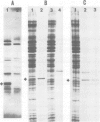
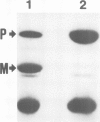
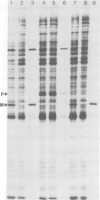
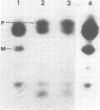
Abstract
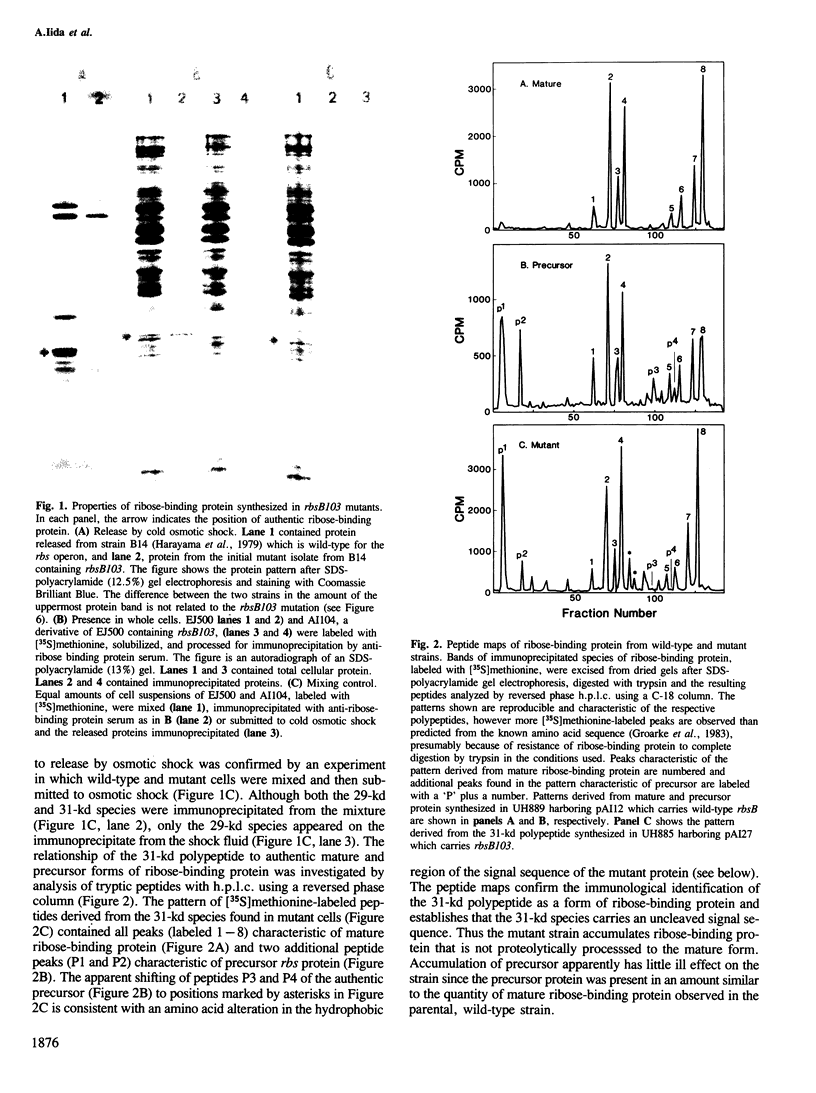
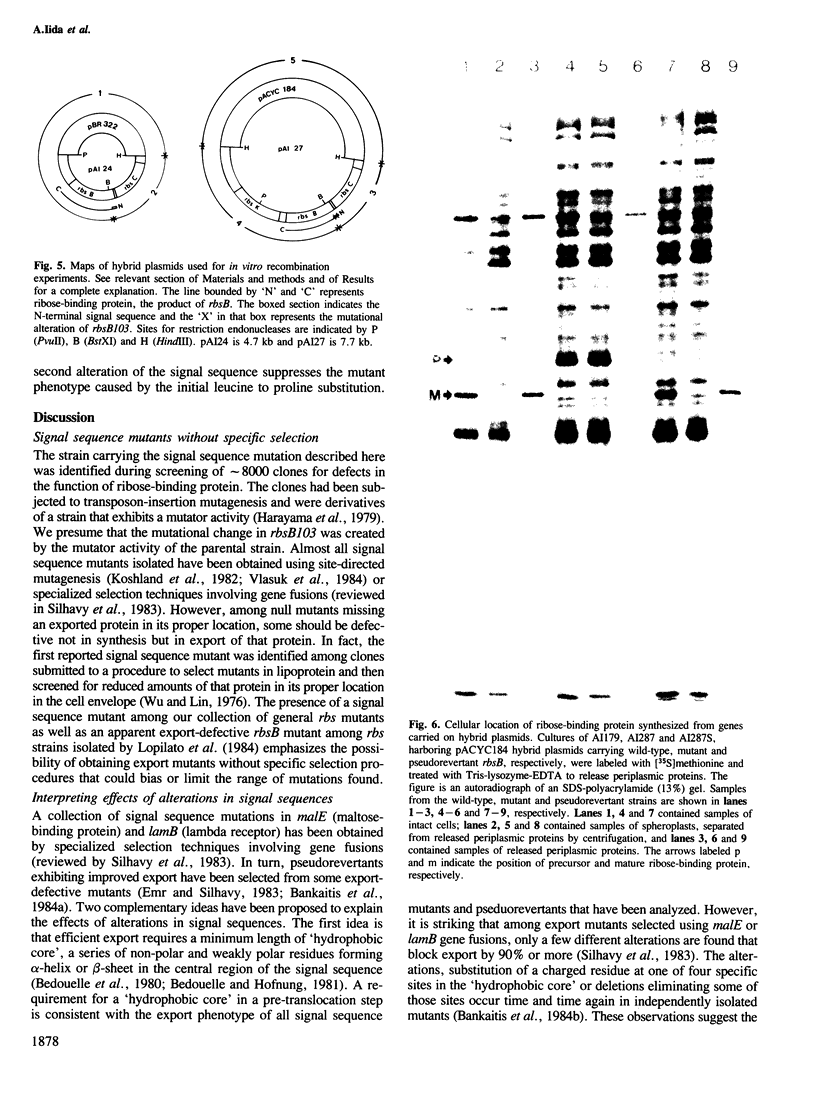
Ribose-binding protein is exported to the periplasmic compartment of Escherichia coli by a process that involves proteolytic cleavage of an amino-terminal extension of amino acids from the precursor form of the protein. In a collection of mutants isolated as defective in the Rbs transport system, a strain was identified that contained only precursor ribose-binding protein, none of which was exported to its normal location in the periplasm. The mutated rbsB contained a base substitution that results in a change of leucine to a proline at position-17 in the signal sequence. A pseudorevertant of the mutant contained proteolytically processed, active ribose-binding protein in the periplasm. The pseudorevertant rbsB carried a second mutation: serine at position-15 in the signal sequence was changed to phenylalanine. Isolation of a signal sequence mutant and a corresponding pseudorevertant without specific selection or site-directed mutagenesis emphasizes the possibility of obtaining export mutants without the use of procedures that could bias or limit the range of mutations found. Explanation of the extreme phenotype of the mutant and the effective correction of that phenotype in the pseudorevertant requires extension of current notions of critical features of signal sequences.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aksamit R. R., Koshland D. E., Jr Identification of the ribose binding protein as the receptor for ribose chemotaxis in Salmonella typhimurium. Biochemistry. 1974 Oct 22;13(22):4473–4478. doi: 10.1021/bi00719a001. [DOI] [PubMed] [Google Scholar]
- Bankaitis V. A., Rasmussen B. A., Bassford P. J., Jr Intragenic suppressor mutations that restore export of maltose binding protein with a truncated signal peptide. Cell. 1984 May;37(1):243–252. doi: 10.1016/0092-8674(84)90320-9. [DOI] [PubMed] [Google Scholar]
- Bedouelle H., Bassford P. J., Jr, Fowler A. V., Zabin I., Beckwith J., Hofnung M. Mutations which alter the function of the signal sequence of the maltose binding protein of Escherichia coli. Nature. 1980 May 8;285(5760):78–81. doi: 10.1038/285078a0. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Clarke L., Carbon J. A colony bank containing synthetic Col El hybrid plasmids representative of the entire E. coli genome. Cell. 1976 Sep;9(1):91–99. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- Emr S. D., Bassford P. J., Jr Localization and processing of outer membrane and periplasmic proteins in Escherichia coli strains harboring export-specific suppressor mutations. J Biol Chem. 1982 May 25;257(10):5852–5860. [PubMed] [Google Scholar]
- Emr S. D., Silhavy T. J. Importance of secondary structure in the signal sequence for protein secretion. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4599–4603. doi: 10.1073/pnas.80.15.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway D. R., Furlong C. E. The role of ribose-binding protein in transport and chemotaxis in Escherichia coli K12. Arch Biochem Biophys. 1977 Dec;184(2):496–504. doi: 10.1016/0003-9861(77)90459-3. [DOI] [PubMed] [Google Scholar]
- Groarke J. M., Mahoney W. C., Hope J. N., Furlong C. E., Robb F. T., Zalkin H., Hermodson M. A. The amino acid sequence of D-ribose-binding protein from Escherichia coli K12. J Biol Chem. 1983 Nov 10;258(21):12952–12956. [PubMed] [Google Scholar]
- Harayama S., Engström P., Wolf-Watz H., Iino T., Hazelbauer G. L. Cloning of trg, a gene for a sensory transducer in Escherichia coli. J Bacteriol. 1982 Oct;152(1):372–383. doi: 10.1128/jb.152.1.372-383.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama S., Palva E. T., Hazelbauer G. L. Transposon-insertion mutants of Escherichia coli K12 defective in a component common to galactose and ribose chemotaxis. Mol Gen Genet. 1979 Mar 20;171(2):193–203. doi: 10.1007/BF00270005. [DOI] [PubMed] [Google Scholar]
- Hazelbauer G. L., Adler J. Role of the galactose binding protein in chemotaxis of Escherichia coli toward galactose. Nat New Biol. 1971 Mar 24;230(12):101–104. doi: 10.1038/newbio230101a0. [DOI] [PubMed] [Google Scholar]
- Hazelbauer G. L., Harayama S. Mutants in transmission of chemotactic signals from two independent receptors of E. coli. Cell. 1979 Mar;16(3):617–625. doi: 10.1016/0092-8674(79)90035-7. [DOI] [PubMed] [Google Scholar]
- Icho T., Iino T. Isolation and characterization of motile Escherichia coli mutants resistant to bacteriophage chi. J Bacteriol. 1978 Jun;134(3):854–860. doi: 10.1128/jb.134.3.854-860.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida A., Harayama S., Iino T., Hazelbauer G. L. Molecular cloning and characterization of genes required for ribose transport and utilization in Escherichia coli K-12. J Bacteriol. 1984 May;158(2):674–682. doi: 10.1128/jb.158.2.674-682.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D., Sauer R. T., Botstein D. Diverse effects of mutations in the signal sequence on the secretion of beta-lactamase in Salmonella typhimurium. Cell. 1982 Oct;30(3):903–914. doi: 10.1016/0092-8674(82)90295-1. [DOI] [PubMed] [Google Scholar]
- Lopilato J. E., Garwin J. L., Emr S. D., Silhavy T. J., Beckwith J. R. D-ribose metabolism in Escherichia coli K-12: genetics, regulation, and transport. J Bacteriol. 1984 May;158(2):665–673. doi: 10.1128/jb.158.2.665-673.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Randall L. L., Hardy S. J. Export of protein in bacteria. Microbiol Rev. 1984 Dec;48(4):290–298. doi: 10.1128/mr.48.4.290-298.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall L. L., Hardy S. J. Synthesis of exported proteins by membrane-bound polysomes from Escherichia coli. Eur J Biochem. 1977 May 2;75(1):43–53. doi: 10.1111/j.1432-1033.1977.tb11502.x. [DOI] [PubMed] [Google Scholar]
- Randall L. L., Josefsson L. G., Hardy S. J. Processing in vitro of precursor periplasmic proteins from Escherichia coli. Eur J Biochem. 1978 Dec;92(2):411–415. doi: 10.1111/j.1432-1033.1978.tb12761.x. [DOI] [PubMed] [Google Scholar]
- Randall L. L. Translocation of domains of nascent periplasmic proteins across the cytoplasmic membrane is independent of elongation. Cell. 1983 May;33(1):231–240. doi: 10.1016/0092-8674(83)90352-5. [DOI] [PubMed] [Google Scholar]
- Silhavy T. J., Benson S. A., Emr S. D. Mechanisms of protein localization. Microbiol Rev. 1983 Sep;47(3):313–344. doi: 10.1128/mr.47.3.313-344.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. C., Lin J. J. Escherichia coli mutants altered in murein lipoprotein. J Bacteriol. 1976 Apr;126(1):147–156. doi: 10.1128/jb.126.1.147-156.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]