Significance
Bacterial dynamins were discovered ∼10 y ago and the explosion in genome sequencing has shown that they radiate throughout the bacteria, being present in >1,000 species. In eukaryotes, dynamins play critical roles in the detachment of endocytic vesicles from the plasma membrane, the division of chloroplasts and peroxisomes, and both the fusion and fission of mitochondria. However, in evolutionary terms, dynamins are of bacterial origin, and yet the biological functions of bacterial dynamins remain poorly understood. Here we demonstrate a critical role for dynamins in bacterial cytokinesis, reminiscent of the essential role of eukaryotic dynamins in the division of chloroplasts and mitochondria.
Keywords: cell division, FtsZ, bacterial dynamins, sporulation, Streptomyces
Abstract
During sporulation, the filamentous bacteria Streptomyces undergo a massive cell division event in which the synthesis of ladders of sporulation septa convert multigenomic hyphae into chains of unigenomic spores. This process requires cytokinetic Z-rings formed by the bacterial tubulin homolog FtsZ, and the stabilization of the newly formed Z-rings is crucial for completion of septum synthesis. Here we show that two dynamin-like proteins, DynA and DynB, play critical roles in this process. Dynamins are a family of large, multidomain GTPases involved in key cellular processes in eukaryotes, including vesicle trafficking and organelle division. Many bacterial genomes encode dynamin-like proteins, but the biological function of these proteins has remained largely enigmatic. Using a cell biological approach, we show that the two Streptomyces dynamins specifically localize to sporulation septa in an FtsZ-dependent manner. Moreover, dynamin mutants have a cell division defect due to the decreased stability of sporulation-specific Z-rings, as demonstrated by kymographs derived from time-lapse images of FtsZ ladder formation. This defect causes the premature disassembly of individual Z-rings, leading to the frequent abortion of septum synthesis, which in turn results in the production of long spore-like compartments with multiple chromosomes. Two-hybrid analysis revealed that the dynamins are part of the cell division machinery and that they mediate their effects on Z-ring stability during developmentally controlled cell division via a network of protein–protein interactions involving DynA, DynB, FtsZ, SepF, SepF2, and the FtsZ-positioning protein SsgB.
The active organization and remodeling of cellular membranes is a fundamental process for all organisms. Many of these remodeling events involve the reshaping, fission, or fusion of lipid bilayers to generate new organelles, release transport vesicles, or stabilize specific membrane structures. In eukaryotes, key cellular processes, such as the fission and fusion of mitochondria, the division of chloroplasts, endocytosis, and viral resistance, are mediated by members of the dynamin superfamily (1). Dynamins and dynamin-like proteins are mechanochemical GTPases that polymerize into helical scaffolds at the surface of membranes. GTP hydrolysis is coupled to a radical conformational change in the protein structure that forces the underlying lipid layer into an energetically unstable conformation that promotes membrane rearrangements, probably via a hemifusion intermediate (2, 3).
Dynamin-like proteins are found in many bacterial species, yet the precise roles of these proteins are still largely unknown. They share a conserved domain architecture with the canonical human Dynamin 1, including the N-terminal GTPase domain, a neck domain involved in dynamin dimerization, and a trunk domain involved in stimulation of GTPase activity (4, 5). In contrast, bacterial dynamins lack the pleckstrin homology motif and proline-rich sequences found in classical dynamins and contain instead other lipid- and protein-binding motifs (5). Structural and biochemical studies on Nostoc punctiforme BLDP1, Bacillus subtilis DynA, and Escherichia coli LeoA have provided clear insight into the mechanism of protein oligomerization and lipid binding (2, 3, 6, 7), but unlike those of their eukaryotic counterparts the biological functions of bacterial dynamins have remained largely unclear. Recent reports suggest that dynamins might function in diverse cellular processes in bacteria, including chromosome replication (8), membrane stress responses (9, 10), and outer membrane vesicle release (7), indicating that they have perhaps evolved to fulfill a range of different functions in bacteria.
Here we show that two bacterial dynamin-like proteins play an important role in sporulation-specific cell division in Streptomyces venezuelae. Streptomycetes are filamentous, antibiotic-producing soil bacteria that have a multicellular life cycle with two distinct modes of cell division: vegetative cross-wall formation and sporulation septation (11) (Fig. 1A). Sporadic cross-walls divide the growing vegetative mycelium into long multinucleoid compartments that remain physically connected. In contrast, dozens of sporulation septa are deposited in a ladder-like pattern between the segregating chromosomes in sporogenic hyphae. These septa constrict, leading to cell–cell separation and the release of equally sized, unigenomic spores. Both forms of cell division require the highly conserved tubulin-like GTPase FtsZ (12, 13), which polymerizes into short dynamic filaments close to the cytoplasmic membrane, forming the so-called Z-ring. The Z-ring provides the spatiotemporal signal for the recruitment of additional cell division proteins to form a multiprotein machine (the divisome) and coordinates peptidoglycan synthesis at the nascent division septum with other cellular processes (14, 15). Two actinomycete-specific proteins, SsgA and SsgB, positively control the spatial distribution of Z-rings in sporogenic hyphae. At the onset of sporulation, FtsZ is recruited to future division sites through a direct interaction with SsgB, which in turn interacts with SsgA (16). However, what determines the positioning of SsgA and SsgB is not known and streptomycetes lack homologs of the canonical septum placement control proteins identified in other bacteria, such as Noc (17), SlmA (18), and the Min system (19).
Fig. 1.
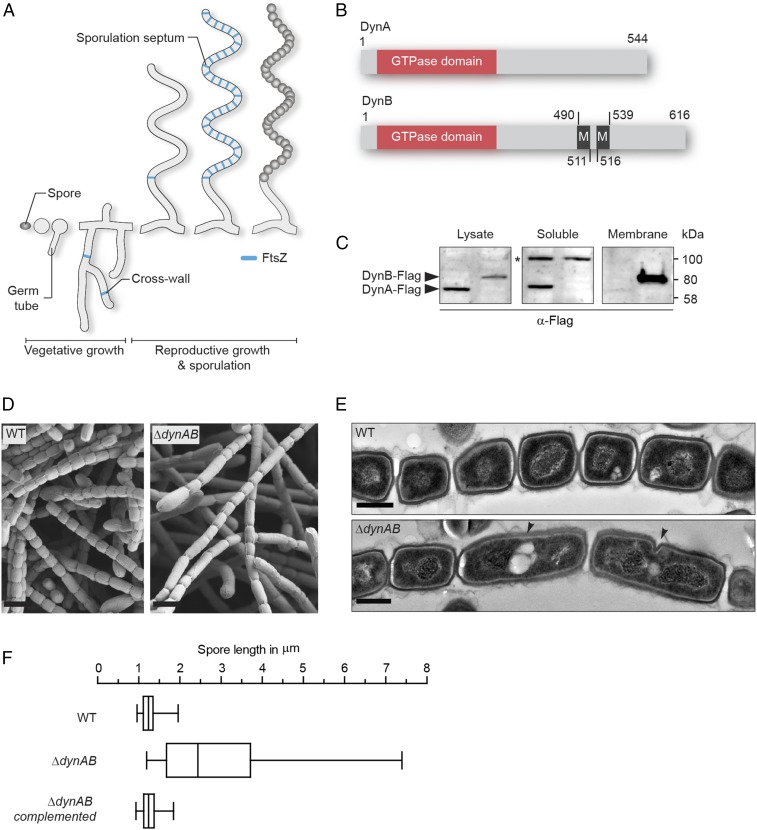
Two dynamin-like proteins are important for sporulation-specific cell division. (A) Schematic depicting the Streptomyces life cycle starting with a spore that germinates and grows into a vegetative mycelium, followed by the formation of a reproductive hypha that differentiates into a chain of equally sized spores. FtsZ associated with vegetative cross-walls and sporulation septa is shown in blue. (B) Schematic showing the predicted domain organization of DynA and DynB. The GTPase domain is shown in orange and transmembrane helices (M) are shown in black. Numbers indicate corresponding amino acid positions. (C) Fractionation experiment using S. venezuelae strains expressing either a functional dynA-3xFLAG (SS93, left lane) or a dynB-3xFLAG (SS140, right lane) fusion from the ΦBT1 attachment site. Whole-cell lysates were separated into soluble and membrane fractions and probed with anti-FLAG antibody. The asterisk denotes a nonspecific signal in the soluble protein fraction. Shown are representative results of biological replicate experiments. (D) Scanning electron micrographs of sporogenic hyphae from wild-type S. venezuelae (WT) and the dynamin mutant (ΔdynAB). (Scale bars: 2 μm.) (E) Transmission electron micrographs of sporogenic hyphae from the WT and ΔdynAB mutant. Black arrowheads indicate failed and asymmetric constrictions in the dynamin mutant. (Scale bars: 500 nm.) (F) Box plot showing the length distribution of spores produced by the WT (n = 1,113), the ΔdynAB mutant (n = 676) and the complemented dynamin mutant (SS23, n = 612). Whiskers denote the 5th and 95th percentile.
To ensure regular septum formation and efficient cell–cell separation, the early stages of divisome assembly requires the stabilization of FtsZ protofilaments on the cytoplasmic membrane, but FtsZ does not interact with the membrane directly. Instead, in other bacterial systems FtsZ filaments are tethered to the membrane through interaction with membrane-anchoring proteins such as FtsA, ZipA, and SepF (20–24). Additional factors, such as the ZapC and ZapD, are critically involved in the stabilization of preformed Z-rings to ensure normal cell division (25–27). S. venezuelae lacks FtsA, ZipA, and the Zap proteins but encodes three SepF homologs, although the functions of these proteins have not been investigated to date.
In filamentous bacteria such as Streptomyces, sporulation-specific cell division represents a unique challenge in which each sporogenic hypha coordinates the almost synchronous placement of dozens of septa. During this process, helical FtsZ filaments tumble along the hypha and then coalesce into long ladders of regularly spaced Z-rings (28, 29). However, the molecular mechanisms that control the stability and functionality of these multiple division-competent Z-rings are unknown. Here we show that two sporulation-specific dynamin-like proteins interact directly with the divisome to stabilize FtsZ rings during Streptomyces sporulation.
Results and Discussion
Two Dynamin-Like Proteins Are Required for Normal Sporulation Septation.
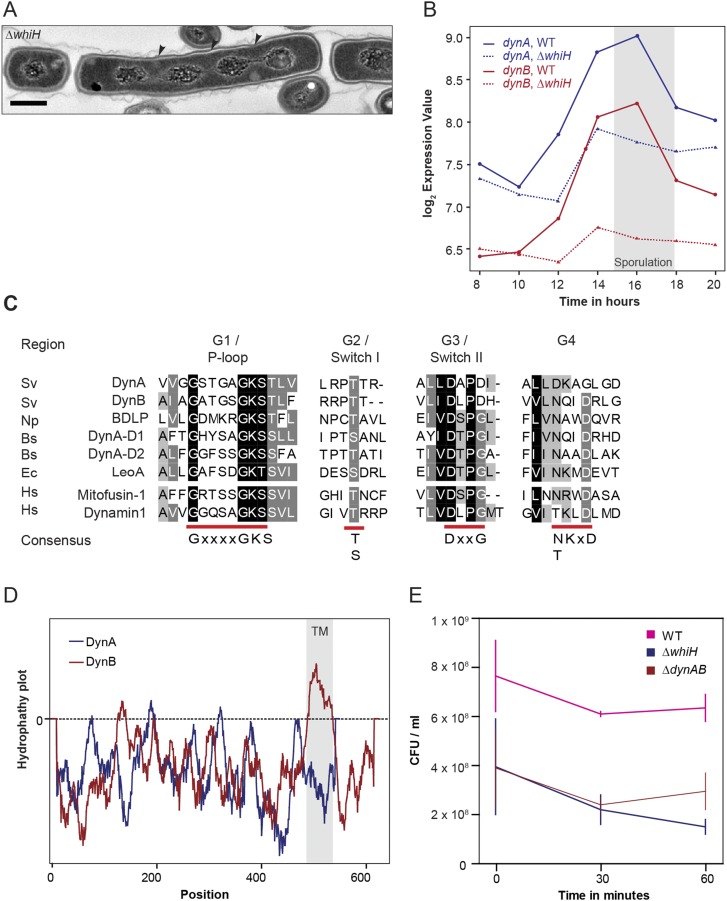
One of the transcriptional regulators critical for the differentiation of sporogenic hyphae into chains of spores is WhiH (30). In whiH mutants, individual sporulation septation events frequently fail, resulting in the creation of long spore compartments with multiple copies of the chromosome (Fig. S1A). To further understand the role of WhiH in sporulation-specific cell division, we screened transcriptional profiling data for genes that showed an altered expression profile in a ΔwhiH background compared with the WT (Fig. S1B). This analysis led to the identification of an operon encoding two dynamin-like proteins that we designated DynA (Sven2472) and DynB (Sven2471) (Fig. 1B). The dynamin genes are induced at the onset of sporulation in the WT and this induction is heavily dependent on whiH (Fig. S1B). Bioinformatic analyses showed that DynA and DynB are structural homologs of the bacterial dynamin-like protein (BDPL1) from the cyanobacterium N. punctiforme (3). Sequence alignments of diverse members of the dynamin superfamily confirmed that DynA and DynB share the highly conserved residues for GTP binding and hydrolysis in the signature N-terminal GTPase domain (Fig. S1C). In addition, DynB carries two predicted transmembrane helices, whereas DynA seems to lack the hydrophobic residues required for a direct interaction with the cytoplasmic membrane (Fig. S1D). Their predicted subcellular locations were confirmed by fractionation experiments, which showed that DynA is a soluble protein whereas DynB cosediments with the membrane (Fig. 1C).
Fig. S1.
S. venezuelae encodes two dynamin-like proteins that are developmentally regulated. (A) TEM image showing the phenotype of the ΔwhiH mutant (SV8). WhiH-deficient cells deposit sporulation septa irregularly, leading to the formation of long spore-like compartments with more than one chromosome. Black arrowheads point to missing sporulation septa. (Scale bar: 500 nm.) (B) Expression profile of dynA and dynB in the WT (solid line) and the ΔwhiH (SV8) mutant (dashed line) over the entire Streptomyces life cycle. The gray box indicates the period of sporulation-specific cell division. (C) Sequence alignment of GTPase domains from selected dynamin family members. Highly conserved residues involved in GTP binding and hydrolysis are underlined in red. Protein sequences: Bs, Bacillus subtilis; Ec, Escherichia coli; Hs, Homo sapiens; Np, Nostoc punctiforme; and Sv, Streptomyces venezuelae. (D) Output from TMPred server predicting two transmembrane helices for DynB (gray box). The corresponding amino acid positions for the predicted transmembrane domains are shown in the schematic in Fig. 1B. (E) Survival of spores from WT, ΔwhiH (SV8), and ΔdynaB (LUV001) cells after heat stress (55 °C) over a time period of 60 min. The experiment was carried out in duplicate and results are shown as means ± SD.
To investigate whether DynA and DynB play a role in developmentally controlled cell division, we generated a dynAB null mutant and imaged sporulating hyphae of WT S. venezuelae and the ΔdynAB::apr (ΔdynAB) mutant by cryo-scanning electron microscopy and transmission electron microscopy (TEM). Strikingly, microscopic analyses revealed that DynAB-deficient hyphae fail to deposit regularly spaced sporulation septa, leading to the formation of long spore compartments (Fig. 1 D and E). This phenotype was fully complemented by expressing dynAB in trans, restoring normal sporulation (Fig. 1F). In addition, transmission electron micrographs of ΔdynAB mutant hyphae showed that the longer spore compartments had multiple chromosomes and often carried asymmetric and incomplete constrictions of the cell envelope, suggesting that cell division had initiated but then aborted at an early stage during septum formation (Fig. 1E). All these phenotypes are strikingly reminiscent of the whiH mutant phenotype in S. venezuelae (Fig. S1A), suggesting that the dynamins largely mediate the effect of WhiH on developmentally controlled cell division. Although DynAB-deficient hyphae frequently fail to complete sporulation septation, the spores that are made by the ΔdynAB mutant seem to be mature, producing the characteristic green spore pigment and showing WT levels of heat resistance (Fig. S1E).
DynA and DynB Colocalize with FtsZ During Sporulation-Specific Cell Division.
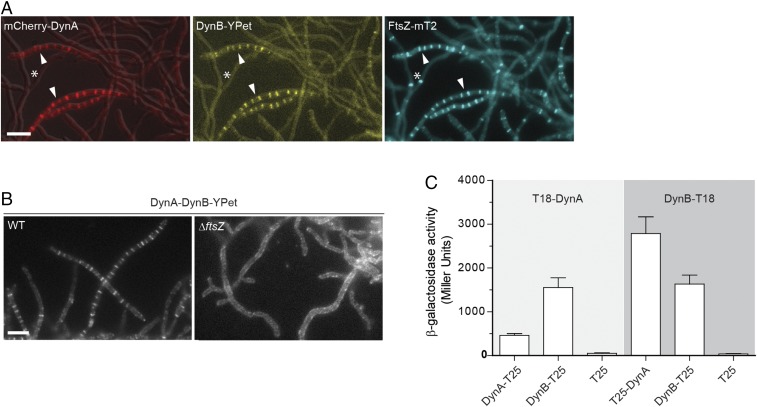
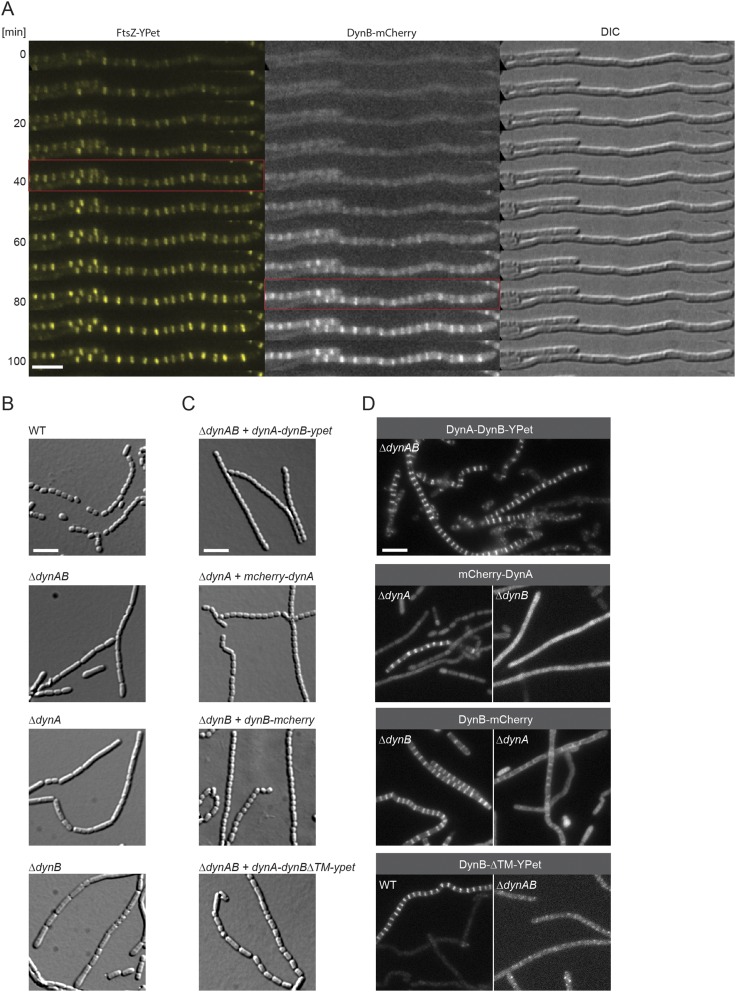
We reasoned that if DynA and DynB play a role in sporulation septation they should accumulate at future division sites. To address this hypothesis, we generated a merodiploid strain in which an mcherry-dynA dynB-ypet operon, expressed from the native dynamin promoter, was integrated into the chromosome at the ΦBT1 integration site. In addition, the same strain was engineered to produce an FtsZ-mTurquoise2 fusion (FtsZ-mT2) to fluorescently label sites of vegetative- and sporulation-specific cell division. Microscopic analysis of the triply labeled S. venezuelae strain revealed that DynA and DynB colocalized with Z-rings specifically at sporulation septa, supporting the idea that DynA and DynB are involved in sporulation-specific cell division (Fig. 2A). Note that DynAB do not localize to vegetative cross-walls (see asterisks in Fig. 2A), in line with the transcriptomic data showing that dynAB transcription is activated at the onset of sporulation (Fig. S1B). An advantage of studying cell division in Streptomyces is that ftsZ null mutants are viable, generating colonies devoid of both vegetative cross-walls and sporulation septa (31). To determine whether DynAB can localize independently of FtsZ, we expressed a functional dynAB-ypet fusion from a constitutive promoter (PermE*) in an S. venezuelae ΔftsZ mutant. Fluorescence microscopy showed that the constitutively expressed DynB-YPet accumulated along the cytoplasmic membrane, and that the distinct ladder-like DynA-DynB-YPet localization seen in the WT was absent in the ΔftsZ mutant (Fig. 2B), indicating that FtsZ is required for DynAB recruitment and placement. This result was further supported by time-lapse imaging showing the appearance of fluorescent FtsZ ladders ∼30–40 min before DynB-mCherry accumulation at the Z-ladders became visible (Fig. S2A).
Fig. 2.
DynA–DynB complexes colocalize with FtsZ at nascent division sites. (A) Subcellular colocalization of fluorescent fusions to DynA (mCherry-DynA) and DynB (DynB-YPet) with FtsZ-mTurquoise2 (FtsZ-mT2). The asterisk denotes vegetative cross-walls and arrowheads point to sporulation septa. Microscopy images of the triply labeled strain (SS206) are representative of at least two independent experiments. (Scale bar: 5 μm.) (B) Localization of DynB-YPet in the WT (SS142) and in the ftsZ null mutant (ΔftsZ, SS238). The dynAB-ypet construct was ectopically expressed from a constitutive promoter (PermE*). (Scale bar: 5 μm.) (C) β-galactosidase activities demonstrating an interaction between DynA and DynB in E. coli BTH101. Positive interaction is detected when DynA and DynB protein fusions to the “T18” and “T25” domains of adenylate cyclase reunite the enzyme, resulting in the synthesis of LacZ. Strains expressing only the T25 domain were used as a negative control. Results are the average of three independent experiments. Error bars represent the SEM.
Fig. S2.
DynA and DynB are codependent and function downstream of FtsZ. (A) Montage of a fluorescent time-lapse sequence showing the localization of FtsZ-YPet and DynB-mCherry (SS196). The red box indicates the appearance of FtsZ-ladders (Left) and the colocalization of DynB (Middle). Corresponding DIC images are shown (Right). Imaging interval was 10 min. (Scale bar: 5 μm.) (B) Sporulating aerial hyphae showing the sporulation septation defect in the ΔdynAB (LUV001), ΔdynA (SS255), and ΔdynB (SS2) mutants compared with the WT. Cells were grown for 4 d on MYM and imaged. (Scale bar: 5 μm.) (C) Images of sporulating aerial hyphae showing the ability of the fluorescent dynamin fusions to complement the ΔdynAB phenotype. All constructs were integrated at the ΦBT1 attachment site and expression of the genes was driven either by the dynamin promoter (SS90, SS266) or by the ermE* promoter (SS263, SS89). No complementation was observed when dynBΔTM-ypet was expressed (Bottom, SS263). (Scale bar: 5 μm.) (D) Sporulating aerial hyphae showing the localization codependency of functional fluorescent fusions to full-length DynA or DynB in the ΔdynA (SS266), ΔdynB (SS89, SS190), and ΔdynAB (SS90, SS95) mutants. The fluorescent DynB protein fusion lacking the two transmembrane domains (DynBΔTM-YPet, Bottom) was produced either in the WT (SS262) or the dynamin mutant (SS263). (Scale bar: 5 μm.)
DynA–DynB Interaction Is Required for Regular Septation.
To gain further mechanistic insight into DynAB action in vivo, we asked whether each dynamin depends on the other for function. First, we generated dynA and dynB single mutants and analyzed their sporogenic hyphae by light microscopy (Fig. S2B). Deletion of either dynA or dynB impaired regular sporulation septation in the same way as removal of both genes, implying their functions are not redundant, and the single mutants could only be complemented by providing the missing gene in trans (Fig. S2C). Next, we examined the subcellular localization dependency of functional fluorescent fusions to DynA or DynB. Although DynA localization to nascent sporulation septa depended on DynB, DynB was still able to accumulate to some extent at septa in the absence of DynA (Fig. S2D). This finding implies that DynB functions in localizing DynA. DynB targeting to future division sites requires the direct interaction with the membrane because a mutant version of DynB lacking the two transmembrane domains (DynBΔTM-YPet) failed to accumulate in the typical FtsZ-like pattern (Fig. S2D). Moreover, removal of the membrane anchor in DynB renders the protein nonfunctional and leads to irregular septation (Fig. S2C).
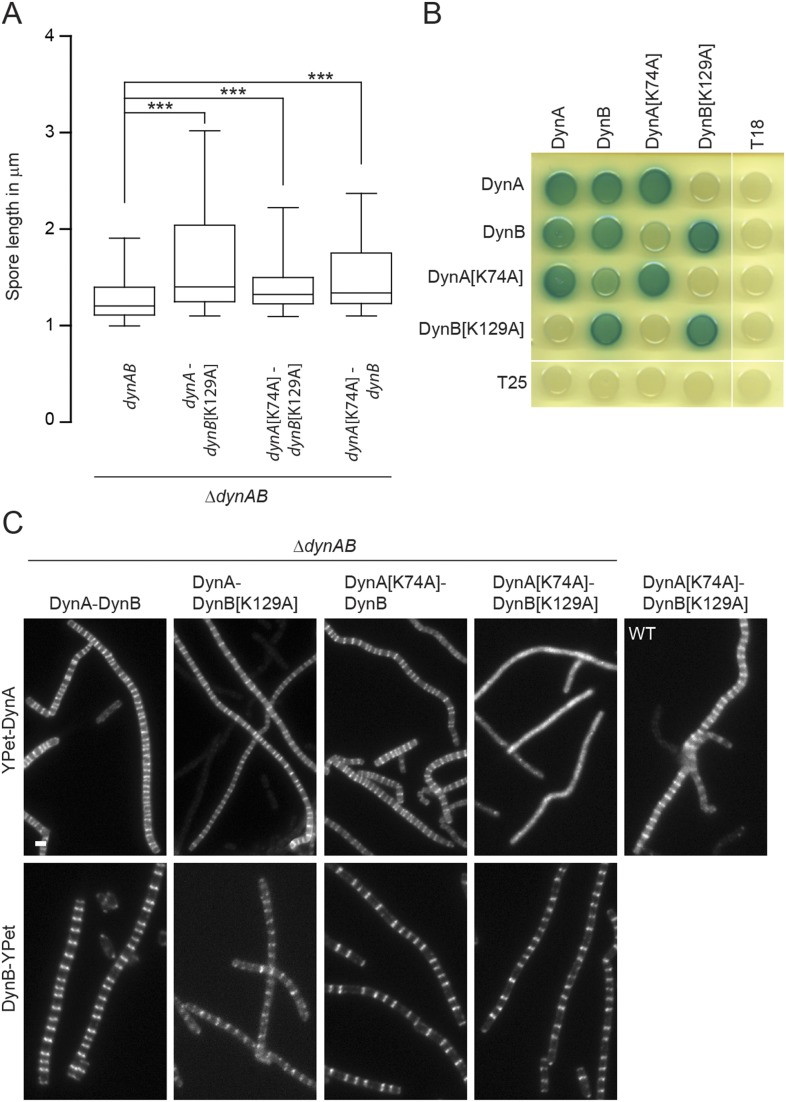
Previous work on the dynamin-like proteins from N. punctiforme and B. subtilis showed that mutation of a highly conserved lysine in the P-loop, a sequence motif in the GTPase domain essential for nucleotide binding (Fig. S1C), significantly reduces GTPase activity (3, 6). We therefore generated the same mutation in the GTPase domains of DynA[K74A] and DynB[K129A]. To assess the importance of GTP binding, we expressed the mutated dynAB operon in trans in a ΔdynAB background and examined whether the corresponding gene products could rescue the dynamin phenotype. Expression of WT dynAB fully complemented the sporulation septation defect, but any combination carrying a P-loop mutation in dynA, dynB, or in both genes failed to restore the WT phenotype (Fig. S3A). Given their subcellular colocalization, we speculated that DynA and DynB might form a heterodimer and that assembly might depend on the nucleotide status of both dynamins. To test this hypothesis, we used a bacterial two-hybrid assay. The results of this assay showed that DynA and DynB self-interact and also bind each other (Fig. 2C and Fig. S3B). Self-interaction was not affected when one of the binding partners carried a P-loop mutation. However, the interaction between DynA and DynB were considerably weaker when one of the partners carried a P-loop mutation compared with the interaction between the WT proteins (Fig. S3B). To further corroborate these results, we examined the subcellular localization of DynA and DynB P-loop mutants in the ΔdynAB mutant (Fig. S3C). As expected from the two-hybrid experiments, DynA[K74A] fails to accumulate at nascent sporulation septa in the presence of DynB[K129A], confirming that defective GTP binding in both DynA and DynB prevents direct interaction. However, WT DynA still localized to sporulation septa when coexpressed with DynB[K129A], suggesting that DynA can still associate with the divisome under these conditions, presumably via interaction with DynB[K129A] and other divisome components. Introducing a P-loop mutation in DynB did not affect its subcellular accumulation at future division sites. Taken together, these results suggest that DynA and DynB form a complex that requires GTP binding for efficient interaction and in vivo function.
Fig. S3.
GTP binding is important for dynamin function and interaction. (A) Spore length analysis showing the extent to which dynamin P-loop mutants are functional. The expression of the dynamin operon containing either WT dynAB (SS142, n = 1,401), a dynA[K74A] (SS211, n = 1,933), dynB[K129A] (SS164, n = 1,933), or both (SS165, n = 1,933) P-loop mutations was driven from the constitutive ermE* promoter from the ΦBT1 attachment site. Spore counts are the result of triplicate experiments. Box plot whiskers represent the 5th and 95th percentile. All P values were calculated using a one-sided t test (***P < 0.001). (B) Bacterial two-hybrid analysis showing representative results of the interaction between WT DynA and DynB and the corresponding P-loop mutant proteins. Cells were grown overnight in LB and spotted on LB agar containing Xgal. Plate was incubated for 2 d at 30 °C in the dark and imaged. (C) Sporulating aerial hyphae showing the subcellular localization of WT YPet-DynA/DynB-YPet and the corresponding P-loop mutant protein derivatives in the ΔdynAB mutant (SS142, SS164, SS165, SS210, SS242, SS244, SS248, and SS250). Note that YPet-DynA[K74A] can localize to division septa in the WT (SS224). (Scale bar: 2 μm.)
FtsZ Rings Are Destabilized in DynAB-Deficient Hyphae.
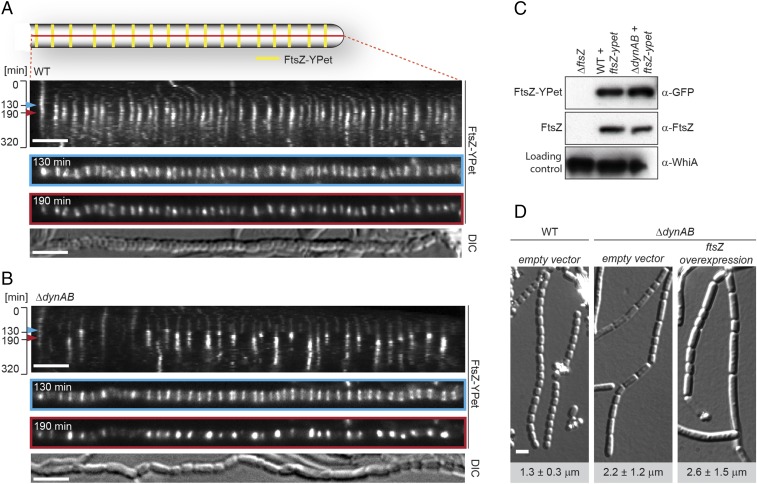
To further investigate the cell division defect in the ΔdynAB mutant we recorded time-lapse images of sporulating WT and ΔdynAB hyphae expressing a fluorescently tagged copy of FtsZ (Fig. 3 A and B, Fig. S4, and Movies S1 and S2). Kymographs of FtsZ-YPet localization during sporulation in the WT showed that Z-rings assemble almost synchronously and are regularly distributed along the sporogenic hyphae (Fig. 3A, t = 130 min and Fig. S4A), giving a ladder-like appearance. Over time, single Z-rings increase in fluorescence intensity, indicating their maturation and ongoing constriction (t = 190 min). After about 2 h, FtsZ-YPet fluorescence decreases and the Z-ladders disappear. This is accompanied by the coordinated constriction of the cell envelope and the completion of septum formation to produce a chain of equally sized spores. Strikingly, kymographs of the ΔdynAB mutant (Fig. 3B and Fig. S4B) reveal a different pattern. Initially, the Z-rings seen in the mutant are of uniform intensity and are deposited at regular intervals, just like in the WT (Fig. 3A, t = 130 min and Fig. S4A), supporting the hypothesis that Z-ring placement is independent of DynAB. However, as cell division progresses, many Z-rings become destabilized and disassemble prematurely in a sudden and synchronous event (Fig. 3B, t = 190 min and Fig. S4B). Thus, the lack of septa or the partial, asymmetric constrictions of the cell envelope seen by TEM in the ΔdynAB mutant (Fig. 1E) can be explained by a reduced processivity of the cytokinetic Z-rings during the early stages of cell division, such that cell division is initiated but not completed. This leads to the formation of longer spore-like compartments carrying more than one copy of the chromosome (Fig. 1E) and is in line with the significant increase in spore length, as determined by measurements of spores produced by the WT and the ΔdynAB mutant (Fig. 1F). The sudden disassembly of constriction-competent Z-rings seen in the dynamin mutant is not caused by reduced FtsZ protein stability because Western blot analysis confirmed that FtsZ and FtsZ-YPet levels are similar in the WT and ΔdynAB strains (Fig. 3C). Moreover, increasing intracellular FtsZ levels through chromosomal integration of an additional copy of ftsZ under the control of a constitutive promoter (PermE*) did not restore normal sporulation to the dynamin mutant (Fig. 3D).
Fig. 3.
DynA and DynB stabilize Z-rings during sporulation-specific cell division. Kymograph analysis of FtsZ-YPet dynamics during sporulation-specific cell division in WT (A) and ΔdynAB cells (B), expressing an additional copy of ftsZ-ypet (strains SS12 and SS14). Blue and red arrowheads denote time points of initial Z-ring assembly (130 min) and maturation (190 min), shown in separate images below. Differential interference contrast (DIC) images show the corresponding spore chain at the end of cell division. Additional kymographs can be found in Fig. S4. (Scale bars: 4 μm.) (C) Immunoblot analysis showing FtsZ and FtsZ-YPet levels in WT (SS12), ΔdynAB (SS14), and ΔftsZ cells. Equal protein concentrations of crude cell lysate were loaded for each lane and samples were probed with anti-FtsZ, anti-GFP, and anti-WhiA antibodies. WhiA is a transcriptional regulator that is present at constant levels throughout the developmental life cycle (42) and was used as a loading control. (D) DIC images of sporulating WT and ΔdynAB hyphae carrying the empty vector (SS4, SS10) or ΔdynAB hyphae constitutively expressing ftsZ from the ermE* promoter (SS11). Numbers indicate mean spore lengths ± SD (n > 460 spores per strain). (Scale bar: 2 μm.)
Fig. S4.
Additional representative kymographs showing the dynamics of fluorescent FtsZ ladders in (A) the WT (SS12) and (B) the dynamin mutant (SS14). Experiments were performed by growing the respective strains in the CellASIC microfluidic system and images were acquired every 8 min. Kymographs show FtsZ-YPet dynamics over 180 min. (Scale bar: 5 μm.)
DynA and DynB Interact with the Divisome.
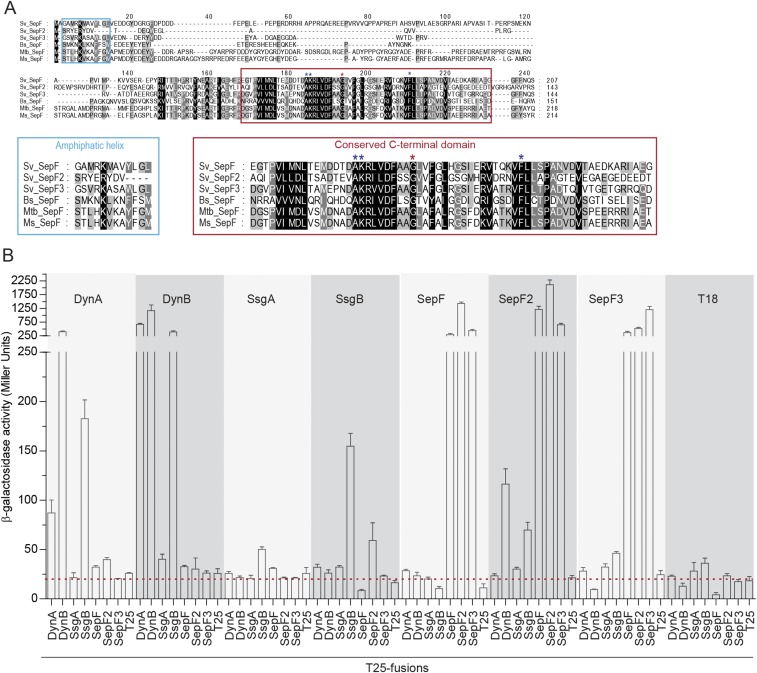
Next we asked whether the dynamins stabilize cytokinetic Z-rings directly or via other proteins. S. venezuelae lacks homologs of ZipA and FtsA, the FtsZ-stabilizing proteins that anchor the Z-ring to the membrane in many other bacteria, but it encodes the actinomycete-specific proteins SsgA and SsgB, involved in FtsZ-ring positioning (16) and three SepF-like proteins: Sven1372, Sven1734, and Sven5776. Sven1734 is encoded in the division and cell wall (dcw) gene cluster, like the single SepF protein found in B. subtilis and Mycobacterium tuberculosis (23, 32). Based on this synteny, we named Sven1734 SepF and the additional Streptomyces SepF-like proteins SepF2 (Sven5776) and SepF3 (Sven1372). Sequence alignments showed that all three Streptomyces SepF homologs have the conserved C-terminal domain found in canonical SepF proteins, including the residues essential for protein dimerization and interaction with FtsZ (20, 24, 33) (Fig. S5A). Notably, SepF2 lacks most of the conserved N-terminal residues that have previously been shown to fold into the amphipathic helix that functions as a lipid anchor in B. subtilis SepF (20) (Fig. S5A), suggesting that SepF2 may not interact directly with the membrane. To test the possibility that DynA and DynB might interact with SsgA, SsgB, or the SepF-like proteins, we performed two-hybrid experiments in E. coli and found that both DynA and DynB interact with SsgB (Fig. 4A). In addition, this assay indicates that both DynB and SsgB bind SepF2 (Fig. 4A) and shows that all three SepF proteins bind themselves and each other (Fig. S5B). Because Streptomyces FtsZ did not show the expected self-interaction in this assay, we complemented our analysis by using the yeast two-hybrid system (Fig. 4B). These experiments showed the expected self-interaction of FtsZ and confirmed the SepF–SepF2 interaction seen in the E. coli system. Importantly, they also showed that SepF binds FtsZ. No interaction was seen between FtsZ and the dynamins. In addition, fluorescence microscopy revealed that both SepF-mCherry and mCherry-SepF2 colocalize with FtsZ-YPet (Fig. 4C), supporting the idea that both proteins are involved in cell division and thus providing an additional functional link between the dynamins and FtsZ.
Fig. S5.
Dynamins interact with the divisome via SepF2. (A) Sequence alignment of selected SepF homologs. The conserved residues in the N terminus (blue box) and C terminus (red box) of the proteins are shown below in larger font. (Left) Sequence alignment of the residues predicted to form an amphipathic helix in the N terminus (20). Note that S. venezuelae SepF2 lacks most of the residues predicted to function in lipid binding. (Right) Sequence alignment of the C-terminal domain, containing the conserved residues responsible for SepF dimer contacts (red asterisk) and interaction with FtsZ (blue asterisks) (20, 24, 33). Bs, B. subtilis; Ms, Mycobacterium smegmatis; Mtb, M. tuberculosis; and Sv, S. venezuelae. (B) β-galactosidase activities showing the relative strengths of the interactions between the dynamins, SsgA, SsgB, and the SepF proteins. Results are the average of three independent experiments. Error bars represent the SEM. Dashed red line indicates background β-galactosidase activity as measured in the negative control (T18–T25).
Fig. 4.
DynA and DynB interact with the cell division machinery. (A) Bacterial two-hybrid analysis of DynA and DynB with SsgA, SsgB, and the three SepF-like proteins. E. coli BTH101 cells carrying plasmids with protein fusion to the T18 and the T25 domain were spotted onto LB agar plates supplemented with Xgal, incubated at 30 °C for 24 h, and imaged. Corresponding β-galactosidase activities of three replicate experiments for each interaction are shown in Fig. S5B. (B) Yeast two-hybrid analysis showing the interaction between SepF and FtsZ. Interaction between proteins fused to the GAL4-activation domain (AD) and the GAL4 DNA-binding domain (BD) allows growth of yeast AH102 on minimal medium, lacking leucine, tryptophan, histidine, and alanine (−LWHA). In parallel, viability of the yeast cells carrying the respective hybrid proteins was confirmed by spotting cells on minimal medium without leucine and tryptophan (−LW). Representative results of three experiments are shown. (C) Colocalization of SepF-mCherry (SS208) and mCherry-SepF2 (SS213) with FtsZ-YPet. Microscopy results are representative of at least two independent experiments. (Scale bar: 2 μm.) (D) The protein interaction wheel between the dynamins and other divisome components based on two-hybrid results. (E) Proposed model of DynA and DynB function in Streptomyces. During sporulation-specific cell division, DynA and DynB form a complex at nascent division sites and interact with the division machinery via binding to SsgB and SepF2. In WT cells, Z-ring formation and cell envelope constriction leads to regular sporulation septum formation, resulting in equally sized, unigenomic spores. In the dynamin mutant, many Z-rings disassemble before completion of division septum synthesis, leading to failed or incomplete and asymmetric cell envelope constrictions and spores of irregular size with variable chromosome number.
Conclusions.
Taken together with the previously demonstrated direct interaction between SsgB and FtsZ (16), our results show that DynA and DynB are part of the divisome (Fig. 4 D and E). In this multiprotein complex the dynamins mediate their effects on Z-ring stability during sporulation-specific cell division via a network of protein–protein contacts involving DynA, DynB, SepF, SepF2, SsgB, and FtsZ (Fig. 4 D and E). Furthermore, 92% of available Streptomyces genomes (n = 139) carry two dynamins and at least two sepF genes, suggesting that they likely play a similar role across the genus. Future studies should attempt to determine whether the dynamins also play a role in remodeling the membrane as it invaginates around the ingrowing cell wall annulus and in catalyzing membrane fusion as the septum eventually closes.
Dynamins have been directly linked to cell division in plant and animals cells (1, 34, 35), where they play important roles in cell plate formation (36, 37), chloroplasts division (38), and vesicle budding from the cleavage furrow (39, 40). The work presented here demonstrates that the involvement of dynamins in cell division is conserved in both eukaryotic cells and bacteria.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions.
All bacterial strains, plasmids, and oligonucleotides used in this study are described in Tables S1 and S2 and Dataset S1. E. coli strains were grown in LB or on LB agar at 37 °C. When required, the following antibiotics were added to the growth medium: 100 μg⋅mL−1 carbenicillin (Carb100), 50 μg⋅mL−1 kanamycin (Kan50), 25 μg⋅mL−1 hygromycin (Hyg25), 50 μg⋅mL−1 apramycin (Apr50), or 25 μg⋅mL−1 chloramphenicol (Cam25). S. venezuelae cells were cultured in maltose-yeast extract-malt extract medium (MYM) made with 50% tap water and and 50% reverse osmosis water and supplemented with R2 trace element solution at 1:500 (41). Liquid cultures were grown under aeration at 30 °C at 250 rpm. When required, MYM agar contained 5 μg⋅mL−1 kanamycin, 25 μg⋅mL−1 hygromycin, or 50 μg⋅mL−1 apramycin. Conjugations between E. coli and S. venezuelae were performed as described in Bush et al. (42).
Table S1.
Strains used in this study
| Strain | Genotype | Construction/description | Source |
| AH109 | MATa, trp1-901, leu2-3, 112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2::GAL1UASGAL1TATA- HIS3, GAL2UAS-GAL2TATA-ADE2, URA3::MEL1UAS-MEL1TATA-lacZ | Host strain for yeast-two hybrid analysis | Clontech |
| TOP10 | F– mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu) 7697 galU galK rpsL (StrR) endA1 nupG | Cloning | Invitrogen |
| ET12567/pUZ8002 | F– dam13::Tn9 dcm6 hsdM hsdR recF143::Tn10 galK2 galT22 ara-14 lacY1 xyl-5 leuB6 thi-1 tonA31 rpsL hisG4 tsx-78 mtl-1 glnV44 | ET12567 containing helper plasmid pUZ8002 | 53 |
| BW25113/pIJ790 | Δ(araD-araB)567 ΔlacZ4787(::rrnB-4) lacIp-4000(lacIQ), λ−rpoS369(Am) rph-1 Δ(rhaD-rhaB)568 hsdR514 | BW25113 containing λ RED recombination plasmid | 54 |
| BTH101 | F– cya-99 araD139 galE15 galK16 rpsL1 (Strr) hsdR2 mcrA1 mcrB1 | Host strain for bacterial two-hybrid analysis | 55 |
| Streptomyces strains | |||
| WT | S. venezuelae ATCC 10712 | Laboratory strain | 52 |
| SV8 | ΔwhiH::apr | M. Bibb, unpublished | |
| ΔftsZ | ΔftsZ::apr | J. McCormick, unpublished | |
| LUV015 | attBϕC31::PftsZ-ftsZ-ypet | pKF351 integrated at the ΦC31 attachment site of S. venezuelae WT | This work |
| LUV001 | ΔdynAB::apr | Chromosomal dynAB locus was replaced with apra-oriT from pIJ773 by ‟Redirect” PCR targeting using oligo KF843 and KF844 | This work |
| SS2 | ΔdynB::apr | Chromosomal dynB locus was replaced with apra-oriT from pIJ773 by ‟Redirect” PCR targeting using oligo 8 and 9 | This work |
| SS4 | attBϕBT1::pIJ10257 | pIJ10257 integrated at the ΦBT1 attachment site of S. venezuelae WT | This work |
| SS10 | ΔdynAB::apr attBϕBT1::pIJ10257 | pIJ10257 integrated at the ΦBT1 attachment site of LUV001 | This work |
| SS11 | ΔdynAB::apr attBϕBT1::PermE*-ftsZ | pSS3 integrated at the ΦBT1 attachment site of LUV001 | This work |
| SS12 | attBϕBT1::PftsZ-ftsZ-ypet | pSS5 integrated at the ΦBT1 attachment site of S. venezuelae WT | This work |
| SS14 | ΔdynAB::apr attBϕBT1::PftsZ-ftsZ-ypet | pSS5 integrated at the ΦBT1 attachment site of LUV001 | This work |
| SS23 | ΔdynAB::apr attBϕBT1::Pdyn-dynAB | pKF337 integrated at the ΦBT1 attachment site of LUV001 | This work |
| SS39 | attBϕC31::PftsZ-ftsZ-mcherry | pSS61 integrated at the ΦC31 attachment site of S. venezuelae WT | This work |
| SS89 | ΔdynB::apr attBϕBT1::PermE*-dynB-mcherry | pSS87 integrated at the ΦBT1 attachment site of SS2 | This work |
| SS90 | ΔdynAB::apr attBϕBT1::Pdyn-dynA-dynB-ypet | pSS89 integrated at the ΦBT1 attachment site of LUV001 | This work |
| SS93 | attBϕBT1::PermE*-dynA-3xFLAG | pSS91 integrated at the ΦBT1 attachment site of LUV001 | This work |
| SS95 | ΔdynAB::apr attBϕBT1::PermE*-dynB-mcherry | pSS87 integrated at the ΦBT1 attachment site of LUV001 | This work |
| SS140 | ΔdynAB::apr attBϕBT1::PermE*-dynA-dynB-3xFLAG | pSS130 integrated at the ΦBT1 attachment site of LUV001 | This work |
| SS142 | ΔdynAB::apr attBϕBT1::PermE*-dynA-dynB-ypet | pSS134 integrated at the ΦBT1 attachment site of LUV001 | This work |
| SS164 | ΔdynAB::apr attB ϕBT1::PermE*-dynA-dynB[K129A]-ypet | pSS141 integrated at the ΦBT1 attachment site of LUV001 | This work |
| SS165 | ΔdynAB::apr attB ϕBT1::PermE*-dynA[K74A]-dynB[K129A]-ypet | pSS142 integrated at the ΦBT1 attachment site of LUV001 | This work |
| SS190 | ΔdynB::apr attBϕBT1::Pdyn-mcherry-dynA | pSS188 integrated at the ΦBT1 attachment site of SS2 | This work |
| SS196 | attBϕC31::PftsZ-ftsZ-ypet | pSS180 integrated at the ΦBT1 attachment site of LUV015 | This work |
| attBϕBT1::Pdyn-dynA-dynB-mcherry | |||
| SS205 | attBϕBT1::Pdyn-mcherry-dynA-dynB-ypet | pSS192 integrated at the ΦBT1 attachment site of S. venezuelae WT | This work |
| SS206 | attBϕC31::PftsZ-ftsZ-mturquoise2 attBϕBT1::Pdyn-mcherry-dynA-dynB-ypet | pSS192 integrated at the ΦBT1 attachment site of SS39 | This work |
| SS208 | attBϕC31::PftsZ-ftsZ-ypet attBϕBT1::PsepF-sepf-mcherry | pSS196 integrated at the ΦBT1 attachment site of LUV015 | This work |
| SS211 | ΔdynAB::apr attB ϕBT1::PermE*-dynA[K74A]-dynB-ypet | pSS208 integrated at the ΦBT1 attachment site of LUV001 | This work |
| SS213 | attBϕC31::PftsZ-ftsZ-ypet attBϕBT1::PsepF2-mcherry-sepf2 | pSS234 integrated at the ΦBT1 attachment site of LUV015 | This work |
| SS224 | attB ϕBT1::PermE*–ypet-dynA[K74A]-dynB[K129A] | pSS317 integrated at the ΦBT1 attachment site of WT S. venzuelae | This work |
| SS238 | ΔftsZ::apr attBϕBT1::PermE*-dynA-dynB-ypet | pSS134 integrated at the ΦBT1 attachment site of S. venezuelae ΔftsZ::apr | This work |
| SS242 | ΔdynAB::apr attB ϕBT1::PermE*-ypet-dynA-dynB | pSS310 integrated at the ΦBT1 attachment site of LUV001 | This work |
| SS244 | ΔdynAB::apr attB ϕBT1::PermE*-ypet-dynA-dynB[K129A] | pSS311 integrated at the ΦBT1 attachment site of LUV001 | This work |
| SS248 | ΔdynAB::apr attB ϕBT1::PermE*–ypet-dynA[K74A]-dynB | pSS316 integrated at the ΦBT1 attachment site of LUV001 | This work |
| SS250 | ΔdynAB::apr attB ϕBT1::PermE*–ypet-dynA[K74A]-dynB[K129A] | pSS317 integrated at the ΦBT1 attachment site of LUV001 | This work |
| SS255 | ΔdynA | pSS107 integrated at dynA locus of S. venezuelae WT via double homologous recombination | This work |
| SS262 | attBϕBT1::PermE*-dynA-dynBΔTM-ypet | pSS353 integrated at the ΦBT1 attachment site of S. venezuelae WT | This work |
| SS263 | ΔdynAB::apr attBϕBT1::PermE*-dynA-dynBΔTM-ypet | pSS353 integrated at the ΦBT1 attachment site of LUV001 | This work |
| SS266 | ΔdynAB::apr attBϕBT1::Pdyn-dynA-mcherry | pSS12 integrated at the ΦBT1 attachment site of LUV001 | This work |
| SS270 | ΔdynA attBϕBT1::PermE*-dynB-mcherry | pSS87 integrated at the ΦBT1 attachment site of SS255 | This work |
Table S2.
Oligonucleotides used in this study
| Oligo | Sequence (5′ → 3′) |
| KF843 | GGCGCGCAGGGCCGCAATAGGCTTCAAAAACACTCATAAATTGGGCATTCATTCCGGGGATCCGTCGACC |
| KF844 | GGAAATGTGGACAACTCCCCCACCCGGGAGGGTGAACCGGAGGGGGAGTTGTGTAGGCTGGAGCTGCTTC |
| KF1114 | TGCATTATGCATCTACCTGCCCCGTACCCC |
| KF1115 | TGCATTATGCATCCTGCCCCGTACCCC |
| KF1116 | TGCATTGGTACCGCCGACCGGGGCTGAAAAC |
| KF1117 | GCGGCCGCGCGCGATATC |
| KF1118 | GCTCCCTGCCCGCTATGGTG |
| KF1119 | TGCATTGGATCCTTACTTATAGAGCTCGTTCATGC |
| KF1120 | TGCATTATGCATGGCTCCGGCTCGGGCAGCATGGTGAGCAAAGGCGAAG |
| KF956 | CCACCGGGACCACCACATATGGCCACCGCCCCCCTTCAGGAAGTCCGGGACGT |
| KF743 | GAGGTGCGGCGTCTAGACGGACCGGTTGCTCCAG |
| 82-fw | TATCACCGCAGATGGTTACC |
| 82-rev | GATGTCATCAGCGGTGGAGT |
| Int-fw | CCCGCCGCTGCTTTTGCACGTGAG |
| ftsZ_pIJ10257_FWD | GGGAATTCCATATGGCAGCACCGCAGAACT |
| ftsZ_pIJ10257_REV | CCCAAGCTTTCACTTCAGGAAGTCCGG |
| ssgA-BACTH F | GCTCTAGAGATGAGCTTCCTCGTCTCGGAGG |
| ssgA-BACTH R | CCGGTACCCGGGCGGTCGGGCCCGCG |
| ssgB-BACTH F | CTGAGGATCCCATGAACACCACGGTCAGCTG |
| ssgB-BACTH R | CCGGTACCCGGCTGTCGGCCAGGATGTG |
| 8 | AGTGACGTCGTGGTGGATGTGGACGTGGAAGCGGGCGGGATTCCGGGGATCCGTCGACC |
| 9 | CGAGGCCGGCTACCTGCCCCGTACCCCCGTCCGCGTCGCTGTAGGCTGGAGCTGCTTC |
| 12 | ATTAATTCATATGTGGAGCCACCCGCAGTTCGAAAAGGGTGCAGTGGTGGATGTGGACGTGGAAGCG |
| 13 | ATAGGTACCCTACCTGCCCCGTACCCCCGTCC |
| 19 | ATTAATTCATATGGTGACCTTGGACGAACGGCC |
| 21 | ATTAATTCATATGGTGGATGTGGACGTGGAAGCG |
| 23 | CTAGAAAGCTTAGATTCTCTCATATGATCATCGATTCGCGACTTAAGCCTAGGTGTACATCTCTCGAGTTAATTAAGGTAC |
| 24 | CTTAATTAACTCGAGAGATGTACACCTAGGCTTAAGTCGCGAATCGATGATCATATGAGAGAATCTAAGCTTT |
| 29 | TAAAGCTTGGACTCGAACCTGCGGCCAAGTGC |
| 30 | ATCCTAGGCTTCTCCTTCTGCAGTACGGAGAGCGCG |
| 32 | TTAATTCCTAGGCCTGCCCCGTACCCCCGTCCGC |
| 63 | AATTAAAGCTTGATGGTGACCTTGGACGAACGGCC |
| 64 | TTAAGGATCCTCCTTCTCCTTCTGCAGTACGGAGAGCG |
| 65 | TTAAGGATCCCATGGTGACCTTGGACGAACGGCC |
| 66 | AATGAATTCTCACTTCTCCTTCTGCAGTACGGAGAGC |
| 67 | AATTAAAGCTTGATGGTGGATGTGGACGTGGAAGCG |
| 68 | AATTAAGGATCCCTCCTGCCCCGTACCCCCGTCCG |
| 69 | AATTAAGGATCCCATGGTGGATGTGGACGTGGAAGCG |
| 70 | AATTAATGAATTCCTACCTGCCCCGTACCCCCGTC |
| 71 | TCCACCGGAGCCGGCGCCTCCACGCTCGTCAAC |
| 72 | GTTGACGAGCGTGGAGGCGCCGGCTCCGGTGGA |
| 73 | GCCACCGGCAGCGGCGCCTCCACCCTCTTCAAC |
| 74 | GTTGAAGAGGGTGGAGGCGCCGCTGCCGGTGGC |
| 98 | TTAATTCCTAGGTCACTTCTCCTTCTGCAGTACGGAGAGC |
| 105 | ATCGGGGTACCGTGGTGGATGTGGACGTGGAAGCG |
| 106 | ATCCGCTCGAGCCTGCCCCGTACCCCCGTCCGCG |
| 107 | ATCGGGGTACCGTGGTGACCTTGGACGAACGGCC |
| 108 | ATCCGCTCGAGCTTCTCCTTCTGCAGTACGGAGAGCGC |
| 112 | TTAATTAACATATGGTCTCCAAGGGCGAGGAGCTG |
| 113 | TTAATTAGATCTTCACTTGTACAGCTCGTCCATGCCC |
| 159 | GACCTCTATTCACAGGGTACGGGCCGG |
| 160 | CCACCGCTGGATCCTACCAACCGGCACGATTGTG |
| 161 | ATCCAGCGGTGGTGACCTTGGACGAACGGCCTC |
| 162 | GGAGTTCGTCGGCGGTGCTGTCG |
| 179 | TATAAAGCTTTCAAGCTCCCGAACGGCAACCTCC |
| 182 | ATAGGTACCGGACGGGGAGGGAAATGTGGACAACTC |
| 191 | ATATAACTAGTGTCGATCAGCTGAGGCCGTTCGTCC |
| 192 | ATATAACTAGTATCGCCGCGCTCTCCGTACTGCAG |
| 270 | TTAATTAACATATGTCCCGCCTGAAGCCGCGCTGCACCTCCCTGGAGATGGTCTCCAAGGGCGAGGAGCTG |
| 271 | ATATACTCGAGCTCCGGGCCCGGCAGCTCCGGGCCCGGCAGCTTGTACAGCTCGTTCATGCCCTCGG |
| 304 | ATATAAGCTTGCCGACCGGGGCTGAAAACTTC |
| 305 | GGAGACCACATGCGAAGAATGCCCAATTTATGAGTG |
| 308 | TCTTCGCATGTGTCCAAGGGCGAGGAGCTGTTCACC |
| 309 | ATATACTCGAGCTCCGGGCCCGGCAGC |
| 310 | ATATACTCGAGGTGGTGACCTTGGACGAACGGCC |
| 311 | ATAGGTACCTCACTTCTCCTTCTGCAGTACGGAGAGCG |
| 332 | ATATATGCATGGCTCCGGCTCGGGCAGCATGGTCTCCAAGGGCGAGGAGGAC |
| 333 | ATATATGCATGGCTCCGGCTCGGGCAGCTCACTTGTACAGCTCGTCCATGCCGCCG |
| 336 | ATTAAAGCTTAGCTCGGCTGCCTCCTGCAGGTC |
| 337 | TTAATTCCTAGGGCTCTGGTTGAAGAATCCGCCCTCTG |
| 338 | ATTAAAGCTTGCTACTGGAGCGGCCTCGCGCAC |
| 395 | GATGGCCCCATGGTCTCCAAGGGCGAGGAGGACAAC |
| 396 | ATATACTCGAGATGAGTAGGTACGAGAGGTACGACGTCAC |
| 397 | AATTAAGATATCCTACGATCGAGGGACCCTGGCC |
| 398 | TTAAGGATCCCATGGGATCGGTGCGTAAGGCGAG |
| 399 | AATTAAGAATTCTCAGCTCTGGTTGAAGAAACCGTCCTG |
| 400 | AATTAAGAATTCGAGCTCTGGTTGAAGAAACCGTCCTGCTG |
| 401 | TTAAGGATCCCATGGCCGGCGCGATGCGCAAG |
| 402 | AATTAAGAATTCTCAGCTCTGGTTGAAGAATCCGCCCTC |
| 403 | AATTAAGAATTCGAGCTCTGGTTGAAGAATCCGCCCTCTGC |
| 404 | TTAAGGATCCCATGAGTAGGTACGAGAGGTACGACGTC |
| 405 | AATTAAGAATTCCTACGATCGAGGGACCCTGGC |
| 442 | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGAGTAGGTACGAGAGGTACGACGTCACC |
| 443 | GGGGACCACTTTGTACAAGAAAGCTGGGTGCTACGATCGAGGGACCCTGGCCC |
| 449 | AATTAAGAATTCATGGCAGCACCGCAGAACTACCTCG |
| 450 | TTAATTCTGCAGATCACTTCAGGAAGTCCGGGACGTCC |
| 464 | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGGTGACCTTGGACGAACGGCC |
| 465 | GGGGACCACTTTGTACAAGAAAGCTGGGTGTCACTTCTCCTTCTGCAGTACGGAG |
| 466 | GGGGACCACTTTGTACAAGAAAGCTGGGTGCTTCTCCTTCTGCAGTACGGAGAGC |
| 467 | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGGTGGATGTGGACGTGGAAGCG |
| 473 | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGGCCGGCGCGATGCGCAAG |
| 474 | GGGGACCACTTTGTACAAGAAAGCTGGGTGTCAGCTCTGGTTGAAGAATCCGCC |
| 476 | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGGGATCGGTGCGTAAGGCG |
| 477 | GGGGACCACTTTGTACAAGAAAGCTGGGTGTCAGCTCTGGTTGAAGAAACCGTCC |
| 619 | GCGACGCCCGCACCCGCTGGCGCGCCTACCCCCGCGACAGCACCGC |
| 620 | GCCCGGCCCCTGCCCCGAGCCTTCGAGGGCGAGGACCGCCGCGGGCCAC |
| 621 | CAGGGGCCGGGCTCCGGCCAGGGGAGCGGCGTGAAGGGGCCTGCCAAGCGG |
| 622 | ATGGCGAAAAGCCGAGAACCCTAGGGGATCCTTACTTATAGAGCTCGTTCATG |
The ΔdynAB::apr (LUV001) and the ΔdynB::apr (SS2) mutant strain were generated using the “Redirect” PCR targeting protocol (43, 44). The resulting mutant strains were confirmed by PCR analysis. The markerless ΔdynA (SS255) mutant strain was generated using I-SceI Meganuclease-mediated gene deletion as described by Fernández-Martínez and Bibb (45).
Preparation of S. venezuelae Crude Cell Lysates.
S. venezuelae cells were grown in MYM overnight, harvested by centrifugation at 5,000 × g for 10 min, and washed once in ice-cold 20 mM Tris⋅HCl, pH 8, and 0.5 mM EDTA. Cell extracts were prepared by resuspending the final pellet in one-fourth of the volume in 20 mM Tris⋅HCl, pH 8, and 0.5 mM EDTA with 1× EDTA-free protease inhibitors (Roche). Cells were lysed by sonication and cell debris was removed by centrifugation at 16,000 × g for 20 min at 4 °C. Protein concentration of cell lysates was determined using the Bradford assay (Bio-Rad) and total protein concentration of each sample was adjusted to 10 mg⋅mL−1.
Immunoblot Analysis.
Protein samples were mixed with 5× SDS sample buffer and boiled for 10 min. Samples containing DynB were incubated with 4× LDS buffer (Expedeon) supplemented with 200 mM DTT for 1 h at 37 °C. Proteins were resolved by SDS/PAGE on 12% polyacrylamide gels, electroblotted on a nitrocellulose membrane. Blocked membranes were probed with anti-WhiA (42) (1:2,500), anti-Flag (F4725, 1:10,000; Sigma), anti-FtsZ (46) (1:30,000), and anti-GFP (ab137827, 1:5,000; Abcam) antibodies. Primary antibodies were detected using anti-rabbit IgG conjugated to HRP (1:10,000; GE Healthcare) and blots were developed using the ECL system (GE Healthcare).
Cellular Fractionation.
Soluble and membrane protein fractions were collected by ultracentrifugation. S. venezuelae strains were grown in 30 mL yeast extract-malt extract mixed with tryptic-soy-broth medium (mixed at a 4:6 ratio). Bacteria were harvested by centrifugation at 5,000 × g for 10 min at 4 °C and washed once with 0.2 M Tris⋅HCl. Cell pellets were resuspended in 1/10 volume of lysis buffer (0.2 M Tris⋅HCl, pH 8, 10 mg⋅mL−1 lysozyme, and 1× EDTA-free protease inhibitors; Roche) and incubated for 30 min at 37 °C and then briefly cooled on ice before lysed by sonication. Cell debris was removed by centrifugation at 16,000 × g for 20 min and cleared cell lysate was ultracentrifuged for 1 h at 100,000 × g at 4 °C to dissociate the soluble protein fraction (supernatant) and membrane proteins (pellet). The soluble fraction was stored at −80 °C. The membrane pellet was washed once with wash buffer (60 mM Tris⋅HCl, pH 8, 0.2 mM EDTA, and 0.2 M sucrose) and sedimented at 100,000 × g at 4 °C for 1 h. The final pellet was dissolved in 1/10 of the initial volume with wash buffer and analyzed by immunoblotting. Experiments were performed in duplicate.
Two-Hybrid Analysis.
Competent E. coli strain BTH101 were transformed with two 100-ng aliquots of “T25” and “T18” protein fusion plasmids in one step. Transformants were selected on LB agar containing Carb100 and Kan50. To test for protein–protein interaction, three individual colonies per interaction were grown overnight in LB with antibiotics. The resulting cultures were spotted (4 μL) onto LB agar containing Carb100, Kan50, 500 μg⋅mL−1 isopropyl β-d-1-thiogalactopyranoside, and 40 μg⋅mL−1 Xgal. Plates were incubated in the dark at 30 °C for 1 d and imaged. Images were processed in Adobe Photoshop CS6. Assays of β-galactosidase activity were performed in triplicates as described by Griffith and Wolf (47) and Slavny et al. (48). Experiments were performed in biological triplicates and technical duplicates and Miller Units were calculated using Graph Prism (version 5.04).
The yeast two-hybrid assays were performed in strain AH109 (Clontech). AH109 was transformed with 100-ng aliquots of bait and prey protein fusion plasmids in one step using the cotransformation technique (49). Transformants were selected on selective yeast synthetic dropout (YSD) medium, lacking leucine and tryptophan (−LW). Single colonies from each transformation plate were resuspended in 100 μL sterile water and 5 μL of each strain was spotted on YSD agar, lacking leucine, tryptophan, adenine, and histidine (−LWAH) to screen for protein interactions and on YSD agar (−L/W) agar to verify growth. Plates were incubated for 4–7 d at 30 °C before growth was analyzed and plates were scanned. Each interaction was tested in biological triplicates.
Electron Microscopy.
Cryo-scanning electron microscopy and TEM were performed as previously described (42, 50).
Widefield Microscopy and Image Analysis.
All images were acquired using a Zeiss Axio Observer Z.1 inverted epifluorescence microscope, using either a Zeiss Alpha Plan-Apo 100×/1.46 Oil DIC M27 or a Plan Apochromat 100×/1.4 Oil Ph3 objective. Snapshots of fluorescent protein localization were taken of cells grown in liquid MYM or from coverslip impression of cells grown on solid MYM medium. For liquid cell samples, 2 μL of an overnight culture was spotted on top of a thin agarose pad on a microscope slide. For imaging of sporulating areal hyphae, a coverslip was placed on the surface of a colony grown on MYM agar for 2 d (fluorescent protein localization) or for 4 d (phenotypic characterization). The coverslip was then moved on top of a thin agarose pad. Fluorescent time-lapse imaging was essentially performed as described in Schlimpert et al. (29). Spores were loaded into B04A microfluidic plates (ONIX; CellASIC) and allowed to germinate and grow by perfusing MYM for 3 h. Sporulation was induced by switching the media flow channel and incubating the growing hyphae in spent MYM, which was derived by filter-sterilizing the growth medium from a sporulating culture. Throughout the experiment, the media flow rate and temperature was maintained at 2 psi and 30 °C. Time-lapse imaging was started 9 h after spores had germinated and images were acquired every 8 min until sporulation was completed. Images, kymographs, and movies were generated in ImageJ. For kymograph analysis, a time window of 40 frames (320 min) was chosen, starting with the cessation of tip extension and a concomitant increase in FtsZ-YPet fluorescence. Selected hyphae were “straightened” in ImageJ and FtsZ-YPet fluorescence was plotted along the length of the hyphae over time using a manually drawn line (width 5).
Spore Size Measurements.
A single colony of sporulating S. venezuelae was spread onto MYM agar to grow into a lawn and the plate was incubated for 3–4 d at 30 °C. Green-pigmented spores were washed off the agar using 20% glycerol and a sterile cotton pad through which spores were collected using a sterile 2-mL syringe. A small aliquot of each spore suspension (1.5 μL) was mounted on a microscope slide on top of a thin agarose pad (1% agarose dissolved in water) and imaged by phase-contrast microscopy. Spore lengths were determined manually using the ZenBlue software (Zeiss) or using the ImageJ plugin MicrobeJ (51). Each experiment was performed in triplicate and data were analyzed using GraphPad.
Microarray Transcriptional Profiling.
Microarray transcriptional profiling experiments of S. venezuelae WT and ΔwhiH cells were performed as described in Bibb et al. (52) and results have been deposited in the ArrayExpress database (accession no. E-MTAB-5853).
Supplementary Material
Acknowledgments
We thank Joe McCormick for the gift of the ftsZ null mutant and the anti-FtsZ antiserum, Matt Bush for sharing bacterial two-hybrid plasmids, Sara Simonini for advice and materials for yeast two-hybrid analysis, Georgia Squyres for helpful discussions, and Grant Calder for technical assistance with the microscope. This work was supported by a Leopoldina Postdoctoral Fellowship (to S.S.), Biotechnology and Biological Sciences Research Council (BBSRC) Grants BB/P001041/1 (to S.S. and M. J. Buttner) and BB/L019825/1 (to M. J. Buttner), BBSRC Institute Strategic Programme Grant BB/J004561/1 to the John Innes Centre, and by grants from the Swedish Research Council (2010-4463 and 2015-05452) and the Crafoord foundation (to K.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the ArrayExpress database (accession no. E-MTAB-5853).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1704612114/-/DCSupplemental.
References
- 1.Ferguson SM, De Camilli P. Dynamin, a membrane-remodelling GTPase. Nat Rev Mol Cell Biol. 2012;13:75–88. doi: 10.1038/nrm3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Low HH, Sachse C, Amos LA, Löwe J. Structure of a bacterial dynamin-like protein lipid tube provides a mechanism for assembly and membrane curving. Cell. 2009;139:1342–1352. doi: 10.1016/j.cell.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Low HH, Löwe J. A bacterial dynamin-like protein. Nature. 2006;444:766–769. doi: 10.1038/nature05312. [DOI] [PubMed] [Google Scholar]
- 4.Bramkamp M. Structure and function of bacterial dynamin-like proteins. Biol Chem. 2012;393:1203–1214. doi: 10.1515/hsz-2012-0185. [DOI] [PubMed] [Google Scholar]
- 5.Bohuszewicz O, Liu J, Low HH. Membrane remodelling in bacteria. J Struct Biol. 2016;196:3–14. doi: 10.1016/j.jsb.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bürmann F, Ebert N, van Baarle S, Bramkamp M. A bacterial dynamin-like protein mediating nucleotide-independent membrane fusion. Mol Microbiol. 2011;79:1294–1304. doi: 10.1111/j.1365-2958.2011.07523.x. [DOI] [PubMed] [Google Scholar]
- 7.Michie KA, Boysen A, Low HH, Møller-Jensen J, Löwe J. LeoA, B and C from enterotoxigenic Escherichia coli (ETEC) are bacterial dynamins. PLoS One. 2014;9:e107211. doi: 10.1371/journal.pone.0107211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ozaki S, et al. A replicase clamp-binding dynamin-like protein promotes colocalization of nascent DNA strands and equipartitioning of chromosomes in E. coli. Cell Rep. 2013;4:985–995. doi: 10.1016/j.celrep.2013.07.040. [DOI] [PubMed] [Google Scholar]
- 9.Sawant P, Eissenberger K, Karier L, Mascher T, Bramkamp M. A dynamin-like protein involved in bacterial cell membrane surveillance under environmental stress. Environ Microbiol. 2016;18:2705–2720. doi: 10.1111/1462-2920.13110. [DOI] [PubMed] [Google Scholar]
- 10.Colangeli R, et al. The Mycobacterium tuberculosis iniA gene is essential for activity of an efflux pump that confers drug tolerance to both isoniazid and ethambutol. Mol Microbiol. 2005;55:1829–1840. doi: 10.1111/j.1365-2958.2005.04510.x. [DOI] [PubMed] [Google Scholar]
- 11.Bush MJ, Tschowri N, Schlimpert S, Flärdh K, Buttner MJ. c-di-GMP signalling and the regulation of developmental transitions in streptomycetes. Nat Rev Microbiol. 2015;13:749–760. doi: 10.1038/nrmicro3546. [DOI] [PubMed] [Google Scholar]
- 12.Bi EF, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 13.Löwe J, Amos LA. Crystal structure of the bacterial cell-division protein FtsZ. Nature. 1998;391:203–206. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- 14.Haeusser DP, Margolin W. Splitsville: Structural and functional insights into the dynamic bacterial Z ring. Nat Rev Microbiol. 2016;14:305–319. doi: 10.1038/nrmicro.2016.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao J, Goley ED. Redefining the roles of the FtsZ-ring in bacterial cytokinesis. Curr Opin Microbiol. 2016;34:90–96. doi: 10.1016/j.mib.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willemse J, Borst JW, de Waal E, Bisseling T, van Wezel GP. Positive control of cell division: FtsZ is recruited by SsgB during sporulation of Streptomyces. Genes Dev. 2011;25:89–99. doi: 10.1101/gad.600211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu LJ, Errington J. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell. 2004;117:915–925. doi: 10.1016/j.cell.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 18.Bernhardt TG, de Boer PA. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol Cell. 2005;18:555–564. doi: 10.1016/j.molcel.2005.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Boer PA, Crossley RE, Rothfield LI. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell. 1989;56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- 20.Duman R, et al. Structural and genetic analyses reveal the protein SepF as a new membrane anchor for the Z ring. Proc Natl Acad Sci USA. 2013;110:E4601–E4610. doi: 10.1073/pnas.1313978110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szwedziak P, Wang Q, Freund SM, Löwe J. FtsA forms actin-like protofilaments. EMBO J. 2012;31:2249–2260. doi: 10.1038/emboj.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hale CA, de Boer PA. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 23.Gola S, Munder T, Casonato S, Manganelli R, Vicente M. The essential role of SepF in mycobacterial division. Mol Microbiol. 2015;97:560–576. doi: 10.1111/mmi.13050. [DOI] [PubMed] [Google Scholar]
- 24.Gupta S, et al. Essential protein SepF of mycobacteria interacts with FtsZ and MurG to regulate cell growth and division. Microbiology. 2015;161:1627–1638. doi: 10.1099/mic.0.000108. [DOI] [PubMed] [Google Scholar]
- 25.Hale CA, et al. Identification of Escherichia coli ZapC (YcbW) as a component of the division apparatus that binds and bundles FtsZ polymers. J Bacteriol. 2011;193:1393–1404. doi: 10.1128/JB.01245-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Durand-Heredia J, Rivkin E, Fan G, Morales J, Janakiraman A. Identification of ZapD as a cell division factor that promotes the assembly of FtsZ in Escherichia coli. J Bacteriol. 2012;194:3189–3198. doi: 10.1128/JB.00176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Durand-Heredia JM, Yu HH, De Carlo S, Lesser CF, Janakiraman A. Identification and characterization of ZapC, a stabilizer of the FtsZ ring in Escherichia coli. J Bacteriol. 2011;193:1405–1413. doi: 10.1128/JB.01258-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grantcharova N, Lustig U, Flärdh K. Dynamics of FtsZ assembly during sporulation in Streptomyces coelicolor A3(2) J Bacteriol. 2005;187:3227–3237. doi: 10.1128/JB.187.9.3227-3237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schlimpert S, Flärdh K, Buttner MJ. Fluorescence time-lapse imaging of the complete S. venezuelae life cycle using a microfluidic device. J Vis Exp. 2016:53863. doi: 10.3791/53863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flärdh K, Findlay KC, Chater KF. Association of early sporulation genes with suggested developmental decision points in Streptomyces coelicolor A3(2) Microbiology. 1999;145:2229–2243. doi: 10.1099/00221287-145-9-2229. [DOI] [PubMed] [Google Scholar]
- 31.McCormick JR, Su EP, Driks A, Losick R. Growth and viability of Streptomyces coelicolor mutant for the cell division gene ftsZ. Mol Microbiol. 1994;14:243–254. doi: 10.1111/j.1365-2958.1994.tb01285.x. [DOI] [PubMed] [Google Scholar]
- 32.Hamoen LW, Meile JC, de Jong W, Noirot P, Errington J. SepF, a novel FtsZ-interacting protein required for a late step in cell division. Mol Microbiol. 2006;59:989–999. doi: 10.1111/j.1365-2958.2005.04987.x. [DOI] [PubMed] [Google Scholar]
- 33.Gündoğdu ME, et al. Large ring polymers align FtsZ polymers for normal septum formation. EMBO J. 2011;30:617–626. doi: 10.1038/emboj.2010.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konopka CA, Schleede JB, Skop AR, Bednarek SY. Dynamin and cytokinesis. Traffic. 2006;7:239–247. doi: 10.1111/j.1600-0854.2006.00385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyagishima SY, Kuwayama H, Urushihara H, Nakanishi H. Evolutionary linkage between eukaryotic cytokinesis and chloroplast division by dynamin proteins. Proc Natl Acad Sci USA. 2008;105:15202–15207. doi: 10.1073/pnas.0802412105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu X, Verma DP. Phragmoplastin, a dynamin-like protein associated with cell plate formation in plants. EMBO J. 1996;15:695–704. [PMC free article] [PubMed] [Google Scholar]
- 37.Kang BH, Busse JS, Bednarek SY. Members of the Arabidopsis dynamin-like gene family, ADL1, are essential for plant cytokinesis and polarized cell growth. Plant Cell. 2003;15:899–913. doi: 10.1105/tpc.009670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyagishima SY, et al. A plant-specific dynamin-related protein forms a ring at the chloroplast division site. Plant Cell. 2003;15:655–665. doi: 10.1105/tpc.009373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng B, Schwarz H, Jesuthasan S. Furrow-specific endocytosis during cytokinesis of zebrafish blastomeres. Exp Cell Res. 2002;279:14–20. doi: 10.1006/excr.2002.5579. [DOI] [PubMed] [Google Scholar]
- 40.Thompson HM, Skop AR, Euteneuer U, Meyer BJ, McNiven MA. The large GTPase dynamin associates with the spindle midzone and is required for cytokinesis. Curr Biol. 2002;12:2111–2117. doi: 10.1016/s0960-9822(02)01390-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. John Innes Foundation; Norwich, UK: 2000. [Google Scholar]
- 42.Bush MJ, Bibb MJ, Chandra G, Findlay KC, Buttner MJ. Genes required for aerial growth, cell division, and chromosome segregation are targets of WhiA before sporulation in Streptomyces venezuelae. MBio. 2013;4:e00684-e13. doi: 10.1128/mBio.00684-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gust B, et al. Lambda red-mediated genetic manipulation of antibiotic-producing Streptomyces. Adv Appl Microbiol. 2004;54:107–128. doi: 10.1016/S0065-2164(04)54004-2. [DOI] [PubMed] [Google Scholar]
- 45.Fernández-Martínez LT, Bibb MJ. Use of the meganuclease I-SceI of Saccharomyces cerevisiae to select for gene deletions in actinomycetes. Sci Rep. 2014;4:7100. doi: 10.1038/srep07100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwedock J, McCormick JR, Angert ER, Nodwell JR, Losick R. Assembly of the cell division protein FtsZ into ladder-like structures in the aerial hyphae of Streptomyces coelicolor. Mol Microbiol. 1997;25:847–858. doi: 10.1111/j.1365-2958.1997.mmi507.x. [DOI] [PubMed] [Google Scholar]
- 47.Griffith KL, Wolf RE., Jr Systematic mutagenesis of the DNA binding sites for SoxS in the Escherichia coli zwf and fpr promoters: Identifying nucleotides required for DNA binding and transcription activation. Mol Microbiol. 2001;40:1141–1154. doi: 10.1046/j.1365-2958.2001.02456.x. [DOI] [PubMed] [Google Scholar]
- 48.Slavny P, Little R, Salinas P, Clarke TA, Dixon R. Quaternary structure changes in a second Per-Arnt-Sim domain mediate intramolecular redox signal relay in the NifL regulatory protein. Mol Microbiol. 2010;75:61–75. doi: 10.1111/j.1365-2958.2009.06956.x. [DOI] [PubMed] [Google Scholar]
- 49.Egea-Cortines M, Saedler H, Sommer H. Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 1999;18:5370–5379. doi: 10.1093/emboj/18.19.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bush MJ, Chandra G, Bibb MJ, Findlay KC, Buttner MJ. Genome-wide chromatin immunoprecipitation sequencing analysis shows that WhiB is a transcription factor that cocontrols its regulon with WhiA to initiate developmental cell division in Streptomyces. MBio. 2016;7:e00523-e16. doi: 10.1128/mBio.00523-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ducret A, Quardokus EM, Brun YV. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat Microbiol. 2016;1:16077. doi: 10.1038/nmicrobiol.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bibb MJ, Domonkos A, Chandra G, Buttner MJ. Expression of the chaplin and rodlin hydrophobic sheath proteins in Streptomyces venezuelae is controlled by σ(BldN) and a cognate anti-sigma factor, RsbN. Mol Microbiol. 2012;84:1033–1049. doi: 10.1111/j.1365-2958.2012.08070.x. [DOI] [PubMed] [Google Scholar]
- 53.Paget MS, Chamberlin L, Atrih A, Foster SJ, Buttner MJ. Evidence that the extracytoplasmic function sigma factor sigmaE is required for normal cell wall structure in Streptomyces coelicolor A3(2) J Bacteriol. 1999;181:204–211. doi: 10.1128/jb.181.1.204-211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.