Abstract
The first-line treatment for non-Hodgkin’s lymphoma is chemotherapy. While generally well tolerated, off-target effects and chemotherapy-associated complications are still of concern. To overcome the challenges associated with systemic chemotherapy, we developed a biology-inspired, nanoparticle drug delivery system (nanoDDS) making use of the nucleoprotein components of the tobacco mosaic virus (TMV). Virus-based nanoparticles, including the high-aspect ratio soft nanorods formed by TMV, are growing in popularity as nanoDDS due to their simple genetic and chemical engineerability, size and shape tunability, and biocompatibility. In this study, we used bioconjugation to modify TMV as a multivalent carrier for delivery of the antimitotic drug valine-citrulline monomethyl auristatin E (vcMMAE) targeting non-Hodgkin’s lymphoma. We demonstrate successful synthesis of the TMV-vcMMAE; data indicate that the TMV-vcMMAE particles remained structurally sound with all of the 2130 identical TMV coat proteins modified to carry the therapeutic payload vcMMAE. Cell uptake using Karpas 299 cells was confirmed with TMV particles trafficking to the endolysosomal compartment, likely allowing for protease-mediated cleavage of the valine-citrulline linker for the release of the active monomethyl auristatin E component. Indeed, effective cell killing of non-Hodgkin’s lymphoma in vitro was demonstrated; TMV-vcMMAE was shown to exhibit an IC50 of ∼250 nM. This study contributes to the development of viral nanoDDS.
Impact statement
Due to side effects associated with systemic chemotherapy, there is an urgent need for the development of novel drug delivery systems. We focus on the high-aspect ratio nanotubes formed by tobacco mosaic virus (TMV) to deliver antimitotic drugs targeted to non-Hodgkin’s lymphoma. Many synthetic and biologic nanocarriers are in the development pipeline; the majority of systems are spherical in shape. This may not be optimal, because high-aspect ratio filaments exhibit enhanced tumor homing, increased target cell interactions and decreased immune cell uptake, and therefore have favorable properties for drug delivery compared to their spherical counterparts. Nevertheless, the synthesis of high-aspect ratio materials at the nanoscale remains challenging; therefore, we turned toward the nucleoprotein components of TMV as a biologic nanodrug delivery system. This work presents groundwork for the development of plant virus-based vehicles for use in cancer treatment.
Keywords: Tobacco mosaic virus, nanoparticle, drug delivery, valine-citrulline monomethyl auristatin E, chemotherapy, lymphoma
Introduction
According to the National Cancer Institute’s Surveillance, Epidemiology, and End Results program’s most recent report, over 800,000 Americans are currently living with or in remission from lymphoma.1 Lymphoma is a neoplastic disorder of the lymphatic system and occurs when lymphocytes, or white blood cells, grow and multiply uncontrollably. Lymphoma can be divided into two categories: Hodgkin’s lymphoma and non-Hodgkin’s lymphoma (NHL). NHL is more common, and patients with this disease have a poorer prognosis.2 While the treatment regimen varies with stage of the disease at the time point of diagnosis, the first-line treatment strategy is chemotherapy.3 Therefore, there is a need for the development of delivery systems to increase efficacy and mitigate off-target toxicities.
One avenue of disease-specific drug targeting is found in the application of antibodies; more specifically antibody-drug conjugates (ADCs).4,5 Antibodies can be selected to virtually any molecular target, and the antibody itself can be therapeutic or can carry a therapeutic cargo. One notable example is the monoclonal antibody therapy rituximab. Rituximab targets CD20, a protein expressed on most B-cell NHL and not normally found in circulation, therefore making this therapy highly specific.6 The addition of rituximab to chemotherapy regimens is the standard for patients diagnosed with B-cell NHL based on early clinical successes.7–9 However, response rates can vary widely from 50% to 95% depending on disease subtype,10,11 indicating there is still room for improvement.
The development of nanoparticles is another approach towards targeted therapies. Nanoparticles typically measure between 10 and 500 nm and are thus small enough to efficiently navigate circulation, traffic through tissues, and target and enter cells.12,13 Nanoparticles are much larger than antibodies (typically measuring 5 × 7 nm) and offer multivalency, i.e. while an IgG antibody offers two antigen-binding sites, a nanoparticle has the potential to present hundreds to thousands of copies of targeting ligands. The multivalency provides a mechanism to increase target specificity through added avidity effects. At the same time, antibodies offer limited drug-loading capacity, while nanoparticle drug delivery systems (nanoDDS) can deliver large payloads of small molecule therapies. Furthermore, multifunctional designs are possible, where toxic payloads and/or contrast agents are loaded into nanoparticles decorated with targeting ligands that enable tissue-specific delivery with increased payloads.
In this work, we turned toward the development of the high-aspect-ratio soft-matter nanoparticles using the nucleoprotein components of tobacco mosaic virus (TMV) as a nanoDDS platform technology. TMV is a plant virus offering attractive properties for development and application in nanoscale biomaterials. TMV is a stable, monodisperse, and biocompatible protein-based scaffold that presents an unparalleled opportunity for engineering.14–18 The TMV structure is known to atomic resolution,19 which allows for the identification of specific reactive groups to be targeted for bioconjugation of medical cargo, such as drugs, contrast agents, and/or targeting ligands.20 Specifically, TMV is a 300 nm by 18 nm rigid hollow nanotube with a 4 nm wide interior channel. Each particle consists of 2130 identical copies of a coat protein (CP) unit, with each CP offering a chemically addressable tyrosine 139 on its exterior surface and glutamic acids 97 and 106 on its interior, solvent-exposed surface.21 Furthermore, genetic engineering allows the introduction of new functionality. One example of this is the lysine-added mutant ‘TMV-lys’ that displays reactive lysine groups on the exterior protein shell and is easily modifiable using well known N-hydroxysuccinimide (NHS) chemistry.17 In addition to the chemical and genetic engineering capabilities, precision shape-engineering, e.g. tuning the aspect ratio of TMV, self-assembling star-shapes or other higher order hierarchical assembles, or even switching shapes from rod-to-sphere, are also possible iterations of the TMV platform technology.16–18,22–24
In this work, we bioconjugated TMV with a valine-citrulline monomethyl auristatin E (vcMMAE) pro-drug. Once targeted to the endolyosomal compartment of cancer cells, the dipeptide valine-citrulline (vc) is cleaved to release MMAE, an antimitotic and cytotoxic drug that inhibits polymerization of tubulin and thus interferes with cell replication, halts cancer growth, and induces apoptosis.25 We describe the synthesis and characterization of the TMV-vcMMAE complex and demonstrate cell killing using an in vitro model of NHL.
Materials and methods
Propagation and purification of TMV-lysine
TMV-lys was propagated using Nicotiana benthamiana plants (a tobacco plant species). The virus was isolated via established purification procedures yielding up to 100 mg of TMV-lys per 100 g of infected tobacco plant leaves.15
UV/VIS spectroscopy
A Thermo Scientific NanoDrop 2000 Spectrophotometer was used to measure the concentration of TMV using the TMV-specific extinction coefficient (1.33 M−1cm−1), the known path length (0.1 cm−1), and the Beer-Lambert equation.
Bioconjugation of TMV-lys with vcMMAE
TMV-lys was reacted with 15-fold molar excess of N-succinmidyl-S-acetylthiopropionate (SATP) (Thermo Fisher) with 10% (v/v) dimethyl sulfoxide (DMSO) in 10 mM potassium phosphate buffer (pH 7.0) at room temperature overnight. The resulting TMV-SATP complex was purified via ultracentrifugation at 42,000 rpm over a 40% (w/v) sucrose cushion, reacted with a deacetylation solution (0.5 M hydroxylamine, 25 mM EDTA in PBS, pH 7.4) for 2 h, and then purified with a PD-10 desalting column (GE Healthcare). Next, the TMV-SATP solution was reacted with 10-fold molar excess of vcMMAE (Med Chem Express) overnight and purified via ultracentrifugation. Recovered yields ranged from 70% to 90% after ultracentrifugation, and yields after desalting column ranged from 30% to 50%.
SDS-PAGE
CPs were analyzed using 4–12% NuPAGE gels (Invitrogen) using 1 × MOPS running buffer (Invitrogen). Twenty micrograms of protein with 4× loading LDS dye was denatured through heating at 100℃ for 5 min. After separation, the gel was first placed in destain solution (10% acetic acid, 50% methanol, and 40% H2O) for 30 min. Ten milliliters of the used destain solution was then diluted with 40 mL H2O and 50 µL of Coomassie Blue R250. The gel was placed in this solution for 30 min to stain for protein, then immediately photographed using an AlphaImager (Biosciences) imaging system.
Transmission electron microscopy
Two microliter drops of 0.1 mg/mL TMV-vcMMAE in H2O were placed onto transmission electron microscopy (TEM) grids and allowed to dry. The following TEM grids were used: Formvar/Carbon 400 mesh, Copper approx. grid hole size: 42 µm, TED PELLA prod. no. 01754-F. The grid was then washed in DI water and stained with 2% (w/v) uranyl acetate for 2 min. After drying, samples were examined using a Zeiss Libra 200FE transmission electron microscope operated at 200 kV.
Cell culture
Karpas 299 cells (ATCC) were maintained in RPMI-1640 at 37℃ in a 5% CO2-humidified atmosphere. The medium was supplemented with 20% (v/v) heat-inactivated fetal bovine serum (FBS), and 1% (v/v) penicillin–streptomycin. All reagents were obtained from Gibco (Thermo Fisher).
Cell viability assay
Cells at 10,000 cells/mL were seeded in a sterile, tissue culture-treated, 96-well clear bottom plate for 24 h at 37℃ in a 5% CO2-humidified atmosphere. Cells were then incubated with fresh media with 0.02 nM to 200 nM vcMMAE for 72 h. At 92 h, Alamar Blue (Thermo Fisher) was added, and cell viability was measured at 96 h according to the manufacturer’s instructions using a fluorescence plate reader with excitation at 540 nm and emission at 610 nm.
Confocal microscopy
Karpas 299 cells were seeded at a density of 500,000 cells/well in a 96-well plate in fresh RPMI and incubated with TMV at 1,000,000 particles/cell for 8 h at 37℃ in a 5% CO2-humidified atmosphere. Cells were spun down at 500 g, washed two times with CELL buffer (0.1 mL 0.5 M EDTA, 0.5 mL FBS, 1.25 mL 1 M HEPES pH 7.0, 48.15 mL PBS), fixed using 2% (v/v) paraformaldehyde in CELL buffer, then washed an additional two times in CELL buffer. Cells were washed in PB buffer (0.2% (v/v) Triton X-100 in DPBS) twice, and incubated with rabbit anti-TMV (1:500) (Pacific Immunology) and mouse anti-human LAMP-1 (1:500) (Sigma Aldrich) in CELL buffer for an hour. The cells were then washed two more times with PB buffer and spun down on cover slips at 2000 r/min for 5 min. Then, the cells were incubated with secondary antibodies using Alexa Fluor 488-labeled goat anti-mouse antibody and Alexa Fluor 555 anti-rabbit antibody (Invitrogen) in PB with 5% (v/v) goat serum for 1 h. Lastly, the slips were washed two more times with CELL buffer, stained with DAPI (Sigma Aldrich), and imaged at 40× on an Olympus FluoView™ FV1000 laser scanning confocal microscope.
Results and discussion
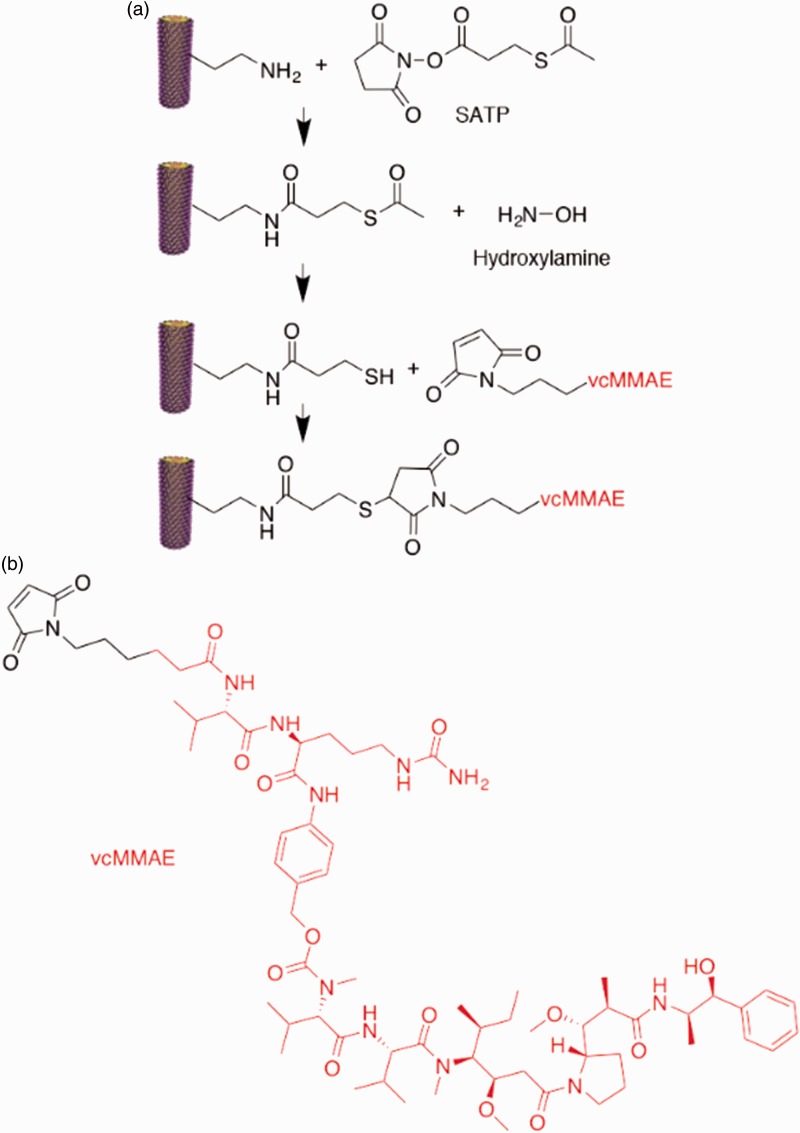
TMV-lys was first propagated in N. benthamiana plants. TMV-lys displays a lysine residue instead of a serine at amino acid position 158; thus, the amine-functional lysine group is solvent-exposed and located at the C-terminus of the CP.17 TMV-lys was extracted in yields of 1 mg of pure virus per gram of infected leaves. To obtain vcMMAE-loaded TMV, the antimitotic drug was bioconjugated using a three-step reaction (Figure 1). First, the amine handle of the TMV-lys was converted into a acetate-protected sulfhydryl group using the bi-functional linker SATP, using a ratio of SATP:TMV CP of 15:1. The reaction was purified via ultracentrifugation over a 40% (w/v) sucrose cushion. The terminating sulfhydryl groups of TMV-SATP were released using hydroxylamine and purified using a PD-10 desalting column, yielding a free thiol handle for conjugation with a maleimide-functionalized vcMMAE through a thiol-Michael reaction using a vcMMAE:TMV CP ratio of 10:1. The reaction mix was again purified by ultracentrifugation over a sucrose cushion, and the final formulation was characterized by UV/VIS spectroscopy, SDS-PAGE, and TEM (Figure 2).
Figure 1.
Chemical structures and bioconjugation reaction. (a) Bioconjugation steps: TMV-lysine was first reacted with SATP via NHS chemistry, TMV-SATP’s thiol group was then deprotected using hydroxylamine, and finally TMV-SH was reacted with the maleimide group of vcMMAE, resulting in TMV-vcMMAE. (b) Chemical structure of vcMMAE. (A color version of this figure is available in the online journal.)
vcMMAE: valine-citrulline monomethyl auristatin E.
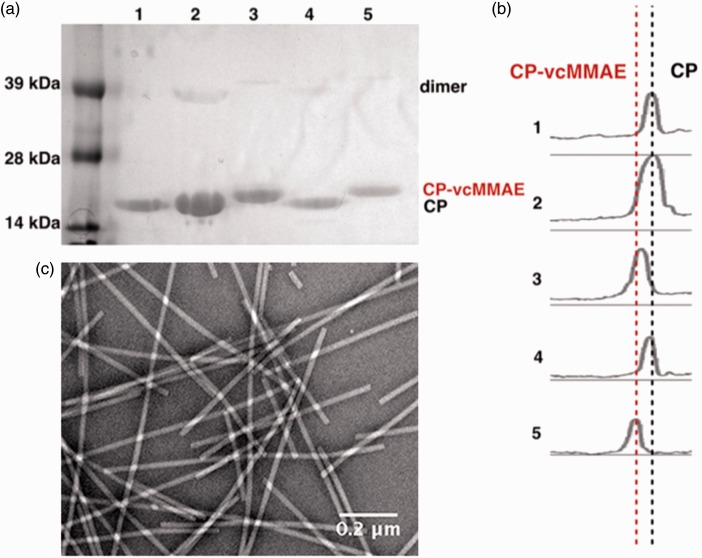
Figure 2.
Biochemical characterization of the vcMMAE-loaded TMV. (a) SDS-PAGE gel of TMV-lys conjugation process. Lane 1: TMV-lys, Lanes 2 and 4: TMV-SATP, and Lanes 3 and 5: TMV-vcMMAE. (b) Densitometry analysis was performed using band analysis tool and ImageJ software showing two distinct bands: CP vs. CP-vcMMAE. (c) TEM micrograph of negatively stained TMV-vcMMAE. (A color version of this figure is available in the online journal.)
CP-vcMMAE: coat protein-valine-citrulline monomethyl auristatin E
UV/VIS spectroscopy was conducted to analyze the amount of protein recovered. Recovered yields ranged from 70% to 90% after ultracentrifugation, and yields after desalting column ranged from 30% to 50% of TMV-vcMMAE.
After each conjugation step, the particles were analyzed using SDS-PAGE (Figure 2(a) and (b)). The TMV CP measures 17.5 kDa and was detectable on the SDS-PAGE (Lane 1, Figure 2(a)). A small percentage of CP dimers was also detectable near the 39-kDa molecular weight standard. The dimers result from inter-CP crosslinking and are often observed, especially when analyzing modified TMV CPs. Shifts in the band pattern toward higher molecular weight bands indicated successful conjugation of vcMMAE (Lanes 3 + 5, Figure 2(a) and (b)). The drug, vcMMAE, has a molecular weight of 1316.64 g/mol, and a shift towards the higher molecular weight band indicated that conjugation was successful; the CP-vcMMAE band was detectable at 20 kDa. Densitometry analysis conducted using ImageJ software indicated ∼100% band separation (Figure 2(b)). In other words, the uniform shift of the band indicated all of the 2130 CPs of each TMV particle, and hence all 2130 available lysine sites per TMV were labeled with the therapeutic cargo.
Finally, TEM revealed that the TMV-vcMMAE formulations remained intact; imaging of negatively stained TEM grids showed structurally sound TMV nanoparticles (Figure 2(c)).
To conclude the synthesis of the vcMMAE-conjugated TMV, we developed a protocol allowing efficient drug loading to TMV. The chemical reaction reached completion with each of the 2130 CPs being modified with a drug molecule. On a %-molecular weight basis, this translates to a drug-loading efficiency of ∼7 wt% per TMV. This is comparable to immunotherapies which have drug loading of up to ∼7 wt% per IgG and up to ∼10 wt% per diabody in antibody conjugated MMAE studies.26,27 Nevertheless, ADCs are limited to only carrying up to eight drug molecules per antibody, which is far fewer than the load which TMV can carry, namely ∼2000 copies of the payload.
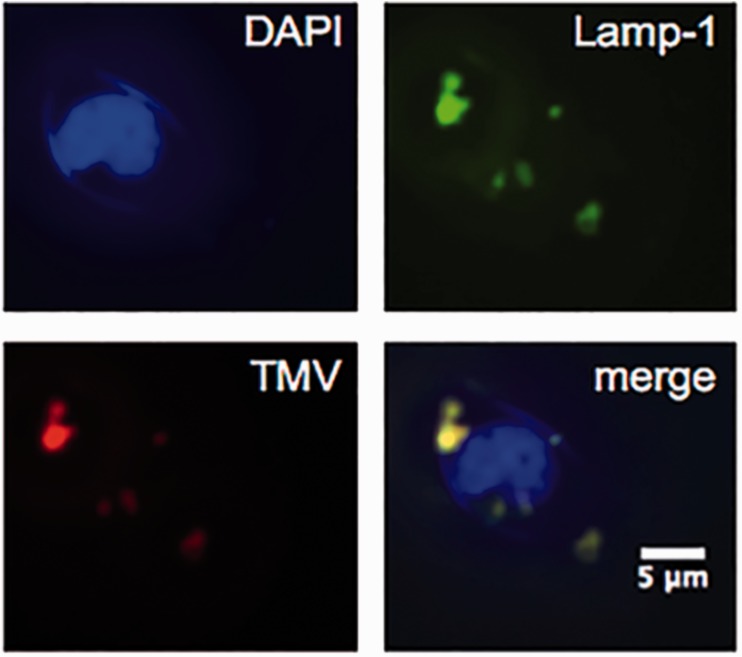
Next, we set out to study the in vitro properties of the drug candidate using Karpas 299 cells, a human-derived large cell NHL. First, we studied the fate of the TMV formulation in these cells: fluorescence imaging indicates that TMV is indeed taken up by these cells and that the nanoparticles target the endolysosomal compartment (Figure 3). These results are as expected, as we and others have previously shown that TMV is internalized by cancer cells through a combination of endocytosis and macropinocytosis and that the particles then co-localize with endolysosomal markers.28–30 We hypothesize that within the protease-rich environment of the endolyosome, the pro-drug TMV-vcMMAE is cleaved at the protease-cleavable vc linker, resulting in release of the active MMAE component. In fact, in our previous studies, we have shown that TMV-conjugated cargos are cleaved and released in endolysosomal extracts even when conjugated via amide bonds.30 Therefore, we hypothesize that the therapeutic cargo MMAE is released from the TMV carrier upon cell uptake and trafficking to the endolysosomal compartment, in which the protein carrier is likely degraded and cleared over time.
Figure 3.
Karpas 299 cell interactions with TMV. Nuclei were stained with DAPI (blue), endolysosomes were stained with mouse anti-human Lamp-1 antibody followed by secondary Alexa Fluor 488 goat anti-mouse antibody (green), and TMV particles were stained with primary rabbit anti-TMV antibody followed by secondary Alexa Fluor 555 goat anti-rabbit antibody (red). Merging of all three channels indicated co-localization of TMV with the endolysosomes (yellow). (A color version of this figure is available in the online journal.)
DAPI: 4',6-diamidino-2-phenylindole; TMV: tobacco mosaic virus
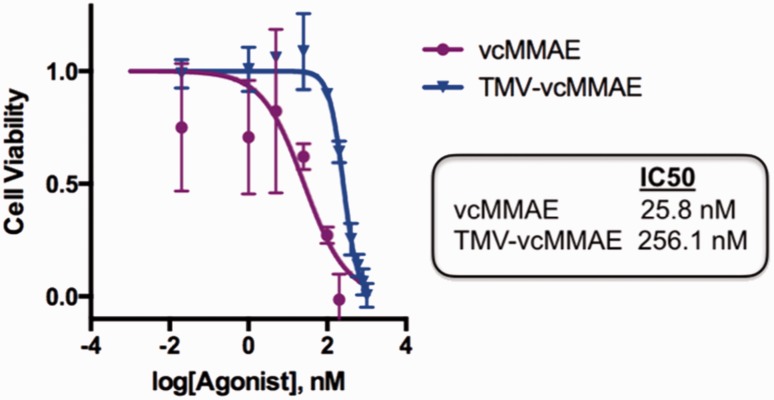
Finally, to evaluate the efficacy of TMV-vcMMAE, cell viability assays were performed using the Alamar Blue assay. We found that vcMMAE delivered by TMV remained efficacious and resulted in cell killing, although at a lower efficacy compared with free vcMMAE. The IC50 for free vcMMAE was determined to be 25.8 nM, while the IC50 for TMV-vcMMAE was found to be 256.1 nM (Figure 4). TMV without the drug conjugate was non-toxic to cells (not shown). IC50 values of free vcMMAE drug are in good agreement with data reported in the literature and lie within the low nanomolar range, and it has been shown that efficacy can be somewhat reduced for ADCs as well, with ranges from 2 nM to 42 nM reported.31–33 The lower IC50 values of the antibody and also nanoparticle conjugates may be explained by lower cell uptake kinetics, as has been reported with other systems.34,35 This could potentially be advantageous for reducing off-target effects from non-specific uptake as future developments of these nanoparticles look into incorporating active targeting ligands.
Figure 4.
Cell viability assay. Alamar Blue cell viability assay of Karpas 299 cells after 72 hours of treatment with vcMMAE (purple) and TMV-vcMMAE (blue) at 0.02 nM to 200 nM vcMMAE; non-conjugated TMV was also tested using the protein concentration corresponding to the highest vcMMAE dose (200 nM), toxicity of TMV was not observed. Error bars denote the standard deviation (experiments were done in triplicate). Data were analyzed and graphed using Prism® v6.0 b software. (A color version of this figure is available in the online journal.)
vcMMAE: valine-citrulline monomethyl auristatin E; TMV: tobacco mosaic virus
Conclusions
We demonstrated the development of TMV as a nanoDDS targeting lymphoma. Bioconjugate chemistry allowed effective drug conjugation yielding vcMMAE-TMV pro-drug. Imaging and cell viability assays using an in vitro model of human B-cell NHL validated the approach by demonstrating cell uptake, endolysosomal processing of the drug, followed by cell killing – the IC50 was determined to be ∼250 nM. This work expands the library of plant virus-based nanoDDS approaches. In recent years, a number of drug-loaded plant virus-based formulations have been developed; for example, we demonstrated efficacy of platinum drug candidates28 as well as photodynamic agents36 delivered by TMV. Similarly, we and others have also shown efficacy of doxorubicin-conjugated TMV rods, disks, and spherical nanoparticles.22,37 Beyond the loading of low-molecular weight chemotherapies and photodynamic therapies, the study demonstrates the functionalization of TMV with a peptide therapeutic, thus opening a new avenue for further exploration.
Acknowledgements
Anna E. Czapar (CWRU) is thanked for helpful discussions. This work was funded in part by NIH grant R03-EB020602 (to NFS).
Authors’ contributions
DLK performed all experiments. AMW designed the bioconjugate chemistry experiments and assisted with the characterization of the vcMMAE-TMV. ASP performed the TEM imaging. NFS conceived and oversaw the entire study. DLK and NFS wrote the manuscript. All authors read and approved the manuscript.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Howlader N, Noone AM, Krapcho M, Miller D, Bishop K, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA (eds). In: SEER Cancer Statistics Review, 1975–2013, National Cancer Institute, Bethesda, MD. Available at: https://seer.cancer.gov/csr/1975_2013/ (2016, accessed 29 June 2017).
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. Cancer J Clin 2017; 67: 7–30. [DOI] [PubMed] [Google Scholar]
- 3.Armitage JO, Gascoyne RD, Lunning MA, Cavalli F. Non-Hodgkin lymphoma. The Lancet Epub ahead of print 30 January 2017. DOI: 10.1016/S0140-6736(16)32407-2. [DOI] [PubMed] [Google Scholar]
- 4.Bouchard H, Viskov C, Garcia-Echeverria C. Antibody-drug conjugates – a new wave of cancer drugs. Bioorg Med Chem Lett 2014; 24: 5357–63. [DOI] [PubMed] [Google Scholar]
- 5.Chen J, Jaracz S, Zhao X, Chen S, Ojima I. Antibody–cytotoxic agent conjugates for cancer therapy. Expert Opin Drug Deliv 2005; 2: 873–90. [DOI] [PubMed] [Google Scholar]
- 6.Prevodnik VK, Lavrenčak J, Horvat M, Novakovič BJ. The predictive significance of CD20 expression in B-cell lymphomas. Diagn Pathol 2011; 6: 33–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coiffier B, Thieblemont C, Van Den Neste E, Lepeu G, Plantier I, Castaigne S, Lefort S, Marit G, Macro M, Sebban C, Belhadj K, Bordessoule D, Fermé C, Tilly H. Long-term outcome of patients in the LNH-98.5 trial, the first randomized study comparing rituximab-CHOP to standard CHOP chemotherapy in DLBCL patients: a study by the Groupe d'Etudes des Lymphomes de l'Adulte. Blood 2010; 116:2040–5. [DOI] [PMC free article] [PubMed]
- 8.Maloney DG. Anti-CD20 antibody therapy for B-cell lymphomas. N Engl J Med 2012; 366: 2008–16. [DOI] [PubMed] [Google Scholar]
- 9.Pfreundschuh M, Schubert J, Ziepert M, Schmits R, Mohren M, Lengfelder E, Reiser M, Nickenig C, Clemens M, Peter N, Bokemeyer C, Eimermacher H, Ho A, Hoffmann M, Mertelsmann R, Trümper L, Balleisen L, Liersch R, Metzner B, Hartmann F, Glass B, Poeschel V, Schmitz N, Ruebe C, Feller AC, Loeffler M; German High-Grade Non-Hodgkin Lymphoma Study Group (DSHNHL). Six versus eight cycles of bi-weekly CHOP-14 with or without rituximab in elderly patients with aggressive CD20+ B-cell lymphomas: a randomised controlled trial (RICOVER-60). Lancet Oncol 2008;9:105–16. [DOI] [PubMed]
- 10.Hiddemann W, Kneba M, Dreyling M, Schmitz N, Lengfelder E, Schmits R, Reiser M, Metzner B, Harder H, Hegewisch-Becker S, Fischer T, Kropff M, Reis HE, Freund M, Wörmann B, Fuchs R, Planker M, Schimke J, Eimermacher H, Trümper L, Aldaoud A, Parwaresch R, Unterhalt M. Frontline therapy with rituximab added to the combination of cyclophosphamide, doxorubicin, vincristine, and prednisone (CHOP) significantly improves the outcome for patients with advanced-stage follicular lymphoma compared with therapy with CHOP alone: results of a prospective randomized study of the German Low-Grade Lymphoma Study Group. Blood 2005;106:3725–32. [DOI] [PubMed]
- 11.McLaughlin P, Grillo-López AJ, Link BK, Levy R, Czuczman MS, Williams ME, Heyman MR, Bence-Bruckler I, White CA, Cabanillas F, others. Rituximab chimeric anti-CD20 monoclonal antibody therapy for relapsed indolent lymphoma: half of patients respond to a four-dose treatment program. J Clin Oncol 1998;16:2825–33. [DOI] [PubMed]
- 12.Albanese A, Tang PS, Chan WCW. The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annu Rev Biomed Eng 2012; 14: 1–16. [DOI] [PubMed] [Google Scholar]
- 13.Torchilin V. Tumor delivery of macromolecular drugs based on the EPR effect. Adv Drug Deliv Rev 2011; 63: 131–5. [DOI] [PubMed] [Google Scholar]
- 14.Bruckman MA, Kaur G, Lee LA, Xie F, Sepulveda J, Breitenkamp R, Zhang X, Joralemon M, Russell TP, Emrick T, Wang Q. Surface modification of tobacco mosaic virus with “click” chemistry. Chem Bio Chem 2008;9:519–23. [DOI] [PubMed]
- 15.Bruckman MA, Steinmetz NF. Chemical modification of the inner and outer surfaces of tobacco mosaic virus (TMV). In: Lin B, Ratna B (eds) Virus hybrids as nanomaterials: methods and protocols, Vol. 1108. New York, NY, U.S.A.: Humana Press, Methods in Molecular Biology book series, 2014, pp.173–185. [DOI] [PMC free article] [PubMed]
- 16.Eber FJ, Eiben S, Jeske H, Wege C. Bottom-up-assembled nanostar colloids of gold cores and tubes derived from tobacco mosaic virus. Angewandte Chem Int Ed 2013; 52: 7203–07. [DOI] [PubMed] [Google Scholar]
- 17.Geiger FC, Eber FJ, Eiben S, Mueller A, Jeske H, Spatz JP, Wege C. TMV nanorods with programmed longitudinal domains of differently addressable coat proteins. Nanoscale 2013; 5: 3808–16. [DOI] [PubMed] [Google Scholar]
- 18.Shukla S, Eber FJ, Nagarajan AS, DiFranco NA, Schmidt N, Wen AM, Eiben S, Twyman RM, Wege C, Steinmetz NF. The impact of aspect ratio on the biodistribution and tumor homing of rigid soft-matter nanorods. Adv Healthc Mater 2015; 4: 874–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Namba K, Stubbs G. Structure of tobacco mosaic virus at 3.6 A resolution: implications for assembly. Science 1986; 231: 1401–7. [DOI] [PubMed] [Google Scholar]
- 20.Pokorski JK, Steinmetz NF. The art of engineering viral nanoparticles. Mol Pharma 2010; 8: 29–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlick TL, Ding Z, Kovacs EW, Francis MB. Dual-surface modification of the tobacco mosaic virus. J Am Chem Soc 2005; 127: 3718–23. [DOI] [PubMed] [Google Scholar]
- 22.Bruckman MA, Czapar AE, VanMeter A, Randolph LN, Steinmetz NF. Tobacco mosaic virus-based protein nanoparticles and nanorods for chemotherapy delivery targeting breast cancer. J Controlled Release 2016; 231: 103–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bruckman MA, Hern S, Jiang K, Flask CA, Yu X, Steinmetz NF. Tobacco mosaic virus rods and spheres as supramolecular high-relaxivity MRI contrast agents. J Mater Chem B 2013; 1: 1482–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lam P, Gulati NM, Stewart PL, Keri RA, Steinmetz NF. Bioengineering of tobacco mosaic virus to create a non-infectious positive control for Ebola diagnostic assays. Sci Rep 2016; 6: 23803–23803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doronina SO, Toki BE, Torgov MY, Mendelsohn BA, Cerveny CG, Chace DF, DeBlanc RL, Gearing RP, Bovee TD, Siegall CB, Francisco JA, Wahl AF, Meyer DL, Senter PD. Development of potent monoclonal antibody auristatin conjugates for cancer therapy. Nat Biotechnol 2003;21:778–84. [DOI] [PubMed]
- 26.Sochaj AM, Swiderska KW, Otlewski J. Current methods for the synthesis of homogeneous antibody-drug conjugates. Biotechnol Adv 2015; 33(6 Pt 1): 775–84. [DOI] [PubMed] [Google Scholar]
- 27.Kim KM, McDonagh CF, Westendorf L, Brown LL, Sussman D, Feist T, Lyon R, Alley SC, Okeley NM, Zhang X, Thompson MC, Stone I, Gerber HP, Carter PJ. Anti-CD30 diabody-drug conjugates with potent antitumor activity. Mol Cancer Ther 2008; 7: 2486–97. [DOI] [PubMed] [Google Scholar]
- 28.Czapar AE, Zheng Y-R, Riddell IA, Shukla S, Awuah SG, Lippard SJ, Steinmetz NF. Tobacco mosaic virus delivery of phenanthriplatin for cancer therapy. ACS Nano 2016; 10: 4119–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X, Wu F, Tian Y, Wu M, Zhou Q, Jiang S, Niu Z. Size dependent cellular uptake of rod-like bionanoparticles with different aspect ratios. Sci Rep 2016; 6: 24567–24567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wen AM, Infusino M, De Luca A, Kernan DL, Czapar AE, Strangi G, Steinmetz NF. Interface of physics and biology: engineering virus-based nanoparticles for biophotonics. Bioconjugate Chem 2015; 26: 51–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Francisco JA, Cerveny CG, Meyer DL, Mixan BJ, Klussman K, Chace DF, Rejniak SX, Gordon KA, DeBlanc R, Toki BE, Law CL, Doronina SO, Siegall CB, Senter PD, Wahl AF. cAC10-vcMMAE, an anti-CD30-monomethyl auristatin E conjugate with potent and selective antitumor activity. Blood 2003; 102: 1458–65. [DOI] [PubMed] [Google Scholar]
- 32.Li F, Emmerton KK, Jonas M, Zhang X, Miyamoto JB, Setter JR, Nicholas ND, Okeley NM, Lyon RP, Benjamin DR, Law CL. Intracellular released payload influences potency and bystander-killing effects of antibody-drug conjugates in preclinical models. Cancer Res 2016; 76: 2710–9. [DOI] [PubMed] [Google Scholar]
- 33.Dornan D, Bennett F, Chen Y, Dennis M, Eaton D, Elkins K, French D, Go MA, Jack A, Junutula JR, Koeppen H, Lau J, McBride J, Rawstron A, Shi X, Yu N, Yu SF, Yue P, Zheng B, Ebens A, Polson AG. Therapeutic potential of an anti-CD79b antibody-drug conjugate, anti-CD79b-vc-MMAE, for the treatment of non-Hodgkin lymphoma. Blood 2009; 114: 2721–9. [DOI] [PubMed] [Google Scholar]
- 34.Danhier F, Lecouturier N, Vroman BI, Jérôme C, Marchand-Brynaert J, Feron O, Préat V. Paclitaxel-loaded PEGylated PLGA-based nanoparticles: in vitro and in vivo evaluation. J Controlled Release 2009; 133: 11–17. [DOI] [PubMed] [Google Scholar]
- 35.Panyam J, Labhasetwar V. Biodegradable nanoparticles for drug and gene delivery to cells and tissue. Adv Drug Deliv Rev 2003; 55: 329–47. [DOI] [PubMed] [Google Scholar]
- 36.Lee KL, Carpenter BL, Wen AM, Ghiladi RA, Steinmetz NF. High aspect ratio nanotubes formed by tobacco mosaic virus for delivery of photodynamic agents targeting melanoma. ACS Biomater Sci Eng 2016; 2: 838–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finbloom JA, Han K, Aanei IL, Hartman EC, Finley DT, Dedeo MT, Fishman M, Downing KH, Francis MB. Stable disk assemblies of a tobacco mosaic virus mutant as nanoscale scaffolds for applications in drug delivery. Bioconjugate Chem 2016; 27: 2480–5. [DOI] [PubMed] [Google Scholar]