Significance
In most biological processes, genes have to be activated and/or repressed. In plants, the TOPLESS protein is essential for gene repression through its action as a corepressor bridging transcription factor with chromatin remodeling complexes. Here we combine biochemical and structural studies to describe the structure of TOPLESS, how it tetramerizes, and how it interacts with its protein partners. We show that both the tetramerization interface and the binding site for protein partners have been conserved since algae, highlighting the ancestrality of TOPLESS function. Comparison of this plant protein with one of its animal counterparts also shows how corepressors can use a common domain differently to achieve similar properties, illustrating the tinkering of evolution in transcriptional repression.
Keywords: TOPLESS, corepressor, auxin signaling, crystal structure, tetramerization
Abstract
Transcriptional repression involves a class of proteins called corepressors that link transcription factors to chromatin remodeling complexes. In plants such as Arabidopsis thaliana, the most prominent corepressor is TOPLESS (TPL), which plays a key role in hormone signaling and development. Here we present the crystallographic structure of the Arabidopsis TPL N-terminal region comprising the LisH and CTLH (C-terminal to LisH) domains and a newly identified third region, which corresponds to a CRA domain. Comparing the structure of TPL with the mammalian TBL1, which shares a similar domain structure and performs a parallel corepressor function, revealed that the plant TPLs have evolved a new tetramerization interface and unique and highly conserved surface for interaction with repressors. Using site-directed mutagenesis, we validated those surfaces in vitro and in vivo and showed that TPL tetramerization and repressor binding are interdependent. Our results illustrate how evolution used a common set of protein domains to create a diversity of corepressors, achieving similar properties with different molecular solutions.
The regulation of transcription is central to many biological processes. It involves transcription factors (TFs) that bind to regulatory sequences and either activate or repress gene expression. For repression, eukaryotes have developed various strategies involving proteins known as corepressors (1). These proteins usually do not directly interact with DNA, but are addressed to regulatory sequences by sequence-specific TFs. Many corepressors work as hub proteins, as they are able to interact with a wide variety of TFs. In many cases, corepressors recruit chromatin remodeling factors (such as histone deacetylases) to the regulatory sequences of down-regulated genes. In eukaryotes, the most studied corepressors include human SMRT (silencing mediator of retinoid acid and thyroid hormone receptor) and NCoR (nuclear receptor corepressor) complexes (2–5), the yeast proteins Sin3 and Tup1 (6, 7), or the homologous Drosophila Groucho (Gro) and mammalian transducin-like enhancer protein (TLE) (8).
Corepressors are also present in plants. In Arabidopsis thaliana, the GRO/TUP family of corepressors has about 13 members, including the founding members LEUNIG (LUG) and TOPLESS (TPL) (9, 10). Both genes were initially identified genetically, based on developmental defects occurring in Arabidopsis mutants bearing mutations in the TPL or LUG genes. The tpl-1 dominant mutation causes an abnormal development of the embryo (11), whereas the lug mutant shows floral organ defects (12). The TOPLESS protein was later found to be involved in multiple pathways and to interact with numerous transcriptional repressors (13). Notably, TPL interacts with IAA proteins involved in transcriptional repression in auxin signaling (14, 15), as well as with the WUS protein regulating plant stem cell homeostasis (13, 16).
LUG and TPL contain several domains previously characterized in other corepressors. They possess WD40 repeats (Fig. 1A) (9), which are also present in the above-mentioned Tup1, Groucho, TLE, and TBL1 [transducin (beta)-like 1], a member of the SMRT/NCoR complex in Homo sapiens (3, 9, 17).
Fig. 1.
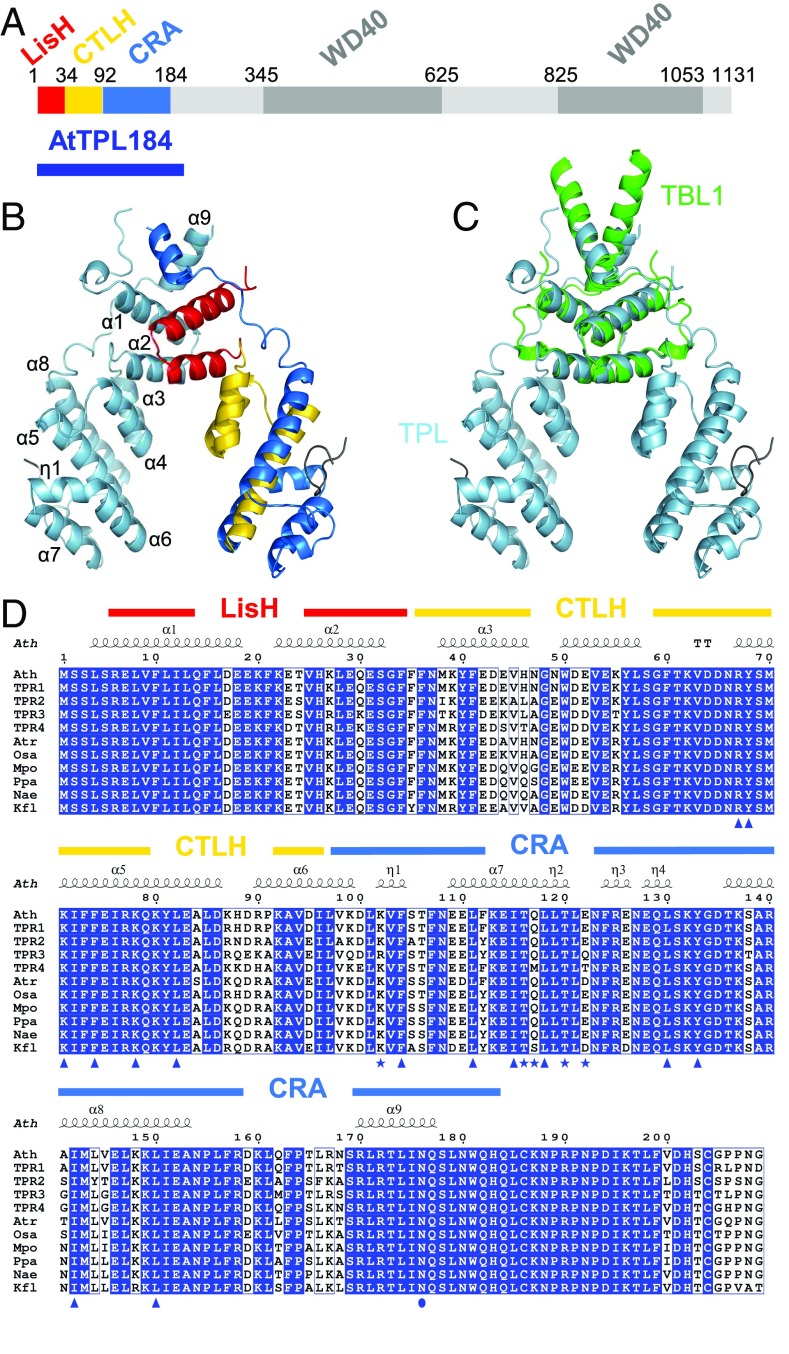
TPL N terminus structure and conservation. (A) AtTPL protein schematic representation. (B) Cartoon representation of AtTPL184 homodimer. LisH, CTLH, and CRA domains are colored in red, gold, and blue in one of the two monomers, respectively. (C) TBL1 (green) and AtTPL184 (blue) structure superimposition. (D) Sequence alignment of the N terminus of TPL and A. thaliana (Ath) TPR proteins. Atr, Amborella trichopoda; Osa, Oryza sativa; Mpo, Marchantia polymorpha; Ppa, Physcomitrella patens; Nae, Nothoceros aenigmaticus; Kfl, K. flaccidum. Triangle, amino acids involved in the interaction with the IAA peptide; star, amino acids involved in tetramerization; circle, N176 residue.
At its N terminus, TPL contains a LIS1 homology (LisH) and a C-terminal to LisH (CTLH) domain. These two domains are often, but not always, found together. LisH domains have been shown to mediate protein dimerization (18, 19). A LisH domain is also present in the TBL1 corepressor; in this case, it confers its capacity both to form tetramers and to interact with hydrophobic peptides from SMRT/NCoR and GPS2 proteins (3). TPL has also been shown to interact with transcriptional regulators through a hydrophobic peptide called the ethylene-responsive element binding factor-associated amphiphilic repression (EAR) motifs domain (13, 20, 21). This interaction was mapped to the LisH CTLH region (14, 22), suggesting TPL and TBL1 might function similarly.
We have solved the crystallographic structure of Arabidopsis TPL, which is structurally similar to the previously determined TPR2 structure from rice (23). Our analyses shed light on this essential plant corepressor; we highlight the presence of a previously unidentified CRA domain, map the tetramerization domain, and reveal the close dependence between peptide binding and tetramerization. We also show that, despite the similarities between TBL1 and TPL, these two tetrameric repressors show remarkably different organizations.
Results
Crystal Structure of A. thaliana TPL N-Terminus.
We focused on the highly conserved N-terminal part of TPL that includes the LisH and CTLH domains plus an additional region with no described structural similarities (Fig. 1A). We solved the structure of the first 184 amino acids from A. thaliana TPL, AtTPL184 and its selenomethionine derivative at 2.61 and 2.92 Å resolution, respectively (SI Appendix, Table S1). Each monomer presents an α-helical topology made by nine α-helices (α1–α9) connected by loops (Fig. 1B): helices α1 and α2 form the LisH domain, α3–α5 the CTLH domain, and α6-α9 are a third domain we identified as a CT11-RanBPM (CRA) domain (24) (see detailed explanation on CRA identification in SI Appendix, Fig. S1). Monomers are arranged into dimers, with the long α9 from the CRA domain bridging the LisH domain (Fig. 1B). Dimerization is thus mediated by two discontinuous regions (the LisH and α9). The configuration adopted by these regions is structurally similar to the LisH-containing domain from the human corepressor TBL1 (3) (Fig. 1C). The AtTPL184 dimers are arranged in tetramers (dimers of dimers), but the crystal packing suggests two possible tetrameric forms (Fig. 2 A and B). The AtTPL184 structure has an rmsd of 0.7 Å for 179 Cα, using DALI with the recently published OsTPR2 structure from rice (23) (SI Appendix, Fig. S2). TPL/TPR protein sequences are indeed highly conserved over several hundreds of My of evolution (from Charophyte algae to angiosperms; Fig. 1D). DALI searches also established that Smu1 (suppressor of mec-8 and unc-52), a splicing factor and replication regulator, shows a high similarity with TPL (rmsd of 0.8 Å for 35 Cα of the LisH domain and of 3.4 Å for 107 Cα of the CTLH-CRA domains), consistent with its described resemblance to TPR2 (25). However, the relative positions of the LisH and CTLH-CRA domains are different, and Smu1 does not tetramerize (SI Appendix, Fig. S3).
Fig. 2.
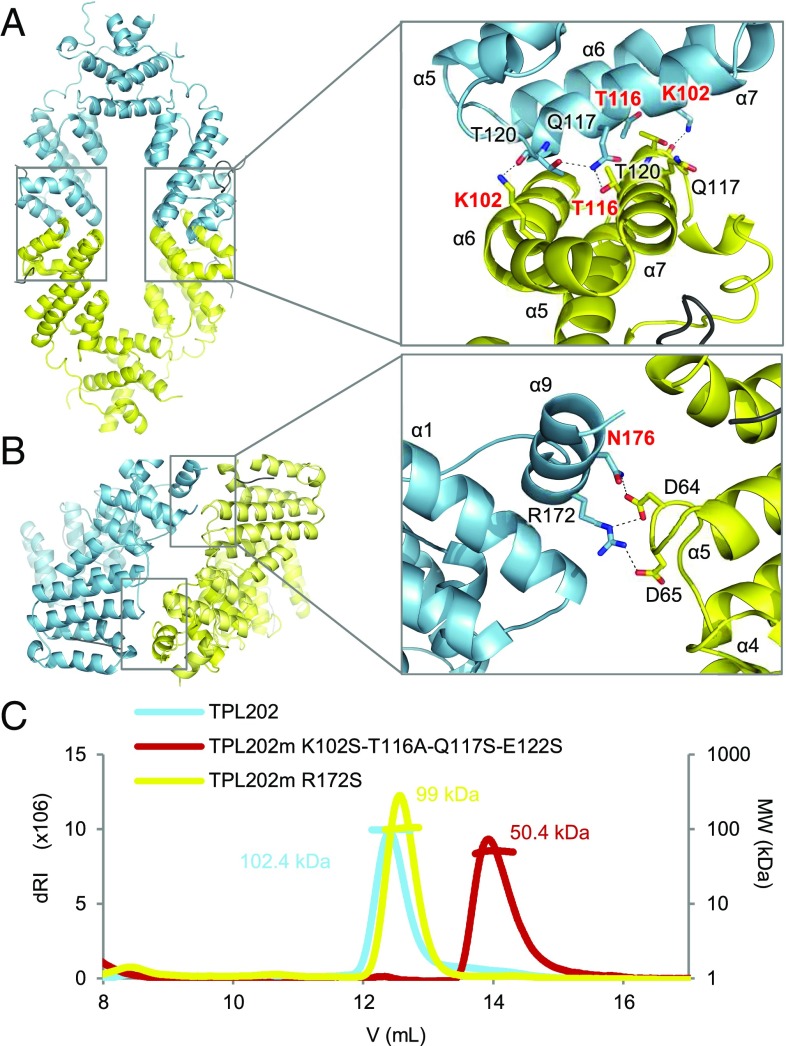
TPL tetramerization. (A) AtTPL184 tetrameric form I (dimers in blue and yellow). (Inset) Close-up view of the tetramerization interface with a rotation of about 90° to better view interacting residues. The interactions between chains C and D (Left) are slightly different from those between A and B (Right), as indicated in SI Appendix, Table S3. Residues mutated in different experiments are shown in red. (B) AtTPL184 tetrameric form II and close-up view of the interface. Main residues shown as sticks; distances <3.4 Å shown as dashes. (C) SEC-MALLS on TPL202 (blue) and TPL202 tetramerization mutants in both tetramerization interfaces I (red) and II (yellow). dRI, differential refractive index; MW, molecular weight.
Characterization of the Tetramerization Interface of AtTPL.
We tested whether the tetrameric arrangement observed in the crystal is valid in solution, using size exclusion chromatography followed by multiple angle laser light scattering (SEC-MALLS). We determined the size of AtTPL184 and AtTPL202, a more soluble construct comprising TPL’s first 202 residues. The measured molecular weights confirmed the tetrameric state of both proteins (SI Appendix, Table S2A). Next, we tested which of the two possible tetrameric conformations mentioned earlier (Fig. 2 A and B) was observed in solution. In the first tetrameric form, also proposed by Ke et al. (23) for OsTPR2, but not experimentally validated, the dimers interact through helices α6 and 7. This arrangement creates two symmetric tetramerization interfaces of 600 Å2 each (tetramerization interface I) (Fig. 2A). The second possible tetramerization interface (tetramerization interface II) involves two symmetric interfaces of 370 Å2 between helix α9 (with residues R172 and N176) and a small loop between helices α5 and α6 (Fig. 2B). This surface is less likely. However, the previously identified dominant tpl-1 point mutation lies within this interface (11). We mutated key residues located in both tetramerization interfaces and determined the molecular weights of resulting proteins using SEC-MALLS (SI Appendix, Table S2 B and C). Mutations of N176 (N176H) or R172 (R172S) from interface II did not affect the complex molecular weight (Fig. 2C and SI Appendix, Table S2C). In contrast, K102S and T116A single mutations in interface I yielded complexes of intermediate molecular weights (3.2–3.5 or 2.6–3.1 monomers, respectively, depending on protein concentrations) (SI Appendix, Table S2B). This behavior is indicative of a fast dimer–tetramer balance that hampers the separation of the two species in the SEC column. We also assayed the effect of other mutations in interface I. TPL202m/Q117M-E122T and TPL202m/Q117S-E122S were still tetrameric, but the quadruple mutation (TPL202m/K102S-T116A-Q117S-E122S) disrupted the tetramer into a dimer (Fig. 2C and SI Appendix, Table S2B). SEC analyses of the oligomerization state of TPL202 and TPL202m/K102S-T116A-Q117S-E122S at different protein concentrations (from 2.25 to 36 µM) showed that the oligomeric state of these proteins is independent of the protein concentration, suggesting TPL likely behaves as a tetramer in vivo (SI Appendix, Fig. S4). K102 and T116 are thus key residues on interface I involved in TPL tetramerization. The central role of K102 is obvious in both AtTPL184 (Fig. 2A) and OsTPR2 structures (SI Appendix, Table S3), as it interacts with T120 of the opposite monomer. The role of T116 is less clear, as it forms a hydrogen bond with Q117 only in the AtTPL184 structure and not in OsTPR2’s. The K102 and T116 residues are, however, both highly conserved among TPL/TPRs from charophytes to angiosperms, suggesting tetramerization is evolutionarily conserved (Fig. 1D).
TPL Interacts with EAR Motifs Through Hydrophobic Groove 3.
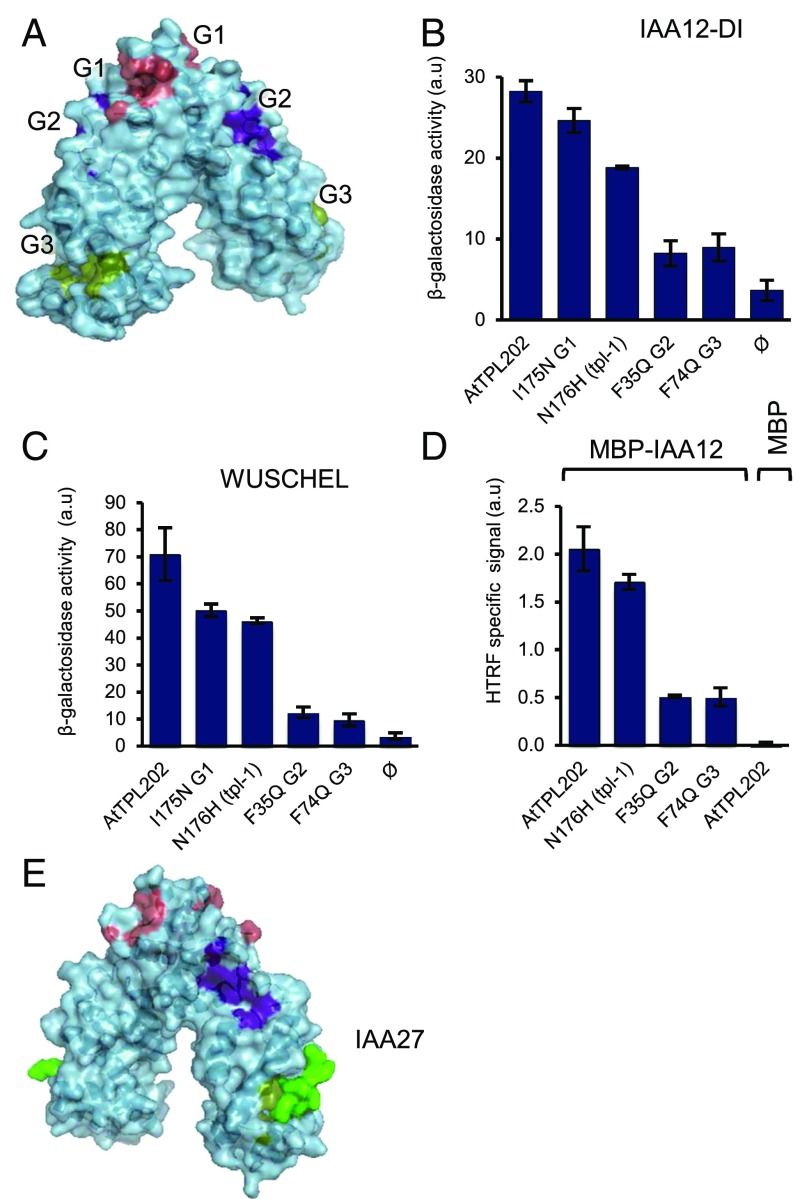
TPL interacts with multiple proteins containing EAR/EAR-like hydrophobic motifs (13). Examination of AtTPL184 monomer structures identified three hydrophobic grooves (G1–G3) as possible candidates to interact with these motifs (Fig. 3A). G1 is formed by the LisH domain and the C-terminal α9 helix of the CRA domain and contains the N176 residue affected by the tpl-1 mutation (11). In TBL1, mentioned earlier, a similar hydrophobic groove is present and interacts with hydrophobic motifs from the SMRT/NCoR corepressor complex (3). G2 is a small hydrophobic groove positioned between the LisH and the CTLH domains. G3 is formed by a three-helix bundle that includes part of the CTLH domain (helix α5) and helices α7 and α8 of the CRA domain.
Fig. 3.
Hydrophobic groove 3 is implicated in the interaction with LxLxL EAR motifs. (A) Hydrophobic grooves (G1, G2, and G3) identified in TPL N terminus. (B and C) Y2H binding assay between TPL-202 (WT and mutants) with IAA12-DI domain (B) and WUSCHEL protein (C). n = 3 for all experiments. Error bars represent SD, ɸ for empty vector. (D) HTRF binding assays between His-TPL202 (WT and mutants) and MBP-IAA12 protein. HTRF-specific signal is reported. n = 3 for all experiments. Error bars represent SD. (E) Structure of AtTPL184 IAA27 peptide IAA27. a.u., arbitrary units.
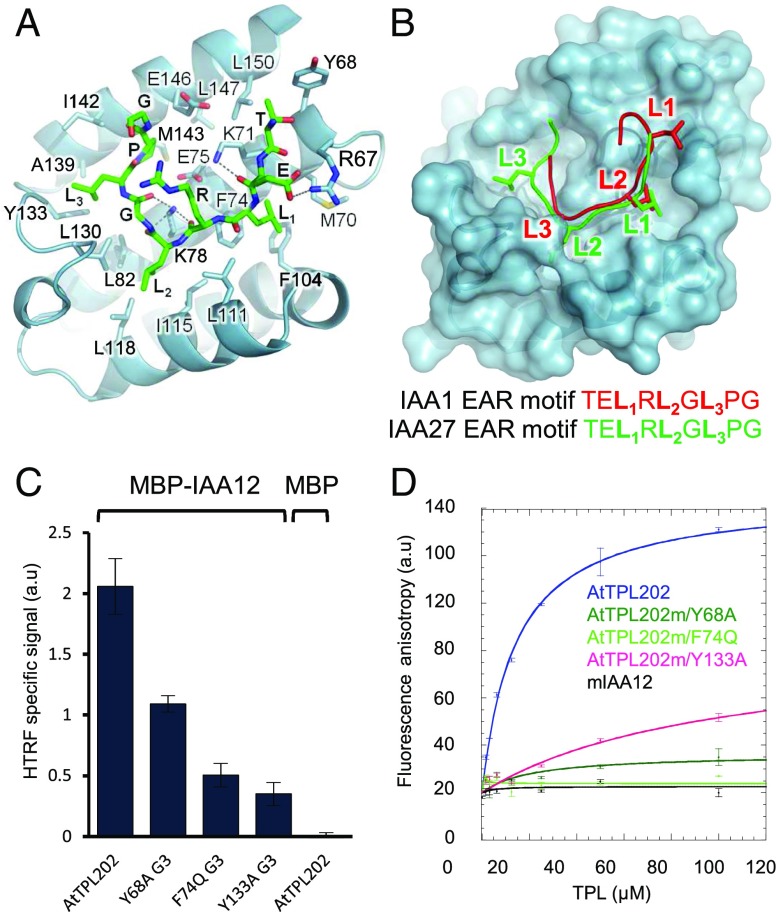
To determine which groove is involved in the binding of EAR/EAR-like motifs, key hydrophobic amino acids were mutated in each of them, and we assessed the interaction between TPL and two of its partners, using yeast two-hybrid (Y2H) and homogeneous time-resolved fluorescence (HTRF) (Fig. 3 B–D). For Y2H assays, we used domain I from the AtIAA12 (IAA12-DI) protein, which contains a LxLxL-type EAR motif known to interact with TPL (14, 23), and the WUSCHEL protein (WUS), which contains a LxLxL-type EAR motif and a TLxLFP WUS-box, both involved in WUS repressor function (16, 26). Both assays showed that mutations in the G1 groove (I175N and N176H) only weakly affected the interaction with IAA12 or WUS (Fig. 3 B–D), in agreement with previous results showing that the TPL N176H mutant still interacts with IAA12 (14) and with the OsTPR2 structure, where G1 is occluded by a Zinc finger and inaccessible to EAR motifs (23). In contrast, mutations in G2 (F35Q) and G3 (F74Q) considerably weakened the interaction with IAA12 or WUS (Fig. 3 B–D), indicating that both G2 and G3 might be important for interacting with EAR/EAR-like motifs. To better characterize these interactions, we cocrystallized AtTPL184 with IAA27 LxLxL-type EAR motif and obtained a high-resolution structure at 1.95 Å of the AtTPL184/EAR complex. No conformational changes are induced by the binding of the peptide, localized in G3 (Figs. 3E and 4A), as observed for OsTPR2 (23). The proline and the three leucine residues of IAA27 peptide (T–E–L1–R–L2–G–L3–P–G) interact with G3 hydrophobic residues (Fig. 4A). This peptide is further positioned in G3 by hydrogen bonds between oxygen atoms of its main chain and lateral chains of K78 and K71 and by additional ionic interactions between the lateral chains of the peptide’s glutamate residue and R67 (Fig. 4A).
Fig. 4.
Interaction of the EAR-motif with the G3 groove of AtTPL. (A) Position of the IAA27 peptide in AtTPL G3. (B) Overlay of IAA1 peptide bound to OsTPR2 on the complex between the IAA27 peptide and AtTPL184. (C) Analysis of AtTPL G3 residues involved in the interaction with IAA12 proteins by HTRF. (D) Analysis of AtTPL G3 residues involved in the interaction with IAA12 peptide by fluorescence anisotropy. a.u., arbitrary units.
Despite the high similarity between AtTPL184 and OsTPR2 structures and the identical sequences of IAA1 and IAA27 peptides, the two peptides are not bound identically: they are shifted by two residues, and the third leucine residue of IAA27 (T–E–L1–R–L2–G–L3–P–G) peptide, L3, interacts with Y133 in the innermost part of G3 in AtTPL184-IAA27, but not in the OsTPR2-IAA1 structure (Fig. 4 A and B). It is possible there is some degeneracy in binding because of the repetitive nature of the EAR peptide with three leucine residues. However, another possibility is that lower resolution of OsTPR2 data did not allow for a precise peptide positioning. We further investigated this binding difference. For this, we mutated key interacting residues in TPL, including Y133, which contacts the peptide only in our configuration. Of the various TPL residues mutated, F74 and Y133 had a great effect on the interaction, supporting the IAA27 peptide positioning as we described it (Fig. 4 C and D). Furthermore, we used the deposited structural data for OsTPR2 (5C7F) and found that our observed register fits these data better than the structure as deposited (SI Appendix, Fig. S5). In particular, the position that is modeled as a glycine in 5C7F shows clear unaccounted-for side chain electron density in a composite omit simulated annealed electron density map. This density fits an arginine side chain well, as in our register (SI Appendix, Fig. S5 C and D). In addition, the 5C7F positioning of the arginine and proline fit the electron density quite poorly, which are both a better fit in our register (SI Appendix, Fig. S5 C and D). Altogether, the peptide register as we describe it is best supported by biochemical and structural evidence.
The G3 groove and the residues contacting the EAR are perfectly conserved throughout TPL evolution (Fig. 1D), even in the green alga Klebsormidium flaccidum (Kf), suggesting ancestral TPL might be able to interact with EAR peptides. We confirmed this hypothesis by showing that KfTPL N terminus interacts with the AtIAA12 protein and the IAA12 EAR motif (SI Appendix, Fig. S6).
Groove 3 and Tetramerization Are Intimately Linked.
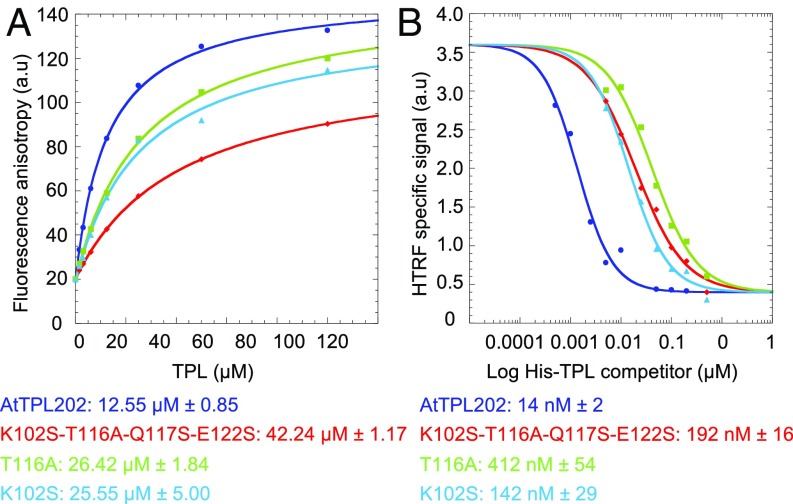
As helices α6 and α7 are part of both the EAR motif binding site and the tetramerization interface of TPL (Figs. 2 and 4), we wondered whether tetramerization and binding of EAR motifs could be interdependent. We tested the TPL oligomeric state in the presence of mutations in G3, using SEC-MALLS, and found that all G3 mutants became dimeric except Y68A (SI Appendix, Table S2D). Conversely, we analyzed whether the previously discussed tetramerization mutants affect TPL binding to EAR motifs or to repressors. Using fluorescence anisotropy with FAM-IAA12 EAR motif, we found that the EC50 (protein effective concentration that gives half maximal response) of TPL202m/K102S-T116A-Q117S-E122S (EC50 = 42.2 µM ± 1.2) was about fourfold higher than that of the wild-type protein (EC50 = 12.6 µM ± 0.8) (Fig. 5A). TPL202m/T116A and TPL202m/K102S also showed an affinity loss in comparison with the wild-type protein.
Fig. 5.
Groove 3 and tetramerization are linked. (A) Characterization of the interaction between IAA12 EAR motif (FAM labeled) and TPL proteins impaired in tetramerization by fluorescence anisotropy assay. EC50 shown below the graph (B) HTRF competition assays performed on an initial GST-IAA12/MBP-AtTPL202 complex by adding increasing amounts of His-tagged AtTPL202 wt and mutant proteins. IC50 shown below the graph. a.u., arbitrary units.
Fluorescence anisotropy results were confirmed by HTRF competition assays. Compared with the wild-type protein (IC50 = 14 nM ± 2), dimeric mutants are 10–30-fold less efficient in competition (IC50 = 192 nM ± 16 for TPL202m/K102S-T116A-Q117S-E122S) (Fig. 5B). Altogether, these results suggest that altering the tetramerization interface cannot be decoupled from peptide binding. This brings up the intriguing possibility that TPL binding to the repressor could alter its tetramerization status. However, an excess of various IAA12 EAR motif peptides did not affect TPL tetramerization (SI Appendix, Tables S2E and S4). TPL thus appears to be tetrameric in the presence and absence of an EAR peptide, but we did not identify any mutation that affects tetramerization without perturbing peptide binding.
TPL N Terminus Mutations Affecting Hydrophobic Grooves or Tetramerization Abolish Auxin-Dependent Transcriptional Repression.
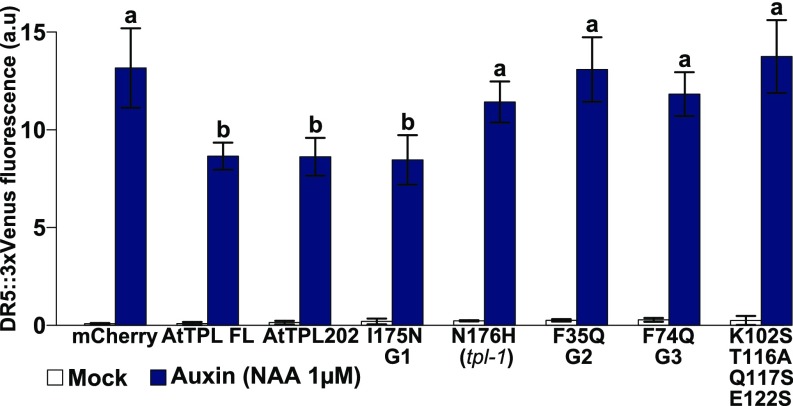
Next we tested the functional importance of the residues involved in peptide binding and TPL tetramerization by analyzing their effect on TPL repressive activity in planta. We focused on auxin signaling, as TPL was previously shown to be recruited by AUX/IAA proteins to repress auxin target genes (11, 14, 27). TPL is differentially expressed in tissues (11, 28), and increasing TPL dosage affects both root and shoot development (29), suggesting TPL expression could be limiting in tissues. We thus generated transgenics overexpressing TPL fused to mCherry under the 35S promoter. Surprisingly, none of the lines tested showed reproducible defects in auxin responses, despite a strong TPL-mCherry expression (SI Appendix, Fig. S7). Conversely, when tested in protoplasts containing an integrated auxin-inducible DR5::VENUS reporter, overexpression of the same TPL-mCherry fusion had a repressive effect of around 40% on the induction of VENUS fluorescence after treatment with 1 μM of the synthetic auxin Naphtalene-1-acetic acid (Fig. 6). Expression of AtTPL202 lacking the two WD40 domains repressed DR5 activity similarly consistent with previous assays in yeast (30, 31). Conversely, the AtTPL202 mutant forms affecting the EAR peptide binding G3 groove (TPL202m/F74Q) and the dimeric mutant TPL202m/K102S-T116A-Q117S-E122S lost their repression activity. A mutant form affected in the G2 groove (TPL202m/F35Q) similarly lost its repressive activity, an observation coherent with the weaker interaction with IAA12 in the Y2H and HTRF assays (Fig. 3). Finally, TPL202m/N176H (tpl-1) also shows a reduced repression capacity despite being still able to interact with IAA12 repressors (Fig. 3) (11). These results confirmed the importance of TPL residues involved in peptide binding in the G3 groove or tetramerization in planta and underline the importance of the AtTPL N terminus for transcriptional repression in auxin signaling. They also demonstrate that intact G1 and G2 grooves are required for repression in planta, despite the apparent absence of direct interaction with EAR domains of IAAs at these grooves.
Fig. 6.
Importance of TPL G3 groove and tetramerization in auxin signaling. In planta repression assay in Col-0 protoplasts containing the integrated DR5::VENUS reporter gene. A loss of repression activity is observable in TPL mutants impaired in their interaction with repressors, their tetramerization, or carrying the tpl-1 mutation. DR5 activity is reported (n = 200 protoplasts). Error bars correspond to the 95% confidence interval. Two variables with different letters are significantly different (corrected P values < 0.05; two-sided Kruskal-Wallis test). a.u., arbitrary units.
Discussion
We have obtained the crystallographic structure of Arabidopsis TPL N terminus, which allowed us to design a series of experiments to study the oligomerization and peptide binding in vitro and in vivo.
EAR Peptide Binding and Tetramerization Are Intimately Related.
As previously shown for OsTPR2 (23), we localized the interaction site of the EAR peptide to one of the three hydrophobic grooves present in TPL-N. The higher resolution of our structure allowed us to refine the interactions between EAR peptide and TPL. Our data suggests a different register than previously proposed (with a two-residue shift), which we validated using mutations in the innermost part of the groove.
Based on gel filtration, TPL-N was previously proposed to form a tetramer (23). Using a combination of SEC-MALLS, mutagenesis of two potential tetramerization interfaces, and an in planta assay, we demonstrated that TPL does indeed tetramerize and that the interface proposed for TPR2 (23) is the same as in AtTPL. It remains to be established whether the full-length TPL protein is indeed tetrameric. Our work also shows that tetramerization and peptide binding are intimately linked: The amino acids responsible for EAR binding and tetramerization are in close proximity, and tetramerization cannot be altered without affecting EAR binding. Nevertheless, EAR peptide binding to TPL does not appear to affect its oligomerization.
Tetramerization is a common feature of corepressors, with a few structural commonalities in tetramers arrangement (3, 7, 17, 32) (SI Appendix, Fig. S8). Why tetramerization is essential for their function is still elusive. We speculate it could be a way to spread chromatin marks along the DNA; for example, to reproduce chromatin domains after mitosis (33). It could also help to recruit multimeric TFs using weak individual interactions that are compensated for by a high avidity (23, 34). To specifically test the importance of tetramerization, mutations are needed that compromise this property without altering other features (such as complex formation with other proteins). However, our work indicates it might be difficult to study the respective contributions of EAR peptide binding and tetramerization in TPL function. We also show that the two other hydrophobic grooves on TPL-N are important for TPL-N function, as introducing mutations in any of them interferes with the interaction with WUS or IAA12 and with TPL-N repressive activity in vivo. Our data suggest these mutations probably do not affect the interaction with the EAR motif itself (SI Appendix, Fig. S9), but TPL-N and rice TPR2-N structures indicate that the G2 groove is accessible and could interact with other protein motifs.
TPL Structure Sheds Light on Other CTLH-CRA Proteins.
The analysis of the TPL structure revealed the presence of a CRA motif. This motif was first identified in RanBP9 (human Ran binding protein 9) for mediating the interaction with FMRP (fragile X mental retardation protein) in the microtubule organizing center (35). CRA motifs are present in thousands of eukaryotic proteins such as RanBPM in human or E3 ligases proteins (SIE3 in plants or Gid in yeast) (35–37). They are often associated with the CTLH domain, but with variable-length spacers between them. Whether CRA and CTLH motifs were forming a single domain or two separate domains had remained elusive because of the lack of structural data. TPL and TPR2 structures show that CTLH and CRA form two distinct but intertwined domains that can mediate tetramerization and hydrophobic peptide binding. Having unambiguously identified the domain next to the CTLH as a CRA domain now allows the modeling of the wealth of proteins possessing the LisH-CTLH-CRA domains, using OsTPR2/AtTPL or Smu1 as models. We inventoried and modeled with high confidence all such Arabidopsis proteins (99.8–100% using Phyre2) (SI Appendix, Table S5), revealing that some possess a TPL-like G3 hydrophobic groove, whereas others are more similar to Smu1, which uses another hydrophobic groove to interact with the RED hydrophobic peptide (SI Appendix, Table S5). Proteins possessing the three domains (LisH, CTLH, and CRA) are found at the very base of the green lineage (including chlorophyte algae such as Chlamydomonas reinhardtii), and also outside the plant kingdom, indicating that those domains have assembled very early in evolution and can now be modeled using TPL/TPR2 structures in many eukaryotic proteins (SI Appendix, Fig. S1).
TBL1 and TPL Use the Same Domain in Different Ways.
TBL1 is a member of the highly studied SMRT/NCoR nuclear receptor complex (3). It functions as a bridge between two members of the complex: GPS2 and SMRT. In TBL1, tetramerization occurs via two interfaces in the LisH domain, forming a dimer of dimers. Binding of partner proteins occurs via a hydrophobic groove on the face of LisH that is distal to the dimerization surfaces (SI Appendix, Fig. S8B). We therefore anticipated that TPL, which also functions as a tetrameric corepressor hub protein and contains a LisH domain, would tetramerize and bind partner proteins in a similar fashion. However, the crystal structures of TPL reveal that it uses a completely different surface for protein–protein interaction that is within the CTLH-CRA domain. Furthermore, unlike TBL1, which uses the “bottom” face of the LisH domain to form a second dimer interface, dimerization in TPL occurs at the base of the CTLH-CRA domain.
Comparing TPL and TBL1 illustrates how corepressors can differently use a common domain (LisH) to achieve similar properties (tetramerization and binding to hydrophobic peptides), and provides an excellent example of the tinkering of evolution (38) that, in this case, used a common toolset to achieve transcriptional repression in different organisms.
Materials and Methods
Material and experimental procedures for vectors construction, protein expression and purification, crystallization, protein structure determination, native molecular mass determination, yeast two hybrid interaction tests, homogeneous-time resolved fluorescence interaction tests, fluorescence anisotropy interaction tests, and repressions assays are described in detail in the SI Appendix.
Supplementary Material
Acknowledgments
We thank R. Miras (Laboratoire Chimie et Biologie des Métaux Grenoble) for help with SEC-MALLS and E. Thevenon and S. Lainé for help. This work was supported by the French National Research Agency programs Auxiflo (ANR-12-BSV6-0005 to T.V. and F.P.) and Gral (ANR-10-LABX-49-01), the Human Frontier Science Program organization (Grant RPG0054-2013 to T.V.), a Université Grenoble Alpes PhD fellowship (R.M.-A.), and the European Community’s Seventh Framework Program (FP7) under grant agreement 283570 (BioStruct-X to R.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 5NQV and 5NQS).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703054114/-/DCSupplemental.
References
- 1.Payankaulam S, Li LM, Arnosti DN. Transcriptional repression: Conserved and evolved features. Curr Biol. 2010;20:R764–R771. doi: 10.1016/j.cub.2010.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: Evolving models of co-repressor action. Nat Rev Genet. 2010;11:109–123. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- 3.Oberoi J, et al. Structural basis for the assembly of the SMRT/NCoR core transcriptional repression machinery. Nat Struct Mol Biol. 2011;18:177–184. doi: 10.1038/nsmb.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watson PJ, Fairall L, Schwabe JWR. Nuclear hormone receptor co-repressors: Structure and function. Mol Cell Endocrinol. 2012;348:440–449. doi: 10.1016/j.mce.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mottis A, Mouchiroud L, Auwerx J. Emerging roles of the corepressors NCoR1 and SMRT in homeostasis. Genes Dev. 2013;27:819–835. doi: 10.1101/gad.214023.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grzenda A, Lomberk G, Zhang JS, Urrutia R. Sin3: Master scaffold and transcriptional corepressor. Biochim Biophys Acta. 2009;1789:443–450. doi: 10.1016/j.bbagrm.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsumura H, et al. Crystal structure of the N-terminal domain of the yeast general corepressor Tup1p and its functional implications. J Biol Chem. 2012;287:26528–26538. doi: 10.1074/jbc.M112.369652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agarwal M, Kumar P, Mathew SJ. The Groucho/Transducin-like enhancer of split protein family in animal development. IUBMB Life. 2015;67:472–481. doi: 10.1002/iub.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Z, Karmarkar V. Groucho/Tup1 family co-repressors in plant development. Trends Plant Sci. 2008;13:137–144. doi: 10.1016/j.tplants.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 10.Lee JE, Golz JF. Diverse roles of Groucho/Tup1 co-repressors in plant growth and development. Plant Signal Behav. 2012;7:86–92. doi: 10.4161/psb.7.1.18377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Long JA, Ohno C, Smith ZR, Meyerowitz EM. TOPLESS regulates apical embryonic fate in Arabidopsis. Science. 2006;312:1520–1523. doi: 10.1126/science.1123841. [DOI] [PubMed] [Google Scholar]
- 12.Sitaraman J, Bui M, Liu Z. LEUNIG_HOMOLOG and LEUNIG perform partially redundant functions during Arabidopsis embryo and floral development. Plant Physiol. 2008;147:672–681. doi: 10.1104/pp.108.115923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Causier B, Ashworth M, Guo W, Davies B. The TOPLESS interactome: A framework for gene repression in Arabidopsis. Plant Physiol. 2012;158:423–438. doi: 10.1104/pp.111.186999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szemenyei H, Hannon M, Long JA. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science. 2008;319:1384–1386. doi: 10.1126/science.1151461. [DOI] [PubMed] [Google Scholar]
- 15.Dinesh DC, Villalobos LIAC, Abel S. Structural biology of nuclear auxin action. Trends Plant Sci. 2016;21:302–316. doi: 10.1016/j.tplants.2015.10.019. [DOI] [PubMed] [Google Scholar]
- 16.Ikeda M, Mitsuda N, Ohme-Takagi M. Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell. 2009;21:3493–3505. doi: 10.1105/tpc.109.069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chodaparambil JV, et al. Molecular functions of the TLE tetramerization domain in Wnt target gene repression. EMBO J. 2014;33:719–731. doi: 10.1002/embj.201387188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim MH, et al. The structure of the N-terminal domain of the product of the lissencephaly gene Lis1 and its functional implications. Structure. 2004;12:987–998. doi: 10.1016/j.str.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 19.Delto CF, et al. The LisH motif of muskelin is crucial for oligomerization and governs intracellular localization. Structure. 2015;23:364–373. doi: 10.1016/j.str.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 20.Kagale S, Links MG, Rozwadowski K. Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol. 2010;152:1109–1134. doi: 10.1104/pp.109.151704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kagale S, Rozwadowski K. EAR motif-mediated transcriptional repression in plants: An underlying mechanism for epigenetic regulation of gene expression. Epigenetics. 2011;6:141–146. doi: 10.4161/epi.6.2.13627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, Kim J, Somers DE. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc Natl Acad Sci USA. 2013;110:761–766. doi: 10.1073/pnas.1215010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ke J, et al. Structural basis for recognition of diverse transcriptional repressors by the TOPLESS family of corepressors. Sci Adv. 2015;1:e1500107. doi: 10.1126/sciadv.1500107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi N, et al. RanBPM, Muskelin, p48EMLP, p44CTLH, and the armadillo-repeat proteins ARMC8alpha and ARMC8beta are components of the CTLH complex. Gene. 2007;396:236–247. doi: 10.1016/j.gene.2007.02.032. [DOI] [PubMed] [Google Scholar]
- 25.Ulrich AKC, Schulz JF, Kamprad A, Schütze T, Wahl MC. Structural basis for the functional coupling of the alternative splicing factors Smu1 and RED. Structure. 2016;24:762–773. doi: 10.1016/j.str.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 26.Zhang F, et al. STENOFOLIA recruits TOPLESS to repress ASYMMETRIC LEAVES2 at the leaf margin and promote leaf blade outgrowth in Medicago truncatula. Plant Cell. 2014;26:650–664. doi: 10.1105/tpc.113.121947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu M-F, et al. Auxin-regulated chromatin switch directs acquisition of flower primordium founder fate. eLife. 2015;4:e09269. doi: 10.7554/eLife.09269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Busch W, et al. Transcriptional control of a plant stem cell niche. Dev Cell. 2010;18:849–861. doi: 10.1016/j.devcel.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 29.Espinosa-Ruiz A, et al. TOPLESS mediates brassinosteroid control of shoot boundaries and root meristem development in Arabidopsis thaliana. Development. 2017;144:1619–1628. doi: 10.1242/dev.143214. [DOI] [PubMed] [Google Scholar]
- 30.Pierre-Jerome E, Jang SS, Havens KA, Nemhauser JL, Klavins E. Recapitulation of the forward nuclear auxin response pathway in yeast. Proc Natl Acad Sci USA. 2014;111:9407–9412. doi: 10.1073/pnas.1324147111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pierre-Jerome E, Moss BL, Lanctot A, Hageman A, Nemhauser JL. Functional analysis of molecular interactions in synthetic auxin response circuits. Proc Natl Acad Sci USA. 2016;113:11354–11359. doi: 10.1073/pnas.1604379113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cerna D, Wilson DK. The structure of Sif2p, a WD repeat protein functioning in the SET3 corepressor complex. J Mol Biol. 2005;351:923–935. doi: 10.1016/j.jmb.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 33.Pirrotta V, Gross DS. Epigenetic silencing mechanisms in budding yeast and fruit fly: Different paths, same destinations. Mol Cell. 2005;18:395–398. doi: 10.1016/j.molcel.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 34.Krishnamurthy VM, Estroff LA, Whitesides GM. 2006. Multivalency in ligand design. Fragment-based Approaches in Drug Discovery, eds Jahnke W and Erlanson DA (Wiley‐VCH, Weinheim, Germany)
- 35.Menon RP, Gibson TJ, Pastore A. The C terminus of fragile X mental retardation protein interacts with the multi-domain Ran-binding protein in the microtubule-organising centre. J Mol Biol. 2004;343:43–53. doi: 10.1016/j.jmb.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 36.Yuan S, et al. A ubiquitin ligase of symbiosis receptor kinase involved in nodule organogenesis. Plant Physiol. 2012;160:106–117. doi: 10.1104/pp.112.199000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Menssen R, et al. Exploring the topology of the Gid complex, the E3 ubiquitin ligase involved in catabolite-induced degradation of gluconeogenic enzymes. J Biol Chem. 2012;287:25602–25614. doi: 10.1074/jbc.M112.363762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacob F. Evolution and tinkering. Science. 1977;196:1161–1166. doi: 10.1126/science.860134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.