Abstract
Despite current standard of care, the average 5-year mortality after an initial diagnosis of heart failure (HF) is about 40%, reflecting an urgent need for new therapeutic approaches. Previous studies demonstrated that the epigenetic reader protein bromodomain-containing protein 4 (BRD4), an emerging therapeutic target in cancer, functions as a critical coactivator of pathologic gene transactivation during cardiomyocyte hypertrophy. However, the therapeutic relevance of these findings to human disease remained unknown. We demonstrate that treatment with the BET bromodomain inhibitor JQ1 has therapeutic effects during severe, preestablished HF from prolonged pressure overload, as well as after a massive anterior myocardial infarction in mice. Furthermore, JQ1 potently blocks agonist-induced hypertrophy in human induced pluripotent stem cell–derived cardiomyocytes (iPSC-CMs). Integrated transcriptomic analyses across animal models and human iPSC-CMs reveal that BET inhibition preferentially blocks transactivation of a common pathologic gene regulatory program that is robustly enriched for NFκB and TGF-β signaling networks, typified by innate inflammatory and profibrotic myocardial genes. As predicted by these specific transcriptional mechanisms, we found that JQ1 does not suppress physiological cardiac hypertrophy in a mouse swimming model. These findings establish that pharmacologically targeting innate inflammatory and profibrotic myocardial signaling networks at the level of chromatin is effective in animal models and human cardiomyocytes, providing the critical rationale for further development of BET inhibitors and other epigenomic medicines for HF.
INTRODUCTION
Heart failure (HF) is a leading cause of mortality, hospitalization, and health care expenditure in the United States (1, 2). Existing pharmacotherapies for systolic HF are β-adrenergic receptor and renin-angiotensin axis antagonists, which dampen the excessive activity of stress neurohormones produced in response to myocardial injury. However, despite the widespread use of these disease-modifying drugs, the average 5-year mortality rate after an initial diagnosis of HF is about 40%, which highlights the urgent need for new therapeutic approaches (1, 2).
In response to persistent hemodynamic and neurohormonal stress, the myocardium undergoes pathological cell state changes characterized by cardiomyocyte hypertrophy, inflammation, myofibroblast activation, and contractile dysfunction (3–6). Although cardiac remodeling may provide short-term adaptation in certain settings, sustained or excessive activation of this process is maladaptive and drives disease progression (3, 4). Studies over the past two decades have established that inhibition of specific signaling pathways that govern stress-induced cardiac remodeling has cardioprotective effects even in the face of persistent stress (4, 7). Notably, ventricular tissue remodeling is a robust predictor of HF severity and death in patients, underscoring the detrimental nature of this process (3, 8–12). Together, these data support the contention that targeting the tissue remodeling process itself can be beneficial without compromising cardiac performance (3, 9,11–13).
The cell state transitions that underlie pathologic cardiac remodeling are rooted in dynamic changes in gene control (14,15) and feature a general state of transcriptional anabolism (3,15). In the stressed heart, multiple upstream signaling pathways converge on the transcription machinery, which integrates these signals by transactivating gene programs that alter cell state (16). Thus, blocking stress-activated signaling cascades at the level of chromatin-dependent signaling represents an attractive therapeutic strategy in HF. In response to cardiac stress, a defined set of transcription factors coordinately binds to specific regulatory regions of the genome. These transcription factors recruit lysine acetyltransferases, which hyperacetylate local chromatin and activate cis-regulatory elements (or enhancers) (16,17). Subsequent transactivation of distal target genes occurs via recruitment of epigenetic “reader” proteins (16), such as bromodomain-containing protein 4 (BRD4) (14,18), that bind acetyllysine via bromodomains and coactivate transcription by assembling complexes that signal to RNA polymerase II (Pol II) (19–22). BRD4 belongs to the bromodomain and extraterminal (BET) family of highly conserved acetyllysine recognition proteins and has been implicated in the maintenance and progression of cancer cell state across a broad range of malignancies (18, 23–25). The recent development of potent, specific, and reversible BET bromodomain inhibitors, such as the first-in-class thienodiazepine small-molecule JQ1 (23), has accelerated mechanistic discovery and translation in this field. JQ1 binds the bromodomains of BET proteins with exquisite shape complementarity and nanomolar affinity, resulting in potent, competitive, and transient displacement of BRD4 from acetylated chromatin (23). Studies across a broad range of cell types demonstrate that JQ1 preferentially blocks transactivation of specific sets of genes in a dose-dependent and context-specific manner (24). Target gene specificity is achieved, in part, because the BRD4 coactivator protein is asymmetrically enriched at massive cell state–specific enhancers (or superenhancers) (26, 27). Transcription of super-enhancer–regulated genes is disproportionately sensitive to depletion of coactivator proteins such as BRD4 (26, 28). Given the tractable therapeutic window of JQ1 in preclinical studies, there has been intense interest in the development of BET bromodomain inhibitors as anticancer drugs. These translational efforts have spawned several ongoing early-phase human cancer trials using BET bromodomain inhibitors, including derivatives of JQ1 (29, 30).
Initial studies from our group demonstrated that BRD4 functions as a critical coactivator of pathologic gene transactivation during cardiomyocyte hypertrophy by a mechanism that involves recruitment of positive transcription elongation factor b (P-TEFb) activity and pause release of Pol II (31, 32). In cultured neonatal rat ventricular myocytes (NRVMs), small-interfering RNA (siRNA)–mediated silencing of BRD4 or chemical inhibition with JQ1 blocks cardinal features of pathologic hypertrophy. In addition, early administration of JQ1 at the very onset of pressure overload in mice prevented the development of cardiac hypertrophy and left ventricular (LV) systolic dysfunction. Although these index studies provided important mechanistic insight, the therapeutic relevance of these early observations remained unknown. In particular, it was not known whether BET bromodomain inhibitors could be used to treat established HF in more clinically relevant experimental settings. Furthermore, it was unknown whether BETs could be therapeutically targeted in human cardiomyocytes and whether there were common transcriptional effects of BET bromodomain inhibitor therapy across pathologic settings and species. Here, we address these major knowledge gaps and identify core mechanisms of therapeutic efficacy that are vital for directing further translation of BET inhibition as a therapeutic strategy for heart disease.
RESULTS
BET bromodomain inhibition is effective in the treatment of preestablished HF
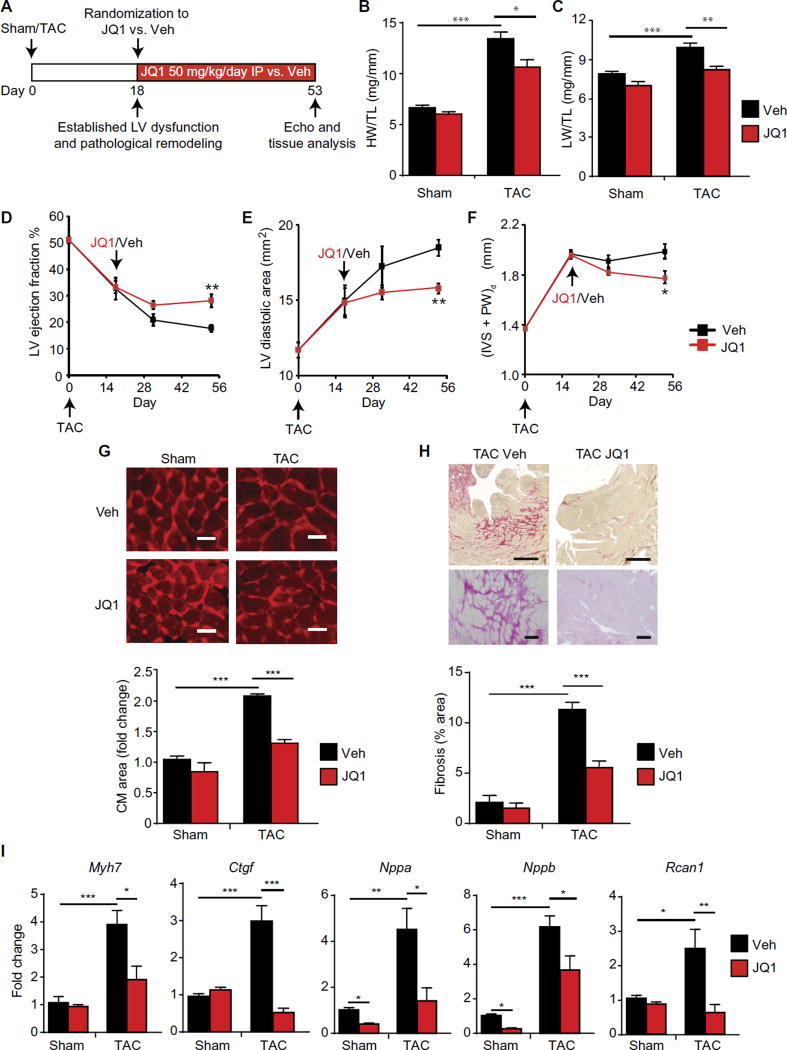
Patients requiring treatment for HF typically present with preestablished disease. Therefore, we first tested whether BET bromodomain inhibition with JQ1 could treat preestablished HF in a mouse model of LV pressure overload. We subjected adult mice to transverse aortic constriction (TAC) or sham surgery. On postoperative day 18, a time point when robust cardiomegaly and LV dysfunction have already developed in this model (31), we randomized mice to receive JQ1 [50 mg/kg daily, intraperitoneally (IP)] versus vehicle and continued out to postoperative week 8 (protocol in Fig. 1A). This dosing scheme of JQ1 has established efficacy and tolerability in previous studies of cancer and cardiovascular disease in mice and does not cause overt toxicity (23, 24, 31–33). In addition, we have previously demonstrated that JQ1 administration at these doses does not affect blood pressure or the pressure gradient across the constricted aorta (31). In this treatment protocol, we found that JQ1 attenuated multiple hallmark features of HF progression in vivo, including cardiomegaly (Fig. 1B), pulmonary edema (Fig. 1C), LV systolic dysfunction, LV cavity dilation, LV wall thickening [Fig. 1, D to F, respectively; echocardiographic data for the final time point (day 53) are shown in fig. S1, A to C], cardiomyocyte hypertrophy (Fig. 1G), and LV fibrosis (Fig. 1H). In addition, JQ1 suppressed the induction of several canonical HF-associated genes (Fig. 1I). Thus, late administration of JQ1 can treat HF even when started at a time when there is robust, preexisting disease.
Fig. 1. BET bromodomain inhibition treats preestablished HF after prolonged pressure overload.
(A) Experimental protocol for preestablished pressure overload model induced by TAC in mice. Veh, vehicle. (B) Ratio of heart weight/tibia length (HW/TL) (n = 10) and (C) lung weight/tibia length (LW/TL) (n = 10). (D) LV ejection fraction, (E) LV diastolic area, and (F) LV wall thickness quantified by echocardiography (n = 10). (IVS + PW)d is the total thickness of the interventricular septum and posterior LV wall at the end-diastole. (G) Representative LV cross sections stained with wheat germ agglutinin (WGA) with quantification of mean cardiomyocyte (CM) cell area shown below (n = 3 to 4). Scale bars, 20 µm. (H) Representative LV cross sections stained with picrosirius red. Scale bars, 400 µm (top) and 100 µm (bottom). Quantification of fibrotic area below (n = 5). (I) Quantitative reverse transcription polymerase chain reaction (qRT-PCR) for indicated genes in mouse heart tissue (n = 5). For (A) to (I), *P < 0.05, **P < 0.01, ***P < 0.001 for indicated comparison. Data are shown as means ± SEM.
BET bromodomain inhibition is effective in the treatment of HF after a large myocardial infarction
Although the TAC model is a widely used method for eliciting hallmark features of HF pathogenesis, the acute onset of severe hemodynamic overload in this model does not represent a common clinical scenario in humans. In contrast, myocardial infarction (MI) is a very common cause of pathologic cardiac remodeling and HF in humans (1). Furthermore, animal models of post-MI HF have served as a major gateway for drug development, as exemplified by studies of angiotensin-converting enzyme inhibitors in rodent preclinical studies (13) and subsequent human clinical trials (11, 12).
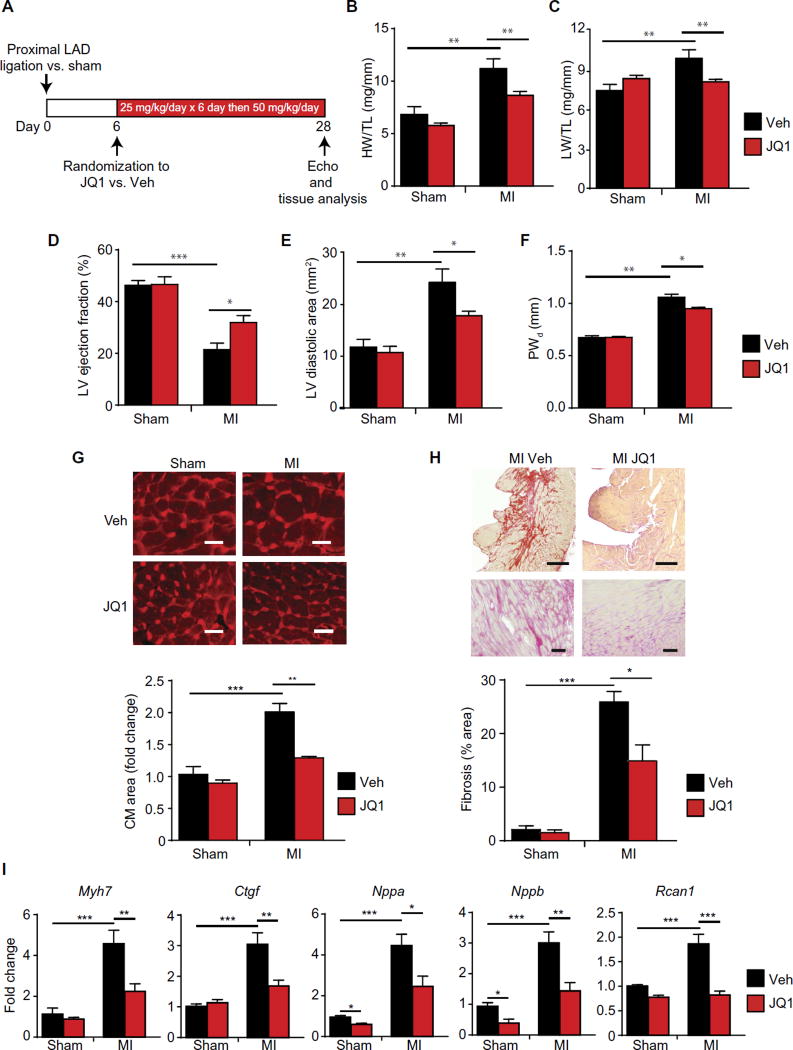
We therefore asked whether JQ1 could be used to treat HF that develops after large, transmural MI in adult mice. We subjected mice to permanent surgical ligation of the proximal left anterior descending (LAD) coronary artery to produce a large, dense, anterior wall MI. As expected for this anatomically proximal level of LAD occlusion (34), ~35% of mice died in the first 5 postoperative days due to LV rupture or overt HF, confirming the severity of this model in our hands. On postoperative day 6, a time point when all infarctions were complete, surviving mice were randomized to receive JQ1 versus vehicle. We initially dosed at 25 mg/kg daily for 6 days (during the period of early surgical recovery) and subsequently ramped up to 50 mg/kg daily until postoperative day 28 (a time point when robust pathologic remodeling of the remote LV myocardium is present in this model) (protocol in Fig. 2A) (35). Similar to our findings in the pressure overload treatment model (Fig. 1), JQ1 attenuated several cardinal features of HF in the post-MI setting, including cardiomegaly (Fig. 2B), pulmonary congestion (Fig. 2C), LV systolic dysfunction (Fig. 2D), LV cavity dilation (Fig. 2E), wall thickening of the remote LV (posterior wall; Fig. 2F), hypertrophy of cardiomyocytes in the remote LV (Fig. 2G), and fibrosis of the remote LV (Fig. 2H). JQ1 also suppressed pathological induction of canonical HF-associated genes in the remote LV myocardium (Fig. 2I). It is possible that therapeutic strategies that attenuate LV remodeling may also prevent normal healing of a transmural infarct, particularly in mouse models, which are predisposed to post-MI rupture (34). However, we did not observe any significant excess in mortality or LV rupture between JQ1 and vehicle treatment groups (two mice died/ ruptured in each group from n = 12 per group after treatment initiation; χ2 = 1), suggesting that this regimen of BET bromodomain inhibition ameliorates HF progression without overt compromise of infarct healing.
Fig. 2. BET bromodomain inhibition ameliorates HF after a massive anterior wall myocardial infarction.
(A) Experimental protocol for MI model induced by surgical ligation of the proximal LAD coronary artery. (B and C) Gravimetric measurements on day 28. Ratios of heart weight/tibia length and lung weight/tibia length were measured (n = 10). (D) LV ejection fraction, (E) LV diastolic area, and (F) LV posterior wall thickness quantified by echocardiography on day 28 [n= 10 (MI) and6 (sham)]. PWd is the thickness of the posterior LV wall (remote LV) at the end-diastole. The mid-LV interventricular septum was transmurally infarcted in this model and was therefore not relevant to the wall thickness assessment. (G) Representative cross sections from remote LV stained with WGA with quantification of mean cardiomyocyte (CM) cell area shown below (n = 3 to 4). Scale bars, 20 µm. (H) Representative cross sections from remote LV stained with picrosirius red. Scale bars, 400 µm (top) and 100 µm (bottom). Quantification of fibrotic area below (n = 5). (I) qRT-PCR for indicated genes in remote LV tissue (n = 5). For (A) to (I), *P < 0.05, **P < 0.01, ***P < 0.001 for indicated comparison. Data are shown as means ± SEM.
BET bromodomain inhibition suppresses transactivation of shared gene regulatory networks across HF models
Given the therapeutic efficacy of JQ1 in two distinct HF models in vivo (Figs. 1 and 2) and our previous observation that BRD4 regulates transcriptional pause release in the heart (31), we hypothesized that there was a common gene-control mechanism mediating these salutary effects across disease settings. We performed transcriptomic profiling from LV tissue using RNA sequencing (RNA-seq) at baseline (sham) and during disease (TAC and MI) from mice treated with vehicle versus JQ1. We first analyzed the transcriptomic effect of chronic JQ1 exposure in the baseline state by comparing the RNA-seq profiles of the sham-vehicle– versus sham-JQ1–treated LV tissue. Differential expression analysis showed that JQ1 reduced expression of 613 genes and increased expression of 125 genes in the left ventricles of sham-treated mice (volcano plot in fig. S2A; complete expression data for all detected transcripts and differential expression calls are provided in table S1). Gene ontology analysis of the JQ1–down-regulated transcripts showed a strong enrichment for innate immune activation, extracellular matrix production, and cell adhesion (fig. S2B). The much smaller set of JQ1–up-regulated genes revealed no major enrichment for functional terms, other than three genes that were cataloged as containing Heat-shock protein 70 binding sites.
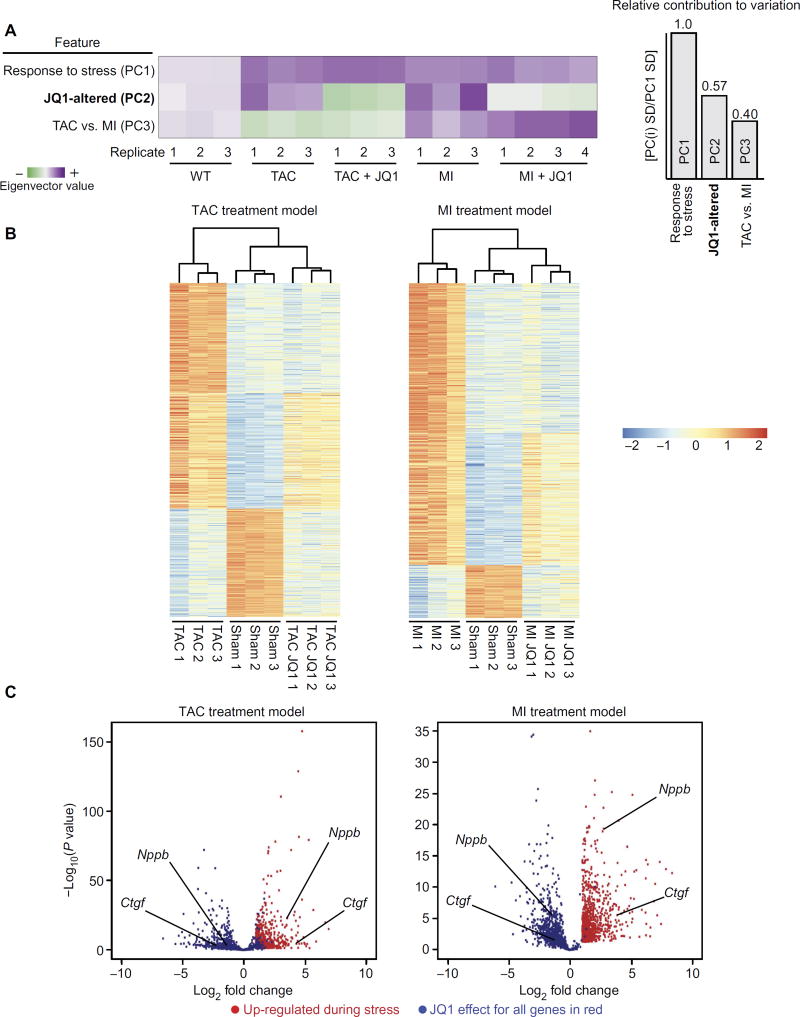
Because JQ1 did not alter baseline cardiac structure or function in the sham group (Figs. 1 and 2 and fig. S1), we next turned our attention to the global set of transcripts that were differentially expressed during disease. Global clustering of transcriptomic profiles by principal components analysis revealed that the two most significant sources of differential expression across all samples were the response to stress (such as TAC or MI) and the effect of JQ1 administration (Fig. 3A). When comparing sham versus stress, the dominant transcriptomic feature was global gene up-regulation (TAC, 725 genes up and 401 genes down; MI, 928 genes up and 205 genes down). These global transcriptomic profiles are consistent with HF as a state of robust transcriptional anabolism (15). For the global set of genes that were differentially expressed during stress, hierarchical clustering of samples (Fig. 3B; shown above the heat maps) demonstrated that the JQ1-treated group clustered more closely with the sham groups, reflecting broad suppression of a pathologic state. The dominant effect of JQ1 across disease models was to dampen the transactivation of a broad subset of stress-inducible genes (Fig. 3B, top third of heat maps). This observation is mechanistically consistent with the function of BRD4 as a key coactivator of enhancer-dependent transcription and effector of Pol II pause release (26–28, 31). The inhibitory effects of JQ1 were specific, as exemplified by a subset of stress-inducible genes that were unaffected by JQ1 (Fig. 3B, middle third of heat maps). Finally, JQ1 had no broad effect in rescuing genes that were down-regulated with stress (Fig. 3B, bottom third of heat maps), a finding that is also consistent with the primary function of BRD4 as a transcriptional coactivator. The volcano plots in Fig. 3C illustrate the potent effects of JQ1 in the dampening of stress-mediated gene induction across the TAC and MI models (differential expression data from RNA-seq are provided in tables S2 and S3). Together, these data demonstrate that these HF models elicit a state of robust transcriptional anabolism and that the dominant role of JQ1 during HF therapy is to dampen the stress-dependent transactivation of a broad but specific set of genes.
Fig. 3. BET bromodomain inhibition suppresses transactivation of a specific stress-inducible gene program during HF pathogenesis.
(A) Global display of information content derived from principal components analysis of RNA-seq data. Each row in the principal components analysis plot is an eigenvector that describes a feature across all samples. The purple to green shading scale reflects the magnitude of the principal components analysis eigenvector for the component of interest. Summary bar plots on the right show that the largest source of variance in the data can be explained by the response to stress (TAC and MI) with the next largest source of variation ascribed to JQ1-associated changes in gene expression. A smaller source of variation can be ascribed to differences in gene expression between the TAC and MI models. (B) Heat map of genes differentially expressed with stress in the TAC (left) and MI model (right). Scale is log2. Statistical criteria from DESeq analysis were ≥2-fold change and a false discovery rate of <0.05. The top one-third of each heat map is the cluster of genes that are up-regulated with stress and dampened by JQ1. The middle one-third of each heat map is the cluster of genes up-regulated with stress and unaffected by JQ1, demonstrating specificity. The bottom one-third of each heat map shows that genes that are down-regulated with stress are not significantly reversed by JQ1. (C) Volcano plot demonstrating magnitude and significance of JQ1 on suppressing induction of stress-inducible genes in both the TAC and MI models. Genes up-regulated by TAC (left) or MI (right) versus sham are plotted in red. Plotted in blue is the effect of JQ1 (stress-JQ1 versus stress-vehicle) for each gene shown in red. Representative genes are called out (black lines) for each volcano plot.
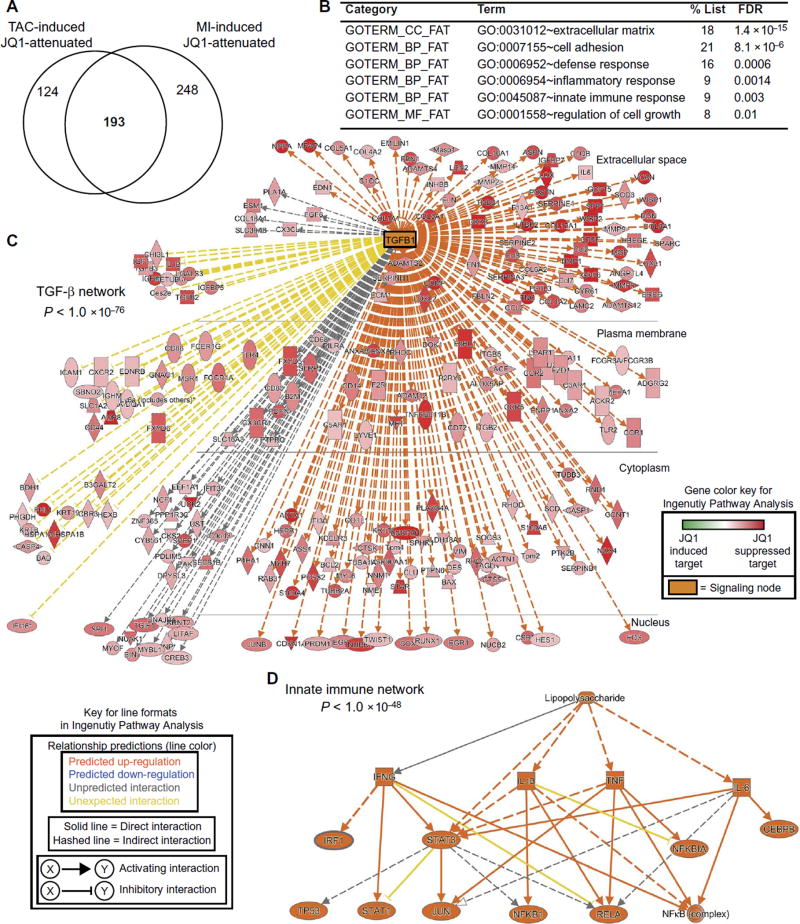
To gain further insight into the therapeutic mechanisms underlying the efficacy of JQ1 in HF, we defined a common set of 193 genes whose transactivation was suppressed by JQ1 in both the TAC and MI models (see Venn diagram; Fig. 4A) (gene list is provided in table S4). Gene ontology analysis of this common set of 193 genes revealed a strong enrichment for specific pathophysiologic processes excessively activated in human HF (6, 36), including extracellular matrix deposition, innate immune and inflammatory responses, and cellular growth (Fig. 4B). We next analyzed this gene set using Ingenuity Pathway Analysis (37, 38) to identify pathologic signaling networks that were specifically blocked by BET bromodomain inhibitor therapy. These unbiased functional analyses revealed striking enrichment for two myocardial signaling networks that were robustly suppressed by JQ1. First, JQ1 was found to block transactivation of genes that form a massive hub centered on transforming growth factor–β (TGF-β) signaling (P < 1.4 × 10−76; Fig. 4C), a central regulator of pathologic cell state transformations in the stressed myocardium (39, 40). Genes affected in this expansive TGF-β network broadly encompassed all major compartments of the cell. The second observation was that the JQ1 inhibitory effect was robustly enriched for genes involved in innate immune responses (signified by the “lipopolysaccharide” term at the top level of the network), which converged upon NFκB/RelA, AP1, and STAT1 as terminal transcriptional effectors (P < 1 × 10−48; Fig. 4D). The robust enrichment for TGF-β and NFκB targets in this set of JQ1-attenuated transcripts was independently corroborated by gene set enrichment analysis (GSEA) (fig. S3A). The GSEA demonstrates that genes up-regulated in our TAC and MI models are statistically enriched for TGF-β and NFκB targets, confirming that these signaling pathways are activated during disease. Consistent with our Ingenuity Pathway Analysis, GSEA demonstrated that the set of genes attenuated by JQ1 in these HF models was statistically enriched for TGF-β and NFκB targets. To gain initial insight into myocardial cell types that might be contributing to the transcriptional response to JQ1 during HF pathogenesis in vivo, we used fingerprinting analysis (fig. S3B) to compare our RNA-seq profiles with curated RNA-seq data sets derived from mouse cardiomyocytes (GSE58453 and GSE68509) (41,42), mouse cardiac fibroblasts (GSE58453) (41), and myeloid cells (GSE57125) (43). These analyses detected a signature for transcriptional activation of fibroblasts in TAC/MI when compared to the sham group. There was a detectable effect of JQ1 in attenuating the transcriptional activation of fibroblast signature. In summary, across in vivo models of HF, BET bromodomain inhibition with JQ1 blocks the transactivation of a shared set of myocardial genes that encompass the coordinated output of TGF-β signaling and proinflammatory transcription factors.
Fig. 4. BET bromodomain inhibition suppresses transactivation of shared transcriptional networks across HF models.
(A) Venn diagram showing significant overlap of gene sets that are stress-induced and JQ1-suppressed across the TAC and MI models (χ2 < 0.001). A common set of 193 genes was identified as being shared targets of BET bromodomain inhibition (gene list is provided in table S4). (B) Gene ontology analysis using DAVID for the Venn overlap genes shown in (A). A false discovery rate (FDR) of <0.05 was considered statistically significant. (C and D) Ingenuity Pathway Analysis. The common gene set was analyzed by Ingenuity Pathway Analysis. These analyses revealed two dominant signaling networks that were highly enriched in the set of genes suppressed by JQ1. (C) A large TGF-β network (P < 1.0 × 10−76). Each target gene is shown in red, with the darkness of red shading designating the relative degree of inhibition by JQ1 of that particular gene. Although the legend includes a green color scale for JQ1–up-regulated genes, there are no green-shaded genes on this diagram because all genes were selected a priori to be those that are suppressed by JQ1. TGFB1 as a central node gene is shown in orange. (D) JQ1 suppressed genes enriched for a network of innate immune signaling nodes with strong convergence on NFκB transcriptional responses (P < 1.0 × 10−48). Keys for the Ingenuity Pathway Analysis color shading and line formatting schemes are provided in the figure.
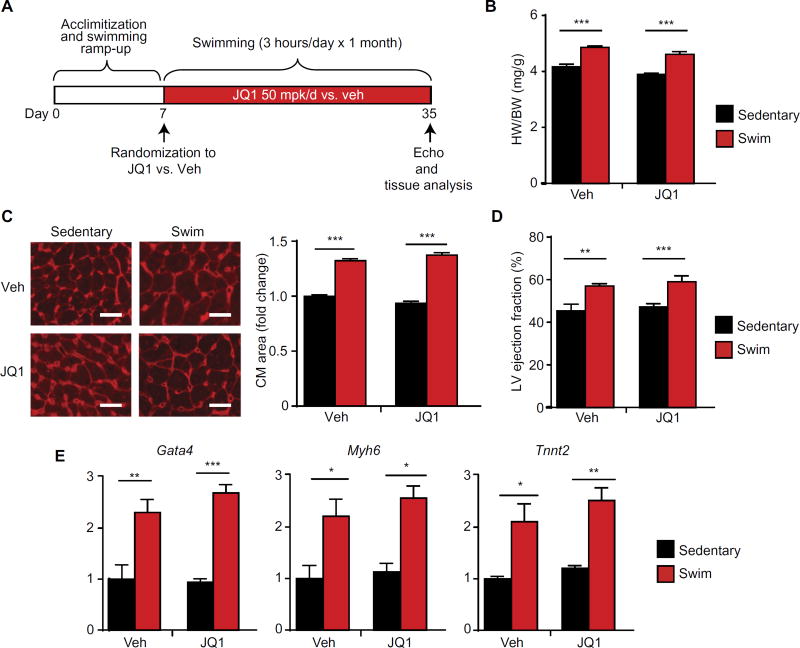
BET bromodomain inhibition does not affect physiological cardiac growth during exercise
Chronic endurance exercise training leads to a physiological form of cardiac growth that has been shown to provide protection against myocardial injury (44, 45). An important distinguishing feature of physiological versus pathological cardiac remodeling is that the former lacks robust induction of innate inflammatory or profibrotic gene programs (45–47). Because our data in murine models showed that the transcriptional effects of JQ1 were specifically enriched for the suppression of proinflammatory and profibrotic gene networks (Figs. 3 and 4), we hypothesized that BET bromodomain inhibition with JQ1 might be permissive for physiological cardiac remodeling during endurance exercise training. To test this hypothesis, adult mice underwent a well-established, high-intensity endurance swimming protocol in which mice are progressively trained to swim for a total of 3 hours/day for 4 weeks (44). Swimming versus sedentary mice were randomized to receive JQ1 (50 mg/kg per day, IP) or vehicle injections (protocol in Fig. 5A). In both the vehicle- and JQ1-treated groups, there were similar increases in cardiac mass (Fig. 5B), cardiomyocyte cross-sectional area (Fig. 5C), LV ejection fraction (Fig. 5D), and expression of transcripts induced with exercise training in this model (such as Gata4 and Tnni3) (Fig. 5E and fig. S4), indicating that JQ1 treatment did not block key features of exercise-induced cardiac plasticity. Hence, JQ1 administration to adult mice at the doses used here can treat HF but does not suppress exercise-induced physiological cardiac growth.
Fig. 5. BET bromodomain inhibition does not suppress physiologic cardiac hypertrophy in response to chronic endurance exercise training.
(A) Schematic of swimming protocol. (B) Gravimetry; heart weight/body weight ratio on day 35 (n = 7). (C) Representative LV cross sections stained with WGA with quantification of mean cardiomyocyte cell area shown at the right (n = 5). Scale bars, 20 µm. (D) Echocardiographic LV ejection fraction on day 35 (n = 7). (E) qRT-PCR for indicated genes in mouse LV tissue (n= 5). For (B) to (E), *P < 0.05, **P < 0.01, ***P < 0.001 for indicated comparisons. No statistically significant JQ1 effect was observed. Data are shown as means ± SEM.
BET bromodomain inhibition blocks agonist-induced hypertrophy in human iPSC-CMs
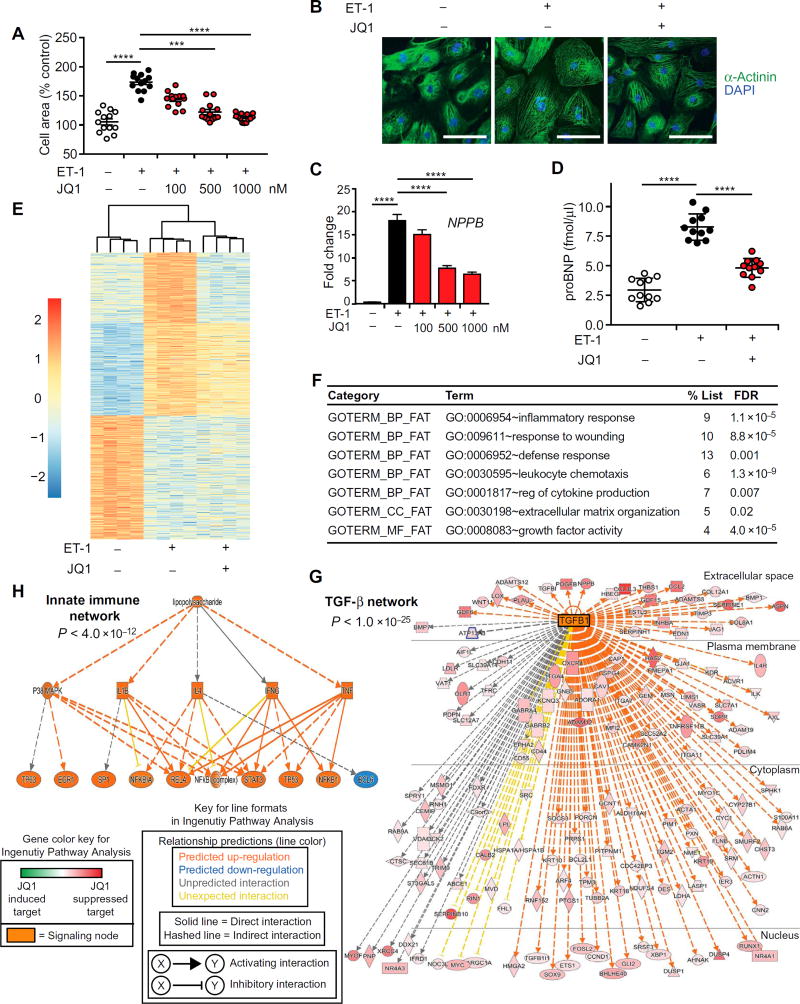
All previous studies of BET bromodomain inhibition in heart disease have been performed in rodent models (31, 32). We therefore asked whether BET bromodomain inhibition could block pathologic cardiomyocyte hypertrophy in human cells. We tested the effects of JQ1 in a widely used human induced pluripotent stem cell–derived cardiomyocyte (iPSC-CM) culture system that originated from an iPSC line derived from a healthy human donor [iCell Cardiomyocytes, Cellular Dynamics International Inc. (CDI)] (48). Previous studies have demonstrated that these cells can mount a hypertrophic response to 10 nM endothelin-1 (ET-1) (48). We found that JQ1 attenuated ET-1–mediated hypertrophic growth in human iPSC-CMs in a dose-dependent manner (Fig. 6, A and B). Consistent with the effects on cellular hypertrophy, qRT-PCR demonstrated that JQ1 suppressed transactivation of NPPB/BNP (Fig. 6C) and other typical marker genes induced in human iPSC-CMs during ET-1 stimulation (fig. S5A) (48). Quantitative enzyme-linked immunosorbent assay (ELISA) confirmed that JQ1 attenuated the ET-1–stimulated increase in BNP protein abundance (Fig. 6D).
Fig. 6. BET bromodomain inhibition blocks agonist-induced hypertrophy and stress-mediated gene transactivation in human iPSC-CMs.
(A) Human iPSC-CMs treated with ET-1 (10 nM), JQ1, or vehicle for 18 hours and stained for α-actinin immunofluorescence for quantification of cell area (n = 12). Bars designate means ± SEM. (B) Representative images of human iPSC-CM stained for α-actinin immunofluorescence. ET-1 (10 nM) and JQ1 (1000 nM); 18 hours. Scale bars, 100 µm. (C) qRT-PCR for NPPB/BNP in iPSC-CMs (n= 12). Data are shown as means ± SEM. (D) ELISA for proBNP protein concentration in medium of iPSC-CM (n = 11). ET-1 (10 nM) and JQ1 (1000 nM); 18 hours. Bars designate means ± SEM. For (A), (C), and (D), ****P < 0.0001, ***P < 0.0005. (E) Heat map of genes differentially expressed in iPSC-CM with ET-1 stimulation. Statistical criteria from DESeq analysis were ≥2-fold change and a false discovery rate of <0.05. The top one-third of each heat map is the cluster of genes that are up-regulated with ET-1 and dampened by JQ1. The middle one-third of each heat map is the cluster of genes up-regulated with ET-1 and unaffected by JQ1, demonstrating specificity. The bottom one-third of each heat map shows that genes that are down-regulated with ET-1 are not significantly reversed by JQ1. Scale is log2. (F) Gene ontology analysis using DAVID for the set of genes that were ET-1–inducible and suppressed by JQ1 (top third of heat map). A false discovery rate of <0.05 was considered statistically significant. (G and H) Ingenuity Pathway Analysis. The set of ET-1–induced genes that were attenuated by JQ1 was analyzed by Ingenuity Pathway Analysis. These analyses revealed two dominant signaling networks that were highly enriched in the set of genes suppressed by JQ1 and paralleled the findings in mouse heart tissue from Fig. 4. (G) A large TGF-β network (P < 1.0 × 10−25). Each target gene is shown in red, with the darkness of red shading designating the relative degree of inhibition by JQ1 of that particular gene. Although the legend includes a green color scale for JQ1–up-regulated genes, there are no green-shaded genes on this diagram because all genes were selected a priori to be those that are suppressed by JQ1. TGFB1 as a central node gene is shown in orange. (H) JQ1 suppressed genes enriched for a network of innate immune signaling nodes with strong convergence on NFκB transcriptional responses (P < 1.0 × 10−48). Keys for the Ingenuity Pathway Analysis color shading and line formatting schemes are provided.
To gain a broader insight into stress-activated gene expression programs affected by JQ1 in human cardiomyocytes, we performed RNA-seq in this experimental system. Similar to the murine RNA-seq profiles (Fig. 3), a heat map of all ET-1–responsive genes in human iPSC-CMs illustrates that the dominant effect of JQ1 was to dampen induction of a subset of ET-1–inducible genes (265 gene subset from a total of 615 ET-1–induced transcripts; Fig. 6E) (differential expression data provided as table S5). Functional annotation of this 265 gene subset revealed strong enrichment for genes involved in inflammatory responses, cellular growth, and extracellular matrix production (Fig. 6F), terms that paralleled the cardiac signatures from murine disease models (Fig. 4). Similar to our murine data, Ingenuity Pathway Analysis revealed that this set of JQ1-responsive human genes robustly enriched for a TGF-β signaling hub (P < 1 × 10−25; Fig. 6G) and an innate immune transcription factor network (signified by the lipopolysaccharide term at the top level of the network) with a strong convergence on NFκB/RelA (P < 4 × 10−12; Fig. 6H). In addition, the robust enrichment for TGF-β and NFκB targets in this set of JQ1-attenuated transcripts in human iPSC-CM was independently corroborated by GSEA (fig. S5B). Together, these data demonstrate that BET bromodomain inhibition with JQ1 blocks agonist-induced hypertrophic responses in human iPSC-CMs in a cell-autonomous manner, with transcriptional effects that parallel our observations in murine models.
DISCUSSION
Our data provide proof of principle that BET bromodomain inhibition, an emerging therapeutic strategy in a variety of human cancers (18, 23–25), can treat preestablished HF in vivo. Our observation that nanomolar concentrations of JQ1 block pathological hypertrophy in human iPSC-CMs suggests that BET inhibition may also exert protective effects in the human heart. Integrated genome-wide analyses across murine models and human cardiomyocytes reveal that BET inhibition suppresses a common pathologic transcriptional program that is highly enriched for inflammatory and profibrotic signaling effectors in the myocardium. Because inflammation and fibrosis are central to HF pathogenesis, molecular mediators of these processes are intently pursued therapeutic targets (6, 49). Our current work demonstrates the efficacy of an epigenomically based therapeutic approach that targets inflammatory and profibrotic cell state transitions at the level of chromatin.
Several BET bromodomain inhibitor drugs are currently being developed in early-phase human cancer trials (29). Our work provides impetus for next-phase testing of this class of therapeutics in largeanimal models of HF, including porcine/ovine MI models. Here, we focused on the ability of JQ1 to ameliorate severe HF that occurs after a complete and dense anterior wall MI. Our findings, which highlight the beneficial effects of JQ1 on the remote/noninfarcted myocardium, are relevant for the treatment of ischemic cardiomyopathy in humans. It will also be interesting to explore whether BET bromodomain inhibition can protect at-risk myocardium in animal models of ischemia-reperfusion injury when delivered at the time of coronary revascularization, although the mechanisms of acute ischemic myoprotection in this setting represent vastly different biological processes than the longer-term beneficial effects on the remote LV shown here. One study has shown that pretreatment with JQ1 in a rat model of permanent coronary artery ligation can reduce biomarkers of infarct severity at 24 hours (50), suggesting that BET bromodomain inhibition may acutely protect ischemic myocytes during an unrevascularized MI. However, it remains unknown whether this strategy provides longer-term benefit or is effective when administered in the perireperfusion phase. In addition to studying acquired forms of HF, it will also be of interest to study the role of BET proteins in the pathogenesis of cardiomyopathies driven by genetic mutations (51), some of which feature extensive activation of profibrotic and proinflammatory pathways in the heart. These lines of investigation, guided by the results of ongoing human cancer trials, will provide the critical foundation for developing BET bromodomain inhibition as a potential therapeutic strategy in human heart disease.
Our RNA-seq data demonstrate that BET bromodomain inhibitors suppress a subset of stress-activated myocardial genes. The mechanisms underlying the selective effects of BET bromodomain inhibition on target genes, a phenomenon which has been observed across multiple cell types (24, 26, 33, 52), are just beginning to be understood. An important contributor to this selectivity is the observation that potent coactivators such as BRD4 are asymmetrically loaded on a critical subset of cell state–defining enhancers that have been termed super-enhancers, which often regulate target genes via Pol II pause release (26–28, 33). Super-enhancer–regulated genes are exquisitely sensitive to local depletion of coactivator proteins, including BRD4, across a number of cell types (26–28, 33, 53). Recent in vitro studies using cultured endothelial cells (33) and NRVMs (54) have demonstrated that during pathological stress, there is a stimulus-coupled flux of BRD4 from baseline regulatory loci to new, stress-activated super-enhancers. In the setting of stress, JQ1 transiently displaces BRD4 from chromatin, prevents the cell’s ability to sustain maximal activity of newly formed super-enhancers, and preferentially dampens transcription of stress-activated gene programs (33). The precise molecular mechanisms governing locus-specific flux of BRD4 to these newly formed super-enhancers is not well understood but may involve recruitment via protein interactions between BRD4 and DNA binding transcription factors (55). Our finding that the myocardial signature of JQ1-suppressed genes is specifically enriched for transcription pathways central to inflammatory and fibrotic responses supports a model in which specific subsets of DNA binding transcription factors may play a key role in driving genome-wide flux of BRD4 during stress. This feature of BRD4 signaling may partially explain why JQ1 did not affect exercise-induced cardiac growth, a form of tissue plasticity that does not typically involve inflammatory or profibrotic transcription programs (45–47).
The preferential inhibitory effect of BET bromodomain inhibitors on stress-dependent enhancer assembly may, in part, contribute to their therapeutic window (55). The pulsatile exposure to JQ1 used here, which has a relatively short half-life in mice (23), can dampen the assembly and net transcriptional output of stress-activated super-enhancers in the injured myocardium. This reversible, phasic, and dose-titratable disruption of super-enhancer–dependent transcription may be homeostatically accommodated in nondiseased tissues in a manner that allows the adult mammalian organism to guard key aspects of basal organ physiology. Consistent with this mode of action, JQ1 does not cause overt toxicity in adult mice under the dosing protocols used in this study (23, 24, 31, 33, 56). Furthermore, JQ1 does not cause major changes in cardiac structure or function in mice subject to sham surgery (31). The general tolerability of JQ1 at the doses used here is further supported by our observation that mice receiving daily JQ1 injections are able to complete a high-intensity, 4-week swimming protocol. However, higher doses of BET inhibitors will bring broader, sustained exposure that may produce a wider range of on-target toxicity (57, 58). A mouse model in which total BRD4 protein abundance is ubiquitously suppressed by an inducible short hairpin RNA (shRNA) demonstrates that excessive levels of total BRD4 depletion result in abnormalities in the intestinal epithelium, bone marrow, and hair follicles—all of which are reversible after the shRNA expression is turned off (56). These observations indicate that the major toxicity profiles of BET inhibitors are likely to be on-target, dose-dependent, and reversible (56). Although our studies demonstrate efficacy and tolerability of JQ1 in mouse models, an important hurdle in next-phase preclinical efforts will be to develop compounds and dosing/delivery schemes that have an appropriate therapeutic index for large-animal models of HF. In addition, the efficacy of BET inhibition when added on to current standard-of-care HF regimens will need to be established. It will also be of interest to test whether adjunctive BET inhibition can protect at-risk ischemic myocardium when administered at the time of coronary reperfusion, although the mechanisms of acute myoprotection in such a setting would likely be different from the chronically beneficial effects on remote myocardium described here. Because several potent BET inhibitor drugs are progressing in early-phase human cancer trials (29), the collective first-in-human experience with this class of therapeutics will serve as an important guide for considering cardiac and other non-cancer applications.
An important area of mechanistic inquiry will be to further dissect which BET family members (BRD2/3/4) and which cell types (for example, cardiac myocytes, cardiac fibroblasts) are mediating the therapeutic effects of small-molecule BET bromodomain inhibition in vivo. In a previous study, specific siRNA-mediated silencing of BRD4 in cultured NRVMs was shown to phenocopy the antihypertrophic effect of JQ1 (31). BRD4 is also the BET family member that allosterically activates the P-TEFb complex via a C-terminal CDK9-interacting domain that is not present in BRD2 or BRD3 (19, 20, 59), which is consistent with our observation that JQ1 suppresses Pol II pause release in the stressed heart (31). Together, these findings strongly implicate cardiac BRD4 as a major target of JQ1 in this context. With regard to which particular cell types in the myocardium are being affected by BET bromodomain inhibition, our RNA-seq studies reveal that JQ1 blocks the induction of a broad program of proinflammatory and profibrotic genes. These transcriptomic signatures encompass genes that are expressed in cardiomyocytes, cardiac fibroblasts, or both. Our data in NRVMs (31) and cultured human iPSC-CMs demonstrate that BET inhibition has cell-autonomous effects on inflammatory and profibrotic gene programs in cardiomyocytes in vitro, suggesting that the cardiomyocyte is a major cell target of BET inhibitors in vivo. Recent studies also demonstrated an important role for BRD4 in hepatic stellate cells (53), suggesting that JQ1 may also block the pathologic activation of resident tissue fibroblasts. Our cell type fingerprinting analysis of mouse left ventricles detected an effect of JQ1 in attenuating the transcriptional activation of fibroblasts. Although this observation is consistent with the ability of JQ1 to attenuate profibrotic and innate inflammatory signaling in the heart, it does not exclude important contributions from cardiomyocytes, immune cells, and other cells that populate the stressed myocardium in mediating the therapeutic response to JQ1. More precise annotation of gene- and tissue-specific effects of BET family members will require the development of conditional genetic models for Brd2/3/4, which will be important resources for this field. Such conditional alleles, when coupled with recently developed Cre drivers (5), will be particularly useful for interrogating the role of BRD4 in activated cardiac myofibroblasts, a cell type that is challenging to manipulate in vivo using viral vectors. It remains important to emphasize that Cre-lox–based conditional gene deletion is permanent and fundamentally very different from the chemical biological approach leveraged here. Chemical probes like JQ1 transiently and reversibly disrupt the interaction of BET proteins with acetylated chromatin in a manner that is temporally precise and dose-titratable (23, 55). These features cannot be recapitulated by current genome-targeting or gene-silencing approaches. Therefore, the ability to dissect gene-specific and cell-autonomous responses in a manner that accurately reflects the biology of these chemical probes would require future development of both gene-specific compounds for individual BET family members and methods for cell type–specific delivery in vivo. The recent development of small molecules that promote reversible degradation of BET proteins may create future opportunities to potently modulate BET function in a more gene- and tissue-restricted manner in vivo, potentially providing an expanded therapeutic window for this pharmacologic strategy (60). Although the delineation of gene- and tissue-specific phenotypes is certainly an important area of investigation, we postulate that pharmacologic BET bromodomain inhibitors, like many other drugs, may be therapeutically effective in vivo because they target multiple cell types.
In broader terms, this work highlights that HF, much like cancer, can be considered to be a disease in which there is aberrant transcriptional control of cell identity (16). In the case of cancer, progressive mutations in the genome drive transcriptional programs that cause uncontrolled cellular expansion and dissemination (16, 18). In HF, excessive exposure to stress signals converges on the gene regulatory apparatus in cardiomyocytes and fibroblasts to drive maladaptive cell state transitions that culminate in progressive organ remodeling and contractile dysfunction. In contrast to several cancer drugs that cause cardiotoxicity (61), our data indicate that BET bromodomain inhibitors may be a privileged class of anticancer therapeutics that has cardioprotective properties. The therapeutic efficacy of BET inhibition in both these settings implies that cancer cells and myocardial cells share common vulnerabilities at the level of chromatin-dependent signal transduction from enhancers. This generalized dependence on stimulus-coupled transcription suggests that in addition to BRD4, the cardiac chromatin signaling machinery may contain other druggable targets that can be exploited for HF therapy.
MATERIALS AND METHODS
Study design
The overall objective of this study was to determine whether BET bromodomain inhibition with the small-molecule JQ1 could treat HF in well-established rodent models (TAC and MI) and attenuate ET-1– induced pathologic remodeling in human iPSC-CMs. RNA-seq studies were performed to assess transcriptomic effects of JQ1 in both murine and human models in an unbiased, genome-wide manner. For murine HF models, mice were randomized to each experimental arm and treatment group. Mouse echocardiograms were analyzed by an individual blinded to the experimental group assignment. For each experiment, sample size reflects the number of independent biological replicates and is provided in the figure legend. The expected effect magnitude and SD for each experiment were guided by examples from published literature for each model used. Using these parameters, sample size was determined by power calculation with α = 0.05 and β = 0.80. Statistical analysis of RNA-seq data was performed in collaboration with M. Lemieux (BioInfo) and the Gladstone Institutes Bioinformatics Core.
Animal models
All protocols concerning animal use were approved by the Institutional Animal Care and Use Committees at the University of California San Francisco and the Case Western Reserve University and conducted in strict accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (62). Studies were conducted with age-matched male C57Bl/6J mice (The Jackson Laboratory, catalog no. 000664). Mice were housed in a temperature- and humidity-controlled pathogen-free facility with 12-hour light/dark cycle and ad libitum access to water and standard laboratory rodent chow.
Preparation of JQ1
JQ1 was synthesized and purified in the laboratory of J. Bradner (Dana-Farber Cancer Institute), as previously published (23). For in vivo experiments, a stock solution [50 mg/ml JQ1 in dimethyl sulfoxide (DMSO)] was diluted to a working concentration of 5 mg/ml in an aqueous carrier (10% hydroxypropyl β-cyclodextrin; Sigma C0926) using vigorous vortexing. Mice were injected at a dose of 50 mg/kg given intraperitoneally once daily. Vehicle control was an equal amount of DMSO dissolved in 10% hydroxypropyl β-cyclodextrin carrier solution. All solutions were prepared and administered using sterile technique. For in vitro experiments, JQ1 was dissolved in DMSO and administered to cells at indicated concentrations using an equal volume of DMSO as control.
Mouse models of cardiac hypertrophy and HF
All mice were male C57Bl/6J mice aged 10 to 11 weeks from The Jackson Laboratory. Mice were placed on a temperature-controlled small-animal surgical table to help maintain body temperature (37°C) during surgery. Mice were anesthetized with ketamine/xylazine, mechanically ventilated (Harvard Apparatus), and subjected to thoracotomy. For TAC surgery, the aortic arch was constricted between the left common carotid and the brachiocephalic arteries using a 7-0 silk suture and a 27-gauge needle, as previously described (31). Intraperitoneal injections of JQ1 (50 mg/kg per day) or vehicle were administered 18 days after TAC surgery and continued until postoperative day 53 (Fig. 1A). For MI surgery, the proximal LAD was permanently ligated at the level of a standard anatomic landmark (left atrial edge) using a 7-0 silk suture, as described previously (63). Occlusion of arterial flow was demonstrated by ischemic color changes in the anterior wall of the heart. Standard three-lead electrocardiography was performed to confirm that left coronary artery ligation has resulted in a myocardial injury current (ST-segment elevations). Intraperitoneal injections of JQ1 were started on postoperative day 6 (25 mg/kg per day), increased to 50 mg/kg per day on postoperative day 12, and continued until postoperative day 28 (Fig. 2A). For sham surgeries, thoracotomy was performed as above, and the aorta (for TAC) or LAD (for MI) was surgically exposed without any further intervention.
Endurance swimming model
Daily swimming exercise was carried out to induce physiological cardiac hypertrophy, as previously described (44) with minor modifications. Swimming exercise started in a ramp-up protocol starting at two sessions of 10 min, with a 10-min increase each day until two sessions of 90 min were reached. Ninety minutes daily of swimming was continued for the next 28 days. Injections of JQ1 (50 mg/kg per day, IP) or vehicle started on the seventh day of this ramp-up protocol (Fig. 5A). The two swimming sessions were separated from each other by at least 4 hours. The temperature of the water was maintained at 31°C with a heating system (JBJ True Temp Titanium Heating System, T3–300), and constant flow of water was maintained by a circulation pump (JBJ OceanStream, OS101). Mice in the sedentary group were subjected to the same water conditions for 5 min weekly. All mice were closely monitored during the training for signs of struggling or discomfort, and no mice died of drowning during the above training protocol.
Transthoracic echocardiography
Mice were anesthetized with 1% inhalational isoflurane and imaged using the Vevo 770 High Resolution Imaging System (FujiFilm VisualSonics Inc.) and the RMV-707B 30-MHz probe. Measurements were obtained from M-mode sampling and integrated electrocardiogram-gated kilohertz visualization (EKV) images taken in the LV short axis at the midpapillary level, as previously described (31, 64). LV areas and ejection fraction were obtained from high-resolution two-dimensional measurements at the end-diastole and end-systole, as previously described (31, 64). For the MI model, measurement of remote LV wall thickness was performed on the inferoposterior wall.
Histological analysis
Short-axis heart sections from the mid–left ventricle were fixed in 10% buffered formalin and embedded in paraffin. Cardiomyocyte cross-sectional area was determined by staining with rhodamine-conjugated WGA (Vector Laboratories, RL-1022) and quantified as previously described (2). Fibrosis was visualized using the picrosirius red staining kit (Polysciences Inc.). Quantification of fibrotic area was performed as previously described (32) with Image-Pro Plus (Media Cybernetics). For the MI model, assessment of remote LV histology was performed on the inferoposterior wall.
Culture, qRT-PCR, and BNP ELISA from human iPSC-CMs
iCell human iPSC-CMs (CDI) were thawed and plated on 96-well plates coated with fibronectin (5 µg/ml) (Sigma), as previously described (48). Cells were plated at a density of 4.1 × 104 cells/cm2. After 48 hours in the iCell Cardiomyocyte Plating Medium (CDI), cells were quiesced by culturing in supplemented William’s E (SWE) medium [cocktail B from the Hepatocyte Maintenance Supplement Pack (Thermo Fisher Scientific) diluted in William’s E medium at a 1:25 dilution] for 4 days. For subsequent hypertrophic stimulation, quiesced iPSC-CMs were treated with or without JQ1 (500 nM) versus DMSO for 3 hours followed by stimulation with or without ET-1 (10 nM) (Sigma) for 18 hours. RNA was purified and reverse-transcribed using the TaqMan Gene Expression Cells-to-CT kit (Life Technologies, AM1278) according to the manufacturer’s instructions. qRT-PCR for human NPPB and B2M (normalizer) was performed with the Life Technologies TaqMan assays (NPPB, Hs00173590_m1; B2M, Hs000984230_m1). qRT-PCR for other human genes was performed using TaqMan chemistry including FastStart Universal Probe Master (Roche), labeled probes from the Universal ProbeLibrary (Roche), and gene-specific oligonucleotide primers (list of qRT-PCR primers and TaqMAN probes are provided in table S6). Relative expression was calculated using the 2ΔΔCt method with normalization to B2M expression. ELISA for BNP protein was performed on 6 µl of medium from each well of a 96-well plate as previously published (48) using the following antibodies: anti-proBNP capture antibody (clone 5B6; Abcam ab13111), anti-proBNP HRP conjugate detection antibody (clone 16F3; Abcam ab13124), and purified NT-proBNP (Phoenix Pharmaceuticals, catalog no. 011–42) as a standard.
Cell area measurements for iPSC-CM
Cells cultured on 96-well plates were fixed in phosphate-buffered saline (PBS) with 2% paraformaldehyde for 20 min, then permeabilized [PBS–Tween 20 (PBST) + 0.1% Triton X-100] for 20 min and blocked (PBST + 5.0% horse serum) for 1 hour at room temperature. Primary antibody against sarcomeric α-actinin (Sigma A7811) was used at a 1:800 dilution in PBST + 5.0% horse serum for 1 hour at room temperature. The anti-mouse secondary antibody tagged with Alexa 488 was used at a dilution of 1:1,000 for 1 hour at room temperature. Wells were scanned on a high-content imaging platform (IN Cell Analyzer, GE Healthcare Systems) at ×200 magnification, and quantified as previously described (48).
RNA purification and qRT-PCR from murine samples
A 10- to 20-mg piece of mouse LV tissue was collected and preserved in RNAlater stabilization reagent (Qiagen) followed by mechanical disruption/homogenization in PureZOL (Bio-Rad) on a TissueLyser II (Qiagen). For the TAC model, a concentric short-axis slice of the LV was prepared for RNA. For the MI model, a short-axis sector of the remote, noninfarcted left ventricle (infero-posterior wall) was prepared for RNA. The RNA-containing aqueous phase was extracted with chloroform. RNA was then purified with Aurum purification kit (BioRad, 732–6830). qRT-PCR was performed using TaqMan chemistry including FastStart Universal Probe Master (Roche), labeled probes from the Universal Probe Library (Roche), and gene-specific oligonucleotide primers on a LightCycler 480 system (Roche). A list of qRT-PCR primers and TaqMAN probes are provided in table S6. Relative expression was calculated using the 2ΔΔCt method with normalization to constitutive genes. Mouse data were normalized to Ppib.
Library preparation and next-generation sequencing
RNA isolated from mouse tissue and human iPSC-CMs were quantified using Qubit fluorometric quantitation and assessed for quality using Agilent Bioanalyzer Nano RNA Chip. For human iPSC-CM samples, the 500 nM JQ1 treatment condition was used for RNA-seq. Samples with RNA integrity number larger than 8 were considered high-quality and suitable for RNA-seq. Library preparation was performed using the Illumina TruSeq Stranded Total RNA kit with Ribo-Zero Gold rRNA depletion, and the library was sequenced on an Illumina HiSeq 2500 at the Genomics Core Facility of Case Western Reserve University (Cleveland, OH) (paired-end, 100 base pairs, >50 M reads per sample; Rapid Run v2 flow cell).
RNA-seq data analysis
Adapters and barcodes were trimmed, and low-quality reads were filtered using fastq-mcf (https://code.google.com/archive/p/ea-utils/). High-quality sequences were aligned to mm10 (mouse data) and hg38 (human data), as appropriate, with TopHat 2.0.13 (65), and assigned to Ensembl genes and counted using “featureCounts” (66), part of the Subread suite (http://subread.sourceforge.net/). (Note: the entry for ENSMUSG00000092329/Gm20388 was removed from the murine Gencode vM4 annotations we used before counting to avoid the artifactual loss of other transcripts, including Acta1, within its ~4-Mb boundaries.) Raw counts were used as inputs to DESeq (v.1.25.0) (67) for differential expression analysis, and Benjamini-Hochberg test was used to control false discovery of differentially expressed genes (68). Heat maps were generated using the Bioconductor package Heatmap (v.1.0.8) using reads per kilobase per million (RPKM) mapped. Values shown are log2-transformed and row-normalized. Samples were clustered using complete linkage and Pearson correlation. Rows were ordered (but not clustered) to highlight patterns between groups. The Venn diagram shows the overlap of transcript IDs. DAVID (v.6.8; https://david.ncifcrf.gov) (69) was used for functional annotation enrichment analyses with the background universe restricted to genes expressed in the species and tissue/cell type being interrogated. Log fold changes were calculated between conditions and imported into the Ingenuity Pathway Analysis (37) server (Qiagen) to identify pathways with enriched differential expression. GSEA (70) was run on expression data preranked by log2 fold change. Curated signatures of TGF-β targets (PLASARI_TGFB1_TARGETS_10HR_UP; Broad Institute C2 data set) and NFκB targets (NFκB_DIRECT_BU; www.bu.edu/nf-kb/gene-resources/target-genes/) were used in the GSEA. Cell type fingerprinting consisted of summing z scores of normalized data sets for each cell type signature (cardiomyocyte: Nppa, Nppb, Myh7, Fstl1, Actc1,Xirp2, and Mybpc2; fibroblast: Postn, Adam12, Wisp2, Col3a1, and Tnc; myeloid: Ly86, Ccr2, Sfpi1/Spi1, Cd80, and Itgam). The following published RNA-seq data sets were fingerprinted as controls: GSE68509 [n = 3 mouse cardiomyocyte (42)], GSE58453 [n = 3 each mouse cardiomyocyte and fibroblast (41)], and GSE57125 [n = 2 cardiac macrophage precursor cells (called “B-type” cells in this publication) (43)]. All RNA-seq data sets were uploaded to Gene Expression Omnibus (GEO) (accession no. GSE96566).
Statistical analysis
Data are reported as means ± SEM unless otherwise indicated in the figure legend. Statistical analyses of murine TAC and MI data were performed using analysis of variance (ANOVA) with Tukey’s honest significance difference test using GraphPad Prism Plus. Statistical analysis of human iPSC-CM area, qRT-PCR, and BNP protein concentration was performed by ANOVA with Holm-Sidak correction for multiple comparisons using GraphPad Prism Plus. For all analyses, P < 0.05 was considered significant. The statistical methods used in the analysis of RNA-seq data are detailed separately above.
Supplementary Material
Fig. S1. Echocardiographic data from sham- and TAC-treated mice at day 53.
Fig. S2. Transcriptomic effects of chronic JQ1 administration in baseline (sham-operated) hearts.
Fig. S3. GSEA and fingerprinting analysis of differentially expressed genes in mouse LV.
Fig. S4. Additional qRT-PCR of exercise-regulated genes in mouse left ventricle.
Fig. S5. qRT-PCR and GSEA in human iPSC-CMs.
Table S1. Normalized RNA-seq expression data for sham-vehicle and sham-JQ1 groups.
Table S2. Normalized RNA-seq expression data for sham-vehicle, TAC-vehicle, and TAC-JQ1 groups.
Table S3. Normalized RNA-seq expression data for sham-vehicle, MI-vehicle, and MI-JQ1 groups.
Table S4. List of 193 genes in the Venn overlap in Fig. 4A.
Table S5. Normalized RNA-seq expression data from human iPSC-CMs for unstimulated, ET-1, and ET-1 + JQ1 groups.
Table S6. List of qRT-PCR primers and TaqMan probes.
Acknowledgments
We thank B. G. Bruneau, C. Y. Lin, and K. S. Pollard for insightful discussions about this work.
Funding: This work was supported by NIH grants DK093821 (S.M.H.), HL127240 (S.M.H., J.E.B., and T.A.M.), and HL116848 (T.A.M.).
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencetranslationalmedicine.org/cgi/content/full/9/390/eaah5084/DC1
Author contributions: Q.D., S.M., J.D.B., D.S., J.E.B., T.A.M., and S.M.H. designed the research and analyzed the data. Q.D., S.M., P.A., H.S., and A.S. performed the research. Q.D., R.Z., Y.H., and H.T.S. performed animal surgeries and echocardiograms. M.E.L. and S.T. assisted with the bioinformatic analyses. J.E.B. provided the key reagents. Q.D., S.M., and S.M.H. wrote the manuscript with critical revisions from J.E.B., J.D.B., D.S., and T.A.M.
Competing interests: S.M.H. and D.S. are scientific co-founders and shareholders of Tenaya Therapeutics and serve as consultants for this entity. J.E.B. is now a shareholder and executive of Novartis Pharmaceuticals. S.M.H., J.E.B., and J.D.B. are co-inventors on patent application US20160095867A1 entitled “BET inhibition therapy for heart disease” held jointly by the Dana-Farber Cancer Institute, the Case Western Reserve University, and the Brigham and Women’s Hospital that covers methods for treating cardiac diseases using BET inhibitors. All other authors declare that they have no competing interests.
Materials and data availability: All RNA-seq data sets are available in GEO (accession no. GSE96566).
REFERENCES AND NOTES
- 1.Braunwald E. Research advances in heart failure: A compendium. Circ. Res. 2013;113:633–645. doi: 10.1161/CIRCRESAHA.113.302254. [DOI] [PubMed] [Google Scholar]
- 2.Roger VL, Go AS, Lloyd-Jones DM, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Makuc DM, Marcus GM, Marelli A, Matchar DB, Moy CS, Mozaffarian D, Mussolino ME, Nichol G, Paynter NP, Soliman EZ, Sorlie PD, Sotoodehnia N, Turan TN, Virani SS, Wong ND, Woo D, Turner MB. Executive summary: Heart disease and stroke statistics—2012 update: A report from the American Heart Association. Circulation. 2012;125:188–197. doi: 10.1161/CIR.0b013e3182456d46. [DOI] [PubMed] [Google Scholar]
- 3.Hill JA, Olson EN. Cardiac plasticity. N. Engl. J. Med. 2008;358:1370–1380. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 4.van Berlo JH, Maillet M, Molkentin JD. Signaling effectors underlying pathologic growth and remodeling of the heart. J. Clin. Invest. 2013;123:37–45. doi: 10.1172/JCI62839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanisicak O, Khalil H, Ivey MJ, Karch J, Maliken BD, Correll RN, Brody MJ, Lin S-JL, Aronow BJ, Tallquist MD, Molkentin JD. Genetic lineage tracing defines myofibroblast origin and function in the injured heart. Nat. Commun. 2016;7:12260. doi: 10.1038/ncomms12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mann DL. Innate immunity and the failing heart: The cytokine hypothesis revisited. Circ. Res. 2015;116:1254–1268. doi: 10.1161/CIRCRESAHA.116.302317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McKinsey TA, Olson EN. Toward transcriptional therapies for the failing heart: Chemical screens to modulate genes. J. Clin. Invest. 2005;115:538–546. doi: 10.1172/JCI24144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Konstam MA, Kramer DG, Patel AR, Maron MS, Udelson JE. Left ventricular remodeling in heart failure: Current concepts in clinical significance and assessment. JACC Cardiovasc. Imaging. 2011;4:98–108. doi: 10.1016/j.jcmg.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 9.St. John Sutton M, Pfeffer MA, Plappert T, Rouleau JL, Moyé LA, Dagenais GR, Lamas GA, Klein M, Sussex B, Goldman S. Quantitative two-dimensional echocardiographic measurements are major predictors of adverse cardiovascular events after acute myocardial infarction. The protective effects of captopril. Circulation. 1994;89:68–75. doi: 10.1161/01.cir.89.1.68. [DOI] [PubMed] [Google Scholar]
- 10.Levy D, Garrison RJ, Savage DD, Kannel WB, Castelli WP. Prognostic implications of echocardiographically determined left ventricular mass in the Framingham Heart Study. N. Engl. J. Med. 1990;322:1561–1566. doi: 10.1056/NEJM199005313222203. [DOI] [PubMed] [Google Scholar]
- 11.Pfeffer MA, Braunwald E, Moyé LA, Basta L, Brown EJ, Jr, Cuddy TE, Davis BR, Geltman EM, Goldman S, Flaker GC, Flaker GC, Klein M, Lamas GA, Packer M, Rouleau J, Rouleau JL, Rutherford J, Wertheimer JH, Morton Hawkins C. Effect of captopril on mortality and morbidity in patients with left ventricular dysfunction after myocardial infarction—Results of the survival and ventricular enlargement trial. N. Engl. J. Med. 1992;327:669–677. doi: 10.1056/NEJM199209033271001. [DOI] [PubMed] [Google Scholar]
- 12.Pfeffer MA, Lamas GA, Vaughan DE, Parisi AF, Braunwald E. Effect of captopril on progressive ventricular dilatation after anterior myocardial infarction. N. Engl. J. Med. 1988;319:80–86. doi: 10.1056/NEJM198807143190204. [DOI] [PubMed] [Google Scholar]
- 13.Pfeffer JM, Pfeffer MA, Braunwald E. Hemodynamic benefits and prolonged survival with long-term captopril therapy in rats with myocardial infarction and heart failure. Circulation. 1987;75:149–155. [PubMed] [Google Scholar]
- 14.Haldar SM, McKinsey TA. BET-ting on chromatin-based therapeutics for heart failure. J. Mol. Cell Cardiol. 2014;74:98–102. doi: 10.1016/j.yjmcc.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sano M, Abdellatif M, Oh H, Xie M, Bagella L, Giordano A, Michael LH, DeMayo FJ, Schneider MD. Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat. Med. 2002;8:1310–1317. doi: 10.1038/nm778. [DOI] [PubMed] [Google Scholar]
- 16.Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152:1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Salvo TG, Haldar SM. Epigenetic mechanisms in heart failure pathogenesis. Circ. Heart Fail. 2014;7:850–863. doi: 10.1161/CIRCHEARTFAILURE.114.001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dawson MA, Kouzarides T, Huntly BJP. Targeting epigenetic readers in cancer. N. Engl. J. Med. 2012;367:647–657. doi: 10.1056/NEJMra1112635. [DOI] [PubMed] [Google Scholar]
- 19.Jang MK, Mochizuki K, Zhou M, Jeong H-S, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 20.Yang Z, Yik JHN, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 21.Bhagwat AS, Roe J-S, Mok BYL, Hohmann AF, Shi J, Vakoc CR. BET bromodomain inhibition releases the mediator complex from select cis-regulatory elements. Cell Rep. 2016;15:519–530. doi: 10.1016/j.celrep.2016.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen C, Ipsaro JJ, Shi J, Milazzo JP, Wang E, Roe J-S, Suzuki Y, Pappin DJ, Joshua-Tor L, Vakoc CR. NSD3-short is an adaptor protein that couples BRD4 to the CHD8 chromatin remodeler. Mol. Cell. 2015;60:847–859. doi: 10.1016/j.molcel.2015.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, Chesi M, Schinzel AC, McKeown MR, Heffernan TP, Vakoc CR, Bergsagel PL, Ghobrial IM, Richardson PG, Young RA, Hahn WC, Anderson KC, Kung AL, Bradner JE, Mitsiades CS. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shu S, Lin CY, He HH, Witwicki RM, Tabassum DP, Roberts JM, Janiszewska M, Huh SJ, Liang Y, Ryan J, Doherty E, Mohammed H, Guo H, Stover DG, Ekram MB, Peluffo G, Brown J, D’Santos C, Krop IE, Dillon D, McKeown M, Ott C, Qi J, Ni M, Rao PK, Duarte M, Wu S-Y, Chiang C-M, Anders L, Young RA, Winer EP, Letai A, Barry WT, Carroll JS, Long HW, Brown M, Liu XS, Meyer CA, Bradner JE, Polyak K. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature. 2016;529:413–417. doi: 10.1038/nature16508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MGM, Qi J, Rahl PB, Sun HH, Yeda KT, Doench JG, Reichert E, Kung AL, Rodig SJ, Young RA, Shipp MA, Bradner JE. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von Schaper E. Roche bets on bromodomains. Nat. Biotechnol. 2016;34:361–362. doi: 10.1038/nbt0416-361. [DOI] [PubMed] [Google Scholar]
- 30.Berthon C, Raffoux E, Thomas X, Vey N, Gomez-Roca C, Yee K, Taussig DC, Rezai K, Roumier C, Herait P, Kahatt C, Quesnel B, Michallet M, Recher C, Lokiec F, Preudhomme C, Dombret H. Bromodomain inhibitor OTX015 in patients with acute leukaemia: A dose-escalation, phase 1 study. Lancet Haematol. 2016;3:e186–e195. doi: 10.1016/S2352-3026(15)00247-1. [DOI] [PubMed] [Google Scholar]
- 31.Anand P, Brown JD, Lin CY, Qi J, Zhang R, Artero PC, Alaiti MA, Bullard J, Alazem K, Margulies KB, Cappola TP, Lemieux M, Plutzky J, Bradner JE, Haldar SM. BET bromodomains mediate transcriptional pause release in heart failure. Cell. 2013;154:569–582. doi: 10.1016/j.cell.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spiltoir JI, Stratton MS, Cavasin MA, Demos-Davies K, Reid BG, Qi J, Bradner JE, McKinsey TA. BET acetyl-lysine binding proteins control pathological cardiac hypertrophy. J. Mol. Cell. Cardiol. 2013;63:175–179. doi: 10.1016/j.yjmcc.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown JD, Lin CY, Duan Q, Griffin G, Federation AJ, Paranal RM, Bair S, Newton G, Lichtman AH, Kung AL, Yang T, Wang H, Luscinskas FW, Croce KJ, Bradner JE, Plutzky J. NF-κB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol. Cell. 2014;56:219–231. doi: 10.1016/j.molcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Unsöld B, Kaul A, Sbroggiò M, Schubert C, Regitz-Zagrosek V, Brancaccio M, Damilano F, Hirsch E, Van Bilsen M, Munts C, Sipido K, Bito V, Detre E, Wagner NM, Schäfer K, Seidler T, Vogt J, Neef S, Bleckmann A, Maier LS, Balligand JL, Bouzin C, Ventura-Clapier R, Garnier A, Eschenhagen T, El-Armouche A, Knöll R, Tarone G, Hasenfuβ G. Melusin protects from cardiac rupture and improves functional remodelling after myocardial infarction. Cardiovasc. Res. 2014;101:97–107. doi: 10.1093/cvr/cvt235. [DOI] [PubMed] [Google Scholar]
- 35.Xu L, Yates CC, Lockyer P, Xie L, Bevilacqua A, He J, Lander C, Patterson C, Willis M. MMI-0100 inhibits cardiac fibrosis in myocardial infarction by direct actions on cardiomyocytes and fibroblasts via MK2 inhibition. J. Mol. Cell. Cardiol. 2014;77:86–101. doi: 10.1016/j.yjmcc.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hannenhalli S, Putt ME, Gilmore JM, Wang J, Parmacek MS, Epstein JA, Morrisey EE, Margulies KB, Cappola TP. Transcriptional genomics associates FOX transcription factors with human heart failure. Circulation. 2006;114:1269–1276. doi: 10.1161/CIRCULATIONAHA.106.632430. [DOI] [PubMed] [Google Scholar]
- 37.Krämer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uosaki H, Cahan P, Lee DI, Wang S, Miyamoto M, Fernandez L, Kass DA, Kwon C. Transcriptional landscape of cardiomyocyte maturation. Cell Rep. 2015;13:1705–1716. doi: 10.1016/j.celrep.2015.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koitabashi N, Danner T, Zaiman AL, Pinto YM, Rowell J, Mankowski J, Zhang D, Nakamura T, Takimoto E, Kass DA. Pivotal role of cardiomyocyte TGF-β signaling in the murine pathological response to sustained pressure overload. J. Clin. Invest. 2011;121:2301–2312. doi: 10.1172/JCI44824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dobaczewski M, Chen W, Frangogiannis NG. Transforming growth factor (TGF)-β signaling in cardiac remodeling. J. Mol. Cell. Cardiol. 2011;51:600–606. doi: 10.1016/j.yjmcc.2010.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matkovich SJ, Edwards JR, Grossenheider TC, de Guzman Strong C, Dorn GW., II Epigenetic coordination of embryonic heart transcription by dynamically regulated long noncoding RNAs. Proc. Natl. Acad. Sci. U.S.A. 2014;111:12264–12269. doi: 10.1073/pnas.1410622111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou H, Dickson ME, Kim MS, Bassel-Duby R, Olson EN. Akt1/protein kinase B enhances transcriptional reprogramming of fibroblasts to functional cardiomyocytes. Proc. Natl. Acad. Sci. U.S.A. 2015;112:11864–11869. doi: 10.1073/pnas.1516237112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leinonen JV, Korkus-Emanuelov A, Wolf Y, Milgrom-Hoffman M, Lichtstein D, Hoss S, Lotan C, Tzahor E, Jung S, Beeri R. Macrophage precursor cells from the left atrial appendage of the heart spontaneously reprogram into a C-kit+/CD45− stem cell-like phenotype. Int. J. Cardiol. 2016;209:296–306. doi: 10.1016/j.ijcard.2016.02.040. [DOI] [PubMed] [Google Scholar]
- 44.Boström P, Mann N, Wu J, Quintero PA, Plovie ER, Panáková D, Gupta RK, Xiao C, MacRae CA, Rosenzweig A, Spiegelman BM. C/EBPβ controls exercise-induced cardiac growth and protects against pathological cardiac remodeling. Cell. 2010;143:1072–1083. doi: 10.1016/j.cell.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maillet M, van Berlo JH, Molkentin JD. Molecular basis of physiological heart growth: Fundamental concepts and new players. Nat. Rev. Mol. Cell Biol. 2013;14:38–48. doi: 10.1038/nrm3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song HK, Hong S-E, Kim T, Kim DH. Deep RNA sequencing reveals novel cardiac transcriptomic signatures for physiological and pathological hypertrophy. PLOS ONE. 2012;7:e35552. doi: 10.1371/journal.pone.0035552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu X, Xiao J, Zhu H, Wei X, Platt C, Damilano F, Xiao C, Bezzerides V, Boström P, Che L, Zhang C, Spiegelman BM, Rosenzweig A. miR-222 is necessary for exercise-induced cardiac growth and protects against pathological cardiac remodeling. Cell Metab. 2015;21:584–595. doi: 10.1016/j.cmet.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carlson C, Koonce C, Aoyama N, Einhorn S, Fiene S, Thompson A, Swanson B, Anson B, Kattman S. Phenotypic screening with human iPS cell-derived cardiomyocytes: HTS-compatible assays for interrogating cardiac hypertrophy. J. Biomol. Screen. 2013;18:1203–1211. doi: 10.1177/1087057113500812. [DOI] [PubMed] [Google Scholar]
- 49.Travers JG, Kamal FA, Robbins J, Yutzey KE, Blaxall BC. Cardiac fibrosis: The fibroblast awakens. Circ. Res. 2016;118:1021–1040. doi: 10.1161/CIRCRESAHA.115.306565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun Y, Huang J, Song K. BET protein inhibition mitigates acute myocardial infarction damage in rats via the TLR4/TRAF6/NF-κB pathway. Exp. Ther. Med. 2015;10:2319–2324. doi: 10.3892/etm.2015.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fatkin D, Seidman CE, Seidman JG. Genetics and disease of ventricular muscle. Cold Spring Harb. Perspect. Med. 2014;4:a021063. doi: 10.1101/cshperspect.a021063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, Taylor MJ, Johns C, Chicas A, Mulloy JC, Kogan SC, Brown P, Valent P, Bradner JE, Lowe SW, Vakoc CR. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ding N, Hah N, Yu RT, Sherman MH, Benner C, Leblanc M, He M, Liddle C, Downes M, Evans RM. BRD4 is a novel therapeutic target for liver fibrosis. Proc. Natl. Acad. Sci. U.S.A. 2015;112:15713–15718. doi: 10.1073/pnas.1522163112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stratton MS, Lin CY, Anand P, Tatman PD, Ferguson BS, Wickers ST, Ambardekar AV, Sucharov CC, Bradner JE, Haldar SM, McKinsey TA. Signal-dependent recruitment of BRD4 to cardiomyocyte super-enhancers is suppressed by a MicroRNA. Cell Rep. 2016;16:1366–1378. doi: 10.1016/j.celrep.2016.06.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol. Cell. 2014;54:728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bolden JE, Tasdemir N, Dow LE, van Es JH, Wilkinson JE, Zhao Z, Clevers H, Lowe SW. Inducible in vivo silencing of Brd4 identifies potential toxicities of sustained BET protein inhibition. Cell Rep. 2014;8:1919–1929. doi: 10.1016/j.celrep.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee DU, Katavolos P, Palanisamy G, Katewa A, Sioson C, Corpuz J, Pang J, DeMent K, Choo E, Ghilardi N, Diaz D, Danilenko DM. Nonselective inhibition of the epigenetic transcriptional regulator BET induces marked lymphoid and hematopoietic toxicity in mice. Toxicol. Appl. Pharmacol. 2016;300:47–54. doi: 10.1016/j.taap.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 58.Matzuk MM, McKeown MR, Filippakopoulos P, Li Q, Ma L, Agno JE, Lemieux ME, Picaud S, Yu RN, Qi J, Knapp S, Bradner JE. Small-molecule inhibition of BRDT for male contraception. Cell. 2012;150:673–684. doi: 10.1016/j.cell.2012.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bisgrove DA, Mahmoudi T, Henklein P, Verdin E. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc. Natl. Acad. Sci. U.S.A. 2007;104:13690–13695. doi: 10.1073/pnas.0705053104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winter GE, Buckley DL, Paulk J, Roberts JM, Souza A, Dhe-Paganon S, Bradner JE. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348:1376–1381. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bellinger AM, Arteaga CL, Force T, Humphreys BD, Demetri GD, Druker BJ, Moslehi JJ. Cardio-oncology: How new targeted cancer therapies and precision medicine can inform cardiovascular discovery. Circulation. 2015;132:2248–2258. doi: 10.1161/CIRCULATIONAHA.115.010484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.National Research Council. Guide for the Care and Use of Laboratory Animals. 8. National Acadmies Press; 2011. [PubMed] [Google Scholar]
- 63.Yang F, Liu YH, Yang XP, Xu J, Kapke A, Carretero OA. Myocardial infarction and cardiac remodelling in mice. Exp. Physiol. 2002;87:547–555. doi: 10.1113/eph8702385. [DOI] [PubMed] [Google Scholar]
- 64.Haldar SM, Lu Y, Jeyaraj D, Kawanami D, Cui Y, Eapen SJ, Hao C, Li Y, Doughman Y-Q, Watanabe M, Shimizu K, Kuivaniemi H, Sadoshima J, Margulies KB, Cappola TP, Jain MK. Klf15 deficiency is a molecular link between heart failure and aortic aneurysm formation. Sci. Transl. Med. 2010;2:26ra26. doi: 10.1126/scitranslmed.3000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liao Y, Smyth GK, Shi W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 67.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Roy. Stat. Soc. Series B. 1995;57:289–300. [Google Scholar]
- 69.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 70.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Echocardiographic data from sham- and TAC-treated mice at day 53.
Fig. S2. Transcriptomic effects of chronic JQ1 administration in baseline (sham-operated) hearts.
Fig. S3. GSEA and fingerprinting analysis of differentially expressed genes in mouse LV.
Fig. S4. Additional qRT-PCR of exercise-regulated genes in mouse left ventricle.
Fig. S5. qRT-PCR and GSEA in human iPSC-CMs.
Table S1. Normalized RNA-seq expression data for sham-vehicle and sham-JQ1 groups.
Table S2. Normalized RNA-seq expression data for sham-vehicle, TAC-vehicle, and TAC-JQ1 groups.
Table S3. Normalized RNA-seq expression data for sham-vehicle, MI-vehicle, and MI-JQ1 groups.
Table S4. List of 193 genes in the Venn overlap in Fig. 4A.
Table S5. Normalized RNA-seq expression data from human iPSC-CMs for unstimulated, ET-1, and ET-1 + JQ1 groups.
Table S6. List of qRT-PCR primers and TaqMan probes.