Abstract
Increased expression of sirtuins lowers the risk of age-related diseases, while their role in the regulation of longevity is not firmly established. Since aging is associated with immunosenescence, we tested whether sirtuin expression was modified in peripheral blood mononuclear cells (PBMC) in an age-related manner and whether this might result from altered expression of the selected miRNAs. The expression of seven SIRT genes and of SIRT1 mRNA-interacting miR-9, miR-34a, miR-132, and miR-199a-5p was evaluated by real-time PCR in PBMC originating from young (Y, n = 57, mean age 27 ± 4.3 years), elderly (E, n = 52, 65 ± 3.4 years), and long-lived (L, n = 56, 94 ± 3.5 years) individuals. Older age was associated with a decreased expression of the majority of the SIRT genes. Most severely affected were median expressions of SIRT1 (P = 0.000001 for the whole studied group, Y vs. E: P < 0.000001, Y vs. L: P < 0.000001), and of SIRT3 (P = 0.000001, Y vs. E: P = 0.000004, Y vs. L: P = 0.000028). Older age was also associated with the increased median expression of miR-34a (P = 0.000001, Y vs. E: P = 0.001, Y vs. L: P = 0.000004) and of miR-9 (P = 0.05, Y vs. L: P = 0.054). In functional studies, miR-9 interacted with the 3′UTR of SIRT1 mRNA. The SIRT1 mRNA level negatively correlated with the expression of miR-34a (r = −0.234, P = 0.003). In conclusion, age-related decrease of SIRT1 expression in PBMC might in part result from overexpression of miR-34a and miR-9. In addition, the sustained expression of the SIRT genes in PBMC is not a prerequisite to longevity in humans but might be one of the reasons for the immune system dysfunction in the elderly.
Impact statement
High expression of sirtuins, particularly SIRT1, lowers the risk of age-related diseases and probably slows down the rate of aging; therefore, their sustained expression should be one of the features of longevity. However, in this work we show that in peripheral blood mononuclear cells (PBMC) of long-lived individuals, expression of majority of the SIRT genes is significantly lower than in cells of young study subjects. In long-lived individuals, downregulation of SIRT1 coexists with upregulation of SIRT1 mRNA-interacting miR-34a and miR-9, indicating the role of epigenetic drift in age-dependent deregulation of SIRT1 expression. Such constellation of SIRT1, miR-34a, and miR-9 expression in PBMC of successfully aging long-lived individuals indicates that, at least in these individuals, it is not a risk factor for morbidity and mortality. It might however affect the function of the immune system and, therefore, aging individuals can profit from interventions increasing the level of SIRT1.
Keywords: SIRT, peripheral blood mononuclear cells, epigenetic drift, aging, longevity
Introduction
Aging and its clinical course depend on multiple factors.1–3 A hundred or so genes have so far been implicated in the regulation of human aging, including SIRT genes (SIRT1–SIRT7) encoding NAD+-dependent deacetylases that control metabolism, production of the reactive oxygen species (ROS), response to oxidative stress and to DNA damage, and inflammatory responses.4–6 Enhanced function and/or overexpression of sirtuin orthologs have been reported to extend the lifespan of Caenorhabditis elegans, yeast, and Drosophila melanogaster; this view, however, is not unanimous.7–10 The lifespan of male mice overexpressing SIRT6 is increased, but the lifespan of SIRT1 transgenic mice is not increased.11,12 In humans, only select SIRT1 and SIRT3 polymorphisms are associated with longevity.13,14 Thus, it is still not firmly established whether or not sirtuins extend the length of life. In contrast, it is generally accepted that a higher activity of sirtuins is associated with protection against the majority of age-related diseases such as type 2 diabetes, cardiovascular disease, and Alzheimer’s disease.15–18 Notably, a calorie restriction diet (CR), the only intervention known so far to extend healthspan and lifespan, leads to SIRT1 and SIRT3 overexpression.19,20 It has been also shown that SIRT1 transgenic mice present features similar to those of animals fed a CR: they are lean, resistant to cancer, as well as to the diet-induced metabolic syndrome.12,21 In SIRT3 transgenic mice, antioxidant mechanisms are enhanced.22 Conversely, CR leads to SIRT1 and SIRT3 overexpression.19,23 In primates, implementation of CR decreased morbidity and mortality,24,25 but this was not replicated by another study.26 Such inconsistency could result from a different composition and source of diet and a slight calorie restriction in the control group of the latter study. In humans, the Comprehensive Assessment of Long-term Effects of Reducing Intake of Energy (CALERIE) trials showed that mild CR reduces weight and improves cardiometabolic risk factors, while self-imposed severe CR with optimal nutrition (CRON) lasting for up to 15 years resulted metabolic, hormonal, and molecular effects previously detected in long-lived CR animals and reduced metabolic and cardiovascular risk factors. However, effects of CR on survival in humans are still not known (reviewed in Most et al.27).
Gene expression is controlled by numerous mechanisms, including epigenetic modifications such as DNA methylation, covalent histone modifications, and the action of miRNAs. Notably, one of the characteristics of aging is epigenetic drift, a subtle but progressive change of the epigenome.28–30
With this in mind, we decided to determine whether successful aging is associated with changes in the expression of sirtuin genes in peripheral blood mononuclear cells (PBMC). We show that even in individuals whose aging is not complicated by age-related diseases, older age is associated with a decreased expression of the majority of the SIRT genes in these cells. We also wished to elucidate whether an altered expression of certain miRNAs might be responsible for the decreased expression of SIRT1 and we found that age-related overexpression of miR-34a and miR-9 might be involved in its downregulation.
Materials and methods
Study subjects
Polish Caucasians were divided into young (Y, n = 57, age range 19–42 years, mean age 27 ± 4.3 years), elderly (E, n = 52, 60–73 years, 65 ± 3.4 years), and long-lived (L, n = 56, 90–102 years, 94 ± 3.5 years). Study subjects were non-obese (body mass index [BMI] < 30 kg/m2) and healthy/relatively healthy: moderate hypertension was allowed for older age groups, and a mild degree physical (the Activities of Daily Living score ≥ 3) and cognitive disability (the Mini Mental State Evaluation score ≥ 20) were also allowed for the long-lived group.31,32 The study protocol was approved by the Bioethics Committee of the Medical University of Warsaw. All participants gave a written informed consent for participation in the study. The anonymity of patients has been preserved at all stages of this investigation.
Isolation of RNA from PBMC
PBMC and RNA were isolated as previously described.33
Reverse transcription, real-time quantification of gene and miRNA expression
Reverse transcription, analysis of SIRT1–SIRT7 mRNA expression, and miRNA expression were performed as previously described, with one modification regarding different annealing temperatures (65℃ for SIRT1, 2, 3, 6; 67℃ for SIRT4, and 60℃ for SIRT5 and 7).34 The list of primers can be found in Table 1.
Table 1.
Primers used for analysis of the expression of SIRT1-SIRT7 in human PBMC
| Primers | Exons | |
|---|---|---|
| SIRT1 | F: 5′ACAGGTTGCGGGAATCCAAAGG3′ R: 5′CCTAGGACATCGAGGAACTACCTG3′ | E5 E7 |
| SIRT2 a | F: 5′CCTCGCCTGCTCATCAACA3′ R: 5′TCCTCCGAGGCCCATAATC3′ | E13 E14 |
| SIRT3 | F: 5′GCTGACGTGATGGCAGACA3′ R: 5′AACCACATGCAGCAAGAACCT3′ | E5 E5 |
| SIRT4 | F: 5′ACAGGGTCCTGTGCTTGGATTG3′ R: 5′TTCAGGACTTGGAAACGCTCTTGC3′ | E2/3 E3 |
| SIRT5 | F: 5′AAGGCTGGCACCAAGAAC3′ R: 5′GCCACAACTCCACAAGAG3′ | E5 E6 |
| SIRT6 | F: 5′TGGTCTCCAGCTTAAACAG3′ R: 5′AAGGCAGTGCAAGCCTCT3′ | E8 E8 |
| SIRT7 a | F: 5′CGTCCGGAACGCCAAATAC3′ R: 5′GACGCTGCCGTGCTGATT3′ | E3 E3/4 |
| ACTB | F: 5′TTCTACAATGAGCTGCGTGTG3′ R: 5′CAGCCTGGATAGCAACGTACA3′ | E3 E4 |
F: forward; PBMC: peripheral blood mononuclear cells; R: reverse.
Primers as in Ashraf et al.35
Functional analysis of miRNA
Candidate miRNAs were searched for with the TargetScanHuman, the miRanda-mirSVR, and the Pictar programs and verified using the rules indicated by Bartel.36–39 We selected miR-9, miR-34a, miR-132, and miR-199a-5p for analysis regarding SIRT1.
DNA corresponding to the full-length 3′UTR of SIRT1 mRNA was amplified from 50 ng of PBMC-originating cDNA with Dream Taq polymerase (Thermo Scientific, Vilnius, Lithuania) using the forward 5′ACTAGAGCTCTAGTGTAATAATTGTGCAGG3′ (added SacI restriction site shown in bold; the STOP codon is underlined) and reverse 5′CTAACTCGAGAACAGAAAAAAGTCAAATGAC3′ (additional XhoI restriction site shown in bold) primers. The PCR reaction was: 3 min at 94℃, 40 cycles of: 30 s at 94℃, 30 s at 62℃, and 2 min 30 s at 72℃, and final extension for 10 min at 72℃. The PCR product was cloned into the pmirGLO reporter vector (Promega, Madison, WI) and sequenced (pmirGLO_SIRT1).
Human embryonic kidney (HEK) 293 cells were cultured and transfected as previously described.34 Forty nanograms of the reporter plasmid and 5 pmol or 10 pmol (30 nmol/L or 60 nmol/L final concentration, respectively) of pre-miRNA (pre-miR-9 or pre-miR miRNA Precursor Negative Control #1; Ambion, Life Technologies, Carlsbad, CA) were used. Cells were then cultured, lysed, and the luminescence was assessed as previously described.34 Each experiment was repeated nine times.
Immunoblot
HEK 293 cells were cultured and transfected with pre-miR-9 or with a negative control miRNA precursor (each at 30 nmol/L or 60 nmol/L final concentration) in a six-well dish. Cells were harvested 24 h after transfection. Total protein was isolated with radioimmunoprecipitation assay (RIPA) buffer and an aliquot of 20 µg was resolved using 10% polyacrylamide gel and transferred onto a nitrocellulose membrane. The membrane was incubated with rabbit anti-SIRT1 primary antibody (1:250, sc-15404, Santa Cruz Biotechnology, TX) and with goat anti-rabbit secondary antibody (1:10,000, 401393-2ML, Merck Milipore, Germany). The control reaction was performed with rabbit anti-GAPDH primary antibody (1:500, sc-25778, Santa Cruz Biotechnology). Proteins were visualized using the SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, IL) and the GeneGnome XRQ bio imaging system (Syngene, UK). Densitometric measurements of three independent immunoblots were performed with the Image Studio Lite software (LI-COR Biosciences, NE).
Statistical analysis
Statistical calculations were performed using Statistica v. 10. To assess normality of the distribution, the Shapiro-Wilk test was used. For SIRT1 expression, statistical analysis was performed with the analysis of variance (ANOVA) and post hoc Fisher’s tests. Since distribution of expressions of the remaining SIRTs and of miRNAs was not normal, statistical analyses were performed with the Kruskal–Wallis test. Correlation between the SIRT1 mRNA and miRNA expressions was calculated by the Spearman’s rank correlation coefficient. The effect of miRNA interaction with the SIRT1 mRNA on the reporter protein activity was analyzed by the two-sided Student’s t-test. For all tests the level of significance was established at 0.05.
Results
Expression of the SIRT1-SIRT7 mRNA in PBMC of young, elderly, and long-lived individuals
We have previously established that age did not affect the mean Cp values for the ACTB control gene in PBMC.36 We also determined that the expression of SIRTs was similar in PBMC of women and men and, therefore, further analyses were performed for all study subjects together.
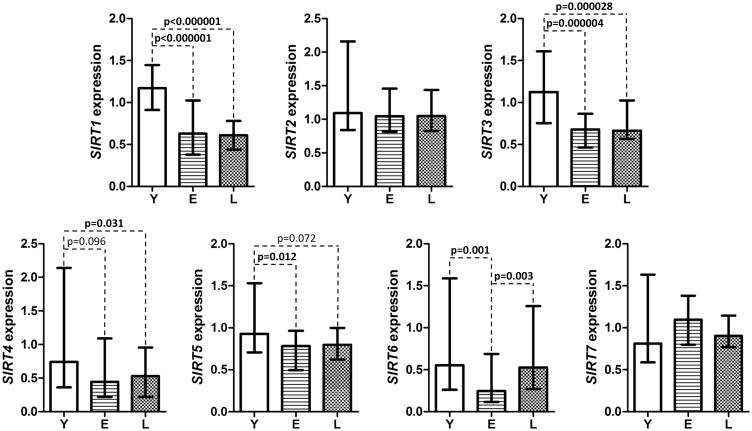
The median expressions of the SIRT genes, presented in arbitrary units (Figure 1) showed that the expressions of SIRT1 and SIRT3 were most severely affected by age (P = 0.000001 and P = 0.000001, respectively) and were reduced approximately two-fold in the E and L groups compared to young controls. The median expression of SIRT4 was significantly lower in the L group than in the Y group, while the median expression of SIRT5 was significantly lower in the E than in the Y group. The expression of SIRT6 was significantly lower in the E group than in the Y and L groups. Finally, age did not affect the expressions of SIRT2 and SIRT7.
Figure 1.
The median expressions of the SIRT1-7 genes in PBMC of individuals of various ages. Results are presented in arbitrary units (specific for each gene) as median (25th, 75th percentile) values. For SIRT1 expression, statistical analysis was performed with the ANOVA and post hoc Fisher’s tests, while for other genes the Kruskal–Wallis test was used. The level of significance was established at 0.05. Y: young; E: elderly; L: long-lived study participants; PBMC: peripheral blood mononuclear cells
Interaction of miR-9 with the 3′UTR of SIRT1 mRNA
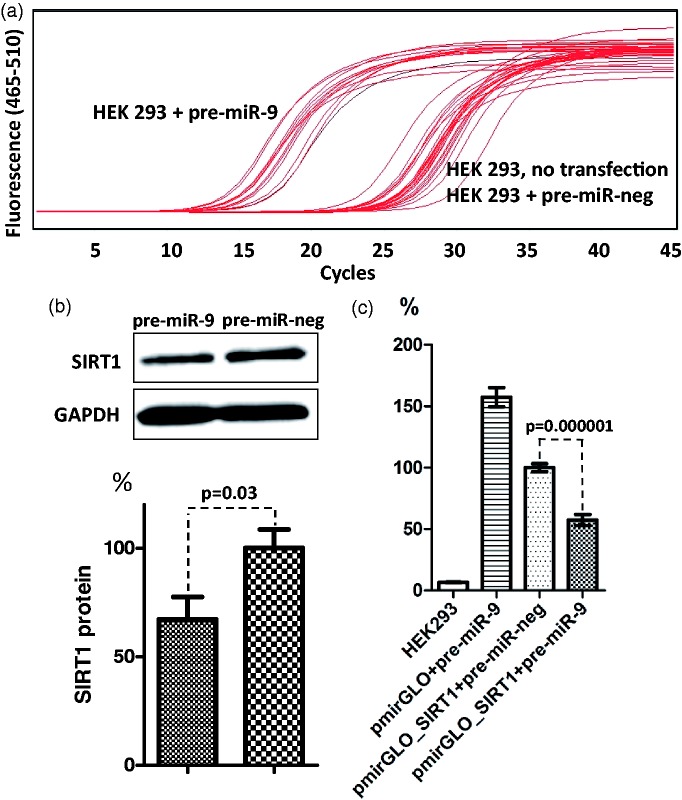
Of all sirtuins, SIRT1 is the one most clearly associated with the phenotype of aging; therefore, we selected this sirtuin for further analysis. We assessed the association of its expression with age-related changes of miR-9, miR-34a, miR-132, and miR-199a-5p levels. The interaction between human SIRT1 mRNA and miR-34a, miR-132 and miR-199a-5 p was demonstrated earlier; therefore, we tested the interaction with only miR-9.40–42 We found that transfection of pre-miR-9 increased the expression of miR-9 in HEK 293 cells while transfection of a negative control miRNA precursor did not, as Cp values for miR-9-overexpressing cells were lower by approximately 10 than those for not transfected cells and cells overexpressing negative control miRNA (Figure 2(a)). Overexpression of miR-9 was associated with the decrease of endogenous SIRT1 protein (by 18% at 30 nmol/L final pre-miR-9 concentration, P = 0.05 and by 33% at 60 nmol/L final concentration, P = 0.03, 70% transfection efficiency, Figure 2(b)). The mean relative luminescence induced by firefly luciferase expressed from the pmirGLO_SIRT1 in the presence of negative control miRNA was normalized to 100%. Co-transfection of pmirGLO_SIRT1 with pre-miR-9 decreased luminescence by 42.6% (P = 0.000001) (Figure 2(c)), suggesting that this miRNA interacted with its binding site present in the 3′UTR of SIRT1 mRNA and decreased the translation of the reporter protein.
Figure 2.
miR-9 interaction with the 3′UTR of SIRT1 mRNA. (a) Amplification curves for RT-PCR analysis of miR-9 expression in HEK 293 cells transfected with miR-9 precursor or with a negative miRNA precursor (each at 30 nmol/L final concentration). Expression of miR-9 was evaluated 24 h after the initiation of transfection. (b) Representative immunoblot of total protein isolated from HEK 293 cells 24 h after the initiation of transfection with miR-9 precursor or with a negative control miRNA precursor (each at 60 nmol/L final concentration), and the result of quantification of densitometric measurements of three independent immunoblots. (c) HEK 293 cells were co-transfected with the pmirGLO reporter vector with or without the cloned sequence corresponding to the 3′UTR of SIRT1 mRNA and with miRNA precursors. The mean relative luminescence induced by firefly luciferase in the presence of negative control miRNA was normalized to 100%. Each experiment was repeated nine times. HEK: human embryonic kidney. (A color version of this figure is available in the online journal.)
Age-dependent changes of expression of miRNAs in PBMC
As a first step, we wished to establish whether the mean U6 snRNA Cp values changed with age in PBMC. Since they did not (Y: 23.3 ± 2.4, E: 23 ± 2.4, L: 23.7 ± 2.2), we used U6 snRNA as a control for evaluating the expression of miR-9, miR-34a, miR-132, and miR-199a-5p. The expression of these miRNAs was similar in women and men and further analyses were performed for both genders together.
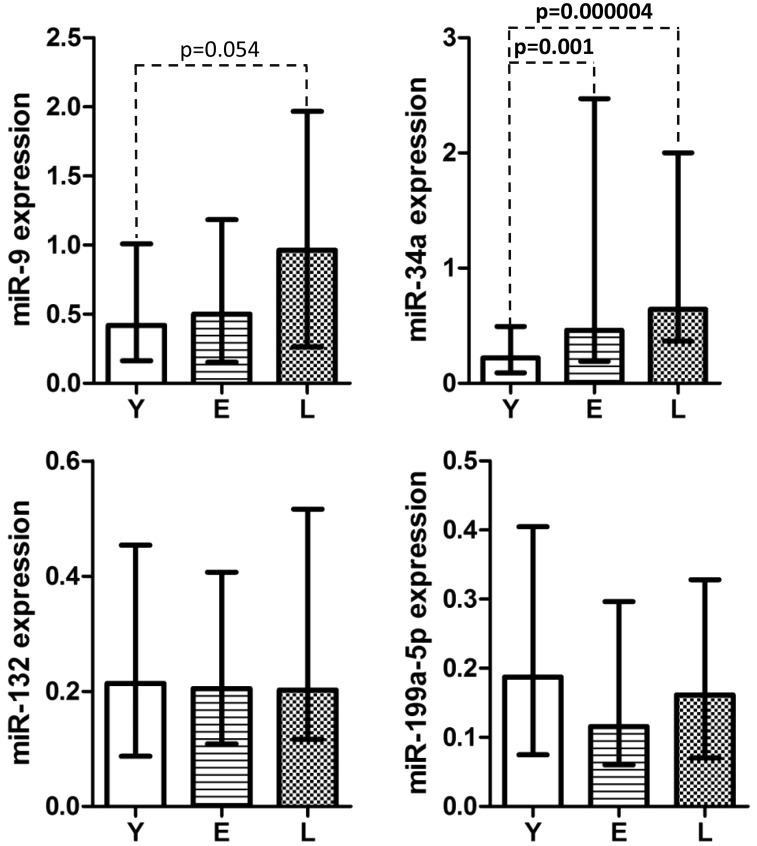
The median expression of miR-34a increased with age (P = 0.000001) and was significantly higher in the E and L groups than in young controls. In addition, the median expression of miR-9 was also affected by age (P = 0.05) and was higher in the L than in the Y group, but this difference did not reach the level of significance. The median expressions of miR-132 and miR-199a-5p were not affected by age in PBMC of the examined individuals (Figure 3).
Figure 3.
The median miR-9, miR-34a, miR-132, and miR-199a-5p expressions in PBMC of individuals of various ages. Results are presented in arbitrary units (specific for each miRNA) as median (25th, 75th percentile) values. Statistical analyses were performed with the Kruskal–Wallis test. The level of significance was established at 0.05. Y: young, E: elderly, L: long-lived study participants. PBMC: peripheral blood mononuclear cells
The SIRT1 mRNA levels negatively correlated with the expression of miR-34a and miR-132 (r = −0.234, P = 0.003 and r = −0.156, P = 0.049, respectively), but not with the expression of miR-9 and that of miR-199a-5p in the whole group of study subjects. There was no correlation between SIRT1 and the studied miRs in any of the age groups while analyzed separately.
Discussion
The available data regarding physiological roles of sirtuins suggest that the preservation of their function should be one of the features of longevity. However, contrary to expectation, in this work we found that in PBMC of aging individuals, older chronological age is associated with a significantly lower expression of the majority of the SIRT genes at the mRNA level, with the expression of SIRT1 and SIRT3 being most severely affected.
SIRT1, mostly located to the nucleus, by deacetylating FOXO transcription factors stimulates the expression of ROS-deactivating enzymes catalase and manganese superoxide dismutase,44–46 and by deacetylating proliferator-activated receptor coactivator-1α increases mitochondrial biogenesis and decreases ROS production.43–47 By deacetylating the Lys310 residue of RelA/p65 component of nuclear factor-κB (NF-κB), SIRT1 inhibits the activity of this transcription factor, supresses inflammation, and enhances the resolution phase of inflammatory response.4,48 In addition, by deacetylating Agt5 and Agt7, SIRT1 induces autophagy, a mechanism removing damaged cellular organelles.49 Alltogether, SIRT1 is a strong inhibitor of the oxidative stress and inflammatory response.48,50 Therefore, aging-associated decrease of SIRT1 can contribute to the occurence of chronic oxidative stress and chronic inflammation, both being hallmarks of inflammaging, and can be associated with the accumulation of damaged organelles (including mitochondria), further impeding the function of immune cells.
SIRT3, the major mitochondrial deacetylase, by deacetylating a number of key mitochondrial proteins promotes urea cycle, fatty acid β-oxidation, and oxidative phosphorylation, as well as increases expression and activity of manganese superoxide dismutase, thus reducing ROS production and promoting ROS scavenging.19,22,43,51–54 Thus, SIRT3 reduces oxidative stress, increases stress defenses, and counteracts the development of aging-associated diseases.22,55,56 Therefore, its decreased expression in immune cells of aged humans can result in an increase in the level of ROS, and in oxidative stress and damage. This, in turn, can activate the NF-κB system.
In conclusion, the decreased expression of SIRT1 and SIRT3 might contribute to the occurence of aging-associated chronic oxidative stress and inflammaging, also present in successfully aging individuals.19,50,56–58 Notably, the fact that the decreased expression of SIRT genes was detected in PBMC of our oldest study subjects indicates that the effect of their downregulation on the function of immune cells is not dominant, could be overcome by other factors and, therefore, does not contribute to the increased morbidity and mortality of successfully aging individuals who are genetically predisposed to longevity.59 It would be highly relevant, however, to establish whether downregulation of SIRTs in PBMC of individuals with less favorable genetic/environmental context is also of minor importance, or indeed contributes to the increased morbidity and mortality of the aging population.
Aging is associated with epigenetic drift.2,3 One of its features is the overexpression of miR-34a: this was detected in the cardiac muscle and endothelial cells of aged mice, as well as in the liver and kidneys of aged rats, among others.60,61 Therefore, the overexpression of this miRNA in PBMC of aging humans described in this work complements existing data. miR-34a exerts its function by downregulating SIRT1 exression, which leads to the described above alterations in the function of immune cells. It has also been shown that miR-34a knockout results in the reduced proliferation and activation of lymph nodes T cells.62 On the other hand, overexpression of miR-34a blocked programmed cell death 1 (PD1) and programmed cell death 1 ligand 1 (PD-L1)-specific apoptosis of T lymphocytes.63 In macrophages, miR-34a promotes M2 polarization linked to immunosuppression.64 Therefore, age-related overexpression of miR-34a seems to be an important pro-inflammatory immunoaging-contributing factor. However, our findings also show that the negative effect of excess miR-34a in PBMC of aged humans does not depend mainly on its inhibitory effect on SIRT1, as downregulation of SIRT1 was detected also in long-lived study subjects who did not suffer from aging-related diseases. Therefore, the genetic/environmental context and/or the inhibition of other factors by miR-34a might play a negative role in human aging.
Microarray analysis showed that miR-9 regulates the expression of a significant number of genes associated with the function of the immune system. It increases the expression of the interferon-induced genes and major histocompatibility complex class I molecules genes, as well as increases the IL-7 and IL-2 expression, and decreases IL-1 and IL-6 expression.65,66 In activated monocytes, NF-κB rapidly increases the expression of miR-9, which in turn reduces the expression of NF-κB, thus closing the negative feedback loop.67 In contrast to miR-34a, miR-9 induces M1 polarization of macrophages, which is associated with the production of pro-inflammatory cytokines.64,68 Therefore, in addition to its role in immunoaging exerted via SIRT1, other miR-9 mechanisms are involved in this process. Nevertheless, its overexpression in PBMC of long-lived humans suggests that it is not detrimental, at least in individuals who age successfully.
PBMC is a mixture of various T and B lymphocyte sub-types, natural killer cells, and monocytes. In humans, even though healthy aging is not associated with significant changes in the absolute numbers of cell types, it is associated with changes in the percentage of cell sub-types, such as a decrease in the number of naive T and B cells and an increase in the number of memory and effector cells, as well as with alterations of their function.69,70 Since aging of various immune cells might be characterized by different expression profiles of SIRTs and miRNAs, it is plausible that the observed expression changes reflect age-related changes in the PBMC composition. Therefore, the analysis of each cell type separately might yield more probing results. According to the literature data, other potential cause for differences in the expression of SIRT1 and miR-34a may be a severe cognitive impairment.71–73 However, we did not test if there were any differences in the levels of SIRT1, miR-34a, and miR-9 in PBMC of our study subjects since our young and elderly study subjects were cognitively normal, and long-lived group was composed of individuals with largely normal cognition or MCI. In addition, we have not examined age-related changes in the expression of SIRTs at the protein level. Such an examination would support the reliability of our results, especially in the case of miR-9. Possibly due to the lack of full complementarity between miR-9 and its binding site, blockage and sequestration of the SIRT1 mRNA were much more likely than degradation, resulting in the lack of correlation between the miR-9 and SIRT1 mRNA levels.74 However, there may be a negative correlation between this miRNA and SIRT1 protein. Unfortunately, performing both fractionation and protein analyses in PBMC was not possible in this study due to the limited amount of blood obtained from the elderly and long-lived study participants.
To sum up, in individuals who healthily age, the sustained expression of the SIRT genes in PBMC is not a prerequisite to longevity. It might however affect the process of immunosenescence.
Conclusion
The fact that downregulation of SIRTs and overexpression of miR-34a and miR-9 were detected in PBMC of long-lived individuals suggests that, at least in these individuals, they are not risk factors for morbidity and mortality but might affect the function of the immune system by accelerating or exacerbating immunosenescence. Therefore, interventions that increase the level of SIRT1 such as CR or resveratrol supplements could improve the function of the aging immune system.
Acknowledgment
This work was supported by the Polish Ministry of Science and Higher Education grant NN401037338.
Author contributions
All authors analyzed data and participated in interpretation of the studies and review of the manuscript. MO conducted the experiments, performed the statistical analysis, MB, ADS, JB, JP, and MG conducted the experiments, PS performed the statistical analysis, MPK designed the study, wrote manuscript. All authors read and approved the final manuscript.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Deelen J, Beekman M, Capri M, Franceschi C, Slagboom PE. Identifying the genomic determinants of aging and longevity in human population studies: progress and challenges. Bioessays 2013; 35: 386–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heyn H, Li N, Ferreira HJ, Moran S, Pisano DG, Gomez A, Diez J, Sanchez-Mut JV, Setien F, Carmona FJ, Puca AA, Sayols S, Pujana MA, Serra-Musach J, Iglesias-Platas I, Formiga F, Fernandez AF, Fraga MF, Heath SC, Valencia A, Gut IG, Wang J, Esteller M. Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci U S A 2012; 109: 10522–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cosentino C, Mostoslavsky R. Metabolism, longevity and epigenetics. Cell Mol Life Sci 2013; 70: 1525–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J 2004; 23: 2369–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, Wright SM, Mills KD, Bonni A, Yankner BA, Scully R, Prolla TA, Alt FW, Sinclair DA. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell 2008; 135: 907–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol 2012; 13: 225–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 2001; 410: 227–30. [DOI] [PubMed] [Google Scholar]
- 8.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and Sir2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 2000; 289: 2126–2126. [DOI] [PubMed] [Google Scholar]
- 9.Longo VD, Kennedy BK. Sirtuins in aging and age-related disease. Cell 2006; 126: 257–68. [DOI] [PubMed] [Google Scholar]
- 10.Burnett C, Valentini S, Cabreiro F, Goss M, Somogyvári M, Piper MD, Hoddinott M, Sutphin GL, Leko V, McElwee JJ, Vazquez-Manrique RP, Orfila AM, Ackerman D, Au C, Vinti G, Riesen M, Howard K, Neri C, Bedalov A, Kaeberlein M, Soti C, Partridge L, Gems D. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature 2011; 477: 482–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, Bar-Joseph Z, Cohen HY. The sirtuin SIRT6 regulates lifespan in male mice. Nature 2012; 483: 218–21. [DOI] [PubMed] [Google Scholar]
- 12.Herranz D, Muñoz-Martin M, Cañamero M, Mulero F, Martinez-Pastor B, Fernandez-Capetillo O, Serrano M. Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nat Commun 2010; 1: 3–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim S, Bi X, Czarny-Ratajczak M, Dai J, Welsh DA, Myers L, Welsch MA, Cherry KE, Arnold J, Poon LW, Jazwinski SM. Telomere maintenance genes SIRT1 and XRCC6 impact age-related decline in telomere length but only SIRT1 is associated with human longevity. Biogerontology 2012; 13: 119–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rose G, Dato S, Altomare K, Bellizzi D, Garasto S, Greco V, Passarino G, Feraco E, Mari V, Barbi C, BonaFe M, Franceschi C, Tan Q, Boiko S, Yashin AI, De Benedictis G. Variability of the SIRT3 gene, human silent information regulator Sir2 homologue, and survivorship in the elderly. Exp Gerontol 2003; 38: 1065–70. [DOI] [PubMed] [Google Scholar]
- 15.Banks AS, Kon N, Knight C, Matsumoto M, Gutiérrez-Juárez R, Rossetti L, Gu W, Accili D. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab 2008; 8: 333–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng F, Wijaya L, Tang BL. SIRT1 in the brain – connections with aging-associated disorders and lifespan. Front Cell Neurosci 2015; 9: 64–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oellerich MF, Potente M. FOXOs and sirtuins in vascular growth, maintenance, and aging. Circ Res 2012; 110: 1238–51. [DOI] [PubMed] [Google Scholar]
- 18.Maiese K. SIRT1 and stem cells: in the forefront with cardiovascular disease, neurodegeneration and cancer. World J Stem Cells 2015; 7: 235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab 2010; 12: 662–7. [DOI] [PubMed] [Google Scholar]
- 20.Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science 2004; 305: 390–2. [DOI] [PubMed] [Google Scholar]
- 21.Bordone L, Cohen D, Robinson A, Motta MC, van Veen E, Czopik A, Steele AD, Crowe H, Marmor S, Luo J, Gu W, Guarente L. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell 2007; 6: 759–67. [DOI] [PubMed] [Google Scholar]
- 22.Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, Gupta MP. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Invest 2009; 119: 2758–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science 2004; 305: 390–2. [DOI] [PubMed] [Google Scholar]
- 24.Colman RJ, Anderson RM, Johnson SC, Kastman EK, Kosmatka KJ, Beasley TM, Allison DB, Cruzen C, Simmons HA, Kemnitz JW, Weindruch R. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science 2009; 325: 201–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colman RJ, Beasley TM, Kemnitz JW, Johnson SC, Weindruch R, Anderson RM. Caloric restriction reduces age-related and all-cause mortality in rhesus monkeys. Nat Commun 2014; 5: 3557–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mattison JA, Roth GS, Beasley TM, Tilmont EM, Handy AM, Herbert RL, Longo DL, Allison DB, Young JE, Bryant M, Barnard D, Ward WF, Qi W, Ingram DK, de Cabo R. Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature 2012; 489: 318–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Most J, Tosti V, Redman LM, Fontana L. Calorie restriction in humans: an update. Ageing Res Rev 2017; 35: 350–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams J, Smith F, Kumar S, Vijayan M, Reddy PH. Are microRNAs true sensors of ageing and cellular senescence? Ageing Res Rev. Epub ahead of print 27 November 2016. pii:S1568-1637(16)30168-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zampieri M, Ciccarone F, Calabrese R, Franceschi C, Bürkle A, Caiafa P. Reconfiguration of DNA methylation in aging. Mech Ageing Dev 2015; 151: 60–70. [DOI] [PubMed] [Google Scholar]
- 30.Pal S, Tyler JK. Epigenetics and aging. Sci Adv 2016; 2: e1600584–e1600584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katz S, Ford AB, Moskowitz RW, Jackson BA, Jaffe MW. Studies of illness in the aged: the index of ADL: a standardized measure of biological and psychosocial function. JAMA 1963; 85: 94–9. [DOI] [PubMed] [Google Scholar]
- 32.Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res 1975; 12: 189–98. [DOI] [PubMed] [Google Scholar]
- 33.Polosak J, Roszkowska-Gancarz M, Kurylowicz A, Owczarz M, Dobosz P, Mossakowska M, Szybinska A, Puzianowska-Kuznicka M. Decreased expression and the Lys751Gln polymorphism of the XPD gene are associated with extreme longevity. Biogerontology 2010; 11: 287–97. [DOI] [PubMed] [Google Scholar]
- 34.Budzinska M, Owczarz M, Pawlik-Pachucka E, Roszkowska-Gancarz M, Slusarczyk P, Puzianowska-Kuznicka M. miR-96, miR-145 and miR-9 expression increases, and IGF-1R and FOXO1 expression decreases in peripheral blood mononuclear cells of aging humans. BMC Geriatrics 2016; 16: 200–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashraf N, Zino S, Macintyre A, Kingsmore D, Payne AP, George WD, Shiels PG. Altered sirtuin expression is associated with node-positive breast cancer. Br J Cancer 2006;95:1056–61. [DOI] [PMC free article] [PubMed]
- 36.TargetScanHuman 7.1. Available at: http://www.targetscan.org/vert_71 (accessed June 2016).
- 37.miRanda-mirSVR. Available at: http://www.microrna.org/microrna/home.do (accessed June 2016).
- 38.PicTar. Available at: http://pictar.mdc-berlin.de (accessed June 2016).
- 39.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136: 215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A 2008; 105: 13421–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strum JC, Johnson JH, Ward J, Xie H, Feild J, Hester A, Alford A, Waters KM. MicroRNA 132 regulates nutritional stress-induced chemokine production through repression of SirT1. Mol Endocrinol 2009; 23: 1876–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu WH, Yao XY, Yu HJ, Huang JW, Cui LY. Downregulation of miR-199a may play a role in 3-nitropropionic acid induced ischemic tolerance in rat brain. Brain Res 2012; 1429: 116–23. [DOI] [PubMed] [Google Scholar]
- 43.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell 2005; 16: 4623–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 2004; 303: 2011–5. [DOI] [PubMed] [Google Scholar]
- 45.Alcendor RR, Gao S, Zhai P, Zablocki D, Holle E, Yu X, Tian B, Wagner T, Vatner SF, Sadoshima J. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ Res 2007; 100: 1512–21. [DOI] [PubMed] [Google Scholar]
- 46.Tanno M, Kuno A, Yano T, Miura T, Hisahara S, Ishikawa S, Shimamoto K, Horio Y. Induction of manganese superoxide dismutase by nuclear translocation and activation of SIRT1 promotes cell survival in chronic heart failure. J Biol Chem 2010; 285: 8375–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1α. EMBO J 2007; 26: 1913–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salminen A, Kauppinen A, Suuronen T, Kaarniranta K. SIRT1 longevity factor suppresses NF-κB-driven immune responses: regulation of aging via NF-κB acetylation? Bioessays 2008; 30: 939–42. [DOI] [PubMed] [Google Scholar]
- 49.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A 2008; 105: 3374–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kauppinen A, Suuronen T, Ojala J, Kaarniranta K, Salminen A. Antagonistic crosstalk between NF-κB and SIRT1 in the regulation of inflammation and metabolic disorders. Cell Signal 2013; 25: 1939–48. [DOI] [PubMed] [Google Scholar]
- 51.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV, Jr, Weissman S, Verdin E, Schwer B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol 2007; 27: 8807–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, Tanokura M, Denu JM, Prolla TA. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell 2010; 143: 802–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hallows WC, Yu W, Smith BC, Devries MK, Ellinger JJ, Someya S, Shortreed MR, Prolla T, Markley JL, Smith LM, Zhao S, Guan KL, Denu JM. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol Cell 2011; 41: 139–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen Y, Zhang J, Lin Y, Lei Q, Guan KL, Zhao S, Xiong Y. Tumour suppressor SIRT3 deacetylates and activates manganese superoxide dismutase to scavenge ROS. EMBO Rep 2011; 12: 534–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, Vassilopoulos A, Ozden O, Park SH, Singh KK, Abdulkadir SA, Spitz DR, Deng CX, Gius D. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell 2010; 17: 41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jing E, Emanuelli B, Hirschey MD, Boucher J, Lee KY, Lombard D, Verdin EM, Kahn CR. Sirtuin-3 (Sirt3) regulates skeletal muscle metabolism and insulin signaling via altered mitochondrial oxidation and reactive oxygen species production. Proc Natl Acad Sci U S A 2011; 108: 14608–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tseng AHH, Shieh SS, Wang DL. SIRT3 deacetylates FOXO3 to protect mitochondria against oxidative damage. Free Radic Biol Med 2013; 63: 222–34. [DOI] [PubMed] [Google Scholar]
- 58.Minciullo PL, Catalano A, Mandraffino G, Casciaro M, Crucitti A, Maltese G, Morabito N, Lasco A, Gangemi S, Basile G. Inflammaging and anti-inflammaging: the role of cytokines in extreme longevity. Arch Immunol Ther Exp 2016; 64: 111–26. [DOI] [PubMed] [Google Scholar]
- 59.Schoenmaker M, de Craen AJ, de Meijer PH, Beekman M, Blauw GJ, Slagboom PE, Westendorp RG. Evidence of genetic enrichment for exceptional survival using a family approach: the Leiden Longevity Study. Eur J Hum Genet 2006; 14: 79–84. [DOI] [PubMed] [Google Scholar]
- 60.Li N, Muthusamy S, Liang R, Sarojini H, Wang E. Increased expression of miR-34a and miR-93 in rat liver during aging, and their impact on the expression of Mgst1 and Sirt1. Mech Ageing Dev 2011; 132: 75–85. [DOI] [PubMed] [Google Scholar]
- 61.Boon RA, Iekushi K, Lechner S, Seeger T, Fischer A, Heydt S, Kaluza D, Tréguer K, Carmona G, Bonauer A, Horrevoets AJ, Didier N, Girmatsion Z, Biliczki P, Ehrlich JR, Katus HA, Müller OJ, Potente M, Zeiher AM, Hermeking H, Dimmeler S. MicroRNA-34a regulates cardiac ageing and function. Nature 2013; 495: 107–10. [DOI] [PubMed] [Google Scholar]
- 62.Sun YX, Li H, Feng Q, Li X, Yu YY, Zhou LW, Gao Y, Li GS, Ren J, Ma CH, Gao CJ, Peng J. Dysregulated miR34a/diacylglycerol kinase ζ interaction enhances T-cell activation in acquired aplastic anemia. Oncotarget 2017; 8: 6142–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang X, Li J, Dong K, Lin F, Long M, Ouyang Y, Wei J, Chen X, Weng Y, He T, Zhang H. Tumor suppressor miR-34a targets PD-L1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell Signal 2015; 27: 443–52. [DOI] [PubMed] [Google Scholar]
- 64.Essandoh K, Li Y, Huo J, Fan GC. MiRNA-mediated macrophage polarization and its potential role in the regulation of inflammatory response. Shock 2016; 46: 122–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thiele S, Wittmann J, Jäck HM, Pahl A. miR-9 enhances IL-2 production in activated human CD4(+) T cells by repressing Blimp-1. Eur J Immunol 2012; 42: 2100–8. [DOI] [PubMed] [Google Scholar]
- 66.Gao F, Zhao ZL, Zhao WT, Fan QR, Wang SC, Li J, Zhang YQ, Shi JW, Lin XL, Yang S, Xie RY, Liu W, Zhang TT, Sun YL, Xu K, Yao KT, Xiao D. miR-9 modulates the expression of interferon-regulated genes and MHC class I molecules in human nasopharyngeal carcinoma cells. Biochem Biophys Res Commun 2013; 431: 610–6. [DOI] [PubMed] [Google Scholar]
- 67.Bazzoni F, Rossato M, Fabbri M, Gaudiosi D, Mirolo M, Mori L, Tamassia N, Mantovani A, Cassatella MA, Locati M. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc Natl Acad Sci U S A 2009; 106: 5282–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu L, McCurdy S, Huang S, Zhu X, Peplowska K, Tiirikainen M, Boisvert WA, Garmire LX. Time Series miRNA-mRNA integrated analysis reveals critical miRNAs and targets in macrophage polarization. Sci Rep 2016; 6: 37446–37446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu J, Li W, Liu Z, Zhang YY, Peng Y, Feng DG, Li LH, Wang LN, Liu L, Li L, Liu J. Ageing-associated changes in cellular immunity based on the SENIEUR protocol. Scand J Immunol 2012; 75: 641–6. [DOI] [PubMed] [Google Scholar]
- 70.Ponnappan S, Ponnappan U. Aging and immune function: molecular mechanisms to interventions. Antioxid Redox Signal 2011; 14: 1551–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schipper HM, Maes OC, Chertkow HM, Wang E. MicroRNA expression in Alzheimer blood mononuclear cells. Gene Regul Syst Biol 2007; 1: 263–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kumar R, Chaterjee P, Sharma PK, Singh AK, Gupta A, Gill K, Tripathi M, Dey AB, Dey S. Sirtuin1: a promising serum protein marker for early detection of Alzheimer's disease. PLoS One 2013; 8: e61560–e61560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kiko T, Nakagawa K, Tsuduki T, Furukawa K, Arai H, Miyazawa T. MicroRNAs in plasma and cerebrospinal fluid as potential markers for Alzheimer's disease. J Alzheimers Dis 2014; 39: 253–9. [DOI] [PubMed] [Google Scholar]
- 74.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol 2012; 19: 586–93. [DOI] [PubMed] [Google Scholar]