Significance
Glioblastoma is a deadly brain tumor with no cure. Brain tumor-initiating cells (BTICs) have been recognized as the key driver behind the unstoppable malignant growth, therapy resistance, and recurrence. BTICs are exceptionally difficult to target because of heterogeneous genetic and epigenetic aberrations that are challenging to reverse therapeutically using conventional pharmaceuticals or biologics. Here we report a lipopolymeric nanoparticle (LPNP) formulation that demonstrates a surprisingly high affinity for BTICs and the capacity to encapsulate multiple siRNAs for potent and targeted anti-BTIC therapy. We show that direct infusion of LPNP siRNAs to brain tumors effectively impedes tumor growth in mouse and provides encouraging survival benefits. This multiplexed nanomedicine platform carries strong potential for personalized anti-BTIC therapies.
Keywords: siRNA, lipopolymeric nanoparticle, glioblastoma transcription factor, brain tumor-initiating cells, convection-enhanced delivery
Abstract
Brain tumor-initiating cells (BTICs) have been identified as key contributors to therapy resistance, recurrence, and progression of diffuse gliomas, particularly glioblastoma (GBM). BTICs are elusive therapeutic targets that reside across the blood–brain barrier, underscoring the urgent need to develop novel therapeutic strategies. Additionally, intratumoral heterogeneity and adaptations to therapeutic pressure by BTICs impede the discovery of effective anti-BTIC therapies and limit the efficacy of individual gene targeting. Recent discoveries in the genetic and epigenetic determinants of BTIC tumorigenesis offer novel opportunities for RNAi-mediated targeting of BTICs. Here we show that BTIC growth arrest in vitro and in vivo is accomplished via concurrent siRNA knockdown of four transcription factors (SOX2, OLIG2, SALL2, and POU3F2) that drive the proneural BTIC phenotype delivered by multiplexed siRNA encapsulation in the lipopolymeric nanoparticle 7C1. Importantly, we demonstrate that 7C1 nano-encapsulation of multiplexed RNAi is a viable BTIC-targeting strategy when delivered directly in vivo in an established mouse brain tumor. Therapeutic potential was most evident via a convection-enhanced delivery method, which shows significant extension of median survival in two patient-derived BTIC xenograft mouse models of GBM. Our study suggests that there is potential advantage in multiplexed targeting strategies for BTICs and establishes a flexible nonviral gene therapy platform with the capacity to channel multiplexed RNAi schemes to address the challenges posed by tumor heterogeneity.
Glioblastoma (GBM) is one of the most challenging tumors to treat (1, 2). Despite decades of research and maximal clinical combination therapy encompassing surgical resection, chemotherapy, and radiation, the median life expectancy of patients has not been extended beyond 2 y after diagnosis (2). Increasing evidence suggests that the genetic, epigenetic, and signaling heterogeneity of GBM underlies the ineffectiveness of currently available therapeutics (1, 2). Additionally, therapeutic schemes devised to challenge brain tumor cells are frequently thwarted by insufficient delivery caused by pharmacokinetics, the blood–brain barrier (BBB), and an altered tumor microenvironment in which tumor-derived signaling recruits immunomodulatory cells and induces extracellular matrix remodeling to build safe harbors of tumorigenic niches (3–5). These obstacles call for tailored therapeutic strategies to counter tumor heterogeneity and overcome roadblocks in delivery. RNAi targeting drivers of tumorigenesis shows strong potential to supplement the development of traditional small-molecule pharmaceutics (6). However, delivery remains a key obstacle for efficient RNAi against tumor drivers (5, 7, 8). RNA-sequencing analysis of patient tissue combined with histology and in situ hybridization (Ivy Glioblastoma Atlas Project; SI Appendix, Fig. S1) show that brain tumor-initiating cells (BTICs) are found predominantly in the bulk of cellular tumors, although their presence in vascular structures and perivascular niches has also been highlighted in recent research (9–13). This distribution pattern suggests that direct delivery to BTICs in the tumor bulk is likely to be more advantageous than systemic delivery approaches, which rely on BBB penetration to maximize the therapeutic benefits of RNAi targeting BTICs.
A class of lipopolymeric nanoparticles (LPNPs) has been formulated to maximize systemic delivery to vasculature-rich organs for antitumor RNAi therapies, and the combination of ionizable, low-molecular-weight lipopolymers custom synthesized to optimize cell entry facilitates tumor cell targeting when delivered directly (14–22). The polymers were synthesized by conjugating epoxide-terminated lipids to low-molecular-weight polyamines with an epoxide ring-opening reaction (23). The resultant nano constructs then were evaluated for their ability to complex with siRNA and knock down gene expression in vitro. The most effective candidates were selected after in vivo assessment. One compound, named “7C1,” was identified from a library of more than 500 candidate delivery materials as the most effective in silencing target genes in vasculature, with a higher performance per dose ratio than previously reported siRNA nano delivery vehicles (15, 23). The capacity to multitarget and the ease of synthesis make 7C1 an attractive therapeutic vehicle for antitumor RNAi. However, the ability of 7C1 to penetrate the BBB has not been established.
A core set of four transcription factors (TFs), namely SOX2, OLIG2, SALL2, and POU3F2, was recently proposed to drive proneural BTICs (24). These TFs show high expression levels in the bulk of patient tumor tissue (SI Appendix, Fig. S1). To evaluate the feasibility of local delivery to BTICs directly in the core of xenografts in mice, we opted to encapsulate combinations of siRNAs in 7C1 LPNPs targeting the four TFs concurrently. Because other circuits are responsible for maintaining the stem cell-like phenotypes in other GBM subgroups listed in The Cancer Gene Atlas (TCGA), and because additional genetic/epigenetic circuits are being identified (25–28), the current choice of RNAi therapeutic targets is not expected to apply to all GBM subtypes. The primary purpose of our work was to establish a proof-of-principle approach and to demonstrate that tumorigenesis can be antagonized using this vehicle and delivery strategy. Given that TFs and other genetic and epigenetic targets were considered “undruggable” by conventional pharmaceutical approaches, and nanomedicine-mediated RNAi strategies have been formulated primarily against membrane-bound or cytoplasmic effectors (29–35), this direct approach in delivering RNAi therapeutics to target tumorigenic gene circuits is a key step forward in diversifying our strategies against one of the most challenging human cancers.
Results
TF Profiling and Selection of Tumor Cell Lines for Therapeutic Targeting.
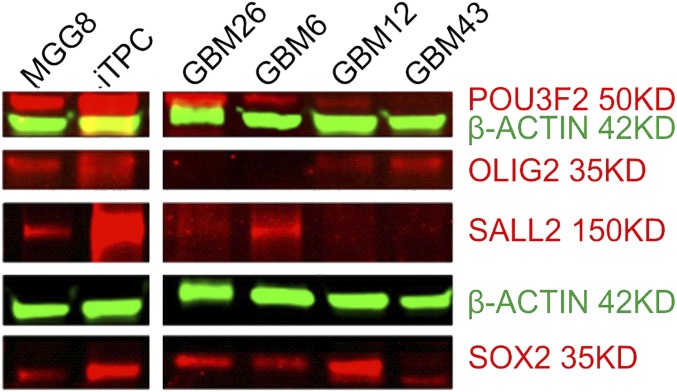
To establish a suitable therapeutic target model using patient-derived GBM BTICs, we cultured GBM cell lines in serum-free Neurobasal medium supplemented for tumorsphere growth conditions (24, 25, 36). Cells then were collected for Western blotting and qRT-PCR analysis of the expression levels of four core TFs (for primer sequences see SI Appendix, Table S1). Both assays indicate that BTICs arising from different cell lines express dramatically variant levels of TFs (Fig. 1 and SI Appendix, Fig. S2); however, MGG8 and GBM43 display the most uniform levels of concurrent expression of the four TFs in both mRNA and protein assays (Fig. 1 and SI Appendix, Figs. S3 and S4), and both cell types have been determined to resemble most closely the proneural subclass of GBM, based on TCGA criteria and genomic analysis (37, 38). Therefore, we chose these two cell lines as TF-dependent BTIC models to examine the feasibility and efficacy of TF RNAi therapy going forward.
Fig. 1.
TF expression profiling of BTIC lines. Western blotting shows distinct TF expression profiles of several patient-derived GBM BTIC lines. MGG8 and GBM43 were selected for modeling of TF-dependent BTICs based on balanced and relatively higher expression of all four TFs.
Validation of Candidate siRNAs Targeting BTIC-Defining TFs.
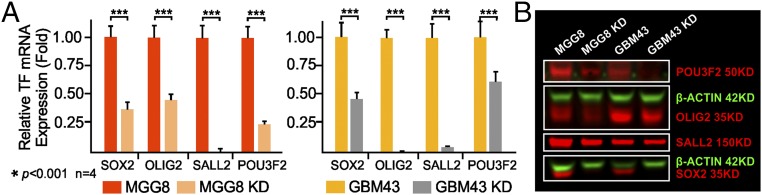
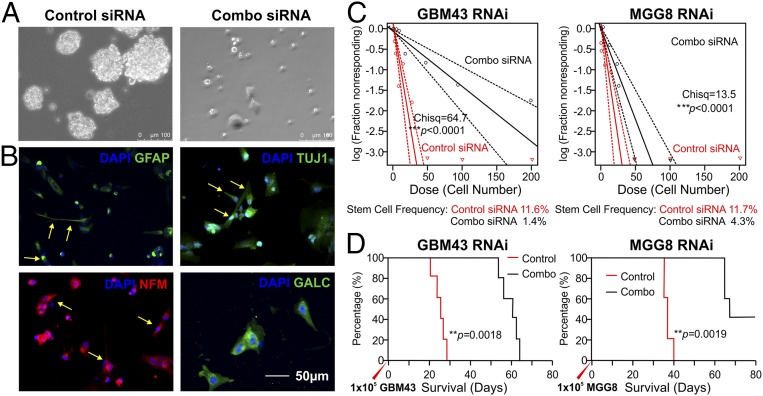
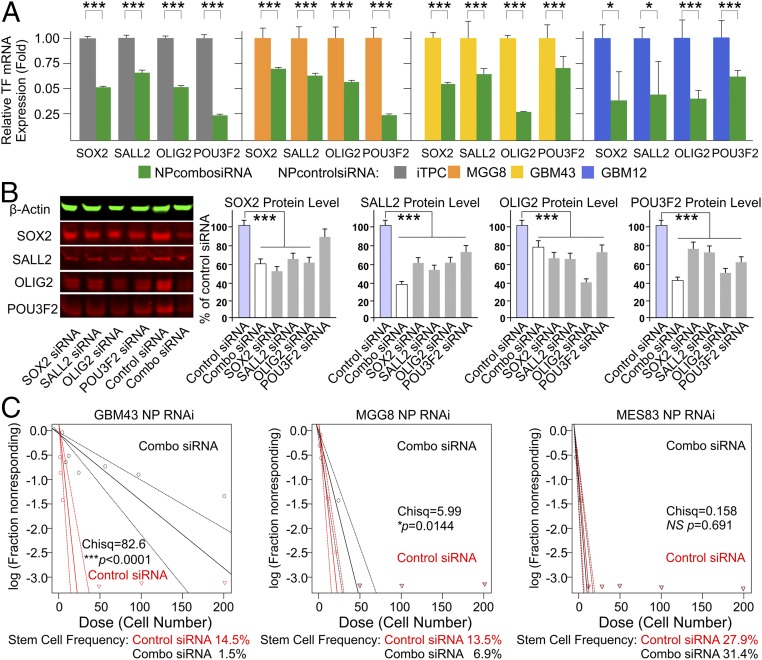
To select the optimal siRNA duplex for TF RNAi, we used sequence-prediction services (Millipore Sigma) and generated three sets of siRNA duplex designs, designated as siRNA1, siRNA2, and siRNA3, for each TF (SI Appendix, Table S2) to test their efficacy in knocking down TF expression. Dilution series were carried out to determine the optimal in vitro TF mRNA knockdown, and combinations of all four finalists (combo siRNAs) were used to treat cells before in vitro and in vivo functional assays to analyze the tumorigenicity of the treated cells. qRT-PCR shows that the delivery of these siRNAs in combination resulted in at least 60% mRNA knockdown by 24 h posttransfection, whereas the delivery of two separate universal nontargeting negative controls did not (Fig. 2). The reduction in protein level became detectable on Western blots and immunocytochemistry by 72 h posttransfection after treatment with 50 nM of siRNA in complete medium (Fig. 2 and SI Appendix, Fig. S3). The biological impact of these four TF knockdowns has been studied through the shRNA strategy (24) in in vitro and in vivo models, but the impact of transient siRNA knockdown, the only clinically translatable approach, has yet to be investigated in formed tumors in situ (39). BTICs treated with combo siRNAs (50 nM for each TF siRNA) for 48 h demonstrated significantly reduced tumor sphere formation and stem cell frequency for MGG8 at cell densities greater than 25 cells per well in 96-well plates (Fig. 3) and abolished single-cell clonal genesis, a hallmark of tumorigenic behaviors (24). BTICs treated with combo siRNAs also display changes in morphology and demonstrate adherent culture with process development (Fig. 3A). Markers of early-stage differentiation along the three neural lineages [e.g., GFAP for astroglia, βIII tubulin (TUJ1) and neurofilament M (NFM) for early-stage neuroprogenitors, and galactocerebroside (GALC) for early-stage oligodendroglia] were observed via immunofluorescence (Fig. 3B). However, cells plated at a higher density of cells in a single well (e.g., more than three cells per well for MGG8, and various increases in cells per well for GBM43) can still give rise to tumor spheres; the dose impact of siRNA combinations needs to be explored in the future (Fig. 3C). The in vitro assay was further supported by the reduced tumorigenicity of MGG8 and GBM43 cells after combo siRNA TF knockdown (50 nM of each TF siRNA for 48 h); when these cells were implanted in athymic nude mice, survival was about twice as long as when tumor cells treated with negative nontargeting control siRNA were implanted (Fig. 3D). The eventual death of all experimental mice indicates that single-dose transient knockdown of master TFs is not a long-term solution in combating the malignant growth of BTICs, and the impact on malignant tumor genome is likely not sustainable. This finding led to the design of repeated or sustained RNAi therapy in vivo to examine whether locally delivered siRNA can maintain the suppression of expression.
Fig. 2.
Validation of candidate siRNAs. Candidate siRNAs were selected for combination therapy against TF expression in BTIC models. (A) qRT-PCR demonstration of effective mRNA knockdown with a combination of the four most effective siRNAs targeting the four TFs in both MGG8 and GBM43 cells (n = 4; ***P < 0.001, t test) (see SI Appendix, Table S2 for these siRNA duplex sequences). (B) Western blotting demonstration of the reduction in the level of TF protein expression with multiplexed RNAi against all four TFs simultaneously. KD, knock down.
Fig. 3.
Functional characterization of TF RNAi in vitro and in vivo. Validation of multiplexed RNAi on tumorigenicity. (A) Representative microscopy image of tumor spheres formed from single BTICs (Left) and differentiated appearance of combo siRNA-treated tumor cells (Right). (B) Immunofluorescence probing of the differentiation states of MGG8 BTICs receiving combo siRNA–LPNP treatment. After combo siRNA treatment for 48 h, BTICs gradually adopted an adherent growth pattern and morphological indications of astroglial (glial fibrillary acid protein, GFAP+), neural (TUJ1+, or neural filament M/NFM+; yellow arrows), and oligodendroglial differentiation (GALC+, green). (C) Extreme limiting dilution assay assessment of the limiting dilution spherogenic potential shows significantly reduced tumor sphere formation at a high cell density per well in 96-well plates. Combo siRNA against TFs abolished single-cell spherogenesis for both the GBM43 and MGG8 BTICs (n = 12; Pearson’s χ2 test). (D) Pretreatment of multiplex RNAi for both GBM43 and MGG8 BTICs significantly prolonged the survival of animals injected with GBM43 or MGG8 cells (n = 5; log-rank test).
Application of 7C1 LPNPs for BTICs.
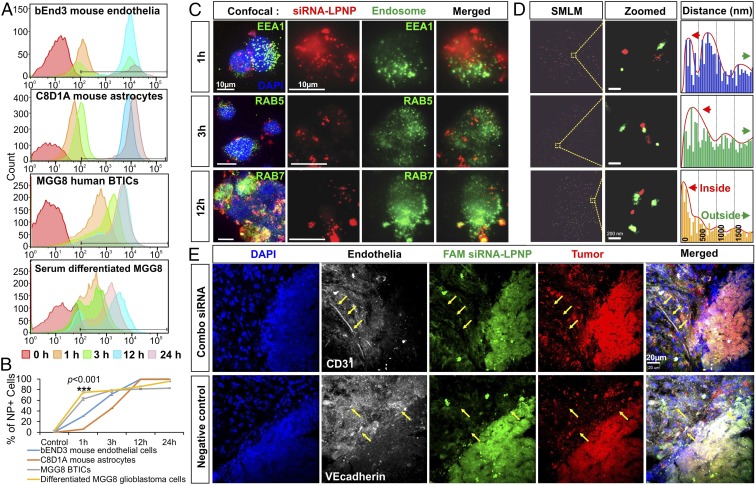
To examine whether the 7C1 nanoparticles (NPs) were suitable for BTIC targeting, we first analyzed the in vitro uptake profiles of MGG8 BTICs using 7C1 LPNPs encapsulated with siRNAs tagged with the Alexa 647 fluorophore. Flow cytometry after an in vitro incubation time series showed surprisingly faster uptake in BTICs than in mouse endothelial cells at 1-h incubation, although by 12–24 h the percentage of BTICs showing NP uptake plateaued at around 80% (Fig. 4A), whereas both the mouse brain endothelial cell line bEnd3 and the astrocyte line C8D1A saturated at nearly 100% (Fig. 4B). This variance might be explained by the growth conditions of these cell lines; BTICs were grown as a tumor sphere suspension, preventing NP uptake in the core of spheres, which form quickly during the incubation of the time series after the initial Accutase dissociation, whereas bEnd3 and C8D1A were grown as adherent cells with flat cell bodies that covered the entire surface area of the culture flasks with maximal exposure to NPs. Serum-containing medium differentiates BTICs (24) and changes the spherogenic growth conditions of BTICs to adherent growth patterns. After 48 h of conditioning in differentiation medium, MGG8 cells demonstrated a higher LPNP uptake rate at 24 h of incubation, similar to that of the adherent mouse brain cells (Fig. 4A). Confocal microscopy indicated that 7C1 NPs were readily endocytosed by BTICs, showing endosomal presence at various stages (Fig. 4C), but more notable is the evidence of endosomal escape at the early stages of LPNP uptake, in that Alexa 647-tagged siRNA in LPNPs can be observed outside the endosomes (Fig. 4C). Heterogeneous intracellular distribution of the endocytosed LPNPs can be observed throughout different time points after the initial treatment within the cell population, because super-resolution nanoscale imaging via single-molecule localization microscopy (SMLM) demonstrated a broad range of distances between antibody-labeled endosomes and the endocytosed LPNPs (Fig. 4D, and see the enlarged view in SI Appendix, Fig. S5), suggesting that perhaps more than one cell-entry mechanism was involved. However, siRNA–LPNP accumulation in late endosomes (RAB7+) may foreshadow a reduced efficiency in gene knockdown. Future investigations to elucidate the exact cell-entry pathways are needed. Interestingly, in vivo delivery of these LPNPs shows strong affinity for BTICs (Fig. 4E), with minimal dispersal to nontumor regions in vivo. Although the cell surface-targeting molecules of the 7C1 lipopolymeric formulation remain unknown, the in vitro and in vivo selectivity for BTIC uptake establishes a solid rationale for BTIC-targeted 7C1 delivery of therapeutic siRNAs.
Fig. 4.
Characterization of siRNA–NP uptake profiles in vitro and in vivo. To characterize the siRNA–NP uptake dynamics in BTICs, in vitro LPNP loading was performed on single-cell suspensions of MGG8 and serum-differentiated MGG8 cells. The bEnd3 mouse brain endothelial cell line and C8D1A mouse astrocytes were used to compare the uptake profiles. (A) Cells were enzymatically digested into single-cell suspensions at various time points for flow cytometry analysis after NP loading was done for different durations: 0, 1, 3, 12, and 24 h. Superimposed flow cytometry graphs collected after cells were loaded with siRNA (Alexa 647-tagged)–NP complexes show swifter uptake of siRNA–NPs in the BTICs than in the bEnd3 mouse brain endothelial cell line and the C8D1A mouse astrocyte line. Serum in the culture medium changes MGG8 cells into adherent phenotypes and alters LPNP uptake dynamics. (B) MGG8 BTICs demonstrate a significantly higher percentage of LPNP uptake by 1 h after seeding (n = 4, ***P < 0.001, t test). The NP+ MGG8 cells plateaued at around 80% of the population by 12 h after NP seeding, the plateauing is reversed by exposure to serum in the culture medium. Mouse bEnd3 and C8D1A cells became nearly saturated by 12 h of NP–siRNA seeding. (C) Confocal microscopy of MGG8 cells shows nearly uniform rapid uptake of FAM-tagged siRNA–NP complexes (green dots) in the cytosol, with some colocalization with the early endosomal marker EEA1 (red signals; yellow indicates colocalization) (Top Row) or RAB5 (red signal) (Middle Row). By 12 h, some of the siRNA–NP complexes were consolidated to RAB7+ late endosomes (yellow signals) (Bottom Row). (D) SMLM super-resolution images of the cells shown in C are analyzed for the relative distances between endosomes (green dots) and the endocytosed Alexa 647-tagged siRNA–LPNPs (red dots). An enlarged view is shown in SI Appendix, Fig. S5. The diverse distribution patterns of distances between LPNPs and endosomes at different time points postloading indicate endosomal escape and potentially additional endocytosis pathways bypassing endosomes. (E) The in vivo distribution of the siRNA–NP complexes (shown in green because of FAM tagging of the siRNAs) is restricted to the BTIC xenograft (shown in red because of mCherry-fLuc lentiviral transduction) at day 4 after intratumoral delivery via an s.c. osmotic pump. Distribution outside the tumor cell xenograft is very limited and is not observed in CD31+ or VE-Cadherin+ mouse brain vasculature (yellow arrows). The border between the tumor xenograft and the FAM-tagged siRNA–NP complexes is clearly visible.
Validation of LPNPs for Combination RNAi.
Instead of using transfection reagents, LPNPs serve as the packaging and delivery vehicle by shielding the negative charge of the siRNA duplex while offering the benefits of low toxicity and decreased liver targeting and kidney clearance when given systemically in the blood stream (23). To examine the feasibility of using LPNPs to deliver RNAi to BTICs and potentially to a tumor-associated microenvironment such as tumor-associated blood vessels, in vitro knockdown of the four master TF mRNAs was performed using 7C1 LPNPs formulated with a combination of siRNAs. Assessment of both the mRNA and protein level confirmed knockdown (Fig. 5), and tumorigenic spherogenesis was suppressed (Fig. 5), albeit to a reduced extent when compared with direct transfection at equivalent siRNA doses. This reduction was likely caused by the endosomal retention of some of the NPs, a well-known long-standing challenge for nanocarrier-mediated siRNA knockdown. In addition, the impact of combo siRNA packaged in LPNPs was different for the distinct GBM subtypes, e.g., the spherogenic potential of MES83, a predominantly mesenchymal BTIC subtype, showed little response to the combo siRNA treatment (Fig. 5C). The predominant biological impact of the LPNP-mediated combo siRNA treatment, based on the Click-iT EdU assay and Ki67 immunostain flow cytometry assays, is the inhibition of cell proliferation (SI Appendix, Figs. S6 and S7). However, different BTICs respond differently when analyzed for cell death (SI Appendix, Fig. S8), with high-dose treatments showing minor toxicity in normal human brain cells, such as astrocytes (SI Appendix, Fig. S8A).
Fig. 5.
Functional characterization of nano RNAi in vitro. (A) Using 7C1 LPNPs to encapsulate siRNAs against master TFs, we show that various degrees of knockdown of TF expression can be observed across different cell lines using 100-nM combination siRNA mixtures (n = 4, *P < 0.05, ***P < 0.001, t test). (B) Single siRNAs were used to compare the relative knockdown of TF protein levels with the combination mixture. There was synergy between TFs in the combination (combo) strategy. Combo siRNAs provided the greatest reduction in protein level by 72 h (***P < 0.001). (C) A functional reduction in BTIC spherogenic potential is demonstrated by a limiting dilution sphere-formation assay that shows significant differences in the ability of various cell lines to form tumor spheres (a significant reduction in stem cell frequency in the treated cells) as a result of the combo nano RNAi therapy (n = 12, ***P < 0.001; Pearson’s χ2 test). This effect is not seen in MES83 cells.
In Vivo Evidence of Tumor Growth Inhibition Mediated by the 7C1 NP–siRNA Complex.
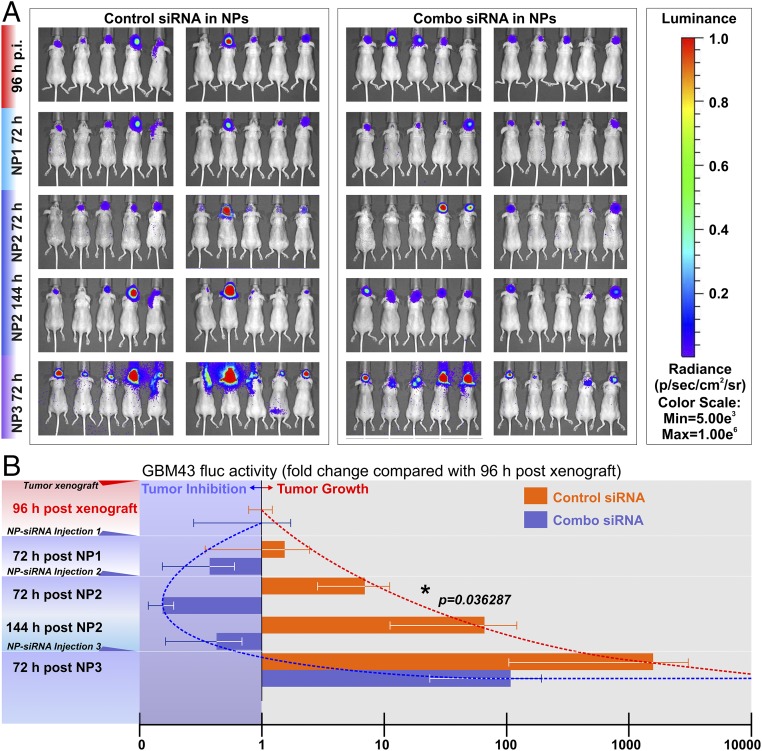
To examine the potential of the7C1 siRNA–NP complex for intratumoral delivery and tumor growth suppression, GBM43 BTIC xenografts were established in mice based on bioluminescence imaging (BLI) signals of firefly luciferase (fLuc)-positive GBM43 tumor mass formation after xenograft implantation. Mice then were given either nontargeting control siRNA–NP treatments or combo siRNA-NP treatment for the TFs via intratumoral injection. BLI signals indicate that the early-stage repeated intratumoral injection of combo siRNA–NP complexes significantly reduced tumor growth as compared with the injection of nontargeting siRNA control NPs at the midpoint of the regimen (72 h after second dosing) (Fig. 6). However, depending on the dosing and frequency of combo siRNA–NP injections, the tumor-inhibiting effects can be short lived (Fig. 6).
Fig. 6.
Functional validation of the antitumor effects of siRNA–NP complexes in vivo. To evaluate the in vivo antitumor impact of the nano RNAi strategy, mice grafted with GMB43-fLuc were given intratumoral combo siRNA–NP complexes against the four TFs. (A) Starting at 96 h post tumor cell xenograft implantation (p.i.), the GBM43 fLuc signals were captured as the baseline activity (one-fold). Interspersed siRNA–NP intratumoral injections were given three times (NP1, NP2, NP3), and the fLuc activities of GBM43 xenografts were captured at 72 h postinjection (an additional time point at 144 h after NP2 was included also). (B) Multiplexed combo TF siRNA–NPs inhibited the fLuc activity and changed the tumor growth trajectory (blue trend line), indicating reduced tumor cell expansion in vivo. Repeated nano RNAi therapy led to a significant reduction of fLuc activity 72 h after the second combo siRNA–NP injection (NP2; n = 10, *P < 0.05, t test), an indicator of in vivo tumor growth arrest. Nontargeting negative control siRNAs in NPs did not stop tumor growth (red trend line). However, the antitumor effect was not sustainable. By the third combo siRNA-NP treatment (NP3), the tumor growth trajectory was restored. However, there was no overall difference between the two groups by ANOVA, indicating that interspersed combo siRNA-NP therapy is insufficient to stop tumor growth entirely.
Contrasting in Vivo Delivery Strategies Resulted in Varying Therapeutic Efficacy.
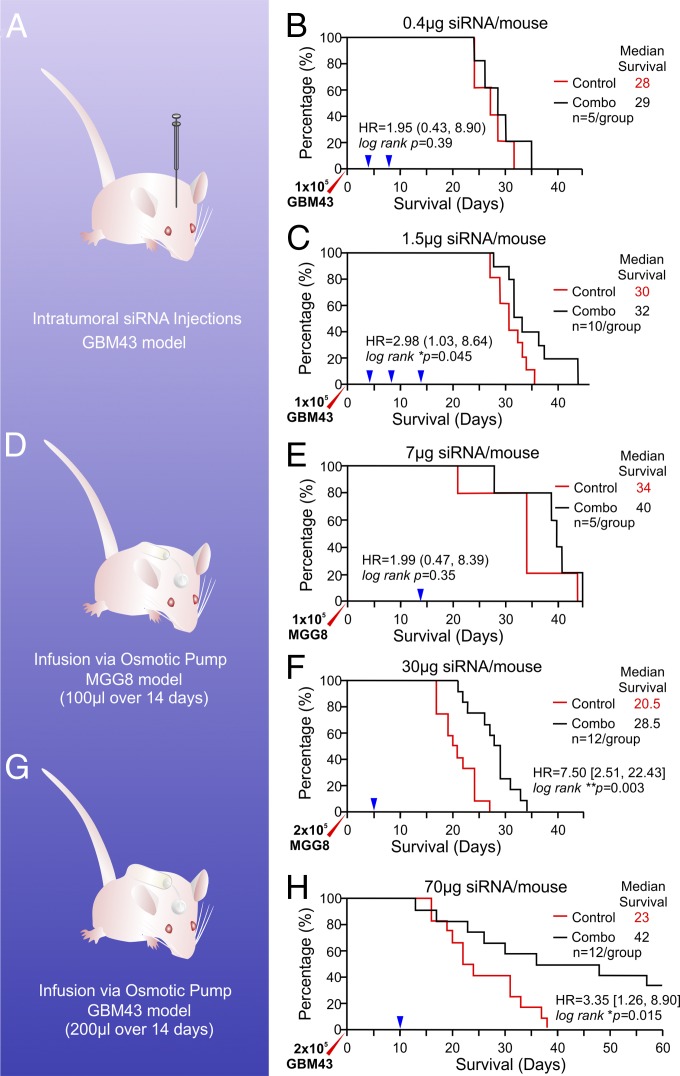
The landmark demonstration of RNAi using double-stranded siRNA in Caenorhabditis elegans (40) and subsequent in vivo demonstration of siRNA therapeutic benefits (41) paved the way for further advances in translational explorations of this technology. Several obstacles with respect to tissue specificity and sustainable potency must be overcome to achieve therapeutic outcomes. Protection against naturally occurring nuclease cleavage, enhanced cellular uptake, and endosomal escape with charge modification and the reduction of kidney clearance by increasing size are key milestones of successful in vivo delivery of siRNAs, and our solution of using a unique class of lipopolymeric nanoconstructs to complex therapeutic siRNA duplexes accomplishes all indices in one fell swoop (14, 15, 22, 42). Overcoming these obstacles helps reduce off-target effects and minimizes toxicity to liver and immune cells in vivo; however, the endothelia-targeting properties of these NPs indicate that systemic delivery of candidate siRNAs is not ideal for targeting brain tumors, even though tumor-induced angiogenesis is prevalent in GBM (43) and cerebral blood flow volume is one of the largest parts of the total cardiac output. Indeed, the biodistribution study of the 7C1 LPNPs used for our study indicates negligible presence in the brain tissue (23). However, with the recent recognition of perivascular niches for BTIC invasive growth (11, 44–47), direct in vivo RNAi delivery via nano vehicles with vasculature affinity may increase the retention and therapeutic efficacy of BTIC-targeting siRNAs in the tumor microenvironment by minimizing off-target distribution. Based on patient data published in the IVY Glioblastoma Atlas Project (IVY GAP; glioblastoma.alleninstitute.org), we identified loci of high TF expression within the bulk tumors of patients diagnosed as having various subtypes of GBM (SI Appendix, Fig. S1). We experimented with two approaches to examine the impact of local direct delivery of the 7C1 siRNA–NP therapy (nano RNAi) to BTIC xenografts. First, we gave two intratumoral injections of combination siRNA–LPNPs (20 µg/kg; 5 µL per injection; 4 d apart) to mice that had been implanted 4 d previously with 1 × 105 xenograft GBM43 cells. There were no survival benefits compared with the control group receiving nontargeting control siRNA–LPNP (P = 0.39, log-rank test; n = 5) (Fig. 7 A and B). We then increased the therapeutic dosing to 75 µg/kg delivered in three separate injections 4–6 d apart starting 4 d (96 h) after tumor cell implantation. There was a significant, but marginal survival benefit (P = 0.045, log-rank test; n = 10; median survival increase = 2 d) (Fig. 7C). Because repeated surgery introduces stress and pain that may impact the survival of the experimental animals, we opted for the convection-enhanced delivery (CED) strategy using an Alzet osmotic pump to deliver a continuous supply of the nano RNAi combination at a rate of 6 µL/24 h for 14 d; this strategy had been successful in our previous application in a rodent model of CNS disorders (48). MGG8 was implanted at a dose of 1 × 105 cells per mouse; then we implanted the pump (Durect Corp.) delivering 350 µg/kg (total average dose, 7 µg per mouse) through a brain infusion kit (Fig. 7D) 10 d after the tumor xenograft to allow sufficient integration of the tumor cells with the host brain microenvironment and to establish brain–blood vessel cooperation and invasive perivascular niches. The implant was left s.c. until the study end point. By design, there is an option to remove the pump upon complete cargo release (by 14 d after implantation) and to replace it with a new pump for continued drug delivery for up to 36 cycles; however, we opted not to do so to reduce surgery-related stress. This single-pump study was designed to establish the feasibility and to document the therapeutic benefit dynamics. The waiting period of 10 d may have enabled further tumor cell invasion of healthy parenchyma, reducing the chance of delivered drug coverage and leading to highly uneven therapeutic outcomes across the five mice per group. As a result, there was a noticeable median survival benefit of 6 d, but the result lacked statistical significance because of variability within the small sample size (n = 5; P = 0.35) (Fig. 7E). Next, we used MGG8 cells implanted at the doubled dose of 2 × 105 cells per mouse to increase the therapeutic threshold, nearly quadrupled the nano RNAi drug dose, and reduced the waiting period after tumor implantation to 5 d. We gave the mice the 1.5 mg/kg nano combination RNAi in a 14-d pump beginning 5 d after tumor implantation. A clear statistical survival benefit was seen when the combination nano RNAi was compared with nontargeting nano siRNA controls (n = 12; P = 0.003, log-rank test) (Fig. 7F), with a median survival extension of 8 d. Because in the nano RNAi treatment group the first animal reached endpoint at 21 d after tumor xenograft, and the last animal reached the end point at 34 d, i.e., 2–15 d after the 14-d nano RNAi supply in the osmotic pump was exhausted, it is conceivable that further survival benefit can be achieved by using either a higher drug concentration or a longer period of drug treatment via replacement pumps. Indeed, using a larger osmotic pump, Alzet model 2002 (delivering 200 µL at a rate of 12 µL/24 h for 14 d) (Fig. 7G) and a GBM43 model established with 2 × 105 cells 10 d before pump implantation, we observed a benefit of 19 d for median survival extension compared with the control group (Fig. 7H). Although there could be a component of high-dose–induced nonspecific toxicity (SI Appendix, Fig. S8), because the control siRNA–LPNP treatment group showed a broadened survival curve ranging over 16–38 d, the overall tolerance of this high-dose delivery is evident in the survival profile of the combo siRNA-treated group (Fig. 7H).
Fig. 7.
In vivo assessment of the survival benefit of nano RNAi on BTIC-driven tumorigenesis. (A) Schematic of the intratumoral bolus injection regimen. (B) Low-dose intratumoral injection (two injections) 4 d after tumor xenograft did not generate survival benefits (n = 5, P = 0.39, log-rank test). (C) A higher drug dose (75 µg/kg) and more redosing (three injections) promoted survival significant benefits for mice with the GBM43 xenograft (*P < 0.05, log-rank test, n = 10), although the medium survival is only 2 d longer than in controls. (D) Schematic of s.c. osmotic pump implantation. (E) A low (7-μg) nano RNAi dose delivered via an s.c.-implanted Alzet pump did not offer significantly longer survival, given the smaller sample size (n = 5), even though there is a median benefit of 6 d. (F) A higher dose (1.5 mg/kg) and larger sample size demonstrated a significant survival benefit for mice with twice as many tumor cell xenografts (n = 12, ***P = 0.003, log-rank test). (G) Schematic of a larger pump implantation. (H) Improved survival profile for xenograft model of GBM43 BTICs (median survival extension = 19 d; log-rank test, *P = 0.015).
Last, it is important to note that the intratumoral delivery of combo siRNA–LPNP indeed inhibits the expression levels of the core TFs based on tissue immunocytochemistry (SI Appendix, Fig. S9) and qRT-PCR following in situ tissue collection via laser-capture microdissection (LCM) (SI Appendix, Fig. S10); however, once the dosing is over, i.e., when the content of the osmotic pumps was consumed, the core TFs may revert back to high expression levels and eventually lead to the demise of the tumor-bearing mice.
Discussion
The recognition of RNAi as a potential anticancer therapeutic strategy coincides with a surging flux of knowledge about the genetic circuits of BTICs and their regulatory impacts on epigenetic states that underscore tumorigenesis. Genome sequencing and transcriptional profiling of contrasting GBM subpopulations have shed light on a large number of genetic events that divide GBM into subclasses of tumors with contrasting hierarchical dependence on disparate genetic aberrations, highlighting the daunting challenge for therapeutics to impede tumor growth through a unified antitumor mechanism. Compounding the complexity of the murky state in the cellular hierarchy within an individual GBM is the recent recognition (based on single-cell RNA sequencing analysis) that cellular heterogeneity is prevalent in individual GBMs (49). The differentiation states of GBM cells have been implicated in the determination of epigenetic plasticity, tumor initiation, and therapy resistance (50, 51), highlighting the potential driving roles of BTICs in GBM malignancy. Because numerous phenotypic markers [e.g., CD133 (52), SSEA-1 (53), CD44 (54), and Integrin α6 (55)] have been used to enrich putative stemlike populations within the GBM, a consensus-oriented therapeutic targeting approach is unlikely to encompass all subtypes of BTICs in a highly heterogeneous GBM. This complexity creates a daunting task for therapeutic targeting of malignant BTICs. Inspired by recent advances in the field of induced pluripotent stem cells (56, 57), efforts have been made to decipher the intrinsic transcriptional network of the cellular hierarchy within the privileged BTIC subpopulations and have resulted in the identification of four master TFs responsible for the maintenance of BTIC phenotypes (24). The expression of these master TFs seems to correlate with the operation of the phenotypic markers established previously, offering hope that similar hierarchical investigations could eventually shed light on common denominators of tumor initiation and enable the consolidation of therapeutic targets.
The concept that forced differentiation abolishes the malignant epigenetic programs underlying tumorigenesis was strengthened further, thanks to efforts in the field of leukemia (58), by the global reset of the epigenetic states to override the tumor genome (59). Although a single factor [e.g., bone morphogenic protein 4 (BMP4) (60)] is capable of inducing BTIC differentiation, complex signaling mixtures, such as serum (61), or a chromatin regulator complex, such as PRC2 (62), demonstrate gradual and at times reversible epigenetic override of the malignant gene circuits of BTICs. Although small-molecule antagonists or inhibitors of the stemness signaling pathways are being explored, the promising safety profile and gene-targeting specificity render siRNAs the ideal therapeutic for modifications of the TF-driven epigenetic state and for prodifferentiation approaches toward malignant BTICs. Given that pro-stemness signaling mechanisms continue to be identified via extensive BTIC epigenetics analysis (63), it is clear that there are no “be-all and end-all” BTIC targets in the context of various tumorigenic genetic subclasses. Combination siRNA targeting of identifiable targets for individual tumors in a personalized fashion becomes a rational goal of therapeutic development. In our experiments, we set out to use patient-derived tumor cells with genetically defined malignancy drivers for siRNA targeting. Given that intratumoral heterogeneity is likely unavoidable (49, 64, 65), subpopulations of BTICs independent of TFs within the tumor would likely overtake the growth even if siRNA combination therapies are effective in suppressing the BTIC populations dependent on TFs (66, 67). This likelihood leads to the prospect of further combined targeting of adaptive malignancy drivers in response to TF suppression as an additional strategy toward personalized precision medicine.
The key limitation to translating the latest genetic/epigenetic discoveries to patient care is drug delivery efficiency. The brain is a privileged organ with broad restrictions on therapeutic penetrance across the BBB. Although promising, RNAi directed at the specialized BTIC populations intertwined within the brain vasculature and normal brain cells face a number of challenges for distinguishable therapeutic benefits. First, siRNA or miRNA as RNAi reagents must survive the RNase activities in the brain tissue microenvironment, so NP encapsulation or conjugation has become a standard approach to shield the therapeutic targeting and in vivo delivery of RNA from degradation. A second challenge is targeting the tumor and administering sufficient accumulated dosing to tumor cells. The third challenge is the requirement for low toxicity and minimal off-targeting and side effects. Last, sustainable dosing at the tumor site is necessary, given the dynamic nature of malignant tumor genetic/epigenetic adaptations to therapy. Numerous nano constructs have been designed to address each of these elements (32, 34, 35, 68, 69); our approach comprehensively combines the favorable benefits of several modalities. It builds on the blood vessel-targeting capacity of the customizable 7C1 lipopolymeric nano vehicle, which has excellent safety profiles (14, 15, 23, 70). The 7C1 delivery system ensures dual strength in direct delivery to the tumor cellular core (where the great majority of BTICs reside) and vasculature-associated invasive niches (11, 44–47) (where residual resistant populations of BTICs accumulate after a standard therapy regimen). Additionally, we maintained continuous focused delivery via a CED brain-infusion catheter that is undergoing clinical trials for both malignant brain tumors and neurodegenerative diseases (71–74). These combined considerations minimize the issues frequently associated with nanomedicine in regards to off-target delivery, imbalanced biodistribution, and the potential for systemic toxicity when high systemic dosing is required to achieve a detectable presence in brain tumor. Concordantly, with this approach we still maintain the capacity to reach the invasive niches that often develop in the intraparenchymal perivascular spaces along the brain vasculature through local distribution and vasculature targeting. Our current data indicate that sustained local delivery significantly extends survival, whereas limited bolus intratumoral injections offer marginal benefits, validating the logic of the delivery strategy. Extending the CED delivery beyond the current dosing period of 14 d may offer additional survival benefit when siRNA dosing limits per unit vehicle are in place with the current nano packaging technologies. Although there could be concerns about the potential for nonmodified siRNA duplexes to induce innate immune response for antitumor effects (75, 76), our use of nu/nu mouse models with human BTIC xenografts and the universal nontargeting negative siRNA controls mitigated this common misinterpretation of antitumor results. For clinical translation, this potential impact will be assessed in immune-competent mouse models, which could reveal the impact of these LPNPs on the immune response of GBM.
In the current report we identify a previously unknown affinity of 7C1 LPNPs for BTICs and describe a CED strategy of 7C1 LPNP-enabled BTIC RNAi therapy in patient-derived xenograft models of GBM. We proved that (i) a multiplexed nanomedicine strategy for RNAi targeting malignant BTIC genomic TF drivers is feasible; (ii) CED-enabled intratumoral delivery offers direct evidence of therapeutic benefits of RNAi therapy against BTICs; and (iii) limited dosing is insufficient to overcome tumor growth despite a clear attenuation of malignant tumor growth. This third finding leads to the prospect of additional multiplexed RNAi therapies against resistant populations of BTICs beyond the four TFs under investigation, once the adaptive malignancy genomic drivers are identified. The study also exposed the need to improve on the current intracranial distribution of nano therapeutics beyond experimental models of CED using Alzet s.c. osmotic pumps. In addition, nontransient RNA-based nanomedicine therapies using new genome-editing tools, such as the rapidly evolving CRISPR technologies, can be delivered via the customizable LPNPs for sustained elimination of tumorigenic molecular markers (14, 70). Furthermore, continued refinement and more customized functionalization of the NPs via surface chemistry engineering may allow antitumor gene editing in multiple cell types present in the tumor microenvironment (e.g., in immune cells and astrocytes) which can be recruited in a locally coordinated antimalignancy campaign to boost therapeutic durability and efficiency.
Materials and Methods
Cell Culture and Reagents.
A panel of patient-derived GBM cells (GBM6, GBM12, GBM26, and GBM43, a gift from C. David James, Northwestern University; MGG8, a gift from M.L.S., Massachusetts General Hospital/Harvard Medical School; and MES83, a gift from Ichiro Nakano, University of Alabama at Birmingham and Shi-Yuan Cheng, Northwestern University) were maintained as neurospheres in serum-free Neurobasal or DMEM F12 medium with B27 and N2 supplements (Thermo Fisher Scientific), broad-spectrum antibiotics, l-glutamine, heparin (Millipore Sigma), EGF, and basic fibroblast growth factor (R&D Systems) (24, 25, 36).
In Vitro Single-Cell and Limiting-Dilution Spherogenic Assays.
Costar polystyrene 96-well plates (Thermo Fisher Scientific) were used to plate single cells per well (24) or in a series of 1, 3, 6, 12, 25, 50, 100, 200 cells per well in 200 µL of serum-free medium (as described above), with 10 µL fresh medium replacements every other day. Cells were imaged under a microscope after 7 d. A BD FACSMelody cell sorter (BD Biosciences) was used to perform the serial dilution plating (SI Appendix, Fig. S11) A Cytation 5 Cell Imaging Multi-Mode Reader (BioTek) was used to collect well images. Results of the limiting dilution assay were assessed using Extreme Limiting Dilution Analysis (ELDA) software (77).
Patient-Derived Xenograft Mouse Model of GBM.
All protocols were approved by the Institutional Animal Care and Use Committees (IACUCs) at The University of Chicago and Northwestern University. The surgical procedures were conducted according to NIH guidelines. Six- to eight-week-old athymic nude male or female mice were obtained from Envigo Research Models and Services and from Charles River Laboratories and were maintained in a pathogen-free facility. Mice were anesthetized with a ketamine HCl (25 mg/mL)/xylazine (2.5 mg/mL) solution, and patient-derived GBM43 or MGG8 cells (without or with TF siRNA knockdown; 1 × 105 or 2 × 105 cells in 2.5 μL sterile saline) were injected in the mouse brain at 2.5 mm depth through a transcranial burr hole created at coordinates 2 mm lateral and 1.5 mm caudal relative to bregma using an established procedure (78). Standard postsurgery care was given following the IACUC-approved protocol. No gender differences were observed in the survival period after tumor cell implantation.
Intratumoral Delivery of Combo siRNA-Encapsulating 7C1 NPs.
All protocols were approved by the IACUC at Northwestern University. Five days after tumor cell implantation, 2–3 separate bolus intratumoral injections of siRNA-complexed 7C1 NPs were given at 4- to 6-d intervals to tumor-bearing mice through the skull burr hole created for tumor cell implantation. Alternatively, Alzet s.c. osmotic pumps (Durect Corporation model 1002, with a capacity of 100 µL over 14 d with a flow rate 6 µL/24 h or model 2002, with a capacity of 200 µL over 14 d with a flow rate 12 µL/24 h) were implanted to supply continuous intratumoral delivery of the siRNA-complexed 7C1 NPs via Brain Infusion Kit 3 (Durect Corporation), which implants a catheter at the depth of tumor cell implantation.
BLI Assessment of Tumor Growth.
Mice were given an i.p. injection of fLuc substrate d-luciferin (GoldBio) before isoflurane anesthesia in an induction chamber for 10 min. BLI signals of tumor growth were captured using a PerkinElmer IVIS Spectrum System with Living Image software 96 h after the tumor cell xenograft, 72 h after the first intratumoral NP injection (NP1), 72 h and 144 h after the second intratumoral NP injection (NP2), and 72 h after the third intratumoral NP injection (NP3). Regions of interest (ROIs) were drawn to cover the entire brain. The sum of the counts within the ROIs was obtained by subtracting the background counts.
Western Blotting.
The proteins were extracted using M-PER reagent (Thermo Fisher Scientific). Equal amounts of proteins were loaded into 4–20% gradient gels (Bio-Rad or Thermo Fisher Scientific) and transferred to an Immobilon FL PVDF membrane (Millipore). The primary antibodies used were against human SOX2, OLIG2 (mouse IgG, R&D Systems, and rabbit IgG, Millipore Sigma), SALL2, POU3F2 (rabbit IgG, Bethyl Laboratories), and β-actin (Cell Signaling Technologies). The secondary antibodies used were anti-rabbit or anti-mouse IgGs conjugated with infrared dyes for multiplex quantitative Western Blot or HRP-conjugated anti-rabbit or anti-mouse IgGs for chemiluminescent blotting. Stained membranes were imaged on a Li-COR Odyssey scanner, and the data were quantified using Image Studio Lite software (Li-COR Biosciences).
Flow Cytometry and Immunocytochemistry Analysis of TF Knockdown.
To analyze TF expression levels, tumor cells were collected after Accutase treatment and were resuspended in Fix/Perm buffer (BioLegend) or were fixed in 2% paraformaldehyde (PFA) followed by permeabilization using 0.03% TritonX-100 in PBS at 2 × 105 cells per 200 µL per well. Cells then were stained with primary antibodies against SOX2, OLIG2, SALL2, POU3F2, or isotype control IgG (eBioscience). Allophycocyanin-labeled secondary antibodies against rabbit IgG were used to incubate cells after washes following primary antibody incubation. Cells were analyzed using a BD LSRFortessa analyzer and FACS Diva software (BD). The flow cytometry data were processed using FlowJo software (FlowJo LLC). Stained cells were fixed in 2% PFA before incubation with Vectashield mounting medium plus DAPI (Vector Labs) for nuclei counterstains in a glass bottom 96-well plate. Confocal microscopy was performed at the University of Chicago Integrated Light Microscopy Facility using a 3i Marianas Yokogawa-type spinning disk confocal microscope with an Evolve EMCCD camera (Photometrics) running SlideBook v5.5 software (Intelligent Imaging Innovations). Digital images then were processed and analyzed for quantitative measurements using Fiji Software.
CellSimple Cell Death and Proliferation Assays.
A CellSimple Cell Analyzer (Cell Signaling Technology) was used to analyze the impact of LPNP–siRNA therapy on BTICs. A CellSimple Annexin V early apoptosis detection kit (Cell Signaling Technology) and a Click-iT EdU Alexa-594 flow cytometry proliferation assay kit (Thermo Fisher Scientific) were used with the CellSimple cassettes for efficient, timely, and accurate live-cell profiling.
qRT-PCR Analysis.
Total RNA was isolated from GBM cells using the RNeasy Plus Kit (Qiagen). One microgram of RNA was reverse-transcribed using the iScript cDNA conversion kit (Bio-Rad). qRT-PCR was conducted using the SYBR Green PCR Kit (Bio-Rad) using the primers indicated in SI Appendix, Table S1. Data analysis was performed using the 2−ΔΔCT method for relative quantification, and all sample values were normalized to the human GAPDH expression value.
Complexing siRNA Formulations into 7C1 NPs.
Purified 7C1 NPs were synthesized and formulated as previously described (23). Specifically, polyethyleneimine with a molecular weight of 600 (PEI600; Millipore Sigma) was combined with 200-proof anhydrous ethanol (Koptec/VWR International) and an epoxide-terminated C15 lipid at a lipid:PEI molar ratio equal to 14:1. The mixture was heated at 90 °C for 48 h before purification was performed with a silica column as previously described (23). To formulate NPs, purified 7C1 was combined with 200-proof ethanol and 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] (Avanti Polar Lipids) at a 7C1:lipid-PEG molar ratio equal to 4:1 in a glass syringe. siRNA was dissolved in pH 3 10 mM citrate solution (Teknova) in a separate syringe. The two syringes were connected to a syringe pump, and the fluid was pushed through a microfluidic device as previously described (22). The resulting NPs were dialyzed in 1× PBS and were sterilized using a 0.22-μm polyethersulfone syringe filter (Genesee Scientific).
NP Characterization.
NP size and structure were analyzed by dynamic light scattering (DLS) (Zetasizer NanoZS; Malvern Instruments) or cryogenic transmission electron microscopy (cryo-TEM) as previously described. DLS samples were measured in sterile 1× PBS at an approximate siRNA concentration of 1.0–3.0 g/mL. Cryo-TEM samples were prepared in a controlled-environment vitrification system at 25 °C and ∼100% relative humidity.
Analysis of NP Uptake Dynamics.
To evaluate the uptake dynamics of the LPNP–siRNA complexes, LPNPs were complexed with Alexa 647-conjugated siRNA sequences and were loaded onto Accutase-dissociated single MGG8 BTICs or control cells (bEnd3 mouse brain endothelial cells and C8D1A mouse astrocytes from ATCC or serum-differentiated MGG8 GBM cells) for incubation for 1, 3, 12, or 24 h. Cells without LPNP incubations were used as baseline controls. The percentage of cells showing uptake of the Alexa 647–siRNA–NP complex was measured using a BD LSRFortessa cell analyzer (BD Biosciences) and FlowJo software. The intracellular locations of the endocytosed LPNPs were determined via coimmunostaining with endosomal marker antibodies such as anti-EEA1, anti-RAB5, and anti-RAB7 antibodies (Cell Signaling Technology) using 2% PFA-fixed BTICs, and the Atto-488–conjugated secondary antibody against rabbit IgG was used to contrast the Alexa 647-tagged siRNA–LPNP complexes. The stained cells then were imaged under a conventional confocal microscope at The University of Chicago Microscopy Core. Images were captured with a 3i Marianas Yokogawa-type spinning disk confocal microscope with an Evolve EMCCD camera (Photometrics) running SlideBook v5.5 software (Intelligent Imaging Innovations).
Super-Resolution SMLM Analysis.
To enhance the optical resolution of subcellular organelles and nanoscale LPNP aggregates further after biological uptake into the cytosol of BTICs, we performed super-resolution imaging of BTICs after LPNP uptake at different time points, using a custom SMLM super-resolution nanoscopy (79, 80). To perform SMLM imaging, 5-μL cell suspensions of each sample were mixed with the imaging buffer and deposited at the center of a freshly cleaned glass slide. The imaging buffer was freshly made and contained TN buffer [50 mM Tris (pH 8.0) and 10 mM NaCl; all chemicals are from Millipore Sigma], an oxygen-scavenging system (0.5 mg/mL glucose oxidase), 40 μg/mL catalase and 10% (wt/vol) glucose, and 143 mM β-mercaptoethanol. A no. 1 coverslip was used to cover the sample and was sealed with dental cement on the glass slide. The samples were placed on the microscope stage and imaged under a total internal reflection fluorescence objective (Nikon CFI apochromat 100×, 1.49 NA). The 473-nm and 645-nm lasers were used to excite fluorescence from Atto-488 and Alexa-647 fluorophores, respectively. Before acquiring SMLM images, we used relatively low-intensity light (∼0.05 W/cm2) to illuminate the sample and recorded the conventional fluorescence image. We then increased the light intensity to ∼2 kW/cm2 to switch off the dyes rapidly for SMLM imaging. We recorded 2,000 images using an EMCCD camera (iXon Ultra 897; Andor) at a frame rate of 50 Hz with field of view of 20 × 20 µm. The super-resolution image was generated using a standard localization algorithm (ThunderSTORM, ImageJ plug-in) (81). To quantify the spatial correlation of the siRNA–NP and endosome, pair correlation functions [g(r)] were calculated using a custom MATLAB code.
Immunocytochemistry Assessment of Tumor Cell Differentiation in Vitro and Growth in Vivo.
Cultured BTIC neurospheres were treated with combo siRNA–LPNPs in vitro for 48 h in 12-well plates containing sterile cover glasses, and cells were fixed on the cover glasses once the morphology became indicative of differentiation using 2% PFA. Cells then were permeabilized and incubated with blocking solution, primary antibodies against differentiation markers such as TUJ1 and NFM for neural lineage, GFAP and S100 for glial lineage, and GalC for oligodendroglial lineage (antibody details are listed in SI Appendix, Table S3). After they were killed, select animals were perfused with 4% ice-cold PFA in 0.1 M PBS to collect tumor-bearing brain tissue. Following postfixation and cryoprotection, brain tissues were embedded in OCT compound (Sakura-Finetek USA Inc.) and were promptly frozen on a mixture of dry ice and isopentane (Thermo Fisher Scientific). Sectioned brain tissue (10 or 20 µm thick) was stained for human cell markers with antibodies against human-specific Nestin (Millipore) to identify human GBM43 and MGG8 tumor cells. TF antibodies then were used to costain for BTIC status. Cy2-, Alexa 555-, or Alexa 647-conjugated secondary antibodies against rabbit IgG or mouse IgG (Thermo Fisher Scientific or Jackson ImmunoResearch) were used to visualize specific binding. Whole-slide scans were performed using a 3DHistech Pannoramic Scan whole-slide scanner (Perkin-Elmer) with a Zeiss Axiocam MRm CCD camera (Carl Zeiss Microscopy) for fluorescence or a Stingray F146C color camera (Allied Vision Technologies) for histology.
LCM and qRT-PCR After Linear RNA Amplification.
To confirm the knockdown impact of siRNAs encapsulated in LPNPs after in vivo administration in xenograft GBM models, freshly harvested mouse brains bearing a GBM xenograft (n = 6) were flash-frozen with OCT compound in isopentane (Thermo Fisher Scientific) chilled with dry ice. These brain tumors were treated with either a single 5-µL LPNP injection (n = 3) 6 d prior or an s.c. osmotic pump via brain infusion kit for 14 d (n = 3). An RNase-free work environment was ensured by wiping the working surfaces of benchtops and the machinery inside the Leica cryostat (model CM1860 UV, Leica Biosystems Nusslock GmbH) with 100% ethanol. The frozen block was then cut into 10-µm-thick sections, collected on MembraneSlide 1.0 PEN (Carl Zeiss Microscopy GmbH), and kept in a freezer at −80 °C until LCM application. A Zeiss Palm LCM system at the Northwestern University Center for Advanced Microscopy Core Facility was used to dissect the brain tumor tissue that was infused with the LPNP–siRNA complexes. The siRNA encapsulated in LPNPs had a FAM (fluorescein) tag; thus the presence of green fluorescence in the tissue was used as a mark for dissection element selection (SI Appendix, Fig. S10). AdhesiveCap 200 PCR tubes (Carl Zeiss Microscopy GmbH) were used to collect dissected tissue from the tissue infused with LPNP–siRNA and from control tissues in the normal parts of the mouse brain (different slides were used for different LCM samples to avoid identity confusion). Samples were kept on dry ice. A MessageBOOSTER cDNA Synthesis kit (catalog no. MBCL90310; Lucigen Corp.) was used for linear amplification of the RNA samples for cDNA conversion, and an RNA Clean & Concentrator kit (catalog no. R1015; Zymo Research) was used to purify RNA before cDNA synthesis for qRT-PCR analysis of TF expression levels.
Statistical Analysis.
All statistical analyses were performed as Student’s t test using GraphPad Prism 5 (GraphPad Software Inc.) unless otherwise specified. The sample size for each group was ≥3, where n represents biological replicates. All numerical data are reported as mean ± SEM. A Kaplan–Meier survival curve was generated, and a log-rank test was applied to compare survival distributions. For all survival experiments n represents number of animals per group. All reported P values were two-sided and were considered to be statistically significant at *P < 0.05, **P < 0.01, ***P < 0.001.
Supplementary Material
Acknowledgments
We thank Dr. Thomas J. Hope of Northwestern University for generous support with the LiCOR system; Dr. C. David James and Dr. Shi-Yuan Cheng at Northwestern University and Dr. Ichiro Nakano from the University of Alabama at Birmingham for the generous gift of patient-derived GBM cells; Dr. Vytas Bindokas at The University of Chicago Integrated Light Microscopy Core for guidance and advice in imaging applications; Paul Mehl, Carolina Ostiguin, and Dr. Suchitra Swaminathan at the Robert H. Lurie Comprehensive Cancer Center Flow Cytometry Core and Drs. Wensheng Liu, Joshua Rappoport, and Constadina Arvanitis for microscopy, bioluminescence imaging, and laser-capture microdissection at the Northwestern University Center for Advanced Microscopy (generously supported by National Cancer Institute Cancer Center Support Grant P30 CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center). This work was supported by NIH Grant R35CA197725 (to M.S.L.), a Burroughs Wellcome Collaborative Travel Grant (to D.Y.), an Elsa U. Pardee Foundation Grant (to D.Y.), and a Northwestern University I3 Pilot Grant (to C.S., M.S.L., and D.Y.).
Footnotes
Conflict of interest statement: D.G.A. and R.L. have filed intellectual property protection related to the 7C1 nanoparticle. D.Y. has an evaluation agreement with Cell Signaling Technology regarding the use of the CellSimpleTM Cell Analyzer and reagents. The authors declare that they have no further competing interests.
This article is a PNAS Direct Submission. D.A.N. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1701911114/-/DCSupplemental.
References
- 1.Wen PY, Reardon DA. Neuro-oncology in 2015: Progress in glioma diagnosis, classification and treatment. Nat Rev Neurol. 2016;12:69–70. doi: 10.1038/nrneurol.2015.242. [DOI] [PubMed] [Google Scholar]
- 2.Van Meir EG, et al. Exciting new advances in neuro-oncology: The avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60:166–193. doi: 10.3322/caac.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hambardzumyan D, Bergers G. Glioblastoma: Defining tumor niches. Trends Cancer. 2015;1:252–265. doi: 10.1016/j.trecan.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ellis HP, et al. Current challenges in glioblastoma: Intratumour heterogeneity, residual disease, and models to predict disease recurrence. Front Oncol. 2015;5:251. doi: 10.3389/fonc.2015.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seymour T, Nowak A, Kakulas F. Targeting Aggressive cancer stem cells in glioblastoma. Front Oncol. 2015;5:159. doi: 10.3389/fonc.2015.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JK, Nam DH, Lee J. Repurposing antipsychotics as glioblastoma therapeutics: Potentials and challenges. Oncol Lett. 2016;11:1281–1286. doi: 10.3892/ol.2016.4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan K, Yang K, Rich JN. The evolving landscape of glioblastoma stem cells. Curr Opin Neurol. 2013;26:701–707. doi: 10.1097/WCO.0000000000000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29:1203–1217. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hjelmeland AB, Lathia JD, Sathornsumetee S, Rich JN. Twisted tango: Brain tumor neurovascular interactions. Nat Neurosci. 2011;14:1375–1381. doi: 10.1038/nn.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z, Rich JN. Hypoxia and hypoxia inducible factors in cancer stem cell maintenance. Curr Top Microbiol Immunol. 2010;345:21–30. doi: 10.1007/82_2010_75. [DOI] [PubMed] [Google Scholar]
- 11.Ricci-Vitiani L, et al. Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature. 2010;468:824–828. doi: 10.1038/nature09557. [DOI] [PubMed] [Google Scholar]
- 12.Francescone R, et al. Glioblastoma-derived tumor cells induce vasculogenic mimicry through Flk-1 protein activation. J Biol Chem. 2012;287:24821–24831. doi: 10.1074/jbc.M111.334540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El Hallani S, et al. A new alternative mechanism in glioblastoma vascularization: Tubular vasculogenic mimicry. Brain. 2010;133:973–982. doi: 10.1093/brain/awq044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin H, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xue W, et al. Small RNA combination therapy for lung cancer. Proc Natl Acad Sci USA. 2014;111:E3553–E3561. doi: 10.1073/pnas.1412686111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sager HB, et al. RNAi targeting multiple cell adhesion molecules reduces immune cell recruitment and vascular inflammation after myocardial infarction. Sci Transl Med. 2016;8:342ra80. doi: 10.1126/scitranslmed.aaf1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Platt RJ, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee H, et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat Nanotechnol. 2012;7:389–393. doi: 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan OF, et al. Dendrimer-inspired nanomaterials for the in vivo delivery of siRNA to lung vasculature. Nano Lett. 2015;15:3008–3016. doi: 10.1021/nl5048972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fenton OS, et al. Bioinspired alkenyl amino alcohol ionizable lipid materials for highly potent in vivo mRNA delivery. Adv Mater. 2016;28:2939–2943. doi: 10.1002/adma.201505822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong Y, et al. Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates. Proc Natl Acad Sci USA. 2014;111:3955–3960. doi: 10.1073/pnas.1322937111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen D, et al. Rapid discovery of potent siRNA-containing lipid nanoparticles enabled by controlled microfluidic formulation. J Am Chem Soc. 2012;134:6948–6951. doi: 10.1021/ja301621z. [DOI] [PubMed] [Google Scholar]
- 23.Dahlman JE, et al. In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight. Nat Nanotechnol. 2014;9:648–655. doi: 10.1038/nnano.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suvà ML, et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157:580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rheinbay E, et al. An aberrant transcription factor network essential for Wnt signaling and stem cell maintenance in glioblastoma. Cell Reports. 2013;3:1567–1579. doi: 10.1016/j.celrep.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mao P, et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci USA. 2013;110:8644–8649. doi: 10.1073/pnas.1221478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim SH, et al. Serine/threonine kinase MLK4 determines mesenchymal identity in glioma stem cells in an NF-κB-dependent manner. Cancer Cell. 2016;29:201–213. doi: 10.1016/j.ccell.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng P, et al. FOXD1-ALDH1A3 signaling is a determinant for the self-renewal and tumorigenicity of mesenchymal glioma stem cells. Cancer Res. 2016;76:7219–7230. doi: 10.1158/0008-5472.CAN-15-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han L, et al. Tat-BMPs-PAMAM conjugates enhance therapeutic effect of small interference RNA on U251 glioma cells in vitro and in vivo. Hum Gene Ther. 2010;21:417–426. doi: 10.1089/hum.2009.087. [DOI] [PubMed] [Google Scholar]
- 30.Van Woensel M, et al. Development of siRNA-loaded chitosan nanoparticles targeting Galectin-1 for the treatment of glioblastoma multiforme via intranasal administration. J Control Release. 2016;227:71–81. doi: 10.1016/j.jconrel.2016.02.032. [DOI] [PubMed] [Google Scholar]
- 31.Lee TJ, et al. RNA nanoparticle as a vector for targeted siRNA delivery into glioblastoma mouse model. Oncotarget. 2015;6:14766–14776. doi: 10.18632/oncotarget.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozielski KL, Tzeng SY, De Mendoza BA, Green JJ. Bioreducible cationic polymer-based nanoparticles for efficient and environmentally triggered cytoplasmic siRNA delivery to primary human brain cancer cells. ACS Nano. 2014;8:3232–3241. doi: 10.1021/nn500704t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Danhier F, Messaoudi K, Lemaire L, Benoit JP, Lagarce F. Combined anti-Galectin-1 and anti-EGFR siRNA-loaded chitosan-lipid nanocapsules decrease temozolomide resistance in glioblastoma: In vivo evaluation. Int J Pharm. 2015;481:154–161. doi: 10.1016/j.ijpharm.2015.01.051. [DOI] [PubMed] [Google Scholar]
- 34.Cohen ZR, et al. Localized RNAi therapeutics of chemoresistant grade IV glioma using hyaluronan-grafted lipid-based nanoparticles. ACS Nano. 2015;9:1581–1591. doi: 10.1021/nn506248s. [DOI] [PubMed] [Google Scholar]
- 35.Jensen SA, et al. Spherical nucleic acid nanoparticle conjugates as an RNAi-based therapy for glioblastoma. Sci Transl Med. 2013;5:209ra152. doi: 10.1126/scitranslmed.3006839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wakimoto H, et al. Human glioblastoma-derived cancer stem cells: Establishment of invasive glioma models and treatment with oncolytic herpes simplex virus vectors. Cancer Res. 2009;69:3472–3481. doi: 10.1158/0008-5472.CAN-08-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarkaria JN, et al. Use of an orthotopic xenograft model for assessing the effect of epidermal growth factor receptor amplification on glioblastoma radiation response. Clin Cancer Res. 2006;12:2264–2271. doi: 10.1158/1078-0432.CCR-05-2510. [DOI] [PubMed] [Google Scholar]
- 38.Verhaak RG, et al. Cancer Genome Atlas Research Network Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wittrup A, Lieberman J. Knocking down disease: A progress report on siRNA therapeutics. Nat Rev Genet. 2015;16:543–552. doi: 10.1038/nrg3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 41.Song E, et al. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Med. 2003;9:347–351. doi: 10.1038/nm828. [DOI] [PubMed] [Google Scholar]
- 42.Dahlman JE, et al. Orthogonal gene knockout and activation with a catalytically active Cas9 nuclease. Nat Biotechnol. 2015;33:1159–1161. doi: 10.1038/nbt.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jhaveri N, Chen TC, Hofman FM. Tumor vasculature and glioma stem cells: Contributions to glioma progression. Cancer Lett. 2016;380:545–551. doi: 10.1016/j.canlet.2014.12.028. [DOI] [PubMed] [Google Scholar]
- 44.Cheng L, et al. Glioblastoma stem cells generate vascular pericytes to support vessel function and tumor growth. Cell. 2013;153:139–152. doi: 10.1016/j.cell.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gilbertson RJ, Rich JN. Making a tumour’s bed: Glioblastoma stem cells and the vascular niche. Nat Rev Cancer. 2007;7:733–736. doi: 10.1038/nrc2246. [DOI] [PubMed] [Google Scholar]
- 46.Hovinga KE, et al. Inhibition of notch signaling in glioblastoma targets cancer stem cells via an endothelial cell intermediate. Stem Cells. 2010;28:1019–1029. doi: 10.1002/stem.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seidel S, et al. A hypoxic niche regulates glioblastoma stem cells through hypoxia inducible factor 2 alpha. Brain. 2010;133:983–995. doi: 10.1093/brain/awq042. [DOI] [PubMed] [Google Scholar]
- 48.Yu D, et al. Alleviation of chronic pain following rat spinal cord compression injury with multimodal actions of huperzine A. Proc Natl Acad Sci USA. 2013;110:E746–E755. doi: 10.1073/pnas.1300083110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patel AP, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bao S, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 51.Chen J, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 53.Son MJ, Woolard K, Nam DH, Lee J, Fine HA. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4:440–452. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anido J, et al. TGF-β receptor inhibitors target the CD44(high)/Id1(high) glioma-initiating cell population in human glioblastoma. Cancer Cell. 2010;18:655–668. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 55.Lathia JD, et al. Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell. 2010;6:421–432. doi: 10.1016/j.stem.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 57.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 58.Ito K, et al. PML targeting eradicates quiescent leukaemia-initiating cells. Nature. 2008;453:1072–1078. doi: 10.1038/nature07016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stricker SH, et al. Widespread resetting of DNA methylation in glioblastoma-initiating cells suppresses malignant cellular behavior in a lineage-dependent manner. Genes Dev. 2013;27:654–669. doi: 10.1101/gad.212662.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Piccirillo SG, et al. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444:761–765. doi: 10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- 61.Lee J, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 62.Natsume A, et al. Chromatin regulator PRC2 is a key regulator of epigenetic plasticity in glioblastoma. Cancer Res. 2013;73:4559–4570. doi: 10.1158/0008-5472.CAN-13-0109. [DOI] [PubMed] [Google Scholar]
- 63.Jahani-Asl A, et al. Control of glioblastoma tumorigenesis by feed-forward cytokine signaling. Nat Neurosci. 2016;19:798–806. doi: 10.1038/nn.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Furnari FB, Cloughesy TF, Cavenee WK, Mischel PS. Heterogeneity of epidermal growth factor receptor signalling networks in glioblastoma. Nat Rev Cancer. 2015;15:302–310. doi: 10.1038/nrc3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wei W, et al. Single-cell phosphoproteomics resolves adaptive signaling dynamics and informs targeted combination therapy in glioblastoma. Cancer Cell. 2016;29:563–573. doi: 10.1016/j.ccell.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu F, et al. Olig2-dependent reciprocal shift in PDGF and EGF receptor signaling regulates tumor phenotype and mitotic growth in malignant glioma. Cancer Cell. 2016;29:669–683. doi: 10.1016/j.ccell.2016.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu F, et al. EGFR mutation promotes glioblastoma through epigenome and transcription factor network remodeling. Mol Cell. 2015;60:307–318. doi: 10.1016/j.molcel.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rozhkova EA. Nanoscale materials for tackling brain cancer: Recent progress and outlook. Adv Mater. 2011;23:H136–H150. doi: 10.1002/adma.201004714. [DOI] [PubMed] [Google Scholar]
- 69.Yang XZ, et al. Sheddable ternary nanoparticles for tumor acidity-targeted siRNA delivery. ACS Nano. 2012;6:771–781. doi: 10.1021/nn204240b. [DOI] [PubMed] [Google Scholar]
- 70.Chahal JS, et al. Dendrimer-RNA nanoparticles generate protective immunity against lethal Ebola, H1N1 influenza, and Toxoplasma gondii challenges with a single dose. Proc Natl Acad Sci USA. 2016;113:E4133–E4142. doi: 10.1073/pnas.1600299113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jahangiri A, et al. Convection-enhanced delivery in glioblastoma: A review of preclinical and clinical studies. J Neurosurg. 2017;126:191–200. doi: 10.3171/2016.1.JNS151591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vogelbaum MA, Aghi MK. Convection-enhanced delivery for the treatment of glioblastoma. Neuro-oncol. 2015;17:ii3–ii8. doi: 10.1093/neuonc/nou354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Saucier-Sawyer JK, et al. Distribution of polymer nanoparticles by convection-enhanced delivery to brain tumors. J Control Release. 2016;232:103–112. doi: 10.1016/j.jconrel.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Debinski W, Tatter SB. Convection-enhanced delivery to achieve widespread distribution of viral vectors: Predicting clinical implementation. Curr Opin Mol Ther. 2010;12:647–653. [PubMed] [Google Scholar]
- 75.Robbins M, et al. Misinterpreting the therapeutic effects of small interfering RNA caused by immune stimulation. Hum Gene Ther. 2008;19:991–999. doi: 10.1089/hum.2008.131. [DOI] [PubMed] [Google Scholar]
- 76.Judge AD, et al. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 77.Hu Y, Smyth GK. ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 78.Ahmed AU, et al. A preclinical evaluation of neural stem cell-based cell carrier for targeted antiglioma oncolytic virotherapy. J Natl Cancer Inst. 2013;105:968–977. doi: 10.1093/jnci/djt141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dong B, et al. Super-resolution spectroscopic microscopy via photon localization. Nat Commun. 2016;7:12290. doi: 10.1038/ncomms12290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dong B, et al. Superresolution intrinsic fluorescence imaging of chromatin utilizing native, unmodified nucleic acids for contrast. Proc Natl Acad Sci USA. 2016;113:9716–9721. doi: 10.1073/pnas.1602202113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ovesný M, Křížek P, Borkovec J, Svindrych Z, Hagen GM. ThunderSTORM: A comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 2014;30:2389–2390. doi: 10.1093/bioinformatics/btu202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.