Significance
The health benefits of ω-3 fatty acids are mediated, in part, through metabolic conversion to bioactive epoxides. Here we detail the discovery and initial characterization of naturally occurring ω-3–derived endocannabinoid epoxides that are formed via enzymatic oxidation of ω-3 endocannabinoids by cytochrome P450s. These dual functional ω-3 endocannabinoid epoxides are anti-inflammatory and vasodilatory and reciprocally modulate platelet aggregation. By virtue of their physiological properties, they are expected to play important roles in neuroinflammation and in cerebrovascular diseases such as stroke.
Keywords: cytochrome P450, epoxygenase, neuroinflammation, endocannabinoid, epoxyeicosatrienoic acids
Abstract
Clinical studies suggest that diets rich in ω-3 polyunsaturated fatty acids (PUFAs) provide beneficial anti-inflammatory effects, in part through their conversion to bioactive metabolites. Here we report on the endogenous production of a previously unknown class of ω-3 PUFA–derived lipid metabolites that originate from the crosstalk between endocannabinoid and cytochrome P450 (CYP) epoxygenase metabolic pathways. The ω-3 endocannabinoid epoxides are derived from docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA) to form epoxyeicosatetraenoic acid-ethanolamide (EEQ-EA) and epoxydocosapentaenoic acid-ethanolamide (EDP-EA), respectively. Both EEQ-EAs and EDP-EAs are endogenously present in rat brain and peripheral organs as determined via targeted lipidomics methods. These metabolites were directly produced by direct epoxygenation of the ω-3 endocannabinoids, docosahexanoyl ethanolamide (DHEA) and eicosapentaenoyl ethanolamide (EPEA) by activated BV-2 microglial cells, and by human CYP2J2. Neuroinflammation studies revealed that the terminal epoxides 17,18-EEQ-EA and 19,20-EDP-EA dose-dependently abated proinflammatory IL-6 cytokines while increasing anti-inflammatory IL-10 cytokines, in part through cannabinoid receptor-2 activation. Furthermore the ω-3 endocannabinoid epoxides 17,18-EEQ-EA and 19,20-EDP-EA exerted antiangiogenic effects in human microvascular endothelial cells (HMVEC) and vasodilatory actions on bovine coronary arteries and reciprocally regulated platelet aggregation in washed human platelets. Taken together, the ω-3 endocannabinoid epoxides’ physiological effects are mediated through both endocannabinoid and epoxyeicosanoid signaling pathways. In summary, the ω-3 endocannabinoid epoxides are found at concentrations comparable to those of other endocannabinoids and are expected to play critical roles during inflammation in vivo; thus their identification may aid in the development of therapeutics for neuroinflammatory and cerebrovascular diseases.
Epidemiological evidence suggests that a diet rich in the ω-3 fatty acids (ω-3 FAs) docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA) promotes beneficial cardiovascular (1), neurological (2), and anti-inflammatory (3) health effects. The biochemical mechanisms facilitating these beneficial effects are yet to be fully elucidated. Mounting evidence suggests that these actions are mediated through both oxidative and nonoxidative routes of metabolism that convert ω-3 FAs into bioactive lipid metabolites.
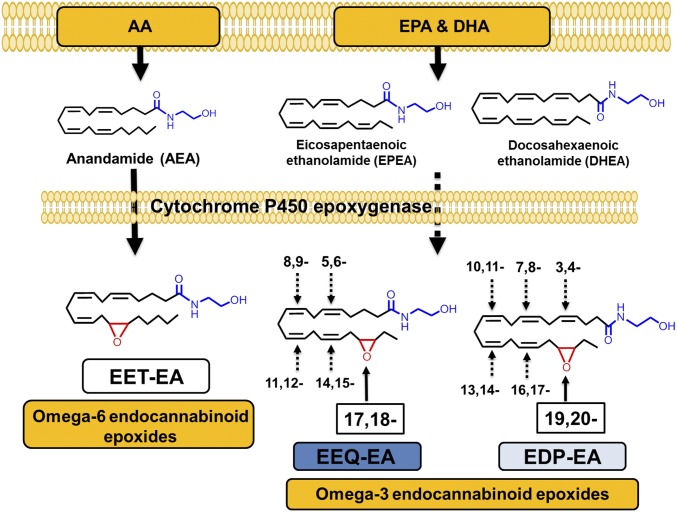
One of the nonoxidative pathways involves the conversion of the ω-3 FAs DHA and EPA into docosahexaenoyl ethanolamide (DHEA) and eicosapentaenoyl ethanolamide (EPEA) (Fig. 1) through the N-acyl ethanolamine synthesis pathway similar to the conversion of arachidonic acid (AA) into arachidonoyl ethanolamine (AEA, anandamide) (4, 5). The endocannabinoids AEA, DHEA, and EPEA exert effects similar to those of Δ9-tetrahydrocannabinol (THC), the active ingredient of Cannabis sativa. Endocannabinoids play important physiological roles that are exerted primarily through the activation of cannabinoid receptor-1 (CB1) and -2 (CB2) (6, 7). CB1 is found predominantly in the CNS, and CB2 is found in both peripheral and CNS immune cells (8).
Fig. 1.
Overview of the CYP epoxygenase-mediated metabolism of endocannabinoids. Both ω-6 and ω-3 dietary fatty acids are stored in plasma membrane and can be converted to the ω-6 and ω-3 endocannabinoids AEA, DHEA, and EPEA. AEA, EPEA, and DHEA are substrates for CYP epoxygenases. The metabolism of AEA produces EET-EA metabolites (the 14,15-EET-EA regioisomer is shown). The metabolism of EPEA and DHEA by CYP epoxygenases leads to the formation of EEQ-EA and EDP-EA, respectively. The terminal endocannabinoid epoxide regioisomer is shown, and other possible epoxides at each double bond are denoted by the numbering system.
AEA signals through both cannabinoid receptors and regulates physiological processes in both the CNS and peripheral tissues (4, 5, 9). DHEA exhibits anticancer (4, 5), anti-inflammatory (10), and synaptogenic (11) properties and has been detected in both the brain and retina at concentrations comparable to those of AEA (12–14). EPEA also has been shown to activate anti-inflammatory pathways (Fig. 1) (10). Interestingly, dietary supplementation with EPA and DHA resulted in increased levels of EPEA and DHEA at the expense of AEA, thereby providing a means for increasing the levels of ω-3 endocannabinoids through dietary consumption (15, 16).
Endocannabinoids are further metabolized by eicosanoid-synthesizing enzymes from the cyclooxygenase (COX), lipoxygenase (LOX), and cytochrome P450 epoxygenase (CYP) pathways to generate complex lipid metabolites with distinct biological functions (17). Recently it was shown that DHEA is a substrate for enzymes of the LOX and COX pathways to yield metabolites with anti-inflammatory properties (18, 19). However, studies examining EPEA or DHEA metabolism by members of the CYP epoxygenase pathway are lacking. CYP epoxygenases convert AA, DHA, and EPA into epoxides that are generally anti-inflammatory, vasodilatory, and antinociceptive in nature (20). Previously, it was shown that AEA is converted to epoxyeicosatrienoic-ethanolamide (EET-EA) by different CYP epoxygenases, including CYP2J2 (Fig. 1) (21). It was further demonstrated that one of the EET-EA regioisomers selectively binds CB2 with 1,000-fold greater affinity than CB1 (22). These results provide a mechanism by which the CYP epoxygenases metabolize endocannabinoids to produce endocannabinoid epoxides that are anti-inflammatory CB2 agonists (22).
Herein, we used targeted lipidomics methods to quantitate the endogenous levels of the ω-3 endocannabinoid epoxides epoxyeicosatetraenoic acid-ethanolamide (EEQ-EA) and epoxydocosapentaenoic acid-ethanolamide (EDP-EA) in rat tissues and human blood. The direct in vitro production of these metabolites was demonstrated in rat brain microsomes, activated BV-2 microglia cells, and recombinant human CYP2J2 (Fig. 1). Additionally, the rates of hydrolysis/inactivation of these metabolites were measured for the epoxide and endocannabinoid motifs via soluble epoxide hydrolase (sEH) and fatty acid amide hydrolase (FAAH), respectively.
In this work we also report the characterization of the bioactivity of ω-3 endocannabinoid epoxides. These bifunctional metabolites contain both ethanolamide and epoxide motifs that function through different signaling pathways with many overlapping functional outcomes. Anti-inflammatory effects are common to both motifs, and, given the presence of these metabolites in the brain, we explored their potential role in a model of neuroinflammation using microglial cells. Additionally, because these epoxides are widely distributed in the periphery and because ω-3 FA epoxides are known to be vasoactive, we studied the “epoxide-like” property of these molecules by measuring vasodilation in bovine coronary artery, angiogenesis in human microvascular endothelial cells (HMVECs), and platelet aggregation in washed human platelets.
Taking these findings together, we identify a class of bioactive lipids that we termed “ω-3 endocannabinoid epoxides,” EDP-EA and EEQ-EA. They are present endogenously and are produced through direct epoxygenation by CYP epoxygenases to yield anti-inflammatory and vasoactive lipid mediators. These metabolites provide targets for the development of therapeutics in the ongoing search for nonaddictive, analgesic, anti-inflammatory, and vasodilatory molecules.
Results
The ω-3 Endocannabinoid Epoxides Are Endogenous Lipids in the Brain and Peripheral Tissues.
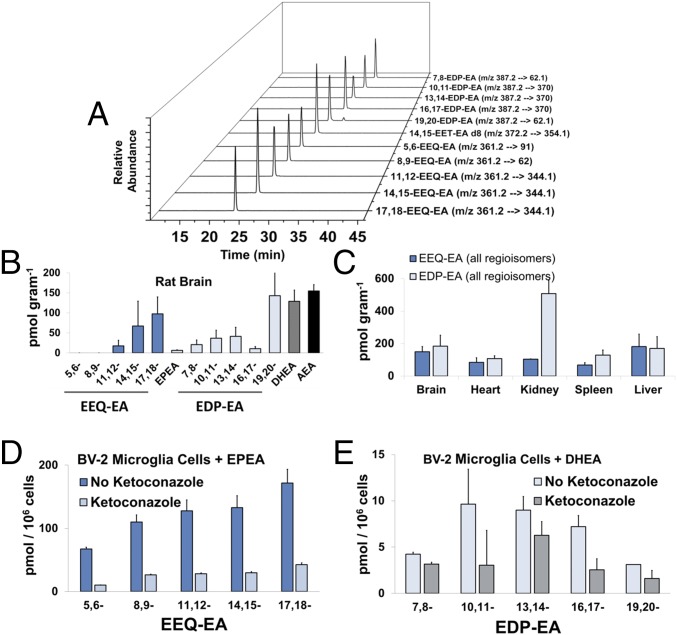
Previously, a wide range of N-acyl amide lipids have been quantified successfully using HPLC coupled with fluorescence detection (23), GC-MS (24), LC-MS (25), and Ag+ coordination LC-MS/MS in the selected reaction monitoring (SRM) mode (26, 27). As shown in Fig. 2A, a LC-MS/MS method in the multiple reaction monitoring (MRM) mode was developed to identify structurally and to quantitate DHEA epoxide (EDP-EA) regioisomers (19,20-, 16,17-, 13,14-, 10,11-, and 7,8-), EEQ-EA regioisomers (17,18-, 14,15-, 11,12-, 8,9-, and 5,6-), EPEA, DHEA, and AEA. The authentic standards of EEQ-EA and EDP-EA regioisomers were prepared in a two-step synthesis process and were purified using HPLC as reported in Experimental Procedures.
Fig. 2.
Endogenous levels of EEQ-EA and EDP-EA regioisomers in rat tissues and their production in BV-2 microglial cells. (A) Authentic standards were used for the development of a LC-MS/MS method in the separation and quantitation of EEQ-EA, EDP-EA, AEA, EPEA, and DHEA lipid mediators using MS/MS fragments and retention times unique to each lipid class. (B and C) Lipid metabolites were extracted and analyzed from pooled Sprague–Dawley rat brain (n = 3) (B) and peripheral organs (C). (D and E) The capacity of LPS-activated BV-2 microglia cells to convert EPEA directly to EEQ-EA regioisomers (D) and DHEA into EDP-EA regioisomers (E) was examined in the absence and presence of the CYP inhibitor ketoconazole (0.5 µM).
The target endocannabinoid metabolites were extracted from the tissues using a two-step extraction method followed by LC-MS/MS (27). To estimate the basal levels of the endogenous EEQ-EA and EDP-EA regioisomers, three sets of pooled (n = 3) Sprague–Dawley rat brain, spleen, heart, liver, and kidneys were extracted and analyzed. As shown in Fig. 2B, both classes of EEQ-EA and EDP-EA metabolites were measured in rat brain, with the two terminal epoxides, 17,18-EEQ-EA and 19,20-EDP-EA, quantified as the predominant metabolites. A similar trend was observed in peripheral organs, with both classes of metabolites being found in all tissues analyzed (Fig. 2C and SI Appendix, Table ST1). Specifically, the values of 17,18-EEQ-EA were relatively consistent among tissues from brain, spleen, heart, liver, and kidney, with levels varying from 60 to 90 pmol/g; the values of 19,20-EDP-EA were more variable, with levels in brain, spleen, heart, liver, and kidney measured at 70–400 pmol/g (Fig. 2C and SI Appendix, Table ST1).
Notably, the levels of DHEA extracted from tissues were comparable to the levels of 19,20-EDP-EA and AEA. Conversely, relatively low levels of the EPEA parent compound were detected, with levels significantly lower than those of 17,18-EEQ-EA. Importantly, the relatively higher DHEA levels and lower EPEA levels are consistent with reports in the literature (10), lending credibility to the quantitative values. Some of the metabolites also were detected at lower levels in porcine brains and in pooled human plasma (SI Appendix, Table ST1). To compare the levels of these metabolites with those of the better-known epoxyeicosanoids, we developed a LC-MS/MS method to monitor all nine comparable metabolites—14,15-EET, 17,18-EEQ, 19,20-EDP, 14,15-EET-EA, 17,18-EEQ-EA, 19,20-EDP-EA, AEA, DHEA, and EPEA—simultaneously. Notably, in rat brain tissues analyzed using this LC-MS/MS method the relative percentages of the epoxyeicosanoids (EETs, EEQs, and EDPs) and endocannabinoid epoxides (EET-EAs, EEQ-EAs, and EDP-EAs) were determined to be 79.2 and 20.8%, respectively. Therefore, the relative levels of the epoxyeicosanoids were approximately fourfold greater than those of the endocannabinoid epoxides. The detection of the ω-3 endocannabinoid epoxides in the different rat tissues warranted further investigation of their production in live microglial cells.
EEQ-EA and EDP-EA Are Naturally Produced by LPS-Stimulated BV-2 Microglial Cells.
Microglial cells are metabolically active brain macrophages that continuously survey the brain parenchyma and maintain homeostasis. If microglia encounter harmful endogenous ligands and microbes, they display proinflammatory activity followed by anti-inflammatory activity to promote defense and healing, respectively. Previously, it was demonstrated that activation of BV-2 microglia resulted in the up-regulation of CYPs that metabolized AEA to form EET-EA regioisomers when supplemented with AEA (22). Given the endogenous presence of EPEA and DHEA in the brain (Fig. 2B), we explored metabolite production by quiescent and activated BV-2 microglial cells in a post-LPS stimulation time course. As shown in SI Appendix, Table ST2, both EEQ-EA and EDP-EA metabolites are naturally produced at 1 h after LPS stimulation and persist throughout a 12-h period. Similarly, EPEA and AEA were observed at 4 h after LPS stimulation, and at 12 h poststimulation the level of these metabolites was more than an order of magnitude greater than that of the ω-3 endocannabinoid epoxides.
To test whether activated BV-2 microglial cells could directly convert the EPEA and DHEA parent compounds into EEQ-EA and EDP-EA metabolites, we analyzed metabolite production in cell medium supplemented with either EPEA or DHEA for 30 min. As shown in Fig. 2 D and E, EPEA and DHEA were rapidly converted to all EEQ-EA and EDP-EA regioisomers, respectively. Total levels of EEQ-EAs were approximately one order of magnitude greater than those of EDP-EAs. Importantly, the production of both the EEQ-EA and EDP-EA metabolites was reduced when cells were preincubated with the CYP inhibitor ketoconazole (0.5 µM), thus showing that the ω-3 endocannabinoid epoxides are produced enzymatically through the CYP epoxygenase pathway. It is known that inflammatory conditions increase the expression levels of CYPs and their metabolites (22). Indeed, our studies show that under inflammatory conditions the levels of all ω-3 endocannabinoid epoxide metabolites are also increased and are synthesized primarily by CYPs.
Epoxygenation of EPEA and DHEA by Rat Brain Microsomes.
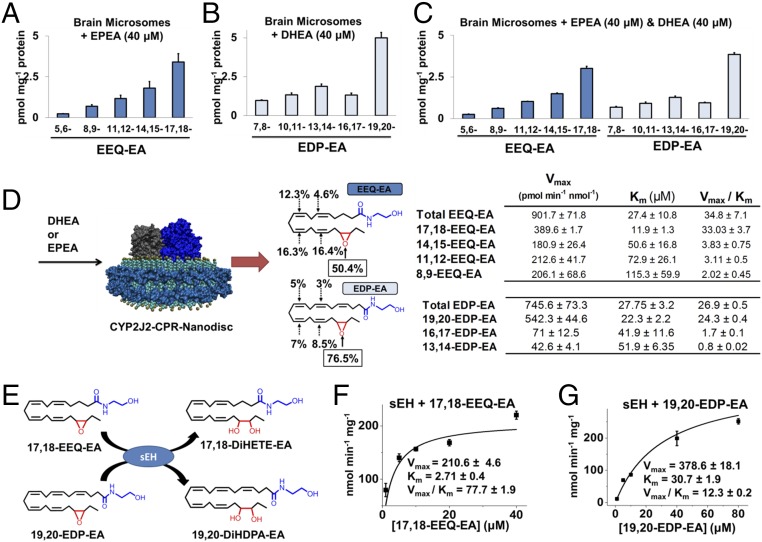
To evaluate the ability of the rat brain CYPs to epoxygenate EPEA and DHEA directly, we performed metabolism studies in which we incubated rat brain microsomes with substrate in the presence of the CYP redox partner cytochrome P450 reductase (CPR) and NADPH. As shown in Fig. 3A, when EPEA was used as the substrate, the microsomal CYP epoxygenases produced all the EEQ-EA regioisomers, with catalytic preference for the terminal olefin (17,18-EEQ-EA), which accounted for 46% of the total epoxide product. Similarly, when DHEA was used in the reactions, epoxygenases produced all the EDP-EA regioisomers, with preference for the terminal olefin (19,20-EDP-EA), which accounted for 48% of total epoxide formed (Fig. 3B).
Fig. 3.
Direct enzymatic production of EEQ-EA and EDP-EA regioisomers and hydrolysis by soluble epoxide hydrolase. Brain microsomes were incubated with substrate in the presence of CPR and NADPH to measure the capacity of endoplasmic reticulum epoxygenases to directly epoxygenate either (A) EPEA (40 μM), (B) DHEA (40 μM), or (C) EPEA (40 μM) + DHEA (40 μM). (D) CYP2J2-CPR was incorporated into nanodiscs. The membrane scaffold protein (cyan) surrounds a lipid bilayer (aquamarine with gold phospholipid head groups) in which both CYP2J2 (dark blue) and CPR (gray) are incorporated. The regioselectivity and kinetics of EPEA and DHEA metabolism by human CYP2J2-CPR nanodiscs was performed leading to the epoxidation of EPEA to EEQ-EA or DHEA to EDP-EA. The production of the total product and each specific EEQ-EA and EDP-EA regioisomers were fit to Michaelis–Menten kinetics with the calculated Km, Vmax and Vmax/Km values listed in the adjacent table. (E) Schematic of soluble epoxide hydrolase (sEH) hydrolyzing 17,18-EEQ-EA and 19,20-EDP-EA. (F–G) Incubations containing sEH ([E] final = 6 nM) in sodium phosphate (100 mM, pH 7.4), bovine serum albumin (0.1 mg/ml) and (F) 17,18-EEQ-EA or (G) 19,20-EDP-EA were performed at 37 °C to measure conversion of epoxides to the corresponding vicinal diols using LC-MS/MS. Enzyme kinetics were estimated using incremental increases of substrate for the generation of kinetic curves that were fitted to the Michaelis–Menten equation for calculation of Vmax and Km. Incubations were performed in triplicate or greater and kinetic parameters were calculated using Origin Pro.
In an effort to understand the substrate specificity and regioselectivity when both substrates are present, we performed rat brain microsomal incubations in the presence of equimolar amounts of EPEA and DHEA (Fig. 3C). Notably, there was some evidence of competition between EPEA and DHEA, because the total EEQ-EA and EDP-EA products were reduced in this reaction. Because the rat CYP epoxygenases are enzymatically different from human CYP epoxygenases, we also demonstrated a similar metabolism profile using porcine brain microsomes (SI Appendix, Fig. S1), which contain a CYP2J isozyme with homology similar to human CYP2J2 (28). Taken together, these findings suggest that similar CYP epoxygenases are implicated in the metabolism of both DHEA and EPEA and that there is competition for the substrates among the various CYP isoforms in rat brain microsomes. Microsomes contain various CYP epoxygenase enzymes; therefore it was important to identify which isoform is responsible for the formation of these metabolites in humans.
Recombinant Human CYP2J2 Incorporated into Nanodiscs Directly Epoxygenates EPEA and DHEA to Form EEQ-EA and EDP-EA Metabolites.
The identification of a human CYP that forms the ω-3 endocannabinoid metabolites was important for the direct translational relevance of our findings to humans. Human CYP2J2 epoxygenates AEA and 2-arachidonoylglycerol (2-AG) (28) and is physiologically relevant, because it is the second most highly expressed P450 in the human brain (29) and is the most highly expressed P450 in human cardiomyocytes (30). However, the cross-species studies are not completely translatable, because BV-2 microglial cells are derived from mice, which have 10 subfamily CYP2J isozymes (CYP2J-5, -6, -7, -8, -9, -11, -12, -13, -14, -15) but lack a comparable CYP2J2 isozyme, making comparative species studies more difficult (31). Importantly, we show that coincubation of BV-2 microglia with a broad inhibitor of CYP enzymes (ketoconazole) significantly reduces the levels of the EEQ-EA and EDP-EA regioisomers (Fig. 2 D and E). These data show that, despite the differences in the CYP isoforms in different animals (mouse vs. rat vs. human), the production of the ω-3 endocannabinoid epoxides is driven by the CYP enzymes. In humans, the epoxygenase pathway is mediated predominantly by CYP2C8, CYP2C9, and CYP2J2. Notably, these isozymes were previously shown to convert EPA and DHA into EEQ and EDP monoepoxides, respectively (32).
CYP2J2 was recombinantly expressed and was incorporated with its redox partner into nanodiscs (28). The first indication that the CYP2J2-CPR nanodiscs could use EPEA and DHEA as substrates was found using LC-electrospray ionization (ESI)-MS (SI Appendix, Fig. S2). Using LC-MS/MS for quantitation, we generated kinetic curves for both EPEA and DHEA metabolism. As shown in Fig. 3D, CYP2J2 nanodisc metabolism of EPEA into EEQ-EA regioisomers followed Michaelis–Menten kinetics (Vmax, Km, and Vmax/Km values are listed in the table in Fig. 3D, Upper Right). Notably, CYP2J2 demonstrated a preference for the EPEA (80 µM) terminal olefin (the average regioselectivity of the different regioisomers is reported in Fig. 3D, Upper Middle). CYP2J2 nanodisc metabolism of DHEA produced EDP-EA regioisomers following Michaelis–Menten kinetics (Vmax, Km, and Vmax/Km are listed in the adjacent table, Fig. 3D, Lower Right). The CYP2J2 enzyme demonstrated a dramatic preference for the DHEA terminal olefin; the average regioselectivity for each regioisomer is reported in Fig. 3D, Lower Middle. Therefore, we demonstrate that CYP2J2 metabolizes EPEA and DHEA with substantially higher efficiency than the rates reported for EPA or DHA (33). Because the terminal epoxides 17,18-EEQ-EA and 19,20-EDP-EA were produced predominantly by the CYP enzymes, their further characterization and biological activity was measured using these epoxides.
Hydrolysis of 17,18-EEQ-EA and 19,20-EDP-EA by sEH and FAAH.
The EEQ-EA and EDP-EA metabolites contain both epoxide and ethanolamide motifs (Fig. 1). Therefore, we evaluated whether these metabolites are targets for both sEH and FAAH. Bioactive fatty acid epoxides are short-lived lipid mediators because they are rapidly inactivated by sEH in vivo. Previous studies have shown that EEQ and EDP are hydrolyzed to their less active di-hydroxy metabolites by sEH (34). To determine if sEH hydrolyzes 17,18-EEQ-EA and 19,20-EDP-EA into their corresponding vicinal diols, 17,18-DiHETE-EA and 19,20-DiHDPA-EA (Fig. 3E), we developed a LC-MS/MS method of quantitating the diols. Incubations containing 17,18-EEQ-EA or 19,20-EDP-EA with sEH were performed. Enzyme kinetics were estimated and fitted to the Michaelis–Menten equation. As shown in Fig. 3F, 17,18-EEQ-EA was hydrolyzed rapidly with a Vmax/Km value of 77.7 ± 1.9 nmol⋅min−1⋅mg−1 sEH. Interestingly, the metabolism of 19,20-EDP-EA was substantially slower, with a Vmax/Km value of 12.3 ± 0.2 nmol⋅min−1⋅ mg−1 sEH (Fig. 3G).
In separate studies, 17,18-EEQ-EA hydrolysis to 17,18-EEQ by rat forebrains containing FAAH was approximately threefold faster than the conversion of 19,20-EDP-EA to 19,20-EDP (SI Appendix, Fig. S3). Taken together, these results show that both 17,18-EEQ-EA and 19,20-EDP-EA are hydrolyzed efficiently by human sEH and FAAH with the 17,18-EEQ-EA metabolite preferred over 19,20-EDP-EA.
Anti-Inflammatory Signaling Actions of 17,18-EEQ-EA and 19,20-EDP-EA.
One of the striking hallmarks of ongoing inflammation in neurodegenerative diseases is chronic microglial activation (35). There is a strong interest in unearthing lipid metabolites that can reduce microglial activation and thereby combat neuroinflammation. Several studies indicate that the endocannabinoids reduce microglial-promoted neuroinflammation through CB2 activation. Consequently, there is a strong interest in the discovery of endocannabinoid derivatives that are CB2-preferring/-selective agonists as means of mitigating inflammatory pathologies (8, 36, 37). The 17,18-EEQ-EA and 19,20-EDP-EA metabolites have N-acyl ethanolamide endocannabinoid functional motifs that also are produced endogenously by activated BV-2 microglia cells (SI Appendix, Table ST2). Moreover the expression of microglial CYPs increases under inflammatory conditions (22) and facilitates the formation of PUFA epoxides that also are involved in anti-inflammatory action in the brain (38). Hence we examined the anti-inflammatory effects of these molecules using LPS-stimulated microglial cells.
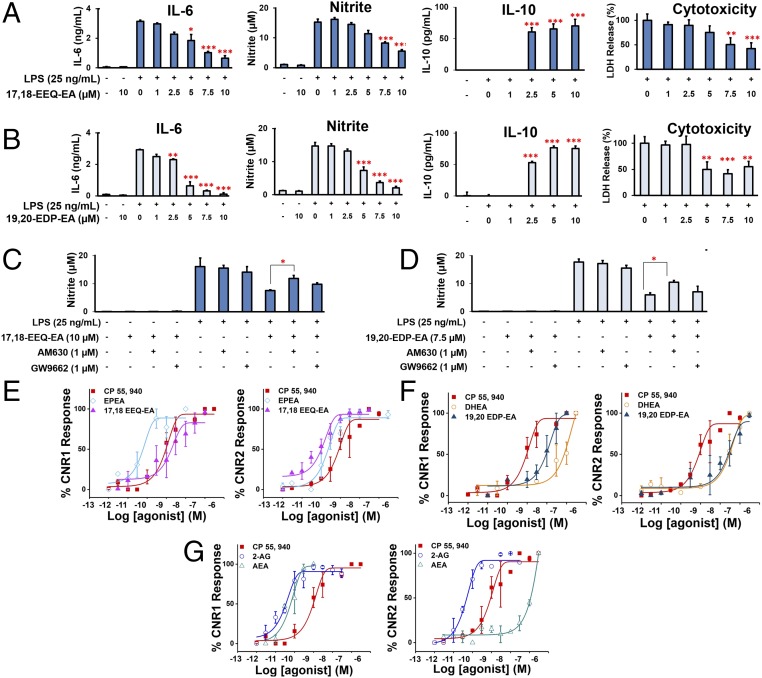
As shown in Fig. 4A, 17,18-EEQ-EA dose-dependently decreased LPS-induced production of IL-6 and nitrite while decreasing LPS-induced cytotoxicity in BV-2 cells, further demonstrating nullification of the proinflammatory effects (39). We also observed significant production of the anti-inflammatory cytokine IL-10 (Fig. 4A). More potent dose-dependent inhibition was measured when 19,20-EDP-EA was used, as evidenced by significant inhibition of IL-6 and nitrite production at concentrations ≥2.5 µM (Fig. 4B). Moreover, the LPS-induced cytotoxicity was significantly decreased at doses ≥5 µM. The 19,20-EDP-EA metabolite also promoted the production of IL-10 in the presence of LPS. Additionally, in BV-2 microglia, 17,18-EEQ-EA and 19,20-EDP-EA were more potent inhibitors of nitrite production than 17,18-EEQ and 19,20-EDP (SI Appendix, Fig. S10). As shown in SI Appendix, Fig. S11, the EPEA parent compound was also found to have some anti-inflammatory effects that were partially reversible when the CYP2J, CYP2C, and CYP2D subfamilies were inhibited by the addition of danazol (40). However, danazol did not reverse DHEA anti-inflammatory effects, possibly because it is a substrate for enzymes of the COX and LOX pathways that also produce anti-inflammatory metabolites (18, 19).
Fig. 4.
Effects of 17,18-EEQ-EA and 19,20-EDP-EA on LPS-stimulated BV-2 microglial cells and signaling properties. (A and B) In dose-response studies, BV-2 microglial cells were pretreated with 17,18-EEQ-EA (A) or 19,20-EDP-EA (B) for 4 h followed by LPS (25 ng/mL) stimulation. The culture medium was collected after 24 h and was analyzed for the proinflammatory cytokines IL-6 and NO and the anti-inflammatory cytokine IL-10. LDH production was measured to assess cell toxicity in LPS in stimulated BV-2 microglia (n = 6). (C and D) The potential targets of 17,18-EEQ-EA (C) and 19,20-EDP-EA (D) were studied using AM630 (a CB2-specific inhibitor) and GW9662 (a PPARγ-specific inhibitor) to gauge the reversal of the anti-inflammatory effects by monitoring nitrite production (n = 6). (E–G) Dose–response curves were generated by monitoring the relative luminescence of cannabinoid receptor 1 (CNR1) and cannabinoid receptor 2 (CNR2) PRESTO-Tango gene-transfected HTLA cells as described in SI Appendix, SI Materials and Methods for 17,18-EEQ-EA, EPEA, and CP 55940 (E), 19,20-EDP-EA, DHEA, and CP 55940 (F), and AEA, 2-AG, and CP 55940 (G). Values shown are the mean ± SEM of experiments performed multiple times (n = 3–7). *P < 0.05, **P < 0.01, and ***P < 0.001.
Both epoxyeicosanoids and endocannabinoids function through multiple receptors and confer complex anti-inflammatory actions (9, 41). Therefore we performed mechanistic studies with selected inhibitors to delineate the mechanism of the anti-inflammatory action (Fig. 4 C and D and SI Appendix, Fig. S4). Using a CB2-selective antagonist (AM630) or a PPARγ-selective antagonist (GW9662), we consistently observed a partial reversal of the anti-inflammatory effects of both metabolites, indicating that CB2 and PPARγ are involved in mediating the anti-inflammatory action of 17,18-EEQ-EA (Fig. 4C) and 19,20-EDP-EA (Fig. 4D). Lastly, the anti-inflammatory effects of the compounds were confirmed at 5 µM using freshly isolated piglet microglia cells stimulated with LPS, which diminished IL-6 production and thus validated the anti-inflammatory actions of these metabolites in primary microglial cells (SI Appendix, Fig. S5).
Determination of the Activation of CB1 and CB2 by AEA, 2-AG, DHEA, EPEA, 17,18-EEQ-EA, and 19,20-EDP-EA Using the β-Arrestin Recruitment PRESTO-Tango Assay.
Previously it was shown that CYP-mediated epoxidation of both AEA and 2-AG produced metabolites with increased CB2 binding and activation (22, 26). Additionally, as shown in Fig. 4 C and D, we determined that the anti-inflammatory action of the ω-3 endocannabinoid epoxides is mediated partly by CB2. In the literature, EPEA and DHEA are reported to be weak agonists of the cannabinoid receptors (10). To explore if oxygenation of EPEA or DHEA by CYPs led to the formation of metabolites with altered cannabinoid receptor activity, we performed receptor functional assays using a β-arrestin recruitment PRESTO-Tango assay (42).
We measured the functional activation of CB1 and CB2 for EDP-EA, EEQ-EA, DHEA, and EPEA at 10 µM and for AEA and 2-AG at 5 µM using the β-arrestin recruitment PRESTO-Tango assay (SI Appendix, Fig. S12). As shown in this figure, the DHEA parent compound was a weak agonist of CB2 with little activation of CB1. However, the DHEA epoxides 19,20-EDP-EA and 16,17-EDP-EA were more potent functional activators of both cannabinoid receptors with a greater preference for CB2. Similarly, the EPEA parent compound was a weak agonist of CB1 with negligible activation of CB2, whereas the EPEA epoxides 17,18-EEQ-EA and 14,15-EEQ-EA exhibited significant CB2 activation. Additionally, to gauge the potency at which these compounds act on CB1 and CB2, we performed functional dose–response studies for AEA, 2-AG (Fig. 4G), DHEA, 19,20-EDP-EA (Fig. 4F), EPEA, and 17,18-EEQ-EA (Fig. 4E) and calculated the EC50 values (SI Appendix, Table ST3). The EC50 value of CP 55940 was similar to those reported in the literature (43). 2-AG activates both receptors with similar potency, whereas AEA activates CB1 with much higher potency than CB2. In the functional dose–response studies, 19,20-EDP-EA binds with a slight preference for CB2, and DHEA also shows a preference for CB2 with very weak activation of CB1. In our studies EPEA and 17,18-EEQ-EA activate both CB1 and CB2 with high potency. Qualitatively, these findings support the hypothesis that the CYP-mediated epoxygenation of ω-6 and ω-3 endocannabinoids alters cannabinoid receptor activation, generally producing CB2-preferring metabolites similar to AEA and 2-AG (22, 26).
The Endocannabinoid Epoxides 17,18-EEQ-EA and 19,20-EDP-EA Are Regulators of Platelet Function.
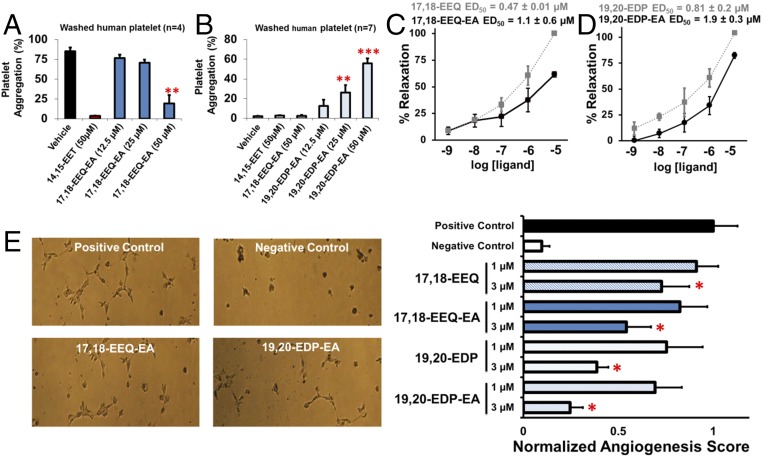
The CYP-mediated epoxygenation of AA produces EETs that are endothelial-derived hyperpolarizing factors (EDHF) that inhibit platelet aggregation and adhesion to endothelial cells (44). Correspondingly, 17,18-EEQ and 19,20-EDP metabolites also inhibit platelet aggregation with IC50 values comparable to those of 14,15-EET (45). Thus we hypothesized that the EEQ-EA and EDP-EA metabolites modulate platelet function. Using washed human platelets, we show that 17,18-EEQ-EA inhibits AA-induced platelet aggregation in a dose-dependent manner (Fig. 5A) (46). These inhibitory effects were not observed in thrombin-, PAR4-AP–, collagen-, or U46619-induced platelet aggregation (SI Appendix, Fig. S6). Additionally, no significant inhibition of platelet aggregation in platelet-rich plasma (PRP) was observed when ADP, collagen, or ristocetin was used to induce aggregation (SI Appendix, Fig. S7). Thus, the anti-platelet aggregatory properties of 17,18-EEQ-EA appear to inhibit the AA-induced platelet aggregation specifically.
Fig. 5.
Biological effects of 17,18-EEQ-EA and 19,20-EDP-EA on platelet aggregation, vasculature tension, and angiogenesis. The physiological functions of 17,18-EEQ-EA and 19.20-EDP-EA regioisomers were examined to characterize their effects on platelet aggregation, vasodilation, and angiogenesis. (A and B) In whole human blood, 17,18-EEQ-EA dose-dependently inhibited AA-induced platelet aggregation (n = 4) (A), whereas 19,20-EDP-EA induced platelet aggregation (B) under stirring conditions in the absence of a platelet agonist (n = 7). (C and D) Dose-dependent relaxation of bovine coronary arteries preconstricted with U-46619 (40 nM) was measured for 17,18-EEQ-EA and 17,18-EEQ (C) and for 19,20-EDP and 19,20-EDP-EA (D) to calculate the ED50. (E) 17,18-EEQ-EA and 19,20-EDP-EA were assessed for their ability to inhibit VEGF-promoted angiogenesis in HMVECs plated on Matrigel (n =7). Compounds were studied in parallel with each of their epoxide and ethanolamide parent compounds. All values are means ± SE; *P < 0.05, **P < 0.01, and ***P < 0.001.
Conversely, 19,20-EDP-EA did not inhibit AA-induced platelet aggregation. Rather a dose-dependent increase in aggregation under stirring conditions, in the absence of any agonist, was observed consistently (Fig. 5B). Because the 19,20-EDP metabolite mediates antiplatelet aggregatory properties, there was a deviation in the behavior of 19,20-EDP-EA, suggesting that the addition of the ethanolamide motif to 19,20-EDP is responsible for the observed proaggregatory effects.
17,18-EEQ-EA and 19,20-EDP-EA Mediate Coronary Artery Vasodilation.
CYP epoxygenases convert the ω-3 fatty acids EPA and DHA into their corresponding epoxides that are vasodilatory in nature (34, 47). These epoxide metabolites are thought to be largely responsible for the observed antihypertensive effects of ω-3 fatty acids (48). Therefore, we compared the vasodilatory effects of 17,18-EEQ-EA and 19,20-EDP-EA with those of the 17,18-EEQ and 19,20-EDP parent compounds by using isometric tension measurements to monitor their ability to relax bovine coronary arteries preconstricted with U-44619. As shown in Fig. 5 C and D, both 17,18-EEQ-EA and 19,20-EDP-EA dose-dependently relaxed constricted bovine coronary arteries. However, the ED50 values were approximately twofold greater than those of 17,18-EEQ and 19,20-EDP. Thus, the endocannabinoid epoxides were vasodilatory in nature, albeit with reduced potency relative to their epoxide parent compounds.
17,18-EEQ-EA and 19,20-EDP-EA Inhibit Angiogenesis in Dermal HMVECs.
Angiogenesis plays many roles both in normal homeostatic functions and in many pathological disease states. Accumulating evidence suggests that consumption of ω-3 fatty acids inhibits angiogenesis in cancer (47). Similarly, endocannabinoids are recognized for their ability to inhibit cancer, in part through their antiangiogenic properties (49, 50). To determine if the EEQ-EAs and EDP-EAs demonstrate a greater, similar, or diminished ability to inhibit angiogenesis, we performed a Matrigel tubulogenesis assay using HMVECs and proangiogenic VEGF, as previously described (51). As shown in Fig. 5E, 17,18-EEQ-EA at 1 and 3 µM dose-dependently inhibited VEGF-mediated tubulogenesis with a decrease of tubulogenesis surface area of 18 and 46%, respectively. Parallel studies were used for 17,18-EEQ, which produced similar, albeit slightly less potent, results with average decreases of 9 and 28% at doses of 1 and 3 µM, respectively. Identical conditions were used to screen 19,20-EDP-EA at 1 and 3 µM; tubulogenesis surface area was decreased by 31 and 75%, respectively. Following a similar but less potent trend, 19,20-EDP at 1 and 3 µM decreased tubulogenesis by 25 and 61%, respectively. Previously, the antiangiogenic actions of the EDP metabolites were shown to be mediated through potent inhibition of VEGF-stimulated cell migration via an as yet unidentified ω-3 epoxyeicosanoid pathway (20).
Discussion
Dysregulated inflammation is a common feature in most human diseases, including several neurodegenerative and cerebrovascular diseases. Lipid mediators play a central role in the regulation of inflammation (52). Therefore, the identification of anti-inflammatory lipid mediators with unique signaling properties is critical for the development of new therapeutics targeting inflammatory disease.
In particular, lipid mediators from the epoxyeicosanoid and endocannabinoid pathways are attractive targets for therapeutic intervention. Epoxyeicosanoid EETs are generated from AA by CYP epoxygenases (41) and have recently been demonstrated to exert potent anti-inflammatory effects in vitro and in vivo (53). Notably, stable EET analogs have been developed for the treatment of cardiovascular disease and inflammation via interaction with the putative EET receptor (54). Endocannabinoids also have been shown to contain anti-inflammatory and anticancer properties. Drugs targeting the endocannabinoid-degrading enzymes such as FAAH are in clinical development to prolong endocannabinoid half-life for the treatment of a wide range of diseases (55). In this study, we report the discovery of anti-inflammatory and vasoactive ω-3 endocannabinoid epoxides that are produced by CYP epoxygenases. These molecules share structural similarity with both their endocannabinoid and epoxide parent compounds and have the ability to exert physiological effects through cross-talk between the endocannabinoid and epoxide signaling pathways.
Studies to establish the biosynthetic routes revealed that the ω-3 endocannabinoids EPEA and DHEA are converted by CYP epoxygenases to generate a class of bioactive lipids, EEQ-EAs and EDP-EAs. In the rat brain, both classes of these mediators were found at levels comparable to those of similar related compounds such as 14,15-EET, AEA, and DHEA. Additionally, cell-culture studies established that both EPEA and DHEA are direct substrates for CYP enzymes in LPS-activated BV-2 microglia, because the epoxygenation was diminished through inhibition of the CYP2, CYP3, and CYP4 families (Fig. 2 D and E) when activated microglia cells were preincubated with ketoconazole (0.5 µM).
To translate these results to the biosynthetic pathway in humans, we studied the metabolism of DHEA and EPEA with CYP2J2 (the major epoxygenase in human brain and heart) and found that EPEA and DHEA are epoxygenated at comparable rates, albeit with a different regioselectivity (Fig. 3D). The total turnover rates of AEA, EPEA, and DHEA are significantly greater than those of AA, EPA, and DHA (28, 32, 56). Specifically, maximal CYP2J2 conversion of DHA to 19,20-EDP was calculated at 22.5 pmol⋅min−1⋅pmol−1 protein (57), whereas the CYP2J2 conversion of DHEA to 19,20-EDP-EA was 542.3 ± 44.6 5 pmol⋅min−1⋅pmol−1 protein, thus demonstrating that the preference for the DHEA substrate is an order of magnitude greater than the preference for DHA. This finding implies that DHEA and EPEA are likely the preferred substrates of CYP2J2 under inflammatory conditions, when CYP and substrate levels both are increased.
Both the epoxides and endocannabinoids are known to exert anti-inflammatory effects in a wide range of tissues, including the brain (8, 38). The spontaneous production of EEQ-EA and EDP-EA regioisomers by LPS-stimulated microglial cells suggested that these metabolites have a role in inflammation (SI Appendix, Table ST2). We show here (Fig. 4 A and B) that both 17,18-EEQ-EA and 19,20-EDP-EA dose-dependently inhibit the production of two prototypic proinflammatory biomarkers, IL-6 and NO, while increasing the production of the anti-inflammatory cytokine IL-10. The change in the cytokine expression levels indicates that these metabolites likely switch microglia states from the proinflammatory phenotype to the repair anti-inflammatory phenotype. Importantly, the direct comparison of the anti-inflammatory properties of EDP-EA and EEQ-EA with those of their respective EEQ and EDP parent compounds demonstrated that the ω-3 endocannabinoid epoxides have enhanced anti-inflammatory properties, most likely because of their interaction with CB2 and with unknown epoxide receptors (Fig. 4E and SI Appendix, Fig. S12). This notion is consistent with recent studies demonstrating that both CB2 ligands and EETs promote the anti-inflammatory macrophage phenotype (58, 59). In this study, the anti-inflammatory effects of the ω-3 endocannabinoid epoxides were partially blocked by AM630 (a CB2 inhibitor), GW99662 (a PPARγ inhibitor) (Fig. 4 C and D), and pertussis toxin (PTX) (SI Appendix, Fig. S4 C and D), suggesting the activation of multiple anti-inflammatory pathways.
Because CB2 is involved in the anti-inflammatory activity of the endocannabinoid epoxides, we used the PRESTO-Tango β-arrestin recruitment assay to demonstrate that the epoxygenation of EPEA and DHEA converts weak CB1/CB2 agonists to partial CB2 agonists (SI Appendix, Fig. S12). Interestingly, 19,20- and 16,17-EDP-EA were shown to be preferential CB2 agonists with CB2:CB1 activation ratios of three- and fourfold, respectively. Additionally, to gauge the potency at which these compounds act on CB1 and CB2, full functional dose-response studies were performed for AEA, 2-AG (Fig. 4G), DHEA, 19,20-EDP-EA (Fig. 4F), EPEA, and 17,18-EEQ-EA (Fig. 4E). The calculated EC50 values demonstrated that the CYP-mediated epoxygenation of ω-6 and ω-3 endocannabinoids changes CB receptor activation, generally producing CB2-preferring metabolites. It is important to note that EPEA and DHEA are converted rapidly to other anti-inflammatory CB2-preferring metabolites in addition to epoxides that can interfere with the CB receptor-activation studies (19). Collectively, these separate studies establish that the CYP-mediated epoxygenation of endocannabinoids such as AEA, 2-AG (22, 26), DHEA, and EPEA produces epoxide metabolites that preferentially target CB2, thereby regulating inflammatory pathologies.
EETs play an important role in neuro-glio–vascular coupling (38). Similar to EETs, EEQs and EDPs are involved in vasodilation and in the inhibition of platelet aggregation; unlike EETs, they inhibit angiogenesis. Thus, we focused further on the actions of 17,18-EEQ-EA and 19,20-EDP-EA in the vasculature. The inhibition of platelet aggregation by 17,18-EEQ-EA (Fig. 5A) is consistent with what is known about the 17,18-EEQ parent compound and other related epoxides. Specifically, EETs, EEQs, and EDPs inhibit platelet aggregation and reduce thrombocyte adhesion (60). Surprisingly, in our studies 19,20-EDP-EA promoted platelet clotting under stirring conditions (Fig. 5B). Because 19,20-EDP, 19,20-DHETE, and DHA are antiaggregatory mediators, our findings reveal a potentially important structure–activity relationship among these lipids. Moreover, our findings afford evidence of a proplatelet aggregatory mechanism of an ω-3–derived metabolite; this mechanism should be taken into consideration when weighing the potential benefits of DHA supplementation.
The physiological relevance of the ω-3 endocannabinoid epoxides is gleaned from the finding that they are bifunctional and are produced by the cross-talk of two important anti-inflammatory pathways—the CYP epoxygenase and endocannabinoid pathways. Although these metabolites exhibit the classical epoxyeicosanoid and endocannabinoid activities, their bifunctional nature imparts the ability to affect multiple signaling pathways (CB2 and PPARγ) to resolve inflammation with higher potency than their parent lipid metabolites (SI Appendix, Figs. S10 and S11). Thus, our results suggest they play an important role in inflammation within the larger context of the epoxyeicosanoid and endocannabinoid lipidomes. Importantly, they are produced at a faster rate by the CYP enzymes and are found in concentrations comparable to those of related lipid mediators. In summary, our findings represent an initial step in evaluating CYP epoxygenase-derived ω-3 endocannabinoid epoxides that, by virtue of their biological properties, can serve as potential therapeutic targets for diseases such as neuroinflammation and cerebrovascular disorders.
Experimental Procedures
Additional details of materials and experimental protocols used are given in SI Appendix, Materials and Methods.
Chemical Synthesis of EEQ-EA and EDP-EA Regioisomers.
EEQ-EA and EDP-EA chemical synthesis was performed using a two-step synthesis. For nonspecific epoxygenation reactions, 10 mg of either EPA or DHA was dissolved in 2 mL of dichloromethane (DCM) with 2 molar equivalents of meta-chloroperoxybenzoic acid (mCPBA) and was reacted for 1 h at room temperature. The reaction was stopped with an equal volume of 10% aqueous NaHCO3 to remove mCPBA from the organic layer, and the aqueous layer was reextracted with equal (3×) volumes of DCM and was dried under vacuum. Purification was achieved using normal-phase HPLC (NP-HPLC) using a Zorbax-NH2 5 µm × 9.4 mm × 250 mm semipreparative column (Agilent, PN880952-208) with an isocratic gradient (hexane/isopropanol/acetic acid; 90:10:0.1) coupled to a HPLC system (SI Appendix, Fig. S8 A and B). For coeluting regioisomers of DHA epoxides (19,20-EDP and 13,14-EDP) and EPA epoxides (17,18-EEQ and 11,12-EEQ), the mixtures were further purified on the same system using reversed-phase HPLC (RP-HPLC), a Sun Fire Prep C18 5 µm × 19 mm × 50 mm column (PN 186002566; Waters), and a mobile system composed of solvent A (H2O/acetonitrile/acetic acid, 95:5:0.1), solvent B (H2O/acetonitrile/acetic acid, 5:95:0.1), and a linear gradient from 50% A to 0% A in 50 min (SI Appendix, Fig. S8 C and D) show similar profile. The synthesis of 17,18-EEQ, 14,15-EEQ, 11,12-EEQ, 8,9-EEQ, 5,6-EEQ, 19,20-EDP, and 16,17-EDP was confirmed by comparing retention times relative to authentic standards purchased from Cayman Chemical and high-resolution mass spectrometry.
In the second step, the coupling of each respective EEQ or EDP regioisomer to ethanolamine was achieved by dissolving the epoxide/1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC)/N-hydroxysuccinimide (NHS)/ethanolamine in acetonitrile in a 1:10:10:10 ratio with rocking at room temperature for 16 h. Last, the resulting EEQ-EA and EDP-EA regioisomers were purified by RP-HPLC using a Sun Fire Prep C18 5 μm × 19 mm × 50 mm column (PN 186002566; Waters). Purity and retention times were assessed using bioanalytical RP-HPLC, and successful synthesis was confirmed by high-resolution mass spectroscopy and proton NMR (SI Appendix, Fig. S9 A and B).
Expression and Purification of CPR.
CPR from Rattus norvegicus was expressed and purified as previously described (61).
LC-MS/MS Analysis.
Samples were analyzed with the 5500 QTRAP LC/MS/MS system (AB Sciex) in the Metabolomics Laboratory of the Roy J. Carver Biotechnology Center, University of Illinois at Urbana–Champaign. Analyst 1.6.2 software (AB Sciex) was used for data acquisition and analysis. The 1200 series HPLC system (Agilent Technologies) includes a degasser, an autosampler, and a binary pump. The LC separation was performed on an Agilent Eclipse XDB-C18 (4.6 mm × 150 mm × 5 μm) with mobile phase A (0.1% formic acid in water) and mobile phase B (0.1% formic acid in acetonitrile). The flow rate was 0.4 mL/min. The linear gradient was as follows: 0–2 min, 90% A; 8 min, 55% A; 13–25 min, 40% A; 30 min, 30% A; 35 min, 25% A; 40–47 min, 20% A; 47.5–54 min, 90% A. The autosampler was set at 5 °C. The injection volume was 10 μL. Positive mass spectra were acquired with the ion spray voltage of 5,500 V under ESI. The source temperature was 450 °C. The curtain gas, ion source gas 1, and ion source gas 2 were 32, 60, and 60 psi, respectively. MRM was used for quantitation: 19,20-EDP-EA and 7,8-EDP-EA, m/z 388.1 → m/z 62.1; 16,17-EDP-EA, 13,14-EDP-EA, and 10,11-EDP-EA, m/z 388.1 → m/z 370.1; 17/18-EEQ-EA, 14/15-EEQ-EA, and 11/12-EEQ-EA, m/z 362.1 → m/z 344.1; 8/9-EEQ-EA, m/z 362.1 → m/z 62.0; 5/6-EEQ-EA, m/z 362.1 → m/z 91.0; EPEA, 346.3 → m/z 62.1; DHEA, 372.4 → m/z 62.1; and anandamide, m/z 348.3 → m/z 62.1. Internal standards were monitored at m/z 350.3 → m/z 66.1 for EPEA-d4, at m/z 376.4 → m/z 66.1 for DHEA-d4, at m/z 352.3 → m/z 66.1 for anadamide-d4, and at m/z 372.1 → m/z 354.1 for 14/15-EET-EA-d8.
LC-MS/MS Method for Quantitation of 17,18-DiHETE-EA and 19,20-DiHDPA-EA.
Samples were analyzed with the 5500 QTRAP LC/MS/MS system (AB Sciex) in the Metabolomics Laboratory of the Roy J. Carver Biotechnology Center, University of Illinois at Urbana–Champaign. Analyst 1.6.2 software was used for data acquisition and analysis. The 1200 series HPLC system (Agilent Technologies) includes a degasser, an autosampler, and a binary pump. The LC separation was performed on an Agilent Eclipse XDB-C18 (4.6 mm × 150 mm × 5 μm) with mobile phase A (0.1% formic acid in water) and mobile phase B (0.1% formic acid in acetonitrile). The flow rate was 0.4 mL/min. The linear gradient was as follows: 0–2 min, 90% A; 8 min, 60% A; 15–22 min, 40% A; 23–29 min, 90% A. The autosampler was set at 10 °C. The injection volume was 1 μL. Positive mass spectra were acquired with the ion spray voltage of 5,000 V under ESI. The source temperature was 450 °C. The curtain gas, ion source gas 1, and ion source gas 2 were 32, 60, and 55 psi, respectively. MRM was used for quantitation: 19,20-DHDPA-EA, m/z 406.3→m/z 370.3; 17,18-DHETE-EA, m/z 380.3→m/z 344.3. Internal standard 14,15-DHET-EA was monitored at m/z 382.3→ m/z 346.3.
Extraction of AEA, EPEA, DHEA, EEQ-EA, and EDP-EAs from Rat Tissue.
The tissue extractions of AEA, DHEA, EPEA, EEQ-EAs, and EDP-EAs were performed similarly as previously reported for the quantitative analysis of the endocannabinoids (27, 62). Sprague–Dawley rats were killed, and whole organs were immediately dissected, pooled (n = 3 per pool), weighed, and mechanically homogenized on ice with a BioHomogenizer (BioSpec) in a 1:1 ratio with PBS supplemented with 12-(3-adamantan-1-yl-ureido)-dodecanoic acid (AUDA) (Cayman Chemical) (30 µm) and PMSF (1 mM) until a homogenous slurry was formed. The equivalent of 1 g of each tissue was homogenized in 40 mL of ethyl acetate/hexane (9:1) with a BioHomogenizer (BioSpec) at room temperature and then was sonicated for 1 min. The resulting solution was washed with a 30% volume of water and centrifuged for layer separation. The organic layer was removed, and the aqueous layer was extracted twice with an equal volume of ethyl acetate/hexane (9:1). The pooled organic supernatant was removed using a Buchi 120 rotary evaporator and was reconstituted in 1 mL of chloroform. The metabolites of interest were isolated by solid-phase extraction (SPE) using a 1-mL silica gel column (no. 214477; Sigma). The reconstituted samples were added to the preconditioned silica column, washed in three column volumes of chloroform, and then eluted with four column volumes of methanol/chloroform (1:9). The eluent was dried, and samples were reconstituted in ethanol for analysis via LC-MS-MS.
To validate the method, the metabolite extraction linearity was examined for each class of lipids from homogenized rat brain tissue. The metabolites 17,18-EEQ-EA, 19,20-EDP-EA, DHEA, EPEA, and AEA were spiked at 20, 60, and 300 ng into homogenized rat brain tissue and extracted as described above. The average percent recovery was calculated by subtracting a nonspiked sample of each metabolite for comparison with authentic standards at low, middle, and high levels on the standard curve. Extraction linearities for 20, 60, and 300 ng of all metabolites (17,18-EEQ-EA, 19,20-EDP-EA, DHEA, EPEA, and AEA) were similar among lipids with average percent recoveries of 89.1 ± 3.1, 84.8 ± 2.8, and 71.5 ± 4.5%, respectively.
Whole-Cell Metabolism Assays.
BV-2 microglial cells were seeded in six-well dishes (5 × 105 cells per well) and grown to 90% confluence in DMEM (Gibco), 10% FBS (Gibco), penicillin (100 U/mL), and streptomycin (0.1 mg/mL) at 37 °C and 5% CO2. At the time of the experiment the supernatant was replaced with serum-free medium, and cells were activated with 100 ng/mL of LPS for 12 h. In one set of experiments the supernatant and cells were collected at 1, 4, and 12 h after LPS stimulation and were examined for the natural production of target lipid mediators. Alternatively, after the 12-h LPS activation, cells were incubated with either EPEA (20 µM) or DHEA (20 µM) in serum-free medium for 30 min. The general P450 inhibitor ketoconazole (0.5 µM) was added in some experiments to inhibit the CYP3A, CYP2C, and CYP2J families. Controls lacking LPS stimulation were run in parallel with the experiment. After the appropriate time period, cells were scraped, and the supernatant was collected and subjected to three rapid freeze–thaw cycles using liquid N2 and a warm water bath (37 °C) for cell lysis. The supernatant and cells were extracted three times with four volumes of ethyl acetate/hexane (9:1).
Incubations of EPEA and DHEA with Brain Microsomes.
Microsomes derived from mouse tissues were prepared using previously described differential centrifugation methods (28). Tissues were dissected from Sprague–Dawley rats, weighed, homogenized, and diluted to 20% (wt/vol) using a buffer containing 250 mM sucrose, 10 mM Tris-Cl (pH 7.5), 1 mM PMSF, and a protease inhibitor mixture for mammalian tissue extraction (no. 25955-11; Nacalai Tesque, Inc.). A series of low (3,000 × g) and medium (10,000 × g) centrifugation speeds was used to remove cellular debris and nuclear and mitochondrial fractions. The resulting supernatant was centrifuged at 100,000 × g for 90 min at 4 °C to pellet the endoplasmic reticulum containing CYPs. The cell pellet was resuspended in 50 mM Tris (pH 7.5), 1 mM DTT, 1 mM EDTA, and 20% glycerol with a Teflon homogenizer. Microsomal protein content was measured using a BCA assay kit (product no. 23233; Thermo Fisher Scientific). Incubations with microsomes were performed at a protein concentration of 1 mg/mL with saturating concentrations of DHEA and EPEA as described in Fig. 3 A–C. Next microsome incubations were extracted using three volumes of ethyl acetate/hexane (9:1), the organic layer was collected and the aqueous layer was reextracted twice more. The organic layer was dried under vacuum and was reconstituted in ice-cold ethanol for LC-MS/MS analysis.
CYP2J2 Nanodisc Kinetic Metabolism of EPEA and DHEA.
Human CYP2J2 was heterologously expressed and incorporated into the lipid bilayers of nanodiscs as previously described (61). Incubation mixtures contained CYP2J2 nanodiscs (0.2 µM), CPR (0.6 µM), butylated hydroxytoluene (BHT) (0.1%), and incremental increases of either EPEA or DHEA (500 µL total volume) in 100 mM phosphate buffer (pH 7.4). Samples were equilibrated at 37 °C for 5 min before the reaction was initiated with the addition of 0.5 mM NADPH. Reactions were stopped after 15 min and were extracted three times with 1 mL of ethyl acetate. The organic layer was dried and reconstituted in ice-cold ethanol for LC-MS/MS analysis.
Kinetic Analysis of 17,18-EEQ-EA and 19,20-EDP-EA Hydrolysis by sEH.
LC-MS/MS was performed to quantitate the conversion of the terminal epoxides to their vicinal diols using LC-MS/MS. Enzymatic linearity over a 10-min period was confirmed. Enzyme kinetics were estimated using incremental increases of substrate for the generation of kinetic curves that were fit to the Michaelis–Menten equation for calculation of Vmax and Km. Incubations were performed with recombinant sEH ([E]final = 6 nM) in NaPO4 (100 mM, pH 7.4), BSA (0.1 mg/mL), and substrate (17,18-EEQ-EA or 19,20-EDP-EA) at 37 °C. After 5 min the reaction was stopped with methanol supplemented with AUDA (0.5 µM), and the solution was centrifuged at 10,000 × g for 10 min to pellet protein. The supernatant was collected and analyzed via LC-MS/MS.
Microglial Neuroinflammation Studies.
BV-2 microglial cells were plated in 24-well dishes (2 × 105 cells per well) and were grown to 80–90% confluence in DMEM (Gibco), 10% FBS (Gibco), penicillin (100 U/mL), and streptomycin (0.1 mg/mL) at 37 °C and 5% CO2. At the time of the experiment, the medium was replaced with serum-free medium, and the cells were pretreated with 19,20-EDP-EA for 4 h before stimulation with 25 ng/mL LPS (Sigma-Aldrich). Medium was collected at 24 h, and NO production was determined by evaluating the nitrite (NO2−) content of the medium using the Griess method. Collected medium (80 µL) was mixed with Griess reagent A (no. 780018; Cayman Chemical) and Griess reagent B (no.780020; Cayman Chemical), and the absorbance at 490 nm was used to quantify NO2− production using a standard curve. Medium supernatant was also examined for the production of IL-6 (no. 583371; Cayman Chemical) and IL-10 (no. 88-7105-22; Affymetrix eBioscience) cytokines. Compound cytotoxicity was assessed by measuring lactate dehydrogenase (LDH) release in the medium using a commercially available kit (no. 601170; Cayman Chemical).
Platelet Aggregation Studies.
The University of Michigan Review Board approved studies, and written informed consent was obtained from all participants before blood collection. Human whole blood was drawn from the antecubital vein of healthy donors. PRP was obtained from whole blood by centrifugation at 200 × g for 10 min. Platelets were pelleted from PRP in the presence of acid citrate dextrose (ACD) (2.5%) and apyrase (0.02 U/mL) by centrifugation at 2,000 × g for 10 min and then were resuspended in Tyrode’s buffer (12 mM NaHCO3, 127 mM NaCl, 5 mM KCl, 0.5 mM NaH2PO4, 1 mM MgCl2, 5 mM glucose, 10 mM Hepes) to a final concentration of 3.0 × 108 platelets/mL. Changes in light transmission were recorded by an eight-channel platelet aggregometer (Chrono-Log) with stirring at 1,200 rpm at 37 °C.
Washed human platelets were incubated with 12.5, 25, and 50 µM 17,18-EEQ-EA for 5 min, and platelet aggregation was induced by 5 µM AA, thrombin (1 nM), PAR4-AP (50 µM), collagen (2 mg/mL), and U46619 (1 µM). DMSO and 14,15-EET were used as controls. No platelet agonists were added for the tests of 19,20-EDP-EA in washed platelets. Experiments with PRP were adjusted to 3.0 × 108 platelets/mL using platelet poor plasma (PPP) from same donor. PRP was incubated with 12.5, 25, and 50 µM 17,18-EEQ EA for 5 min, and platelet aggregation was induced by ADP (1 µM), collagen (2 µg/mL), or ristocetin (1 mg/mL).
Bovine Coronary Artery Vasodilation Assay.
Fresh bovine hearts were procured on ice from the local butcher. The left anterior coronary artery was dissected from the adjacent connective tissue and cut into 2-mm ring sections. The artery lumen then was mounted on two parallel pins attached to a piezoelectric transducer for isometric tension measurements using a Myograph Tissue Bath System (DMT) as previously described (63). Basal tension (100% relaxation) was set at 30 mN, and tissues were equilibrated in Krebs buffer (130 mM NaCl, 4.7 mM KCl, 1.18 mM KH2PO4, 1.17 mM MgSO4, 14.9 mM NaHCO3, 5.5 mM dextrose, 0.026 mM EDTA, 1.6 mM CaCl2) for 1 h. Maximal contractions were determined by washing tissues with Krebs buffer supplemented with 60 mM KCl for 3 min, and then tissues were washed with Krebs buffer to return to basal tension. This step was repeated until consistent contractile measurements were achieved. The thromboxane receptor agonist U46619 (20–40 nM) was used to constrict the rings to ∼40–80% of maximal contraction (0% relaxation). Cumulative additions of EEQ, EDP, EEQ-EA, and EDP-EA (1 nM, 10 nM, 100 nM, 1 µM, and 10 µM, respectively) were added to each chamber, and the resulting percent relaxation was plotted against the corresponding substrate dose.
Inhibition of Human Microvascular Cell Tube Formation.
The EEQ, EEQ-EA, EDP, and EDP-EA inhibition of VEGF-promoted tube formation was assessed in HMVECs (Cell Systems) as previously described (51). Briefly, 35 µL of Matrigel was plated on the bottom of a 48-well plate and allowed to gel for 30 min at 37 °C. Approximately 15,000 HMVECs then were seeded per well in EBM-2 medium (Lonza) supplemented with VEGF at 50% of the concentration used in EGM-2. After the compounds were added at the desired concentration, tube formation by HMVECs was quantified after 8 h. Cells were imaged using a Cannon Rebel digital single lens reflex camera, and the tube area was quantified using ImageJ.
Statistics.
Results are presented as average ± SE unless noted otherwise. Kinetic parameters were determined by nonlinear regression of the Michaelis–Menten equation using OriginPro 8.6 software (OriginLab). Data were analyzed for statistical significance using a two-sided Student’s t test where *P < 0.05, **P < 0.01, and ***P < 0.001.
Supplementary Material
Acknowledgments
We thank Dr. Zhong Li at the Metabolomics Laboratory of the Roy J. Carver Biotechnology Center, University of Illinois at Urbana–Champaign (UIUC) for his contributions; Furong Sun at the School of Chemical Science Mass Spectrometry Facility, UIUC; the Sligar Lab for the gene encoding MSP1D1; Dr. Natasha Snider for helpful discussions; Dr. Shah, the Ko laboratory, and the Campbell laboratory for training in the BCA vasodilation assay; the Roth laboratory and the National Institute of Mental Health’s Psychoactive Drug Screening Program (NIMH PDSP) center for PRESTO-Tango assays; Dan Gilman for the gifts of fresh bovine hearts; and Holly Fairfield for procuring freshly isolated Sprague–Dawley rat tissues. The genes encoding CB1 and CB2 were generous gifts from Professor Ken Mackie. This work was supported in part by American Heart Association (AHA) Scientist Development Grants 15SDG25760064 (to A.D.) and 1R01GM115584-01A1 (to A.D.); AHA Predoctoral Fellowship 14PRE20130015 (to D.R.M.); and NIH Office of Dietary Supplement Grants GM105671 (to M.H.), HL114405 (to M.H.), and HD069899 (to R.W.J.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610325114/-/DCSupplemental.
References
- 1.Kris-Etherton PM, Harris WS, Appel LJ. American Heart Association. Nutrition Committee Fish consumption, fish oil, omega-3 fatty acids, and cardiovascular disease. Circulation. 2002;106:2747–2757. doi: 10.1161/01.cir.0000038493.65177.94. [DOI] [PubMed] [Google Scholar]
- 2.Morris MC, Evans DA, Tangney CC, Bienias JL, Wilson RS. Fish consumption and cognitive decline with age in a large community study. Arch Neurol. 2005;62:1849–1853. doi: 10.1001/archneur.62.12.noc50161. [DOI] [PubMed] [Google Scholar]
- 3.Duda MK, et al. Fish oil, but not flaxseed oil, decreases inflammation and prevents pressure overload-induced cardiac dysfunction. Cardiovasc Res. 2009;81:319–327. doi: 10.1093/cvr/cvn310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown I, et al. Cannabinoids and omega-3/6 endocannabinoids as cell death and anticancer modulators. Prog Lipid Res. 2013;52:80–109. doi: 10.1016/j.plipres.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Brown I, et al. Cannabinoid receptor-dependent and -independent anti-proliferative effects of omega-3 ethanolamides in androgen receptor-positive and -negative prostate cancer cell lines. Carcinogenesis. 2010;31:1584–1591. doi: 10.1093/carcin/bgq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skaper SD, Di Marzo V. Endocannabinoids in nervous system health and disease: The big picture in a nutshell. Philos Trans R Soc Lond B Biol Sci. 2012;367:3193–3200. doi: 10.1098/rstb.2012.0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fonseca BM, Costa MA, Almada M, Correia-da-Silva G, Teixeira NA. Endogenous cannabinoids revisited: A biochemistry perspective. Prostaglandins Other Lipid Mediat. 2013;102-103:13–30. doi: 10.1016/j.prostaglandins.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Ashton JC, Glass M. The cannabinoid CB2 receptor as a target for inflammation-dependent neurodegeneration. Curr Neuropharmacol. 2007;5:73–80. doi: 10.2174/157015907780866884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pacher P, Bátkai S, Kunos G. The endocannabinoid system as an emerging target of pharmacotherapy. Pharmacol Rev. 2006;58:389–462. doi: 10.1124/pr.58.3.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meijerink J, Balvers M, Witkamp R. N-Acyl amines of docosahexaenoic acid and other n-3 polyunsatured fatty acids - from fishy endocannabinoids to potential leads. Br J Pharmacol. 2013;169:772–783. doi: 10.1111/bph.12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim HY, Spector AA. Synaptamide, endocannabinoid-like derivative of docosahexaenoic acid with cannabinoid-independent function. Prostag Leukotr Ess. 2013;88:121–125. doi: 10.1016/j.plefa.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugiura T, et al. Transacylase-mediated and phosphodiesterase-mediated synthesis of N-arachidonoylethanolamine, an endogenous cannabinoid-receptor ligand, in rat brain microsomes. Comparison with synthesis from free arachidonic acid and ethanolamine. Eur J Biochem. 1996;240:53–62. doi: 10.1111/j.1432-1033.1996.0053h.x. [DOI] [PubMed] [Google Scholar]
- 13.Bisogno T, Delton-Vandenbroucke I, Milone A, Lagarde M, Di Marzo V. Biosynthesis and inactivation of N-arachidonoylethanolamine (anandamide) and N-docosahexaenoylethanolamine in bovine retina. Arch Biochem Biophys. 1999;370:300–307. doi: 10.1006/abbi.1999.1410. [DOI] [PubMed] [Google Scholar]
- 14.Sheskin T, Hanus L, Slager J, Vogel Z, Mechoulam R. Structural requirements for binding of anandamide-type compounds to the brain cannabinoid receptor. J Med Chem. 1997;40:659–667. doi: 10.1021/jm960752x. [DOI] [PubMed] [Google Scholar]
- 15.Artmann A, et al. Influence of dietary fatty acids on endocannabinoid and N-acylethanolamine levels in rat brain, liver and small intestine. Biochim Biophys Acta. 2008;1781:200–212. doi: 10.1016/j.bbalip.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 16.Rossmeisl M, et al. Metabolic effects of n-3 PUFA as phospholipids are superior to triglycerides in mice fed a high-fat diet: Possible role of endocannabinoids. PLoS One. 2012;7 doi: 10.1371/journal.pone.0038834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rouzer CA, Marnett LJ. Endocannabinoid oxygenation by cyclooxygenases, lipoxygenases, and cytochromes P450: Cross-talk between the eicosanoid and endocannabinoid signaling pathways. Chem Rev. 2011;111:5899–5921. doi: 10.1021/cr2002799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meijerink J, et al. Inhibition of COX-2-mediated eicosanoid production plays a major role in the anti-inflammatory effects of the endocannabinoid N-docosahexaenoylethanolamine (DHEA) in macrophages. Br J Pharmacol. 2015;172:24–37. doi: 10.1111/bph.12747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang R, et al. Decoding functional metabolomics with docosahexaenoyl ethanolamide (DHEA) identifies novel bioactive signals. J Biol Chem. 2011;286:31532–31541. doi: 10.1074/jbc.M111.237990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang G, Kodani S, Hammock BD. Stabilized epoxygenated fatty acids regulate inflammation, pain, angiogenesis and cancer. Prog Lipid Res. 2014;53:108–123. doi: 10.1016/j.plipres.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snider NT, Walker VJ, Hollenberg PF. Oxidation of the endogenous cannabinoid arachidonoyl ethanolamide by the cytochrome P450 monooxygenases: Physiological and pharmacological implications. Pharmacol Rev. 2010;62:136–154. doi: 10.1124/pr.109.001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snider NT, Nast JA, Tesmer LA, Hollenberg PF. A cytochrome P450-derived epoxygenated metabolite of anandamide is a potent cannabinoid receptor 2-selective agonist. Mol Pharmacol. 2009;75:965–972. doi: 10.1124/mol.108.053439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt A, Brune K, Hinz B. Determination of the endocannabinoid anandamide in human plasma by high-performance liquid chromatography. Biomed Chromatogr. 2006;20:336–342. doi: 10.1002/bmc.568. [DOI] [PubMed] [Google Scholar]
- 24.Muccioli GG, Stella N. An optimized GC-MS method detects nanomolar amounts of anandamide in mouse brain. Anal Biochem. 2008;373:220–228. doi: 10.1016/j.ab.2007.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J, Paudel KS, Derbenev AV, Smith BN, Stinchcomb AL. Simultaneous Quantification of Anandamide and Other Endocannabinoids in Dorsal Vagal Complex of Rat Brainstem by LC-MS. Chromatographia. 2009;69:1–7. doi: 10.1365/s10337-008-0841-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen JK, et al. Identification of novel endogenous cytochrome p450 arachidonate metabolites with high affinity for cannabinoid receptors. J Biol Chem. 2008;283:24514–24524. doi: 10.1074/jbc.M709873200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kingsley PJ, Marnett LJ. Analysis of endocannabinoids by Ag+ coordination tandem mass spectrometry. Anal Biochem. 2003;314:8–15. doi: 10.1016/s0003-2697(02)00643-7. [DOI] [PubMed] [Google Scholar]
- 28.McDougle DR, Kambalyal A, Meling DD, Das A. Endocannabinoids - anandamide and 2-arachidonoylglycerol are substrates for human cytochrome P450 2J2 epoxygenase. J Pharmacol Exp Ther. 2014;351:616–627. doi: 10.1124/jpet.114.216598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dutheil F, et al. Xenobiotic-metabolizing enzymes and transporters in the normal human brain: Regional and cellular mapping as a basis for putative roles in cerebral function. Drug Metab Dispos. 2009;37:1528–1538. doi: 10.1124/dmd.109.027011. [DOI] [PubMed] [Google Scholar]
- 30.Chaudhary KR, Batchu SN, Seubert JM. Cytochrome P450 enzymes and the heart. IUBMB Life. 2009;61:954–960. doi: 10.1002/iub.241. [DOI] [PubMed] [Google Scholar]
- 31.Zhou GL, et al. Deletion of the murine cytochrome P450 Cyp2j locus by fused BAC-mediated recombination identifies a role for Cyp2j in the pulmonary vascular response to hypoxia. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Westphal C, Konkel A, Schunck WH. CYP-eicosanoids-A new link between omega-3 fatty acids and cardiac disease? Prostag Oth Lipid M. 2011;96:99–108. doi: 10.1016/j.prostaglandins.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 33.Arnold C, et al. Arachidonic acid-metabolizing cytochrome P450 enzymes are targets of ω-3 fatty acids. J Biol Chem. 2010;285:32720–32733. doi: 10.1074/jbc.M110.118406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morisseau C, et al. Naturally occurring monoepoxides of eicosapentaenoic acid and docosahexaenoic acid are bioactive antihyperalgesic lipids. J Lipid Res. 2010;51:3481–3490. doi: 10.1194/jlr.M006007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amor S, Puentes F, Baker D, van der Valk P. Inflammation in neurodegenerative diseases. Immunology. 2010;129:154–169. doi: 10.1111/j.1365-2567.2009.03225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benito C, et al. Cannabinoid CB2 receptors in human brain inflammation. Br J Pharmacol. 2008;153:277–285. doi: 10.1038/sj.bjp.0707505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagarkatti P, Pandey R, Rieder SA, Hegde VL, Nagarkatti M. Cannabinoids as novel anti-inflammatory drugs. Future Med Chem. 2009;1:1333–1349. doi: 10.4155/fmc.09.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iliff JJ, et al. Epoxyeicosanoid signaling in CNS function and disease. Prostag Oth Lipid M. 2010;91:68–84. doi: 10.1016/j.prostaglandins.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishio K, et al. Attenuation of lipopolysaccharide (LPS)-induced cytotoxicity by tocopherols and tocotrienols. Redox Biol. 2013;1:97–103. doi: 10.1016/j.redox.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee CA, et al. Identifying a selective substrate and inhibitor pair for the evaluation of CYP2J2 activity. Drug Metab Dispos. 2012;40:943–951. doi: 10.1124/dmd.111.043505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spector AA. Arachidonic acid cytochrome P450 epoxygenase pathway. J Lipid Res. 2009;50:S52–S56. doi: 10.1194/jlr.R800038-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kroeze WK, et al. PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat Struct Mol Biol. 2015;22:362–369. doi: 10.1038/nsmb.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yin H, et al. Lipid G protein-coupled receptor ligand identification using beta-arrestin PathHunter assay. J Biol Chem. 2009;284:12328–12338. doi: 10.1074/jbc.M806516200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krotz F, et al. Membrane potential-dependent inhibition of platelet adhesion to endothelial cells by epoxyeicosatrienoic acids. Arterioscl Throm Vas. 2004;24:595–600. doi: 10.1161/01.ATV.0000116219.09040.8c. [DOI] [PubMed] [Google Scholar]
- 45.VanRollins M. Epoxygenase metabolites of docosahexaenoic and eicosapentaenoic acids inhibit platelet aggregation at concentrations below those affecting thromboxane synthesis. J Pharmacol Exp Ther. 1995;274:798–804. [PubMed] [Google Scholar]
- 46.Ikei KN, et al. Investigations of human platelet-type 12-lipoxygenase: Role of lipoxygenase products in platelet activation. J Lipid Res. 2012;53:2546–2559. doi: 10.1194/jlr.M026385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang G, et al. Epoxy metabolites of docosahexaenoic acid (DHA) inhibit angiogenesis, tumor growth, and metastasis. Proc Natl Acad Sci USA. 2013;110:6530–6535. doi: 10.1073/pnas.1304321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fischer R, et al. Dietary omega-3 fatty acids modulate the eicosanoid profile in man primarily via the CYP-epoxygenase pathway. J Lipid Res. 2014;55:1150–1164. doi: 10.1194/jlr.M047357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Petrocellis L, et al. The endogenous cannabinoid anandamide inhibits human breast cancer cell proliferation. Proc Natl Acad Sci USA. 1998;95:8375–8380. doi: 10.1073/pnas.95.14.8375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pisanti S, et al. Antiangiogenic activity of the endocannabinoid anandamide: Correlation to its tumor-suppressor efficacy. J Cell Physiol. 2007;211:495–503. doi: 10.1002/jcp.20954. [DOI] [PubMed] [Google Scholar]
- 51.Abdeen AA, Weiss JB, Lee J, Kilian KA. Matrix composition and mechanics direct proangiogenic signaling from mesenchymal stem cells. Tissue Eng Part A. 2014;20:2737–2745. doi: 10.1089/ten.tea.2013.0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khanapure SP, Garvey DS, Janero DR, Letts LG. Eicosanoids in inflammation: Biosynthesis, pharmacology, and therapeutic frontiers. Curr Top Med Chem. 2007;7:311–340. doi: 10.2174/156802607779941314. [DOI] [PubMed] [Google Scholar]
- 53.Thomson SJ, Askari A, Bishop-Bailey D. Anti-inflammatory effects of epoxyeicosatrienoic acids. Int J Vasc Med. 2012;2012:605101. doi: 10.1155/2012/605101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Imig JD, et al. Development of epoxyeicosatrienoic acid analogs with in vivo anti-hypertensive actions. Front Physiol. 2010;1:157. doi: 10.3389/fphys.2010.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petrosino S, Di Marzo V. FAAH and MAGL inhibitors: Therapeutic opportunities from regulating endocannabinoid levels. Curr Opin Investig Drugs. 2010;11:51–62. [PubMed] [Google Scholar]
- 56.Walker VJ, Griffin AP, Hammar DK, Hollenberg PF. Metabolism of anandamide by human cytochrome P450 2J2 in the reconstituted system and human intestinal microsomes. J Pharmacol Exp Ther. 2016;357:537–544. doi: 10.1124/jpet.116.232553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arnold WR, Baylon JL, Tajkhorshid E, Das A. Asymmetric binding and metabolism of polyunsaturated fatty acids (PUFAs) by CYP2J2 epoxygenase. Biochemistry. 2016;55:6969–6980. doi: 10.1021/acs.biochem.6b01037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tomar S, Zumbrun EE, Nagarkatti M, Nagarkatti PS. Protective role of cannabinoid receptor 2 activation in galactosamine/lipopolysaccharide-induced acute liver failure through regulation of macrophage polarization and microRNAs. J Pharmacol Exp Ther. 2015;353:369–379. doi: 10.1124/jpet.114.220368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dai M, et al. Epoxyeicosatrienoic acids regulate macrophage polarization and prevent LPS-induced cardiac dysfunction. J Cell Physiol. 2015;230:2108–2119. doi: 10.1002/jcp.24939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fitzpatrick FA, et al. Inhibition of cyclooxygenase activity and platelet aggregation by epoxyeicosatrienoic acids. Influence of stereochemistry. J Biol Chem. 1986;261:15334–15338. [PubMed] [Google Scholar]
- 61.McDougle DR, Palaria A, Magnetta E, Meling DD, Das A. Functional studies of N-terminally modified CYP2J2 epoxygenase in model lipid bilayers. Protein Sci. 2013;22:964–979. doi: 10.1002/pro.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Richardson D, Ortori CA, Chapman V, Kendall DA, Barrett DA. Quantitative profiling of endocannabinoids and related compounds in rat brain using liquid chromatography-tandem electrospray ionization mass spectrometry. Anal Biochem. 2007;360:216–226. doi: 10.1016/j.ab.2006.10.039. [DOI] [PubMed] [Google Scholar]
- 63.Hedegaard ER, et al. KV 7 channels are involved in hypoxia-induced vasodilatation of porcine coronary arteries. Br J Pharmacol. 2014;171:69–82. doi: 10.1111/bph.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.