhesion molecule implicated in leukocyte migration.24 Amica1 (alias Jaml) however, a transmembrane protein involved in leukocyte transendothelial migration25 and T-cell costimulation,26 was downregulated in HS rats, demonstrating that within the same functional process (leukocyte adhesion and migration) different molecules can be specifically regulated.
In HS, we observed the downregulation of genes involved in the adaptive immune response: the immunoglobulin genes Cd79b and Ighm, genes of the rat major histocompatibility complex (RT1-Ba, RT1-Da, RT1-Db1, RT1-N3), Ms4a1 encoding a transmembrane protein, selectively expressed on mature B cells.27 This observation is similar to the downregulation of the adaptive immune functions described in septic shock by Hotchkiss et al.,28 and it also suggests that common patterns can be found in different types of shock.
In inflammation, proresolving lipid mediators are powerful modulators and activators of the resolution mechanisms. For example, Alox5ap and Ltb4r, which encode the arachidonate 5-lipoxygenase-activating protein required for the biosynthesis of leukotrienes and leukotriene B4 receptor, were upregulated in HS. Leukotrienes are lipid mediators produced from arachidonic acid, an essential fatty acid, and are involved in the initial inflammatory response and in leukocyte adhesion and chemotaxis.29 Leukotrienes and 5-lipoxigenase have been identified as mediators of acute lung injury in a murine model of HS.30 Downregulation in HS rats was detected for the prostaglandin receptor Ptger2, whereas the receptors Ffar2 and Ldlr, that bind free fatty acids and low-density lipoproteins, respectively, were upregulated in HS rats. The finding that receptor molecules that bind different classes of lipids are modulated in HS rats underlines the role of lipids as mediators of inflammation and is in line with the changes in lipid metabolism described by D’Alessandro et al.31 in HS. The same authors reported a derangement of the amino acid metabolism in HS,31 as we observed for the upregulation of Bcat1 and Ggt1 involved in amino acid catabolism, of Ass1 participating to arginine biosynthesis and for the downregulation of Prodh involved in proline degradation.
The activation of the complement cascade also plays an essential role in the adverse immune consequences of HS and resuscitation.32 C5aR, whose ligand is the potent and ubiquitous inflammatory stimulator C5a, is upregulated in HS rats. In a murine model of ischemia-reperfusion, Zhang et al.33 demonstrated that C5aR, an important pathogenic factor in shock and myocardial injury, is upregulated in cardiomyocytes after I/R and C5aR blockade inhibits microvascular permeability and myocardial inflammatory response through the suppression of MAP kinase. In HS rats, we observed the upregulation of Cfd gene, the complement factor D that cleaves and activates factor B, the rate-limiting step of the alternative pathway of complement activation. Similarly, D’Alessandro et al.34 and Mittal et al.35 found the upregulation of Cfd protein level in mesenteric lymph from HS rats. The activation of the complement cascade has also been observed in injured patients with a proteomic approach,36 which showed an increase in the levels of proteins involved in complement activation in the mesenteric lymph of trauma patients.
Reperfusion after HS restores blood flow to organs and tissues but it can also damage the microvasculature and particularly the endothelial cells lining the inner surface of blood vessels, which are extremely vulnerable to I/R. The vascular endothelium is covered by the endothelial glycocalyx (EG), a carbohydrate-rich layer that supports the anticoagulant state of the endothelium and maintains intercellular tight junctions.37 Hyaluronic acid (HA), a glycosaminoglycan, is an important constituent of the EG38 and contributes to maintain its integrity. In the setting of tissue injury, HA is degraded into lower molecular weight fragments that promote inflammation.39 In HS rats, we observed the upregulation of Hyal2 gene encoding a hyaluronidase that performs HA degradation. The cleavage of HA by Hyal2 stimulates monocytes to produce proinflammatory cytokines, chemokines and the recruitment of additional inflammatory cells.40
Transcripts encoding inhibitors of neutrophil protease activity were upregulated in our HS rats. Neutrophils are the first responders to inflammation and have the ability to release large amounts of ROS and other cytotoxic contents. Their endogenous serine proteases, in particular elastase and cathepsin G, when released from activated neutrophils, can destroy host proteins and contribute to morbidity and mortality in inflammatory diseases.41 Olfm4 and Cst7, that are upregulated in HS rat, encode Olfactomedin 4 and Cystatin F, respectively, which are inhibitors of neutrophil proteases. Recently, Baumann et al.42 showed that a neutrophil protease inhibitor (SerpinB1) is critical for neutrophil survival through inhibition of cathepsin G and a similar mechanism might also occur in HS, possibly involving Olfm4 and Cst7 as well. Using a proteomic approach, D’Alessandro et al.34 found a time-dependent increase of several serine-protease inhibitors (serpins) in mesenteric lymph of hemorrhagic rats.
The restoration of blood flow during reperfusion in HS is a potent trigger for the generation of ROS, lipid peroxidation by free radicals and further damage to ischemic tissue.7 A major source of ROS during I/R is NADPH oxidase (NOX), which generates superoxide anion, an oxygen-free radical. ROS rapidly react with lipids, proteins, DNA, and activate apoptotic pathways.43 In our model of HS, we observed that genes involved in ROS inactivation were upregulated. Nqo1 is a cellular superoxide scavenger, highly and rapidly inducible after exposure to oxidative stress.44 Mt2A and Mt1, which belong to the family of metallothioneins expressed in most cells and tissues, are potent antioxidants and are inducible by a variety of stress conditions.45 Mt2A is able to inhibit lipid peroxidation, an important source of injury in ischemia-reperfusion.46 In injured patients, the accumulation of proteins involved in detoxification was observed in post shock mesenteric lymph.36
Ass1 gene, which was up-regulated in HS, encodes argininosuccinate synthase 1, the rate-limiting enzyme in arginine biosynthesis that is required for sustained production of nitric oxide (NO) by activated macrophages.47 NO production may activate stress-induced transcription factors like NF-κB and STAT, involved in promotion of inflammation.48 In line with this observation, in HS rats, we found the downregulation of Tnfaip3 and NF-κBIɛ, which are involved in the suppression of NF-κB inflammatory pathway.49
In HS rats, Gpnmb and Trim72 were upregulated as well. They participate in tissue and cell membrane repair. Gpnmb is a transmembrane phagocytic protein that regulates the degradation of internalized debris and dead cells in macrophages. It has been described as necessary in tissue repair of injured kidney50 and reported as a neuroprotective factor in cerebral I/R injury.51 Trim72, tripartite motif containing 72, plays a central role in the acute membrane repair process by mediating a rapid membrane patching after injury.52 It is crucial in membrane repair response in cardiomyocytes and protects the heart from cardiomyocyte loss, showing a cardioprotective function in I/R injury.53 Interestingly, although Trim72 is reported as a muscle-specific gene, we detected its expression and upregulation in the blood of HS rats, suggesting that Trim72 could also have an active functional role also in circulating cell membrane repair.
This study represents, to the best of our knowledge, the first pilot systematic effort at describing whole blood transcriptome in HS. We used paired end RNAseq, a powerful method for the analysis of differential expression that shows high reproducibility and good agreement with other methods including qPCR.54,55 Our study has the limitation of a small sample size. For this reason, to increase the robustness of our findings, we applied two widely used pipelines for RNAseq data analysis (DeSeq2 and EdgeR) and examined in the downstream analyses only the DEGs identified by both methods.
The experimental protocol we used to induce HS includes the storage of blood, after withdrawal, for 2 h at 22℃ before reinfusion to restore circulating volume and blood pressure. We assessed the potential effects of storage on blood gene expression in a transcriptome analysis on blood from an analogous model of swine HS. This analysis showed that the effect of HS and reperfusion on differential gene expression was significantly more relevant (536 DEGs – HS vs. baseline) than the effect of blood storage (18 DEGs – stored blood vs. baseline). Five transcripts are in common and they account for less than 1% of 536 DEGs in swine shock (unpublished data). Therefore, we believe that the effect of blood storing conditions during the hypovolemic phase in our rat experiment was negligible.
In conclusion, using whole blood as RNA source, we were able to provide a systemic picture of gene expression changes that are induced in whole blood by HS and reperfusion. This experiment highlights specific functions activated in blood cells (inflammation and immunity) and other mechanisms of cell adaptation (ROS scavenging and cell membrane repair). Moreover, we were able to confirm previous observations reported in HS using other -omics approaches: a change in expression of genes encoding for lipid receptors and amino acid metabolism is consistent with previous metabolomics studies, as well as the increased expression of genes with antioxidant function and of the complement pathway which is in line with previous proteomic studies. Therefore, blood transcriptome, as well as other -omics methods, could prove useful to investigate the mechanisms of HS and identify potential markers and therapeutic targets of organ damage in hemorrhagic shock.
Supplementary Material
Acknowledgements
“ShockOmics” grant #602706 of the European Union; “CelSys Shock” Marie Curie International Outgoing Fellowship PIOF-GA-2012-328796 of the European Union; Career Development Award (CDA2) 1IK2BX001277-01A1 from the Department of Veterans Affairs, Veterans Health Administration; Office of Research and Development and NIH grant GM 85072.
Authors’ contributions
GWSS, FA, CB, experimental design; MHS, FADL, animal experiments; DB, FDA, FT RNAseq experiment; MB, DB, SL, data analysis; CB, DB, manuscript preparation; CB, DB, FA, GWSS, EBK, GB, manuscript revision. DB and MB contributed equally to this paper. FA and CB are joint senior authors.
References
- 1.Kauvar DS, Wade CE. The epidemiology and modern management of traumatic hemorrhage: US and international perspectives. Crit Care 2005; 9(Suppl 5): S1–S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kochanek KD, Murphy SL, Xu J. Deaths: final data for 2011. Natl Vital Stat Rep 2015; 63: 1–120. [PubMed] [Google Scholar]
- 3.Mongardon N, Dyson A, Singer M. Is MOF an outcome parameter or a transient, adaptive state in critical illness? Curr Opin Crit Care 2009; 15: 431–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carden DL, Granger DN. Pathophysiology of ischaemia-reperfusion injury. J Pathol 2000; 190: 255–66. [DOI] [PubMed] [Google Scholar]
- 5.Kvietys PR, Granger DN. Endothelial cell monolayers as a tool for studying microvascular pathophysiology. Am J Physiol 1997; 273: G1189–99. [DOI] [PubMed] [Google Scholar]
- 6.Rahbar E, Cardenas JC, Baimukanova G, Usadi B, Bruhn R, Pati S, Ostrowski SR, Johansson PI, Holcomb JB, Wade CE. Endothelial glycocalyx shedding and vascular permeability in severely injured trauma patients. J Transl Med 2015; 13: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Granger DN, Kvietys PR. Reperfusion injury and reactive oxygen species: the evolution of a concept. Redox Biol 2015; 6: 524–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liew CC, Ma J, Tang HC, Zheng R, Dempsey AA. The peripheral blood transcriptome dynamically reflects system wide biology: a potential diagnostic tool. J Lab Clin Med 2006; 147: 126–32. [DOI] [PubMed] [Google Scholar]
- 9.Aletti F, Maffioli E, Negri A, Santamaria MH, DeLano FA, Kistler EB, Schmid-Schönbein GW, Tedeschi G. Peptidomic analysis of rat plasma: proteolysis in hemorrhagic shock. Shock 2015; 45: 540–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liao Y, Smyth GK, Shi W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014; 30: 923–30. [DOI] [PubMed] [Google Scholar]
- 12.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15: 550–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26: 139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009; 25: 1091–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fülöp A, Turóczi Z, Garbaisz D, Harsányi L, Szijártó A. Experimental models of hemorrhagic shock: a review. Eur Surg Res 2013; 50: 57–70. [DOI] [PubMed] [Google Scholar]
- 16.Oppenheim JJ, Yang D. Alarmins: chemotactic activators of immune responses. Curr Opin Immunol 2005; 17: 359–65. [DOI] [PubMed] [Google Scholar]
- 17.Mira JC, Szpila BE, Nacionales DC, Lopez M, Gentile LF, Mathias BJ, Vanzant EL, Ungaro R, Holden D, Rosenthal MD, Rincon J, Verdugo PT, Larson SD, Moore FA, Brakenridge SC, Mohr AM, Baker HV, Moldawer LL, Efron PA. Patterns of gene expression among murine models of hemorrhagic shock/trauma and sepsis. Physiol Genomics 2016; 6: 135–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, Iwasaki A, Flavell RA. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci USA 2004; 101: 5598–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Divanovic S, Trompette A, Atabani SF, Madan R, Golenbock DT, Visintin A, Finberg RW, Tarakhovsky A, Vogel SN, Belkaid Y, Kurt-Jones EA, Karp CL. Negative regulation of Toll-like receptor 4 signaling by the Toll-like receptor homolog RP105. Nat Immunol 2005; 6: 571–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lafleur AM, Lukacs NW, Kunkel SL, Matsukawa A, Arbor A, Studies M. Role of CC chemokine CCL6/C10 as a monocyte chemoattractant in a murine acute peritonitis. Mediators Inflamm 2004; 13: 349–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henderson NC, Sethi T. The regulation of inflammation by galectin-3. Immunol Rev 2009; 230: 160–71. [DOI] [PubMed] [Google Scholar]
- 22.Filipe MD, Meijers WC, Rogier van der Velde A, de Boer RA. Galectin-3 and heart failure: prognosis, prediction & clinical utility. Clin Chim Acta 2015; 443: 48–56. [DOI] [PubMed] [Google Scholar]
- 23.Jakob SM, Pick R, Brechtefeld D, Nussbaum C, Kiefer F, Sperandio M, Walzog B. Hematopoietic progenitor kinase 1 (HPK1) is required for LFA-1-mediated neutrophil recruitment during the acute inflammatory response. Blood 2013; 121: 4184–94. [DOI] [PubMed] [Google Scholar]
- 24.Bradfield PF, Nourshargh S, Aurrand-Lions M, Imhof BA. JAM family and related proteins in leukocyte migration (Vestweber series). Arterioscler Thromb Vasc Biol 2007; 27: 2104–12. [DOI] [PubMed] [Google Scholar]
- 25.Alvarez JI, Kebir H, Cheslow L, Chabarati M, Larochelle C, Iv J, Prat A. JAML mediates monocyte and CD8 T cell migration across the brain endothelium. Ann Clin Transl Neurol 2015; 2: 1032–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Witherden DA, Verdino P, Rieder SE, Garijo O, Robyn E, Teyton L, Fischer WH, Wilson IA, Havran WL. The adhesion molecule JAML is a costimulatory receptor for epithelial gammadelta T cell activation. Science 2010; 329: 1205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eon Kuek L, Leffler M, Mackay GA, Hulett MD. The MS4A family: counting past 1, 2 and 3. Immunol Cell Biol 2016; 94: 11–23. [DOI] [PubMed] [Google Scholar]
- 28.Hotchkiss RS, Monneret G, Payen D. Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nat Rev Immunol 2013; 13: 862–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serhan CN. Pro-resolving lipid mediators are leads for resolution physiology. Nature 2014; 510: 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eun JC, Moore EE, Mauchley DC, Johnson CA, Meng X, Banerjee A, Wohlauer MV, Zarini S, Gijón MA, Murphy RC. The 5-lipoxygenase pathway is required for acute lung injury following hemorrhagic shock. Shock 2012; 37: 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D’Alessandro A, Moore HB, Moore EE, Wither M, Nemkov T, Gonzalez E, Slaughter A, Fragoso M, Hansen KC, Silliman CC, Banerjee A. Early hemorrhage triggers metabolic responses that build up during prolonged shock. Am J Physiol – Regul Integr Comp Physiol 2015; 308: 1034–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szebeni J, Baranyi L, Savay S, Götze O, Alving CR, Bünger R, Mongan PD. Complement activation during hemorrhagic shock and resuscitation in swine. Shock 2003; 20: 347–55. [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, Qin G, Liang G, Li J, Barrington RA, Liu D. C5aR-mediated myocardial ischemia/reperfusion injury. Biochem Biophys Res Commun 2007; 357: 446–52. [DOI] [PubMed] [Google Scholar]
- 34.D’Alessandro A, Dzieciatkowska M, Peltz ED, Moore EE, Jordan JR, Silliman CC, Banerjee A, Hansen KC. Dynamic changes in rat mesenteric lymph proteins following trauma using label-free mass spectrometry. Shock 2014; 42: 509–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mittal A, Middleditch M, Ruggiero K, Loveday B, Delahunt B, Jüllig M, Cooper GJS, Windsor JA, Phillips ARJ. Changes in the mesenteric lymph proteome induced by hemorrhagic shock. Shock 2010; 34: 140–9. [DOI] [PubMed] [Google Scholar]
- 36.Dzieciatkowska M, Alessandro AD, Moore EE, Wohlauer M, Banerjee A, Silliman CC, Hansen KC. Lymph is not a plasma ultrafiltrate: a proteomic analysis of injured patients. Shock 2015; 42: 485–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tarbell JM, Cancel LM. The glycocalyx and its significance in human medicine. J Intern Med 2016; 280: 97–113. [DOI] [PubMed] [Google Scholar]
- 38.Weinbaum S, Tarbell JM, Damiano ER. The structure and function of the endothelial glycocalyx layer. Annu Rev Biomed Eng 2007; 9: 121–67. [DOI] [PubMed] [Google Scholar]
- 39.Scheibner KA, Lutz MA, Boodoo S, Fenton MJ, Powell JD, Horton MR. Hyaluronan fragments act as an endogenous danger signal by engaging TLR2. J Immunol 2006; 177: 1272–81. [DOI] [PubMed] [Google Scholar]
- 40.De la Motte C, Nigro J, Vasanji A, Rho H, Kessler S, Bandyopadhyay S, Danese S, Fiocchi C, Stern R. Platelet-derived hyaluronidase 2 cleaves hyaluronan into fragments that trigger monocyte-mediated production of proinflammatory cytokines. Am J Pathol 2009; 174: 2254–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benarafa C. The serpinB1 knockout mouse: a model for studying neutrophil protease regulation in homeostasis and inflammation. Methods Enzymol 2011; 499: 135–48. [DOI] [PubMed] [Google Scholar]
- 42.Baumann M, Pham CTN, Benarafa C. SerpinB1 is critical for neutrophil survival through cell-autonomous inhibition of cathepsin G. Blood 2013; 121: 3900–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nathan C, Ding A. Snapshot: reactive oxygen intermediates (ROI). Cell 2010; 140: 951.e2–951.e2. [DOI] [PubMed] [Google Scholar]
- 44.Siegel D, Gustafson DL, Dehn DL, Han JY, Boonchoong P, Berliner LJ, Ross D. NAD(P)H:quinone oxidoreductase 1: role as a superoxide scavenger. Mol Pharmacol 2004; 65: 1238–47. [DOI] [PubMed] [Google Scholar]
- 45.Chiaverini N, De Ley M. Protective effect of metallothionein on oxidative stress-induced DNA damage. Free Radic Res 2010; 44: 605–13. [DOI] [PubMed] [Google Scholar]
- 46.Wakida K, Shimazawa M, Hozumi I, Satoh M, Nagase H, Inuzuka T, Hara H. Neuroprotective effect of erythropoietin, and role of metallothionein-1 and -2, in permanent focal cerebral ischemia. Neuroscience 2007; 148: 105–14. [DOI] [PubMed] [Google Scholar]
- 47.Qualls JE, Subramanian C, Rafi W, Smith AM, Balouzian L, Defreitas AA, Shirey KA, Reutterer B, Kernbauer E, Stockinger S, Decker T, Miyairi I, Vogel SN, Salgame P, Rock CO, Murray PJ. Sustained generation of nitric oxide and control of mycobacterial infection requires argininosuccinate synthase 1. Cell Host Microbe 2012; 12: 313–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hierholzer C, Harbrecht B, Menezes JM, Kane J, MacMicking J, Nathan CF, Peitzman AB, Billiar TR, Tweardy DJ. Essential role of induced nitric oxide in the initiation of the inflammatory response after hemorrhagic shock. J Exp Med 1998; 187: 917–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marchant DJ, Boyd JH, Lin DC, Granville DJ, Garmaroudi FS, McManus BM. Inflammation in myocardial diseases. Circ Res 2012; 110: 126–44. [DOI] [PubMed] [Google Scholar]
- 50.Li B, Castano AP, Hudson TE, Nowlin BT, Lin S-L, Bonventre JV., Swanson KD, Duffield JS. The melanoma-associated transmembrane glycoprotein Gpnmb controls trafficking of cellular debris for degradation and is essential for tissue repair. FASEB J 2010; 24: 4767–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakano Y, Suzuki Y, Takagi T, Kitashoji A, Ono Y, Tsuruma K, Yoshimura S, Shimazawa M, Iwama T, Hara H. Glycoprotein nonmetastatic melanoma protein B (GPNMB) as a novel neuroprotective factor in cerebral ischemia-reperfusion injury. Neuroscience 2014; 277: 123–31. [DOI] [PubMed] [Google Scholar]
- 52.Cai C, Masumiya H, Weisleder N, Matsuda N, Nishi M, Hwang M, Ko J-K, Lin P, Thornton A, Zhao X, Pan Z, Komazaki S, Brotto M, Takeshima H, Ma J. MG53 nucleates assembly of cell membrane repair machinery. Nat Cell Biol 2009; 11: 56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X, Xie W, Zhang Y, Lin P, Han L, Han P, Wang Y, Chen Z, Ji G, Zheng M, Weisleder N, Xiao RP, Takeshima H, Ma J, Cheng H. Cardioprotection of Ischemia/Reperfusion injury by cholesterol-dependent MG53-mediated membrane repair. Circ Res 2010; 107: 76–83. [DOI] [PubMed] [Google Scholar]
- 54.Fang Z, Cui X. Design and validation issues in RNA-seq experiments. Brief Bioinform 2011; 12: 280–7. [DOI] [PubMed] [Google Scholar]
- 55.Su Z, Łabaj PP, Li SS, Thierry-Mieg J, Thierry-Mieg D, Shi W, Wang C, Schroth GP, Setterquist RA, Thompson JF, Jones WD, Xiao W, Xu W, Jensen RV, Kelly R, Xu J, Conesa A, Furlanello C, Gao H, Hong H, Jafari N, Letovsky S, Liao Y, Lu F, Oakeley EJ, Peng Z, Praul CA, Santoyo-Lopez J, Scherer A, Shi T, Smyth GK, Staedtler F, Sykacek P, Tan XX, Thompson EA, Vandesompele J, Wang MD, Wang J, Wolfinger RD, Zavadil J, Auerbach SS, Bao W, Binder H, Blomquist T, Brilliant MH, Bushel PR, Cai W, Catalano JG, Chang CW, Chen T, Chen G, Chen R, Chierici M, Chu TM, Clevert DA, Deng Y, Derti A, Devanarayan V, Dong Z, Dopazo J, Du T, Fang H, Fang Y, Fasold M, Fernandez A, Fischer M, Furió-Tari P, Fuscoe JC, Caimet F, Gaj S, Gandara J, Gao H, Ge W, Gondo Y, Gong B, Gong M, Gong Z, Green B, Guo C, Guo L, Guo LW, Hadfield J, Hellemans J, Hochreiter S, Jia M, Jian M, Johnson CD, Kay S, Kleinjans J, Lababidi S, Levy S, Li QZ, Li L, Li L, Li P, Li Y, Li H, Li J, Li S, Lin SM, López FJ, Lu X, Luo H, Ma X, Meehan J, Megherbi DB, Mei N, Mu B, Ning B, Pandey A, Pérez-Florido J, Perkins RG, Peters R, Phan JH, Pirooznia M, Qian F, Qing T, Rainbow L, Rocca-Serra P, Sambourg L, Sansone SA, Schwartz S, Shah R, Shen J, Smith TM, Stegle O, Stralis-Pavese N, Stupka E, Suzuki Y, Szkotnicki LT, Tinning M, Tu B, van Delft J, Vela-Boza A, Venturini E, Walker SJ, Wan L, Wang W, Wang J, Wang J, Wieben ED, Willey JC, Wu PY, Xuan J, Yang Y, Ye Z, Yin Y, Yu Y, Yuan YC, Zhang J, Zhang KK, Zhang W, Zhang W, Zhang Y, Zhao C, Zheng Y, Zhou Y, Zumbo P, Tong W, Kreil DP, Mason CE, Shi L. A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium. Nat Biotechnol 2014; 32: 903–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.