Significance
We have coupled mass spectrometry-based phosphoproteomic analyses with treatment using various selective PDE inhibitors to characterize the PDE-regulated phosphoproteome of CD3/CD28-stimulated Jurkat cells. Predictive algorithms were used to identify likely upstream regulatory kinases, metabolic pathways, and biological processes that can be regulated by different PDEs. Here we compare the phosphoproteomes of different functional compartments subserved by combinations of individual PDE isozymes in a T-cell model. We observed unique phosphoproteomes associated with specific combinations of PDEs. These data allow one to prioritize future experiments to understand further how these pathways are regulated by specific PDEs. The results also have substantial implications for the design and use of selective PDE inhibitors in clinical practice.
Keywords: cAMP, phosphodiesterase, phosphoproteomics, PDE, PKA
Abstract
Specific functions for different cyclic nucleotide phosphodiesterases (PDEs) have not yet been identified in most cell types. Conventional approaches to study PDE function typically rely on measurements of global cAMP, general increases in cAMP-dependent protein kinase (PKA), or the activity of exchange protein activated by cAMP (EPAC). Although newer approaches using subcellularly targeted FRET reporter sensors have helped define more compartmentalized regulation of cAMP, PKA, and EPAC, they have limited ability to link this regulation to downstream effector molecules and biological functions. To address this problem, we have begun to use an unbiased mass spectrometry-based approach coupled with treatment using PDE isozyme-selective inhibitors to characterize the phosphoproteomes of the functional pools of cAMP/PKA/EPAC that are regulated by specific cAMP-PDEs (the PDE-regulated phosphoproteomes). In Jurkat cells we find multiple, distinct PDE-regulated phosphoproteomes that can be defined by their responses to different PDE inhibitors. We also find that little phosphorylation occurs unless at least two different PDEs are concurrently inhibited in these cells. Moreover, bioinformatics analyses of these phosphoproteomes provide insight into the unique functional roles, mechanisms of action, and synergistic relationships among the different PDEs that coordinate cAMP-signaling cascades in these cells. The data strongly suggest that the phosphorylation of many different substrates contributes to cAMP-dependent regulation of these cells. The findings further suggest that the approach of using selective, inhibitor-dependent phosphoproteome analysis can provide a generalized methodology for understanding the roles of different PDEs in the regulation of cyclic nucleotide signaling.
Cellular signaling is a complex orchestration of multiple effector molecules making up communication relays that ultimately coordinate cellular responses. Since its original discovery nearly 60 y ago (1, 2), cAMP has been shown to regulate many different biological processes in multiple cell types (3). cAMP coordinates the regulation of these cellular responses by activating cAMP-dependent protein kinases (PKAs) (4), specific guanine nucleotide exchange factors (EPACs) (5), and cyclic nucleotide-gated ion channels (6). It also can directly regulate some of the cyclic nucleotide phosphodiesterases (PDEs). The PDEs comprise a family of enzymes that catalyze the hydrolysis of cAMP or cGMP to 5′-AMP or 5′-GMP and thus regulate the amplitude, duration, and subcellular localization of cyclic nucleotide signaling.
It is thought that the expression and subcellular distribution of cAMP, PDEs (7), adenylyl cyclases (8), effector molecules (9), and target substrates are organized in such a way as to form “functional compartments.” These compartments may correspond to known subcellular (10) regions or may be more diffuse and smaller than can be resolved by light microscopy. For example, PDE3B is localized predominately at membrane surfaces and is thought to regulate cAMP in regions proximal to these surfaces (11). Similarly, in activated T cells, PDE4 is recruited to the T-cell receptor in a β-arrestin–dependent manner, thus lowering cAMP in the vicinity of this part of the T-cell activation pathway (12).
In T lymphocytes, PDE inhibitors and other agents that increase cAMP have in general been shown to reduce cell proliferation and to bias T-helper polarization toward Th2, Treg, or Th17 phenotypes (13, 14). In a few cases increased cAMP may even potentiate the T-cell activation signal (15), particularly at early stages of activation. Recent MS-based proteomic studies have been useful in characterizing changes in the phosphoproteome of T cells under various stimuli such as T-cell receptor stimulation (16), prostaglandin signaling (17), and oxidative stress (18), so much of the total Jurkat phosphoproteome is known. Until now, however, no information on the regulation of phosphopeptides by PDEs has been available in these cells.
Inhibitors of cAMP PDEs are useful tools to study PKA/EPAC-mediated signaling, and selective inhibitors for each of the 11 PDE families have been developed (19–21). The classical pharmacological approach to study cAMP-regulated systems has been to increase global cAMP in vitro or in vivo by means of adenylyl cyclase activation and/or treatment with relatively nonselective PDE inhibitors. Consequently, information about the composition and function of specific subcellular cAMP-signaling compartments was largely masked. We used sets of highly selective PDE inhibitors to perturb putative cAMP compartment(s) combined with unbiased, global MS-based analysis to capture dynamic changes in the phosphoproteomes regulated by these PDEs. We treated Jurkat cells with various combinations of selective inhibitors of PDEs 1, 3, 4, 7, and 8 in the presence or absence of low, physiological concentrations of PGE2. Total phosphorylation changes were quantified by label-free LC-MS/MS (22–24), leading to the identification of 13,589 sites, of which 618 were sensitive to PDE inhibition. In doing so, we identified two distinct functional compartments regulated by two different combinations of PDE inhibitors.
Results
Global cAMP Levels Are Elevated Only by Specific Combinations of PDE Inhibitors.
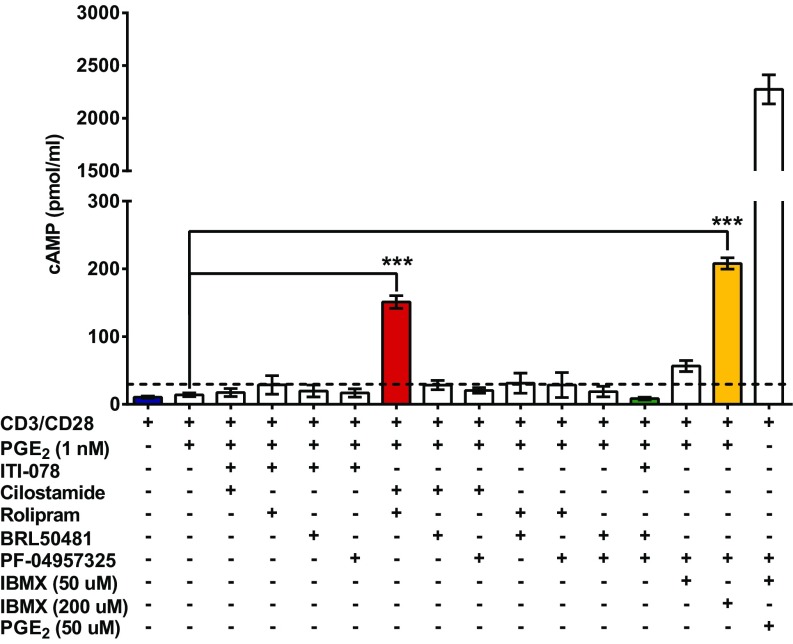
To determine which sets of PDE inhibitors to use in phosphoproteomic studies, we first treated Jurkat cultures with individual isozyme-selective PDE inhibitors or various combinations of these inhibitors and measured the resulting changes in cAMP levels. Previous studies identified PDEs 1B, 1C, 3B, 4A, 7A, and 8A as the predominant cAMP-hydrolyzing PDE mRNA transcripts expressed in Jurkat cells (25–27). In theory, any of these PDEs or any combination of PDEs could regulate different functional cAMP compartments in the cell. We also used 50 µM and 200 µM isobutylmethylxanthine (IBMX), a potent, general, nonselective PDE inhibitor, in combination with 200 nM PF-04957325 (a PDE8-selective inhibitor) to inhibit the majority of cAMP-hydrolyzing PDEs. For these initial studies, we then chose the inhibitor combinations that seemed most likely to influence the greatest number of PDE-regulated functional compartments for follow-up by phosphoproteomic analysis (Fig. 1).
Fig. 1.
Inhibition of multiple phosphodiesterases is necessary to increase intracellular cAMP. Jurkat cells (1 × 107) were treated as indicated for 20 min. Cells were centrifuged briefly, and the supernatant was discarded. Cell pellets were lysed by adding 1 mL of a 1:99 mixture of 11.65 M HCL and 95% EtOH. Pellets were dispersed using a P1000 pipet tip and were vortexed. The lysate was incubated at room temperature for 30 min. Following incubation, the extraction volume was transferred to a fresh microfuge tube and was dried in a speed vacuum. cAMP was resuspended in 150 µL of 0.1 M HCl, acetylated, and assayed using a cAMP ELISA kit according to the manufacturer’s recommendations (Cayman Biochemical). Bars represent the mean of a minimum of three individual experiments; error bars show SD. ITI-078/BRL50481/PF-04957325 and IBMX 50 µM data are from two individual experiments; error bars indicate the range. Statistical analysis was performed using a Student’s t test; ***P < 0.0001. The dashed line indicates the greatest increase of cAMP from a single PDE inhibitor (rolipram) treatment.
Somewhat unexpectedly, treatment with an individual PDE inhibitor alone did not cause a significant increase in total cAMP in either the absence or the presence of low PGE2 (1 nM) (SI Appendix, Fig. S1). However, when combined with the low concentration of PGE2 (1 nM), combined treatment with cilostamide (a PDE3 inhibitor) and rolipram (a PDE4 inhibitor) greatly increased cAMP, from 9.7 to 144 pmol/mL (Fig. 1). Interestingly, the combination of PGE2 plus 50 µM IBMX and 200 nM PF-04957325 increased cAMP to a lesser degree (from 9.7 to ∼50 pmol/mL) than the combination of PGE2 plus cilostamide and rolipram, perhaps because of the known antagonistic effect of IBMX on adenosine-stimulated cAMP. However, a higher concentration of IBMX (200 µM) and 200 nM PF-04957325 plus PGE2 (1 nM) increased cAMP ∼38-fold (from 6.6 to 250 pmol/mL) (Fig. 1). In these conditions, the highest level of cAMP was seen in the presence of a supersaturating concentration of 200 µM PGE2 (Fig. 1). Overall, the results suggested that most putative cAMP compartments would be saturated only, if at all, in the high PGE2 condition, thus implying that it should be feasible to use lower combinations of PDE inhibitors to begin to resolve the phosphorylations that occur in the PDE-specific functional compartments of the cell.
Phosphoproteomic Interrogation of PDE Functional Compartments (the PDE-Regulated Phosphoproteomes).
To identity the constituents of putative PDE-regulated compartments, we designed a phosphoproteomic approach using the combination of PDE inhibitors that caused the greatest increases in global cAMP based on our cAMP assay results. This approach is illustrated in SI Appendix, Fig. S2A. Preliminary analysis of the number of phosphopeptides identified in the basal condition (SI Appendix, Fig. S2B) suggested that nearly maximal identification could be achieved with nine LC-MS/MS runs for each treatment condition (with three or more biological replicates and three analytical replicates each). A single LC-MS/MS run yielded on average 5,146 unique quantified phosphopeptides, and increasing the number of LC-MS/MS runs to nine approached a plateau number of ∼10,000 phosphopeptides per biological condition (SI Appendix, Fig. S2B). The Pearson correlation coefficients between biological replicates and analytical replicates were between 0.5–0.9 and ≥0.9, respectively (SI Appendix, Fig. S2C). In total, we identified 13,589 phosphopeptides and 3,241 proteins. For further downstream functional analyses, we included only phosphopeptides that were observed in at least 60% of the LC-MS/MS runs in the respective treatment groups. Of the 13,589 phosphopeptides identified, we found 618 phosphopeptides distributed among 461 unique proteins that were significantly regulated by the PDE inhibitor treatments [false discovery rate (FDR) ≤0.05, two-tailed t test, permutation-based FDR]. (Please refer to Dataset S1 for the complete list).
Different PDEs Regulate Distinct Functional Compartments.
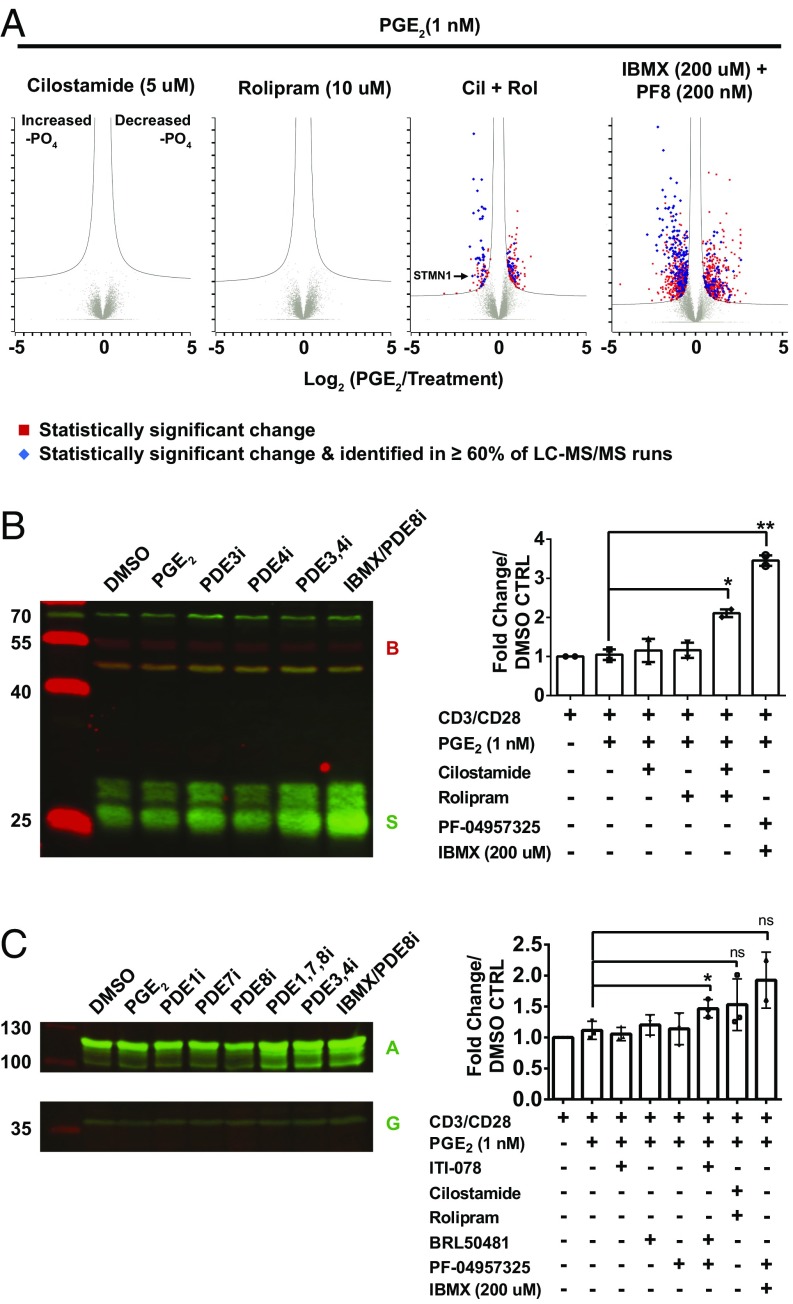
Consistent with the cAMP assays, no phosphosites were significantly altered by individual PDE inhibitor treatments alone under the basal condition (no PGE2) (Dataset S1) or in the 1-nM PGE2-stimulated state (Fig. 2A). However, we observed a synergistic increase in phosphopeptides identified when two or more PDEs were inhibited (Fig. 2A). To corroborate the proteomics data, we used commercially available (Abcam and Cell Signaling) antibodies to probe for changes in phosphorylation in phosphosites identified in our proteomics study, Stathmin1 (STMN1) S63 and Rho/Rac guanine nucleotide exchange factor 2 (ARHGEF2) S858. In agreement with the proteomics study, we found that neither PDE3 or PDE4 inhibitors alone, nor the combination of PDE1, PDE7, and PDE8 inhibitors, caused a change in STMN1 S63 phosphorylation, but inhibition of both PDE3 and PDE4 increased STMN1 S63 phosphorylation (Fig. 2B, see also SI Appendix, Fig. S7). In this case also, treatment with 200 µM IBMX plus the PDE8 inhibitor caused the greatest increase in phosphorylation. The individual PDE1, PDE7, and PDE8 inhibitors alone did not increase ARHGEF2 S858 phosphorylation, but the combination of all three inhibitors did (Fig. 2C).
Fig. 2.
Inhibiting multiple PDEs increases the number and magnitude of phosphorylation changes. (A) Volcano plots of the number of phosphosites significantly modulated compared with DMSO as a subset of the total proteome are presented as the log ratio of the DMSO intensity over the PDE inhibitor intensity plotted against the negative log of the P value. Red squares indicate phosphosites significantly modulated over PGE2. Blue diamonds indicate significantly regulated sites that occur in at least 60% of LC-MS/MS runs. (B) Immunoblot analysis of changes in STMN1phosphorylation at S63. Cells (1 × 107) were treated as previously described. Cells were harvested and boiled in 200 µL Laemmli buffer and transferred to nitrocellulose. Membranes were probed with anti-STMN1 antibody (S) (1:2,000) (Abcam) and anti–β-actin (B) (1:200,000) (GeneTex). Membranes were quantified on the Odyssey Scanner Clx (LI-COR). The blot is shown on the left, and quantification is shown on the right. Error bars show SD. Statistical analysis was performed using a Student’s t test; *P = 0.02, **P = 0.006. (C) Immunoblot analysis of changes in Rho/RAC guanine nucleotide exchange factor 2 phosphorylation at S858. Cells (1 × 107) were treated as previously described, harvested, boiled in 200 µL Laemmli buffer, and transferred to nitrocellulose. Membranes were probed with anti-ARHGEF2 antibody (A) (1:2,000) (Cell Signaling) or anti-GAPDH antibody (G) (1:4,000) (Cell Signaling). Error bars indicate SD. Statistical analysis was performed using a Student’s t test; *P = 0.04; ns, not significant.
In an attempt to define the functional compartments also regulated by PDEs other than PDE3 and PDE4, we performed the same analysis with 50 µM IBMX plus 200 nM PF-04957325 and 200 µM IBMX plus 200 nM PF-04957325. The expectation was that the 200 µM IBMX plus PF-04957325 condition would likely define the total PDE-regulated functional compartments. Each of these conditions was assessed in the absence or presence of a low concentration (1 nM) of PGE2. Indeed, many more regulated phosphosites were modulated when all PDEs were inhibited (Fig. 3). We surmised that the remaining PDEs (i.e., not PDE3 and PDE4), which included PDEs 1, 7, and 8 (or some combination thereof), might also subserve functional compartments different from those regulated by PDE3 and/or PDE4. Therefore, we also treated Jurkat cells with a combination of 200 nM ITI-078 (a PDE1 inhibitor), 30 µM BRL50481 (a PDE7 inhibitor), and 200 nM PF-04957325 (a PDE8 inhibitor), also in the absence or presence of a low (1 nM) level of PGE2.
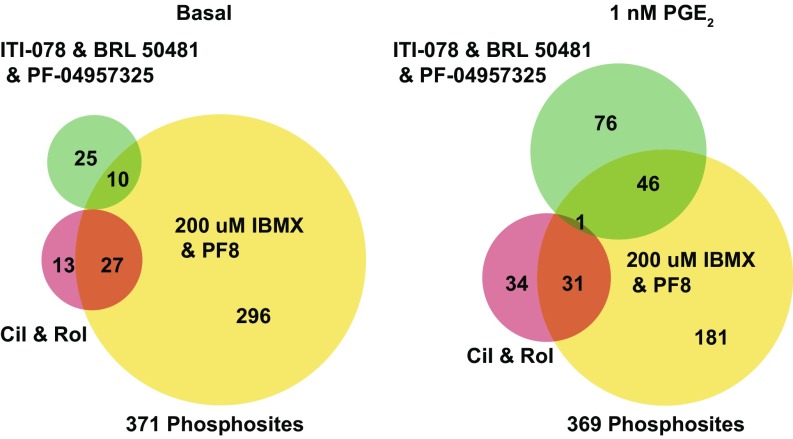
Fig. 3.
Venn diagrams of the number of phosphosites increased by selective PDE inhibition. MaxQuant was used to search mass spectra against the UniProt human reference proteome; 13,589 phosphopeptides were identified. For further analysis, phosphopeptides had to satisfy the following conditions: first, phosphopeptides must have an intensity value in 60% of the total LC-MS/MS runs in that experimental condition; second, they must be statistically significant by a two-tailed, two-sample t test, multiple comparison FDR of 0.05. BioVenn was used to plot the number of unique or common phosphosites identified in each condition (90).
The subset of cilostamide- and rolipram-regulated phosphosites made up a portion of the total IBMX- and PF-04957325-regulated phosphosites (Fig. 3, Left). Forty phosphosites were selectively regulated by inhibiting PDE3 and PDE4 and not by inhibiting the combination of PDEs 1, 7, and 8 (Fig. 3, and SI Appendix, Table S3). Table 1 shows examples of phosphosites increased by the combination of PDE 3 and PDE4 inhibitors in the absence and presence of PGE2 (for the full list, refer to Dataset S1). Under PGE2-stimulated conditions 66 phosphosites were regulated by the PDE3 and PDE4 treatment, and 123 phosphosites were regulated by the PDE1/7/8 treatment (Fig. 3, Right). However, even with increased peptide identification, only one common phosphosite, S811 on Slingshot protein phosphatase 2 (SSH2), was statistically altered by both PDE inhibitor treatment groups. This general absence of overlap strongly suggested that the pool(s) of cAMP regulated by the combination of PDE1/7/8 inhibitors are functionally different from the pool(s) regulated by the combination of PDE3 and PDE4 inhibitors and that perhaps SSH2 is regulated by cAMP in more than one functional compartment (Fig. 3, Right and Table 1).
Table 1.
Top phosphorylated proteins in response to PDE3 plus PDE4 inhibition in the absence (−) and presence (+) of 1 nM PGE2 compared with changes seen in response to the other PDE inhibitors under the same conditions
| Gene | Description | Position no. | Sequence | Fold change over control | |||
| ITI, BRL, PF8 | CIL and ROL | IBMX 50 µM | IBMX 200 µM | ||||
| Response to PDE3 and PDE4 inhibitor (−) PGE2 | |||||||
| CAD | CAD protein | 1,343 | GRRLSSFVT | 1.10 | 3.31 | 2.80 | 2.86 |
| RANBP2 | E3 SUMO-protein ligase RanBP2 | 1,509 | PRKQSLPAT | 1.30 | 3.17 | 3.98 | 4.23 |
| MAP2K2 | Dual specificity mitogen-activated protein kinase kinase 2 | 394 | NQPGTPTRT | 0.37 | 3.01 | 0.34 | 0.44 |
| MAGED2 | Melanoma-associated antigen D2 | 200 | ARRASRGPI | 0.00 | 2.92 | 0.86 | 1.48 |
| HIST1H1E | Histone H1.4; Histone H1.3 | 37 | KRKASGPPV | 1.44 | 2.82 | 3.79 | 4.01 |
| HIST1H1C | Histone H1.2 | 36 | PRKASGPPV | 0.89 | 2.80 | 2.37 | 2.61 |
| UBE2O | E2/E3 hybrid ubiquitin-protein ligase UBE2O | 515 | SRKKSIPLS | 0.66 | 2.52 | 1.93 | 0.95 |
| RCSD1 | CapZ-interacting protein | 108 | ASPKSPGLK | 1.54 | 2.27 | 0.69 | 1.05 |
| ATAD2 | ATPase family AAA domain-containing protein 2 | 1,302 | RARRSQVEQ | 1.24 | 2.23 | 3.53 | 3.92 |
| SEC22B | Vesicle-trafficking protein SEC22b | 137 | RNLGSINTE | 1.21 | 2.16 | 2.68 | 2.89 |
| GAS2L1 | GAS2-like protein 1 | 316 | ERRGSRPEM | 1.25 | 2.15 | 2.32 | 1.67 |
| SSH2 | Protein phosphatase Slingshot homolog 2 | 811 | PKKNSIHEL | 1.41 | 2.13 | 1.87 | 2.19 |
| Response to PDE3 and PDE4 inhibitor (+) PGE2 | |||||||
| RANBP2 | E3 SUMO-protein ligase RanBP2 | 1,509 | PRKQSLPAT | 2.20 | 4.51 | 5.01 | 5.21 |
| HIST1H1C | Histone H1.2 | 36 | PRKASGPPV | 1.23 | 4.05 | 3.23 | 3.88 |
| MAGED2 | Melanoma-associated antigen D2 | 200 | ARRASRGPI | 0.68 | 3.84 | 1.02 | 1.53 |
| SEC22B | Vesicle-trafficking protein SEC22b | 137 | RNLGSINTE | 1.44 | 3.36 | 3.45 | 3.50 |
| PWP1 | Periodic tryptophan protein 1 homolog | 485 | ARNSSISGP | 1.27 | 3.29 | 4.42 | 3.20 |
| STMN1 | Stathmin | 63 | ERRKSHEAE | 0.42 | 3.25 | 1.57 | 1.59 |
| HIST1H1E | Histone H1.4 | 37 | KRKASGPPV | 1.94 | 3.17 | 5.58 | 5.42 |
| NUMA1 | Nuclear mitotic apparatus protein 1 | 1,955 | LRRASMQPI | 1.49 | 2.91 | 3.59 | 3.59 |
| NFRKB | Nuclear factor related to kappa-B-BP | 310 | GRKGSLAAL | 1.28 | 2.88 | 2.17 | 2.33 |
| CAD | CAD protein | 1,343 | GRRLSSFVT | 1.41 | 2.87 | 5.24 | 4.13 |
| MKI67 | Antigen KI-67 | 538 | TKRKSLVMH | 1.14 | 2.53 | 3.25 | 3.02 |
Consensus PKA primary amino acid phosphorylation sequences are shown in bold, underlined, italicized text. Statistically significantly regulated changes in phosphorylation in the other PDE inhibitor conditions (P < 0.05) are shown in bold. BRL, BRL50481; CIL, cilostamide; ITI, ITI-078; PF8, PF-04957325; ROL, rolipram.
Identifying Kinases That Modulate PDE-Regulated Phosphoproteomes.
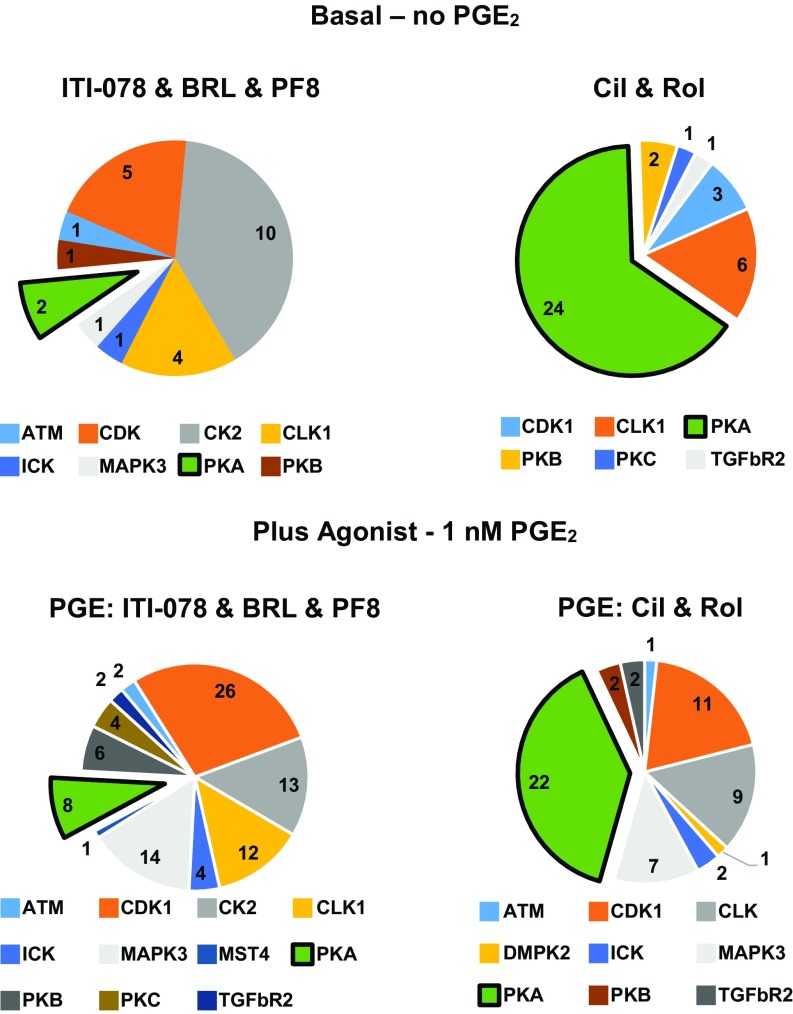
The distinct functional pools of PDE-regulated phosphosites were further characterized by analyzing which regulatory kinases would be most likely to phosphorylate the PDE inhibitor-dependent sites. Analysis of the sequences of the phosphosites with the program NetPhorest (28), a web-based tool for kinase prediction (https://omictools.com), suggested that the majority of phosphosites regulated by combined inhibition of PDEs 1, 7, and 8 are primarily phosphorylated by casein kinase II or by a kinase with a similar substrate recognition motif in both the basal and PGE2-stimulated conditions (Fig. 4, Left). In contrast, the same algorithm suggested that most of the phosphosites increased by inhibiting both PDE3 and PDE4 are primarily phosphorylated by PKA (Fig. 4, Right). We observed a similar trend in PGE2-stimulated cells. The majority of sites predicted to be phosphorylated by PKA contain a modified S/T residue contained in the classic PKA consensus motif R/K, R/K, X, S/T (red bars in SI Appendix, Fig. S3 B and D). However, 132 of 151 phosphorylation sites in the cells treated with combined PDE1/7/8 inhibitors did not contain this predicted PKA consensus site (SI Appendix, Fig. S3 A and C).
Fig. 4.
Predicted kinase utilization for different PDE/agonist combinations. A truncated peptide sequence of four amino acid residues flanking the regulated phosphosite was used as an input sequence into NetPhorest to predict kinases responsible for phosphorylating identified sites. The threshold score was set at 0.21.
Prioritizing Phosphosites with Likely Biological Relevance.
To begin to identify which of the phosphosites were most likely to be biologically relevant, we searched a database of phosphosites annotated by Xiao et al. (29) for predicted regulatory function. This database was compiled using an algorithm that compares the sequences of the phosphosites with a series of criteria including evolutionary conservation, secondary structure, regional disorder, and the degree of sequence similarity to kinase-recognition motifs of known phosphorylation substrates. Of the 618 sites that were significantly modulated by PDE inhibitor treatment, 160 were predicted by this algorithm to be likely to have a regulatory function (refer to Dataset S1 for the full list). In fact, 55 of these identified phosphosites already have annotations in the PhosphositePlus (30) database, indicating that there is empirical evidence demonstrating that phosphorylation at the identified site regulates protein function (examples are given in Table 2).
Table 2.
Sites in our dataset modulated by any PDE inhibitor condition that are identified as regulatory in the PFP database (28)
| Gene | Description | Amino acid | Position no. | Predictive models | RP | |||
| B | L | M | R | |||||
| LASP1 | LIM and SH3 domain protein 1 | S | 146 | — | — | — | — | + |
| ARHGEF2 | Rho guanine nucleotide exchange factor 2 | S | 858 | — | — | + | — | + |
| PTPN7 | Tyrosine-protein phosphatase nonreceptor type 7 | S | 125 | + | + | + | + | + |
| BRAF | Serine/threonine-protein kinase B-raf | S | 446 | + | + | + | + | + |
| NOP58 | Nucleolar protein 58 | S | 502 | — | — | — | — | + |
| NUP50 | Nuclear pore complex prot Nup50 | S | 287 | — | — | — | — | + |
| RAB3IP | Rab-3A-interacting protein | S | 162 | — | — | — | — | + |
| BAD | Bcl2-associated agonist of cell death | S | 74/75 | + | — | — | + | + |
| PGRMC1 | Member-associated progesterone receptor component 1 | S | 57 | — | — | — | — | + |
| STAT1 | Signal transducer and activator of transcription | S | 727 | — | + | + | + | + |
| SLC9A1 | Sodium/hydrogen exchanger 1 | S | 796 | — | — | — | + | + |
| SP1 | Transcription factor Sp1 | S | 7 | + | + | + | + | + |
| PRKCB | Protein kinase C β-type | S | 660 | — | + | + | — | + |
| TBC1D1 | TBC1 domain family member 1 | T | 596 | + | + | + | + | + |
| ETS1 | Protein C-ets-1 | S | 282 | + | — | + | + | + |
| PPP1R2 | Protein phosphatase inhibitor 2 | S | 121 | + | + | + | + | + |
| PPP1R2 | Protein phosphatase inhibitor 2 | S | 122 | + | + | + | + | + |
| HMGA1 | High mobility group protein HMG-I/HMG-Y | T | 53 | — | — | — | — | + |
| STMN1 | Stathmin | S | 63 | + | — | — | — | + |
| CAD | CAD protein | S | 1,343 | + | + | + | + | + |
| CAMKK1 | C++/calmodulin-dependent protein kinase kinase 1 | S | 485 | — | — | — | — | + |
| USP20 | Ubiquitin carboxyl-terminal hydrolase 20 | S | 333 | — | — | — | — | + |
A truncated peptide sequence of four amino acid residues flanking the regulated phosphosite was used to screen the PFP proteomic database for predicted functional phosphosites. Predictive models used by PFP are Bayes (B), logistic (L), multilayer (M), and random (R). Empirically determined regulatory sites (RP) as derived from the PhosphositePlus database (29) are reported in the last column.
This approach also allowed us to identify a series of phosphosites likely to be regulatory but for which a regulatory role had not yet been empirically determined. Several examples of the treatment with PDE1/7/8 inhibitors are shown in Table 3, and examples of treatment with PDE3 and PDE4 inhibitors are shown in Table 4. We found 50 potential regulatory sites in the group of PDE1/7/8 inhibitors (Table 3) and 30 potential regulatory sites in the group of PDE3 and PDE4 inhibitors (examples are given in Table 4). Clearly, these sites are prime candidates for further follow-up with genetic and mutagenesis approaches to corroborate possible new regulatory roles for these PDE-modulated phosphosites.
Table 3.
Examples of sites of unknown function in the dataset modulated by PDE1/7/8 inhibitors that are predicted to be regulatory in the PFP database (28)
| Gene | Description | Amino acid | Position no. | Predicted kinase | Predictive models | |||
| B | L | M | R | |||||
| ETS1 | Protein C-ets-1 | S | 285 | N/A | + | — | + | + |
| HDAC4 | Histone deacetylase 4 | S | 453 | CK2α | — | — | — | + |
| RB1 | Retinoblastoma-associated protein | S | 624 | N/A | + | + | + | + |
| ANAPC2 | Anaphase-promoting complex subunit 2 | S | 314 | MOK | + | + | + | + |
| HIST1H1B | Histone H1.5 | S | 18 | N/A | + | — | — | — |
| NBEAL2 | Neurobeachin-like protein 2 | T | 1,683 | N/A | — | — | + | — |
| TP53BP1 | Tumor suppressor p53-binding protein 1 | S | 552 | MAPK1 | + | + | + | + |
| BCL11B | B-cell lymphoma/leukemia 11B | T | 131 | CDK2 | — | — | + | — |
A truncated peptide sequence of four amino acid residues flanking the regulated phosphosite was used to screen the PFP proteomic database for predicted functional phosphosites (28). Sites were considered positive if at least one of four prediction models suggested function. Predictive models used by PFP are Bayes (B), logistic (L), multilayer (M), and random (R). The same sequence was used in NetPhorest (27) to predict the regulatory kinase. The threshold was set at 0.21. N/A, not applicable.
Table 4.
Examples of sites of unknown function in the dataset modulated by PDE3 and PDE4 inhibitors that are predicted to be regulatory in the PFP database (28)
| Gene | Description | Amino acid | Position no. | Predicted kinase | Predictive models | |||
| B | L | M | R | |||||
| NUMA1 | Nuclear mitotic apparatus protein 1 | S | 1,955 | N/A | + | + | + | + |
| MKI67 | Antigen KI-67 | S | 538 | PKAα | — | — | + | — |
| NUMA1 | Nuclear mitotic apparatus protein 1 | S | 2,033 | N/A | — | — | — | + |
| FLNA | Filamin-A | T | 2,309 | CLK2 | + | + | — | + |
| ATG16L1 | Autophagy-related protein 16-1 | S | 269 | ATM | — | + | + | — |
| PLEKHF2 | Pleckstrin homology domain-containing family F member 2 | S | 16 | PKAα | — | — | + | — |
| TEX2 | Testis-expressed sequence 2 protein | S | 295 | PKAα | — | + | + | — |
| ABL1 | Tyrosine-protein kinase ABL1 | S | 16 | PKAα | — | — | + | — |
| RIF1 | Telomere-associated protein RIF1 | S | 2,205 | PKAα | + | + | — | + |
| SNX1 | Sorting nexin-1 | S | 188 | PKAα | — | + | — | + |
| CUL4A | Cullin-4A | S | 10 | PKAα | + | — | — | — |
| PRKDC | DNA-dependent protein kinase catalytic subunit | S | 893 | PKAγ | + | + | — | + |
| CAMKK2 | Ca++/calmodulin-dependent protein kinase kinase 2 | S | 468 | N/A | — | — | — | + |
| CDCA2 | Cell division cycle-associated protein 2 | S | 962 | PKAα | + | — | — | — |
| MACF1 | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 | S | 7,068 | N/A | — | — | + | — |
A truncated peptide sequence of four amino acid residues flanking the regulated phospho-site was used to screen the PFP proteomic database for predicted functional phosphosites (28). Sites were considered positive if at least one of four prediction models suggested function. Predictive models used by PFP are Bayes (B), logistic (L), multilayer (M), and random (R). The same sequence was used in NetPhorest (27) to predict the regulatory kinase. The threshold was set at 0.21. N/A, not available.
Interaction Networks Defined by STRING Analysis.
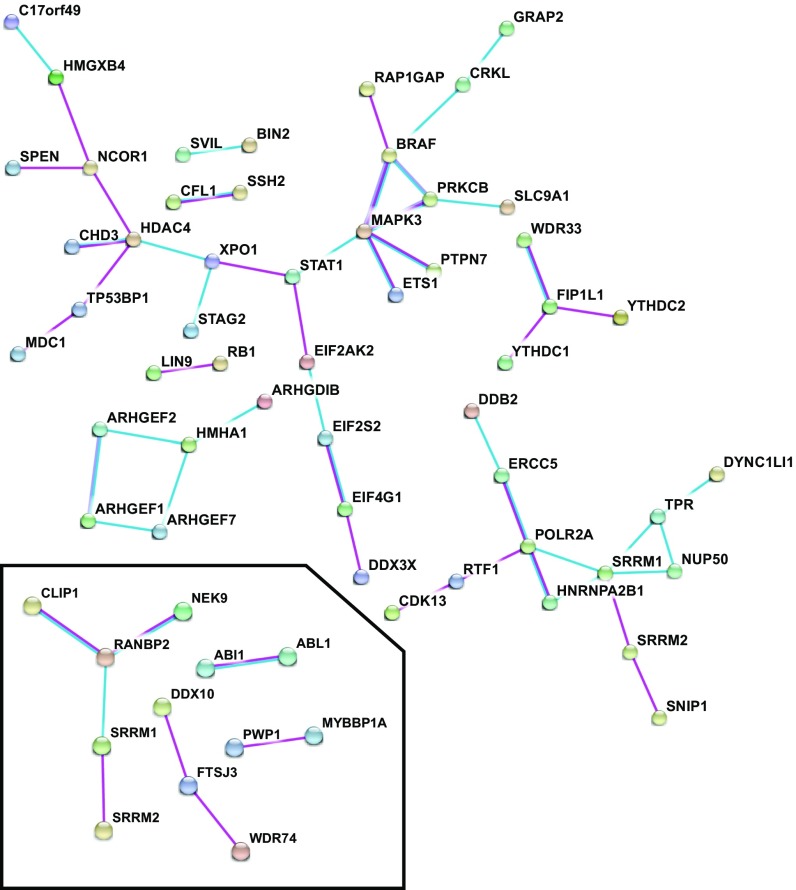
We used STRING analysis (31) as another method to suggest which biological processes or pathways might be regulated in each PDE inhibitor treatment group and to prioritize further which phosphosites warranted further investigation. STRING identifies proteins that have been empirically shown to interact (experiment option) or that are known components of an annotated common pathway (database option). This analysis showed a greater number of interacting proteins in the group treated with PDE1/7/8 inhibitors than in the group treated with PDE3 and PDE4 inhibitors (Fig. 5). As expected, the greatest numbers of interactions were observed in the group treated with 200 µM IBMX plus the PDE8 inhibitor (SI Appendix, Fig. S4). Under default network visualization settings, with the inhibition of PDEs 1, 7, and 8 there are clusters of interacting proteins around MAPK3/ERK1, HDAC4, and POL2RA (Fig. 5). The combined inhibition of PDE3 and PDE4 shows a cluster of interacting proteins around RANBP2 (Fig. 5). Last, when all PDEs were inhibited by IBMX plus the PDE8 inhibitor, additional clusters of proteins interacting with HDAC1, SIN3A, MAPRE1, ARHGEF7, ABL1, LCK, CHEK1, and PRKAB1 are visible (SI Appendix, Fig. S4). Each of these clusters is indicative of proteins associated with particular pathways or functions. The observation that cAMP modulates several members of a cluster most likely suggests that cAMP regulates multiple points of the pathway/process.
Fig. 5.
Interaction analysis of proteins with phosphosites regulated by treatment with inhibitors of PDEs 1, 7, and 8 and with inhibitors of PDE3 and PDE4 (Inset). A gene list was generated from phosphosites statistically significantly regulated by inhibition of PDEs 1, 7, and 8 and by inhibition of PDE3 and PDE4 in both the basal and PGE2-stimulated condition. The lists were submitted to the STRING analysis web portal to query the Experiments and Databases source options, with a minimum interaction score of 0.700. For visual clarity, disconnected nodes have been omitted from the interaction map.
Identification of Biological Processes Regulated by Specific Combinations of PDE Inhibitors.
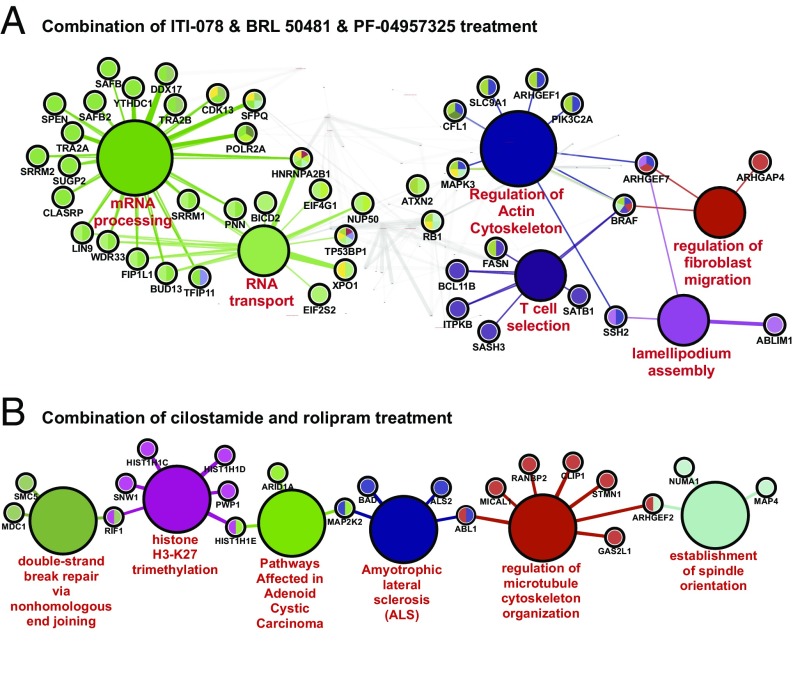
Gene Ontology (GO) analysis of the list of phosphoproteins regulated by PDE1/7/8 inhibitor treatment grouped 90 of 133 genes into 17 functional clusters (SI Appendix, Fig. S5 and Table S1). Fig. 6A shows an example of six identified functional clusters and the genes associated with their respective processes. GO analysis of the combination of PDE3 and PDE4 inhibitor treatment sites resulted in 20 of 74 genes grouped into six functional clusters (Fig. 6B and SI Appendix, Table S2). GO analysis of the 200-µM IBMX treatment data resulted in 317 of 368 genes grouped into 34 clusters. Therefore it is quite likely that these different combinations of PDE inhibitors subserve different functional pools of cAMP and that the different functional pools in turn regulate the different functions identified by the GO analysis. It should be noted that, particularly in the presence of PGE2, several common gene products are identified in both the PDE3 plus PDE4 inhibitor condition and the PDE1/7/8 inhibitor condition. In general, however, different sites are phosphorylated by different combinations of PDE inhibitors. Several possibilities for this observation are discussed in the next section.
Fig. 6.
GO analysis to identify functional processes regulated by PDE inhibitors. GO analysis was performed using the ClueGo Cytoscape plug-in (90). (A) Lists of unique proteins for each series of PDE inhibitor treatment with combined PDE1/7/8 inhibitors, a subset of functions relevant to T-cell biology. The remaining functions are shaded gray. Refer to SI Appendix, Fig. S1 for the full network. (B) Combined PDE3 and PDE4 inhibitors were generated from the statistically significantly regulated phosphosites. Each list was used to query the Kyoto Encyclopedia of Genes and Genomes, GO biological function database, and Wikipathways databases. ClueGo parameters were set as follows: GO Term Fusion selected; display only pathways with P values ≤0.05; GO tree interval, all levels; GO term minimum number of genes, 3; threshold of 4% of genes per pathway; and κ score of 0.42. GO terms are presented as nodes and are clustered together based on the similarity of genes present in each term or pathway. Major biological functions are listed in red text. Smaller circles indicate proteins associated with the adjoining biological process. Circles with multiple colors are associated with more than one process.
Discussion
In this study, we have used a nonbiased MS phosphoproteomics approach coupled with isozyme-selective PDE inhibitors to identify some of the PDE-regulated phosphoproteomes modulated by several different cAMP PDEs in a model T-cell line. According to Ponomarenko (32), there are more than 20,000 different proteins in the human proteome, not including splice variants and posttranslationally modified proteins. No single cell is expected to express all possible proteins at all times. In our datasets, we identified 3,241 phosphorylated proteins containing 13,589 phosphopeptides present in CD3/CD28-stimulated Jurkat T cells. The number of phosphopeptides we identified was comparable to the number of phosphopeptides identified in other MS studies using different MS paradigms: 10,500 by Mayya et al. (33), 11,454 Salek et al. (34), and 12,799 de Graaf et al. (22). Of these 13,589 phosphopeptides, 618 were significantly regulated by specific combinations of selective PDE antagonists compared with DMSO or PGE2 controls. Nine LC-MS/MS runs were sufficient to identify ∼88% of the enriched phosphopeptides per biological condition. We observed a very high correlation (>0.9 Pearson correlation coefficient) between analytical LC-MS/MS replicates. As expected, there was greater variation between biological replicates, which we attribute to inherent variations in culture conditions and sample processing.
We have defined PDE-regulated functional compartments as compartments containing proteins that are acutely modulated by a selective PDE inhibitor treatment. Using the subset of conditions and drugs that we studied, we observed at least three distinct functional compartments regulated by PDEs: first, those regulated by a combination of PDE1/7/8 inhibitors; second, those regulated by the combination of PDE3 and PDE4 inhibitors; and third, those regulated by the combination of IBMX and a PDE8 inhibitor that were not regulated by the other two combinations. Of note, it was necessary to inhibit both PDE3 and PDE4 to elicit any changes in the compartment regulated by the combination of PDE3 and PDE4 inhibitors; individual inhibitors of PDE3 and PDE4 had little or no effect by themselves. Time and resources did not allow testing of all combination of all PDE inhibitors, so it is likely that other distinct functional compartments exist. In these initial studies, we used the combination of PDE1/7/8 inhibitors as a comparison condition to investigate possible compartments not regulated by PDE3 and PDE4 inhibitors. Indeed, this condition potentially represents seven different functional compartments regulated by all the possible combinations of PDEs 1, 7, and 8; therefore, data from the PDE1/7/8 condition should be viewed in this context. However, because it has been previously reported that little or no PDE1 activity or protein is present in Jurkat cells, it is quite likely that the majority of effects observed in the PDE1/7/8-inhibited condition are in fact caused by PDE7 and/or PDE8 regulation (26). For a full understanding of all of the potential PDE synergies present in T cells and to understand fully the different PDE phosphoproteomes, phosphorylation profiles from each PDE inhibitor and PDE inhibitor combination will need to be determined in future studies. For example, both PDE3 and PDE4 inhibitors will need to be tested in all combinations with PDE1/7/8 inhibitors. It is known, for example, that a combination of PDE4 and PDE8 inhibitors is particularly effective in MA-10 cells (35, 36), so it seems quite likely that additional PDE-regulated functional compartments will be identified as different combinations of PDE inhibitors are investigated.
Sensitivity and Phosphopeptide Coverage.
No one yet knows how many phosphosites are regulated by PDEs in T cells. Using multiple (nine or more) LC/MS runs deepened our proteomic coverage. However, we know the complete proteome was not sampled, because key regulatory proteins such as CSK were not identified. This incomplete sampling may explain, in part, why we observed no effects with PDE3 or PDE4 inhibitors alone. Phosphorylation changes in low-abundance proteins are inherently difficult to detect by this method, especially if the phosphopeptides are not enriched by the method or if they coelute with more abundant phosphopeptides. Because the combination of IBMX plus the PDE8 inhibitor yielded many more phosphosites than those identified with the combined total of the PDE3 and PDE4 inhibitors plus the combination of PDE1/7/8 inhibitors, it seems quite likely that other combinations of PDE inhibitors will give different and expanded PDE-regulated phosphoproteome signatures.
In general, the effects of the PDE inhibitor on the phosphoproteome correlated reasonably well with the observed changes in cAMP caused by the same PDE inhibitor treatments, suggesting that the concentrations of cilostamide (5 µM) and rolipram (10 µM) used, even though predicted to elicit ∼90% inhibition of PDE3 and PDE4, respectively, were still quite selective, because neither treatment alone could regulate either cAMP or the phosphoproteomics data. In theory, the nonselective inhibitor IBMX (200 µM) plus PF-04957325 (200 nM) should inhibit all cAMP-PDEs. This combination caused the greatest number of changes in regulated sites, both in the basal and PGE2-stimulated states. However, as mentioned above, this effect also suggests that other functional compartments regulated by different combinations of PDEs remain to be identified and annotated. Both the phosphoproteomic results and the changes in cAMP seen in response to the PDE inhibitor treatments also suggested that major functional compartments in these cells are most likely not regulated by single PDEs. In addition, it is, of course, possible that some functional cAMP compartments may be subserved by three or more PDEs. This possibility was not tested. Dong et al. (26) reported that inhibiting PDE3, PDE4, and PDE7 together maximally potentiated glucocorticoid-mediated apoptosis. Unfortunately, time and resources did not allow phosphoproteomic studies under all the possible combinations of PDEs expressed in the Jurkat cell line. Eventually these studies will need to be done to understand the full PDE-regulated phosphoproteomes.
It is perhaps worth repeating that the largest changes both in the phosphoproteome and cAMP data were seen in the PGE2-stimulated conditions rather than in the basal condition. Thus it appears that these functional compartments are dynamic and can be influenced by the source of cAMP. Bloom et al. (37) reported that pools of cAMP in distinct subcellular locales could be detected in T cells stimulated with different agonists. Therefore, it is plausible, for example, that the functional compartment(s) defined by the inhibition of PDEs 1, 7, and 8 or by the inhibition of PDE3 plus PDE4 could be different in adenosine-stimulated cells versus PGE2-stimulated cells. Additionally, the time-course phosphoproteomic studies by Giansanti et al. (17) and Golkowski et al. (35) suggest that these functional compartments change in time as well, indicating an additional parameter that will need to be explored to understand fully the scope of the functional PDE-regulated phosphoproteomes. Clearly a more comprehensive study of cAMP/PDE functional compartments is needed to investigate all the possible combinations of PDE inhibitors under multiple agonist stimulation paradigms.
Direct Phosphorylation by PKA.
Of the 618 significantly regulated phosphosites, 55 are annotated in PhosphositePlus, indicating that, when phosphorylated, these sites have been empirically determined to regulate protein function directly or indirectly. Of these 55 phosphosites 13 were at canonical PKA consensus sequences and therefore are likely to be directly phosphorylated by PKA. These include ARFIP1 S132 (38, 39), ARHGEF2 S886 (40), BAD S152 (41, 42), BRAF S446 (43), CAD S1406 (44), CAMKK1 S458 (45), KIF3A S687 (46, 47), LASP1 S146(48–50), PTPN7 S44 (51, 52), RCC1 S11(53–55), RPS6 S235 (56), STMN1 S63 (57–59), and TAL1 172 (60, 61). In fact, eight of these sites—in ARHGEF2, BAD, CAD, CAMKK1, LASP1, PTPN7, STMN1, and TAL1—have been reported to be substrates of PKA in other cell types. These results strongly corroborate the combined inhibitor-phosphoproteomic approach outlined in this article. Moreover, these proteins are associated with a wide array of biological functions and thus suggest that cAMP coordinates not only a few rate-limiting steps but rather a large number of sites that in turn regulate multiple processes in the T cells. In fact, this observation may call into question the generally taught concept that only a single rate-limiting step is likely to be the major regulatory site for cAMP/PKA in many pathways.
As indicated above, these regulatory sites should not be interpreted as exclusively PKA substrates. For example, STMN1 S63, RCC1 S11, and PTPN7 S44 also can be phosphorylated by CaMKIV (58), CDK1 (55), and PKCƟ (51), respectively. Therefore it is plausible, and perhaps even likely, that the sequences of these sites have evolved so that different kinases can regulate the same site under specific but differing cellular contexts. Nonetheless, the identification of these phosphosites also tends to validate the approach of using shotgun MS to identify biologically relevant phosphosites in response to treatment with combinations of PDE inhibitors. Therefore, we were encouraged to expand our analysis beyond sites with canonical PKA consensus sequences to identify other PDE-dependent phosphosites with probable regulatory actions, because they could lead to the identification of novel molecular mechanisms by which cAMP and PDEs coordinate cellular responses.
PDE Synergies: Inhibition of Multiple PDEs Is Needed to Regulate Individual Sites and Processes.
The majority of phosphosites were not regulated until more than one PDE was inhibited. This finding was corroborated by Western blots of STMN1 S63 and ARHGEF2 S858 (Fig. 2 B and C). Of note, probing by Western blot revealed increased phosphorylation of ARHGEF2 S858 by combined treatment with PDE3 and PDE4 inhibitors which was not noted in the initial analysis of the phosphoproteomics data because a single statistical outlier confounded the analysis (SI Appendix, Fig. S6). Giembycz et al. (62) have reported the proliferation of primary T cells decreased only when both PDE3 and PDE4 were inhibited. The current data may, in part, provide a molecular explanation for their observations. That is, molecular effectors controlling proliferation are well regulated by PDE inhibition only when more than one PDE is inhibited.
The many examples of PDE synergy in the current datasets at both the cAMP and PDE-regulated phosphoproteome level also likely have substantial implications for drug design. Data from a clinical study by Franciosi et al. (63) showed that RPL554, a dual PDE3 and PDE4 inhibitor, was an effective bronchodilator and reduced inflammation in patients with chronic obstructive pulmonary disease. Although perhaps not yet fully embraced by the pharmaceutical industry, an increasing number of such functional examples are being elucidated in which multiple PDEs need to be inhibited to elicit a pharmacological response. To our knowledge very few such screening studies have been carried out previously. The current data demonstrate this same principle of PDE synergy at a molecular level and suggest that a phosphoproteomic approach could be used as a part of an initial preclinical screen to determine which PDEs need to be inhibited to maximize a therapeutic effect or to minimize an unwanted side effect.
PDE1/7/8-Regulated Functional Compartment(s).
Interestingly, the characteristics of the functional compartment(s) regulated by combined inhibition of PDEs 1, 7, and 8 differ from the characteristics of the compartment(s) regulated by PDE 3 and PDE4. The stark differences between these two functional compartments were not fully expected. We found, for example, that the majority of sites regulated by PDE3 and PDE4 have a PKA consensus sequence, and NetPhorest analysis predicted that the majority of these sites would be phosphorylated by PKA. This finding strongly suggested that they are direct substrates of PKA and/or perhaps also other AGC-type kinases. In contrast, a much greater diversity of kinases was predicted to regulate the phosphosites in the PDE1/7/8 compartment(s), and the majority of sites regulated in these compartments do not have a PKA consensus site. In both the basal and PGE2-stimulated state, CK2 is the predominately predicted kinase for this functional compartment. One possible mechanism that might be operative here is that cAMP might activate PKA or EPAC upstream of CK2 so that the effects of cAMP, although real, are indirect. If so, this activity represents an unexplored mechanism of cAMP action. PDE1/7/8 inhibitors also may have effects on phosphatase activity.
Biological Processes Regulated by Different PDE-Regulated Phosphoproteomes.
GO analysis was performed to provide insight into which biological processes might be regulated by each series of PDE inhibitor combinations. We observed that the biological processes regulated by inhibition of PDEs 1, 7, and 8 were largely distinct from those regulated by inhibition of PDE 3 and PDE4, as might be expected because different proteins are phosphorylated. It is worth noting that, although functional clusters such as spindle organization, regulation of cytoskeleton, and repair of double-stranded breaks are common to both treatment groups, the genes present in each node are different among treatment groups, suggesting that inhibiting different combinations of PDEs can affect the same biological process but likely does so via different mechanisms. A number of the biological processes identified by GO analysis have been previously reported to be regulated by cAMP. These include mRNA splicing (64–66), spindle organization (67, 68), fibroblast migration (69, 70), lamellipodium assembly (71, 72), apoptosis (73–76), ATM signaling (77, 78), gene silencing via microRNA (79–82), T-cell selection (83), and cytoskeletal reorganization (84). This study suggests which molecular substrates might participate in the regulation of these processes. To date the roles of cAMP/PKA signaling are even less well defined for most of the other biological processes implicated, such as chromatin remodeling (85–87) and chromosomal segregation. Again, the phosphopeptides that map to these processes should be a good place to start mechanistic studies of cAMP/PDE effects on these processes.
The combined approach of using selective inhibitors with phosphoproteomic analysis builds on previous classical and phosphoproteomic studies (17) in several key aspects. First, the majority of previous phosphoproteomic studies used high concentrations of agents such as cAMP analogs or receptor agonists to increase cAMP globally. In other studies, relatively nonselective phosphatase inhibitors were used. In general, the investigators in these studies were most interested in defining a maximal cAMP-regulated or phosphatase-regulated phosphoproteome. Our data suggest that a much more nuanced understanding, particularly regarding the physiological roles of different subsets of PDEs, can be achieved by using selective PDE inhibitors at their selective concentrations to interrogate the phosphoproteome of a cell.
Second, using this combination approach, we have identified a number of proteins known to be key regulators of important pathways/processes that have been largely understudied in the context of cAMP regulation. For example, as also seen in our recent description of a PDE-regulated phosphoproteome of MA-10 cells (35), a number of small G protein-regulated pathways were identified. As in the MA-10 system, the regulation of these pathways is likely to be at the level of the guanine nucleotide exchange factors and GTPase-activating proteins that modulate the small Rho-type GTPases rather than a direct phosphorylation of the GTPase itself. In both studies, inhibition of a combination of PDEs increased ARHGEF2 phosphorylation on S886. This site has been previously reported to regulate ARHGEF2 activity (40). Moreover, ARHGEF2-dependent RhoA activity also has been shown to regulate the uropod of migrating T cells (71), in accordance with the report by Vang et al. (88) that PDE8 inhibition caused a decrease in T-cell motility. In the current study, the combination of PDEs 1, 7, and 8 showed the largest increase in ARHGEF2 phosphorylation. In the context of T-cell biology, cytoskeletal reorganization has been intimately linked with T-cell receptor signaling. The increased phosphorylation of ABLIM1, ARHGEF1, ARHGEF7, ARHGAP4, BRAF, CFL1, MAPK3, PIK3C2A, SLC9A1, and SSH2 (all proteins associated with cytoskeletal reorganization), suggested that PDE8, possibly in combination with another PDE (likely PDE7), may regulate T-cell migration at multiple points (see SI Appendix, Supplemental Methods and Materials for further discussion of other PDE-modulated functional compartments). However, further studies will be necessary to prove this hypothesis.
Finally, we have identified a number of PDE-modulated phosphosites on proteins not previously known to be regulated by cAMP/PDEs. For example, RANBP2 has been identified as an EPAC1 interactor. Gloerich et al. (89) showed that EPAC1 bound to RAN-GTP, which in turn bound to the cluster of zinc finger domains of RANBP2. They also showed that phosphatase inhibitor treatment increased the phosphorylation of RANBP2 zinc finger domains and prevented EPAC binding. However, the exact site/zinc finger domain was not identified. Inhibition of PDE3 and PDE4 caused a significant increase in RANBP2-S1509 phosphorylation (Tables 2 and 4) under both basal and PGE2 conditions. S1509 is immediately C terminal of the third zinc finger domain and is within a canonical PKA consensus sequence. It therefore is likely that PKA phosphorylates RANBP2 at S1509 and disrupts RAN-GTP/EPAC binding to the third zinc finger domain. Again, further studies will be necessary to prove this notion.
In summary, we have used MS phosphoproteomic analysis on Jurkat cultures treated with various selective concentrations of inhibitors of PDEs 1, 3, 4, 7, and 8 to begin to characterize the phosphoproteome of the functional pools of cAMP subserved by these PDEs. Predictive algorithms were used to identify upstream regulatory kinases and to prioritize potential regulatory phosphosites for future investigation. The big advantage of this approach lies in the unbiased identification of regulated phosphosites and the sheer number of sites identified. This study compares the phosphoproteomes of two functional compartments subserved by any individual PDE isozyme or by any combinations of PDEs in a T-cell model, not only in the basal state but in an adenylyl cyclase-stimulated state as well. We observed at least two functional pools of cAMP in Jurkat cells that are distinct from one another. The data underscore the need to understand the exact pharmacological response to individual and combinations of PDE inhibitors. Currently we do not know how many functional PDE-regulated cAMP compartments exist in Jurkat cells, nor do we understand if similar functional PDE-regulated cGMP pools exist in these cells. We also need to know if these cyclic nucleotide pools regulate one another and which biological processes each of these functional pools controls. The answers to these questions will undoubtedly be complex and assuredly will be cell-context and time dependent. However, addressing these questions using the combined selective PDE inhibitor approach coupled with phosphoproteomic analyses is well within our current abilities. The volume of data to be gleaned is staggering, but with enough experimental iterations it should be possible, using phosphosites as molecular signatures, to construct a functional atlas of PDE-regulated cyclic nucleotide signaling not only in Jurkat cells but also in any other model cell culture system. Using other agonists and antagonists, the same approach should be possible for the cyclic nucleotide synthetic pathways.
Materials and Methods
The methods and sources of materials can be found in SI Appendix, Supplemental Methods and Materials.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
Data deposition: Phosphoproteomic raw data have been deposited with the Mass Spectrometry Interactive Virtual Environment (MassIVE) maintained by the Center for Computational Mass Spectrometry (CCMS) in the Computer Science and Engineering Department of the University of California, San Diego (accession no. MSV000081115).
See Commentary on page 7741.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703939114/-/DCSupplemental.
References
- 1.Sutherland EW, Rall TW. Fractionation and characterization of a cyclic adenine ribonucleotide formed by tissue particles. J Biol Chem. 1958;232:1077–1091. [PubMed] [Google Scholar]
- 2.Rall TW, Sutherland EW. Formation of a cyclic adenine ribonucleotide by tissue particles. J Biol Chem. 1958;232:1065–1076. [PubMed] [Google Scholar]
- 3.Robison GA, Butcher RW, Sutherland EW. Cyclic AMP. Annu Rev Biochem. 1968;37:149–174. doi: 10.1146/annurev.bi.37.070168.001053. [DOI] [PubMed] [Google Scholar]
- 4.Walsh DA, Perkins JP, Krebs EG. An adenosine 3′,5′-monophosphate-dependant protein kinase from rabbit skeletal muscle. J Biol Chem. 1968;243:3763–3765. [PubMed] [Google Scholar]
- 5.de Rooij J, et al. Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature. 1998;396:474–477. doi: 10.1038/24884. [DOI] [PubMed] [Google Scholar]
- 6.DiFrancesco D, Tortora P. Direct activation of cardiac pacemaker channels by intracellular cyclic AMP. Nature. 1991;351:145–147. doi: 10.1038/351145a0. [DOI] [PubMed] [Google Scholar]
- 7.Chini CC, Grande JP, Chini EN, Dousa TP. Compartmentalization of cAMP signaling in mesangial cells by phosphodiesterase isozymes PDE3 and PDE4. Regulation of superoxidation and mitogenesis. J Biol Chem. 1997;272:9854–9859. doi: 10.1074/jbc.272.15.9854. [DOI] [PubMed] [Google Scholar]
- 8.Cooper DM. Compartmentalization of adenylate cyclase and cAMP signalling. Biochem Soc Trans. 2005;33:1319–1322. doi: 10.1042/BST0331319. [DOI] [PubMed] [Google Scholar]
- 9.Hasler P, Moore JJ, Kammer GM. Human T lymphocyte cAMP-dependent protein kinase: Subcellular distributions and activity ranges of type I and type II isozymes. FASEB J. 1992;6:2735–2741. doi: 10.1096/fasebj.6.9.1319361. [DOI] [PubMed] [Google Scholar]
- 10.Corbin JD, Sugden PH, Lincoln TM, Keely SL. Compartmentalization of adenosine 3′:5′-monophosphate and adenosine 3′:5′-monophosphate-dependent protein kinase in heart tissue. J Biol Chem. 1977;252:3854–3861. [PubMed] [Google Scholar]
- 11.Liu H, Maurice DH. Expression of cyclic GMP-inhibited phosphodiesterases 3A and 3B (PDE3A and PDE3B) in rat tissues: Differential subcellular localization and regulated expression by cyclic AMP. Br J Pharmacol. 1998;125:1501–1510. doi: 10.1038/sj.bjp.0702227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abrahamsen H, et al. TCR- and CD28-mediated recruitment of phosphodiesterase 4 to lipid rafts potentiates TCR signaling. J Immunol. 2004;173:4847–4858. doi: 10.4049/jimmunol.173.8.4847. [DOI] [PubMed] [Google Scholar]
- 13.Boniface K, et al. Prostaglandin E2 regulates Th17 cell differentiation and function through cyclic AMP and EP2/EP4 receptor signaling. J Exp Med. 2009;206:535–548. doi: 10.1084/jem.20082293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao C, et al. Prostaglandin E2-EP4 signaling promotes immune inflammation through Th1 cell differentiation and Th17 cell expansion. Nat Med. 2009;15:633–640. doi: 10.1038/nm.1968. [DOI] [PubMed] [Google Scholar]
- 15.Conche C, Boulla G, Trautmann A, Randriamampita C. T cell adhesion primes antigen receptor-induced calcium responses through a transient rise in adenosine 3′,5′-cyclic monophosphate. Immunity. 2009;30:33–43. doi: 10.1016/j.immuni.2008.10.020. [DOI] [PubMed] [Google Scholar]
- 16.Nguyen TD, et al. The phosphoproteome of human Jurkat T cell clones upon costimulation with anti-CD3/anti-CD28 antibodies. J Proteomics. 2016;131:190–198. doi: 10.1016/j.jprot.2015.10.029. [DOI] [PubMed] [Google Scholar]
- 17.Giansanti P, Stokes MP, Silva JC, Scholten A, Heck AJ. Interrogating cAMP-dependent kinase signaling in Jurkat T cells via a protein kinase A targeted immune-precipitation phosphoproteomics approach. Mol Cell Proteomics. 2013;12:3350–3359. doi: 10.1074/mcp.O113.028456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghesquiere B, et al. Redox proteomics of protein-bound methionine oxidation. Mol Cell Proteomics. 2011;10:M110 006866. doi: 10.1074/mcp.M110.006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan S, Yan C. PDE1 isozymes, key regulators of pathological vascular remodeling. Curr Opin Pharmacol. 2011;11:720–724. doi: 10.1016/j.coph.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nthenge-Ngumbau DN, Mohanakumar KP. Can cyclic nucleotide phosphodiesterase inhibitors be drugs for Parkinson’s disease? Mol Neurobiol. January 6, 2017 doi: 10.1007/s12035-016-0355-8. [DOI] [PubMed] [Google Scholar]
- 21.Bobin P, et al. Cyclic nucleotide phosphodiesterases in heart and vessels: A therapeutic perspective. Arch Cardiovasc Dis. 2016;109:431–443. doi: 10.1016/j.acvd.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 22.de Graaf EL, Giansanti P, Altelaar AF, Heck AJ. Single-step enrichment by Ti4+-IMAC and label-free quantitation enables in-depth monitoring of phosphorylation dynamics with high reproducibility and temporal resolution. Mol Cell Proteomics. 2014;13:2426–2434. doi: 10.1074/mcp.O113.036608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soderblom EJ, Philipp M, Thompson JW, Caron MG, Moseley MA. Quantitative label-free phosphoproteomics strategy for multifaceted experimental designs. Anal Chem. 2011;83:3758–3764. doi: 10.1021/ac200213b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jersie-Christensen RR, Sultan A, Olsen JV. Simple and reproducible sample preparation for single-shot phosphoproteomics with high sensitivity. Methods Mol Biol. 2016;1355:251–260. doi: 10.1007/978-1-4939-3049-4_17. [DOI] [PubMed] [Google Scholar]
- 25.Erdogan S, Houslay MD. Challenge of human Jurkat T-cells with the adenylate cyclase activator forskolin elicits major changes in cAMP phosphodiesterase (PDE) expression by up-regulating PDE3 and inducing PDE4D1 and PDE4D2 splice variants as well as down-regulating a novel PDE4A splice variant. Biochem J. 1997;321:165–175. doi: 10.1042/bj3210165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong H, Zitt C, Auriga C, Hatzelmann A, Epstein PM. Inhibition of PDE3, PDE4 and PDE7 potentiates glucocorticoid-induced apoptosis and overcomes glucocorticoid resistance in CEM T leukemic cells. Biochem Pharmacol. 2010;79:321–329. doi: 10.1016/j.bcp.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 27.Bloom TJ, Beavo JA. Identification and tissue-specific expression of PDE7 phosphodiesterase splice variants. Proc Natl Acad Sci USA. 1996;93:14188–14192. doi: 10.1073/pnas.93.24.14188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bozza PT, Payne JL, Goulet JL, Weller PF. Mechanisms of platelet-activating factor-induced lipid body formation: Requisite roles for 5-lipoxygenase and de novo protein synthesis in the compartmentalization of neutrophil lipids. J Exp Med. 1996;183:1515–1525. doi: 10.1084/jem.183.4.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao Q, Miao B, Bi J, Wang Z, Li Y. Prioritizing functional phosphorylation sites based on multiple feature integration. Sci Rep. 2016;6:24735. doi: 10.1038/srep24735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hornbeck PV, et al. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szklarczyk D, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ponomarenko EA, et al. The size of the human proteome: The width and depth. Int J Anal Chem. 2016;2016:7436849. doi: 10.1155/2016/7436849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayya V, et al. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci Signal. 2009;2:ra46. doi: 10.1126/scisignal.2000007. [DOI] [PubMed] [Google Scholar]
- 34.Salek M, et al. Quantitative phosphoproteome analysis unveils LAT as a modulator of CD3ζ and ZAP-70 tyrosine phosphorylation. PLoS One. 2013;8:e77423. doi: 10.1371/journal.pone.0077423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Golkowski M, Shimizu-Albergine M, Suh HW, Beavo JA, Ong SE. Studying mechanisms of cAMP and cyclic nucleotide phosphodiesterase signaling in Leydig cell function with phosphoproteomics. Cell Signal. 2016;28:764–778. doi: 10.1016/j.cellsig.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimizu-Albergine M, Tsai LC, Patrucco E, Beavo JA. cAMP-specific phosphodiesterases 8A and 8B, essential regulators of Leydig cell steroidogenesis. Mol Pharmacol. 2012;81:556–566. doi: 10.1124/mol.111.076125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bloom FE, Wedner HJ, Parker CW. The use of antibodies to study cell structure and metabolism. Pharmacol Rev. 1973;25:343–358. [PubMed] [Google Scholar]
- 38.Gehart H, et al. The BAR domain protein Arfaptin-1 controls secretory granule biogenesis at the trans-Golgi network. Dev Cell. 2012;23:756–768. doi: 10.1016/j.devcel.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 39.Cruz-Garcia D, et al. Recruitment of arfaptins to the trans-Golgi network by PI(4)P and their involvement in cargo export. EMBO J. 2013;32:1717–1729. doi: 10.1038/emboj.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meiri D, et al. Modulation of Rho guanine exchange factor Lfc activity by protein kinase A-mediated phosphorylation. Mol Cell Biol. 2009;29:5963–5973. doi: 10.1128/MCB.01268-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou XM, Liu Y, Payne G, Lutz RJ, Chittenden T. Growth factors inactivate the cell death promoter BAD by phosphorylation of its BH3 domain on Ser155. J Biol Chem. 2000;275:25046–25051. doi: 10.1074/jbc.M002526200. [DOI] [PubMed] [Google Scholar]
- 42.Lizcano JM, Morrice N, Cohen P. Regulation of BAD by cAMP-dependent protein kinase is mediated via phosphorylation of a novel site, Ser155. Biochem J. 2000;349:547–557. doi: 10.1042/0264-6021:3490547. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Tran NH, Wu X, Frost JA. B-Raf and Raf-1 are regulated by distinct autoregulatory mechanisms. J Biol Chem. 2005;280:16244–16253. doi: 10.1074/jbc.M501185200. [DOI] [PubMed] [Google Scholar]
- 44.Kotsis DH, et al. Protein kinase A phosphorylation of the multifunctional protein CAD antagonizes activation by the MAP kinase cascade. Mol Cell Biochem. 2007;301:69–81. doi: 10.1007/s11010-006-9398-x. [DOI] [PubMed] [Google Scholar]
- 45.Wayman GA, Tokumitsu H, Soderling TR. Inhibitory cross-talk by cAMP kinase on the calmodulin-dependent protein kinase cascade. J Biol Chem. 1997;272:16073–16076. doi: 10.1074/jbc.272.26.16073. [DOI] [PubMed] [Google Scholar]
- 46.Phang HQ, et al. POPX2 phosphatase regulates the KIF3 kinesin motor complex. J Cell Sci. 2014;127:727–739. doi: 10.1242/jcs.126482. [DOI] [PubMed] [Google Scholar]
- 47.Xiong R, et al. Herpes simplex virus 1 US3 phosphorylates cellular KIF3A to downregulate CD1d expression. J Virol. 2015;89:6646–6655. doi: 10.1128/JVI.00214-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Butt E, et al. Actin binding of human LIM and SH3 protein is regulated by cGMP- and cAMP-dependent protein kinase phosphorylation on serine 146. J Biol Chem. 2003;278:15601–15607. doi: 10.1074/jbc.M209009200. [DOI] [PubMed] [Google Scholar]
- 49.Mihlan S, et al. Nuclear import of LASP-1 is regulated by phosphorylation and dynamic protein-protein interactions. Oncogene. 2013;32:2107–2113. doi: 10.1038/onc.2012.216. [DOI] [PubMed] [Google Scholar]
- 50.Vaman V S A, et al. LASP1, a newly identified melanocytic protein with a possible role in melanin release, but not in melanoma progression. PLoS One. 2015;10:e0129219. doi: 10.1371/journal.pone.0129219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saxena M, Williams S, Taskén K, Mustelin T. Crosstalk between cAMP-dependent kinase and MAP kinase through a protein tyrosine phosphatase. Nat Cell Biol. 1999;1:305–311. doi: 10.1038/13024. [DOI] [PubMed] [Google Scholar]
- 52.Nika K, et al. Lipid raft targeting of hematopoietic protein tyrosine phosphatase by protein kinase C theta-mediated phosphorylation. Mol Cell Biol. 2006;26:1806–1816. doi: 10.1128/MCB.26.5.1806-1816.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hood FE, Clarke PR. RCC1 isoforms differ in their affinity for chromatin, molecular interactions and regulation by phosphorylation. J Cell Sci. 2007;120:3436–3445. doi: 10.1242/jcs.009092. [DOI] [PubMed] [Google Scholar]
- 54.Horiike Y, Kobayashi H, Sekiguchi T. Ran GTPase guanine nucleotide exchange factor RCC1 is phosphorylated on serine 11 by cdc2 kinase in vitro. Mol Biol Rep. 2009;36:717–723. doi: 10.1007/s11033-008-9234-3. [DOI] [PubMed] [Google Scholar]
- 55.Hutchins JR, et al. Phosphorylation regulates the dynamic interaction of RCC1 with chromosomes during mitosis. Curr Biol. 2004;14:1099–1104. doi: 10.1016/j.cub.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 56.Wettenhall RE, Morgan FJ. Phosphorylation of hepatic ribosomal protein S6 on 80 and 40 S ribosomes. Primary structure of S6 in the region of the major phosphorylation sites for cAMP-dependent protein kinases. J Biol Chem. 1984;259:2084–2091. [PubMed] [Google Scholar]
- 57.Yip YY, Yeap YY, Bogoyevitch MA, Ng DC. cAMP-dependent protein kinase and c-Jun N-terminal kinase mediate stathmin phosphorylation for the maintenance of interphase microtubules during osmotic stress. J Biol Chem. 2014;289:2157–2169. doi: 10.1074/jbc.M113.470682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marklund U, et al. Serine 16 of oncoprotein 18 is a major cytosolic target for the Ca2+/calmodulin-dependent kinase-Gr. Eur J Biochem. 1994;225:53–60. doi: 10.1111/j.1432-1033.1994.00053.x. [DOI] [PubMed] [Google Scholar]
- 59.Beretta L, Dobránsky T, Sobel A. Multiple phosphorylation of stathmin. Identification of four sites phosphorylated in intact cells and in vitro by cyclic AMP-dependent protein kinase and p34cdc2. J Biol Chem. 1993;268:20076–20084. [PubMed] [Google Scholar]
- 60.Li Y, et al. Dynamic interaction between TAL1 oncoprotein and LSD1 regulates TAL1 function in hematopoiesis and leukemogenesis. Oncogene. 2012;31:5007–5018. doi: 10.1038/onc.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prasad KS, Brandt SJ. Target-dependent effect of phosphorylation on the DNA binding activity of the TAL1/SCL oncoprotein. J Biol Chem. 1997;272:11457–11462. doi: 10.1074/jbc.272.17.11457. [DOI] [PubMed] [Google Scholar]
- 62.Giembycz MA, Corrigan CJ, Seybold J, Newton R, Barnes PJ. Identification of cyclic AMP phosphodiesterases 3, 4 and 7 in human CD4+ and CD8+ T-lymphocytes: Role in regulating proliferation and the biosynthesis of interleukin-2. Br J Pharmacol. 1996;118:1945–1958. doi: 10.1111/j.1476-5381.1996.tb15629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Franciosi LG, et al. Efficacy and safety of RPL554, a dual PDE3 and PDE4 inhibitor, in healthy volunteers and in patients with asthma or chronic obstructive pulmonary disease: Findings from four clinical trials. Lancet Respir Med. 2013;1:714–727. doi: 10.1016/S2213-2600(13)70187-5. [DOI] [PubMed] [Google Scholar]
- 64.Li H, et al. In vivo selection of kinase-responsive RNA elements controlling alternative splicing. J Biol Chem. 2009;284:16191–16201. doi: 10.1074/jbc.M900393200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jarnaess E, et al. Splicing factor arginine/serine-rich 17A (SFRS17A) is an A-kinase anchoring protein that targets protein kinase A to splicing factor compartments. J Biol Chem. 2009;284:35154–35164. doi: 10.1074/jbc.M109.056465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aksaas AK, Eikvar S, Akusjärvi G, Skålhegg BS, Kvissel AK. Protein kinase a-dependent phosphorylation of serine 119 in the proto-oncogenic serine/arginine-rich splicing factor 1 modulates its activity as a splicing enhancer protein. Genes Cancer. 2011;2:841–851. doi: 10.1177/1947601911430226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vandame P, et al. The spatio-temporal dynamics of PKA activity profile during mitosis and its correlation to chromosome segregation. Cell Cycle. 2014;13:3232–3240. doi: 10.4161/15384101.2014.950907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gharibi Sh, et al. Effect of phosphodiesterase type 3 inhibitor on nuclear maturation and in vitro development of ovine oocytes. Theriogenology. 2013;80:302–312. doi: 10.1016/j.theriogenology.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 69.Sinha C, et al. PKA and actin play critical roles as downstream effectors in MRP4-mediated regulation of fibroblast migration. Cell Signal. 2015;27:1345–1355. doi: 10.1016/j.cellsig.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Olmedo I, et al. EPAC expression and function in cardiac fibroblasts and myofibroblasts. Toxicol Appl Pharmacol. 2013;272:414–422. doi: 10.1016/j.taap.2013.06.022. [DOI] [PubMed] [Google Scholar]
- 71.Heasman SJ, Ridley AJ. Multiple roles for RhoA during T cell transendothelial migration. Small GTPases. 2010;1:174–179. doi: 10.4161/sgtp.1.3.14724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen L, Zhang JJ, Huang XY. cAMP inhibits cell migration by interfering with Rac-induced lamellipodium formation. J Biol Chem. 2008;283:13799–13805. doi: 10.1074/jbc.M800555200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang L, et al. Gene expression signatures of cAMP/protein kinase A (PKA)-promoted, mitochondrial-dependent apoptosis. Comparative analysis of wild-type and cAMP-deathless S49 lymphoma cells. J Biol Chem. 2008;283:4304–4313. doi: 10.1074/jbc.M708673200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lalli E, Sassone-Corsi P, Ceredig R. Block of T lymphocyte differentiation by activation of the cAMP-dependent signal transduction pathway. EMBO J. 1996;15:528–537. [PMC free article] [PubMed] [Google Scholar]
- 75.Ivanov VN, Lee RK, Podack ER, Malek TR. Regulation of Fas-dependent activation-induced T cell apoptosis by cAMP signaling: A potential role for transcription factor NF-kappa B. Oncogene. 1997;14:2455–2464. doi: 10.1038/sj.onc.1201088. [DOI] [PubMed] [Google Scholar]
- 76.Gupta M, et al. Presence of pentoxifylline during T cell priming increases clonal frequencies in secondary proliferative responses and inhibits apoptosis. J Immunol. 1999;162:689–695. [PubMed] [Google Scholar]
- 77.Eliezer Y, Argaman L, Kornowski M, Roniger M, Goldberg M. Interplay between the DNA damage proteins MDC1 and ATM in the regulation of the spindle assembly checkpoint. J Biol Chem. 2014;289:8182–8193. doi: 10.1074/jbc.M113.532739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cho EA, Kim EJ, Kwak SJ, Juhnn YS. cAMP signaling inhibits radiation-induced ATM phosphorylation leading to the augmentation of apoptosis in human lung cancer cells. Mol Cancer. 2014;13:36. doi: 10.1186/1476-4598-13-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vo N, et al. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci USA. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li P, Xue WJ, Feng Y, Mao QS. MicroRNA-205 functions as a tumor suppressor in colorectal cancer by targeting cAMP responsive element binding protein 1 (CREB1) Am J Transl Res. 2015;7:2053–2059. [PMC free article] [PubMed] [Google Scholar]
- 81.Huang B, et al. miR-142-3p restricts cAMP production in CD4+CD25- T cells and CD4+CD25+ TREG cells by targeting AC9 mRNA. EMBO Rep. 2009;10:180–185. doi: 10.1038/embor.2008.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Feng G, Yan Z, Li C, Hou Y. microRNA-208a in an early stage myocardial infarction rat model and the effect on cAMP-PKA signaling pathway. Mol Med Rep. 2016;14:1631–1635. doi: 10.3892/mmr.2016.5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vig M, et al. Commitment of activated T cells to secondary responsiveness is enhanced by signals mediated by cAMP-dependent protein kinase A-I. Mol Pharmacol. 2002;62:1471–1481. doi: 10.1124/mol.62.6.1471. [DOI] [PubMed] [Google Scholar]
- 84.Howe AK. Regulation of actin-based cell migration by cAMP/PKA. Biochim Biophys Acta. 2004;1692:159–174. doi: 10.1016/j.bbamcr.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 85.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 86.Lai AY, Wade PA. Cancer biology and NuRD: A multifaceted chromatin remodelling complex. Nat Rev Cancer. 2011;11:588–596. doi: 10.1038/nrc3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hedrich CM, et al. cAMP responsive element modulator (CREM) α mediates chromatin remodeling of CD8 during the generation of CD3+ CD4- CD8- T cells. J Biol Chem. 2014;289:2361–2370. doi: 10.1074/jbc.M113.523605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vang AG, et al. PDE8 regulates rapid Teff cell adhesion and proliferation independent of ICER. PLoS One. 2010;5:e12011. doi: 10.1371/journal.pone.0012011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gloerich M, et al. The nucleoporin RanBP2 tethers the cAMP effector Epac1 and inhibits its catalytic activity. J Cell Biol. 2011;193:1009–1020. doi: 10.1083/jcb.201011126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bindea G, et al. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.