Significance
Type 1 diabetes (T1D) is a major autoimmune disease with increasing incidence in recent years. In this study, we found that the intestinal viromes of cases were less diverse than those of controls. We identified eukaryotic viruses and bacteriophage contigs that are associated with the presence or absence of autoimmunity. These viruses provide targets for future mechanistic studies to differentiate causal and incidental associations between the virome and protection against the development of T1D.
Keywords: virome, microbiome, Circoviridae, type 1 diabetes, bacteriophages
Abstract
Viruses have long been considered potential triggers of autoimmune diseases. Here we defined the intestinal virome from birth to the development of autoimmunity in children at risk for type 1 diabetes (T1D). A total of 220 virus-enriched preparations from serially collected fecal samples from 11 children (cases) who developed serum autoantibodies associated with T1D (of whom five developed clinical T1D) were compared with samples from controls. Intestinal viromes of case subjects were less diverse than those of controls. Among eukaryotic viruses, we identified significant enrichment of Circoviridae-related sequences in samples from controls in comparison with cases. Enterovirus, kobuvirus, parechovirus, parvovirus, and rotavirus sequences were frequently detected but were not associated with autoimmunity. For bacteriophages, we found higher Shannon diversity and richness in controls compared with cases and observed that changes in the intestinal virome over time differed between cases and controls. Using Random Forests analysis, we identified disease-associated viral bacteriophage contigs after subtraction of age-associated contigs. These disease-associated contigs were statistically linked to specific components of the bacterial microbiome. Thus, changes in the intestinal virome preceded autoimmunity in this cohort. Specific components of the virome were both directly and inversely associated with the development of human autoimmune disease.
The microbiome contains multiple types of organisms, including bacteria, archaea, fungi, and eukaryotic organisms, as well as the viruses that infect both the host and other members of the microbiome (1). Changes in various components of the microbiome have been observed in association with multiple human diseases (1–4). Prominent among diseases that are influenced by microorganisms is type 1 diabetes (T1D), a disease involving an autoimmune attack on the insulin-secreting beta cells of the pancreatic islets of Langerhans. Changes in intestinal bacteria have been linked to the development of T1D in some studies (5, 6). Some common observations include increased abundance of the Bacteroides genus (7–9) and reduced abundance of butyrate-producing bacteria (7, 10). In a study that included 11 seropositive children from Finland and Estonia, of whom 4 progressed to T1D, Kostic et al. demonstrated a reduction in bacterial diversity among the cases (11). These changes emerged after the appearance of autoantibodies that are predictive of T1D (11), potentially suggesting that intestinal bacteria might be involved in the progression from beta-cell autoimmunity to clinical disease rather than in the initiation of the disease process (6).
Changes in intestinal viruses have been observed in patients with inflammatory bowel disease and AIDS (2–4, 12), indicating that changes in the virome can be associated with infectious and inflammatory diseases. In some cases, these changes involve eukaryotic viruses, whereas in other cases, disease-associated changes in bacteriophages have been identified. Studies in mice clearly document that virus infection can trigger damage to the islets (13–16). Furthermore, intestinal eukaryotic viruses have been implicated in triggering human T1D, based mostly on seroepidemiologic studies of responses to one or a few candidate viruses (17–19). However, in another study, no changes in the virome were detected for 3–9 mo before the development of autoimmunity (20). In a recent study, CrAssphage, a human bacteriophage, was correlated with Bacteroides dorei, but a direct association of this virus with islet autoimmunity was not observed (21). Importantly, although it generally has been assumed that viruses cause harm by triggering diseases, studies in animal models show that virus infection is not necessarily harmful and can provide symbiotic benefits (22–24), and actually may offer protection from the development of T1D (25, 26). Thus, an evaluation of the virome in children at risk for T1D might have the potential to identify viruses associated with either disease risk or protection from disease. In this study, we defined the RNA and DNA intestinal virome of children before the development of autoimmunity and identified changes in the virome potentially associated with protection against or development of this human autoimmune disease.
Results
Cohort and Sample Sequencing.
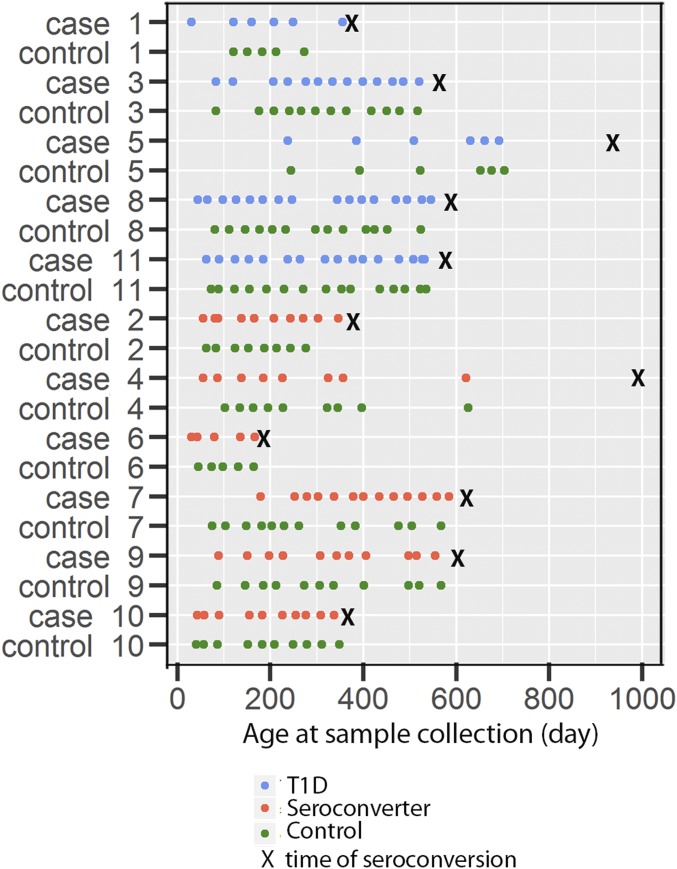
To characterize the intestinal virome and its relationship to T1D disease susceptibility, we studied sequential fecal samples from a well-characterized prospective longitudinal study of infants at risk for T1D (11). Infants from Finland and Estonia were recruited at birth based on their HLA risk genotype. Parents collected infant stools at approximately monthly intervals. The cohort included 11 children who developed serum autoantibodies associated with progression to T1D (defined as positivity for at least two of the five autoantibodies analyzed, referred to as “cases” herein) (11). Of these 11 cases of autoimmunity, 4 developed T1D before the initial publication of the cohort and 1 progressed to T1D at age 77 mo. Therefore, this study involved an analysis of sequential stool samples collected before development of autoimmunity in 11 children. Six of these 11 children exhibited autoantibodies without progression to T1D (the “seroconverter” group), and the other five seroconverted and developed T1D (the “T1D” group) (Fig. 1). These 11 cases were matched with 11 controls for sex, HLA genotype, age, delivery route, and country (Table 1) (11).
Fig. 1.
Cohort and study design. Each point represents a stool sample. “X” represents the time of seroconversion detected.
Table 1.
Patient cohort
| Subject | Subject ID | Country of origin | T1D status | HLA type | Autoantibody positive | Age at SC, d | Age at T1D diagnosis, d | Sex |
| Case 1 | E003251 | Finland | T1D case | DQB1*0302/*0501-DRB1*0401 | IAA, GADA, IA-2A, ZNT8A, ICA | 358 | 1,168 | Female |
| Control 1 | E003061 | Finland | Nonconverter | DQB1*0302/*0502-DRB1*0405 | None | N/A | N/A | Female |
| Case 3 | E006574 | Finland | T1D case | DQB1*0302/*0501-DRB1*0401 | IAA, GADA, IA-2A, ZNT8A, ICA | 533 | 1,340 | Male |
| Control 3 | E006646 | Finland | Nonconverter | DQB1*0302/*0501-DRB1*0401 | None | N/A | N/A | Male |
| Case 5 | E010937 | Finland | T1D case | DQA1*05/*03-DQB1*02/*0302-DRB1*0401 | IAA, IA-2A, ZNT8A, ICA | 905 | 960 | Female |
| Control 5 | E013487 | Finland | Nonconverter | DQA1*05/*03-DQB1*02/*0302-DRB1*0401 | None | N/A | N/A | Female |
| Case 8 | E022137 | Finland | T1D case | DQB1*0302/*0501-DRB1*0401 | IAA, GADA, IA-2A, ZNT8A, ICA | 562 | 2,346 | Male |
| Control 8 | E021406 | Finland | Nonconverter | DQB1*0302/*0302-DRB1*0401/*0401 | None | N/A | N/A | Male |
| Case 11 | T025418 | Estonia | T1D case | DQA1*0201/*03-DQB1*02/*0302-DRB1*0404 | IAA, GADA, IA-2A, ICA | 540 | 880 | Female |
| Control 11 | T025411 | Estonia | Nonconverter | DQB1*0302/*0501-DRB1*0404 | None | N/A | N/A | Female |
| Case 2 | E003989 | Finland | Seroconverter | DQB1*0302/*04-DRB1*0401 | IAA, GADA, ZNT8A, ICA | 347 | N/A | Male |
| Control 2 | E001463 | Finland | Nonconverter | DQB1*0302/*04-DRB1*0401 | None | N/A | N/A | Male |
| Case 4 | E010629 | Finland | Seroconverter | DQB1*0302/*0501-DRB1*0401 | IAA, GADA, ZNT8A, ICA | 945 | N/A | Male |
| Control 4 | E010590 | Finland | Nonconverter | DQB1*0302/*04-DRB1*0401 | None | N/A | N/A | Male |
| Case 6 | E017751 | Finland | Seroconverter | DQA1*05-DQB1*02/*0604 | IAA, ICA | 175 | N/A | Female |
| Control 6 | E016924 | Finland | Nonconverter | DQA1*05-DQB1*02/*04 | None | N/A | N/A | Female |
| Case 7 | E018113 | Finland | Seroconverter | DQB1*0302/*04-DRB1*0401 | IAA, GADA, IA-2A, ZNT8A, ICA | 588 | N/A | Female |
| Control 7 | E018268 | Finland | Nonconverter | DQB1*0302/*0604-DRB1*0404 | None | N/A | N/A | Female |
| Case 9 | E026079 | Finland | Seroconverter | DQB1*0302/*04-DRB1*0401 | IAA, GADA | 580 | N/A | Male |
| Control 9 | E029817 | Finland | Nonconverter | DQB1*0302/*04-DRB1*0404 | None | N/A | N/A | Male |
| Case 10 | T013815 | Estonia | Seroconverter | DQA1*05/*0201-DQB1*02/*02 | IAA, GADA | 350 | N/A | Female |
| Control 10 | T014292 | Estonia | Nonconverter | DQA1*05/*03-DQB1*02/*0301 | None | N/A | N/A | Female |
Delivery route: all subjects had vaginal delivery. N/A, not applicable; SC, seroconversion.
To define longitudinal changes in the intestinal virome, we prepared libraries from DNA and RNA isolated from virus-like particle (VLP) preparations from all fecal samples collected before seroconversion and control samples. Samples were matched for collection time as closely as possible (Fig. 1). Shotgun sequencing was carried out on 220 samples, with a median of 11 samples per individual (range, 5–17) using the paired-end 2 × 250 nt Illumina MiSeq platform (3). We obtained a mean of 1.93 ± 0.92 million sequences per sample, of which a mean of 68.5% ± 14.3% were of high quality (Dataset S1). Deduplicated sequences were analyzed using VirusSeeker (27) to detect bacteriophage and eukaryotic viral sequences, which accounted for 0.04–93.17% of deduplicated sequences. There were no significant differences in the total number of sequences, the number of quality-controlled unique sequences, or the percentage of eukaryotic viral sequences between cases and controls. However, controls had a significantly higher percentage of bacteriophage sequences (P = 0.017, repeated measures ANOVA with permutation test).
Intestinal Virome in Children at Risk for T1D.
We used two complementary approaches to compare intestinal viromes between cases and controls. First, we used read-based analysis to detect bacteriophage and eukaryotic viral sequences based on the best BLAST hit when queried against National Center for Biotechnology Information (NCBI) nucleotide (NT) and protein (NR) databases (27). Viral abundances were normalized to total deduplicated sequences (referred to as “relative abundance” herein) from each sample and used for statistical comparison of viromes.
Because viruses are the fastest-mutating genetic elements on Earth (23), and limitations in the reference viral databases result in the inability to annotate the great majority of sequences present in purified “virus” samples (28, 29), we also performed contig-based analysis. Sequences from each sample were individually assembled to minimize the possibility of chimeric sequence formation (27). The taxonomic identity of each contig was assigned based on the best BLAST hit when queried against the NCBI NT and NR databases (27). Contigs obtained from all samples were pooled, resulting in 10,715 contigs ≥1,000 bp in length, of which 6,550 unique contigs were obtained after deduplication. In total, 3,666 contigs were classified as viral contigs. The longest contig was 108,785 bp, which shared the greatest sequence similarity to bacillus phage BCU4 in the Myoviridae with a genome of 154,371 bp. If the 5′ and 3′ ends of a contig overlapped by at least 10 bases with ≥99% nucleotide identity, then the overlapping sequences were trimmed, and the contig was considered a circular full-length genome. A total of 422 potential complete circular genomes were obtained. The longest of these had a length of 99,507 bp after trimming and shared the highest sequence similarity with Staphylococcus phage pSco-10 in the Siphoviridae with a genome of 101,986 nt. The results suggest that our VLP preparation and sequencing enabled us to obtain abundant viral sequences and complete viral genomes in the stool samples.
A reference contig database was created consisting of the 6,550 unique contigs, the four full-length CRESS DNA virus (circular, rep-encoding single-stranded DNA virus) (30, 31) genomes obtained (see below), and crAssphage (32), a prevalent bacteriophage whose full-length genome was not in the version of the NCBI database downloaded. Deduplicated individual sequencing reads from each sample were aligned to the contig database using FR-HIT (33). A mean of 93.2 ± 13.0% of the deduplicated sequences per sample were mapped to these contigs (sequence identity ≥95% over the length of an individual sequence). A normalized matrix of the number of reads per kbp of contig sequence per million raw reads per sample (RPKM) of all the viral contigs was used to create a viral contig abundance matrix. This viral contig abundance matrix was used to measure α and β diversity of the virome (34) and to perform Random Forests analysis.
We detected 178 genera (145 genera of bacteriophage and 33 genera of eukaryotic virus) out of the 609 total viral genera defined by the International Committee on Taxonomy of Viruses (2015 release). Nearly full-length genomes were obtained for viruses related to six eukaryotic viral families: Anelloviridae, Circoviridae, Bocaviridae, Picobirnaviridae, Picornaviridae, and Reoviridae (Dataset S2). Nine nearly full-length picornavirus genomes with high sequence identity to human parechovirus and human coxsackievirus A4 genomes were identified in seven different subjects (Dataset S2). In subject case 3 (E006574), nearly full-length sequences of 10 of the 11 dsRNA segments of a rotavirus genome most closely related to rotavirus A were obtained (Dataset S2). Although the presence of these sequences does not prove infection with the viruses detected, the children were exposed to a broad range of eukaryotic viruses (12, 23).
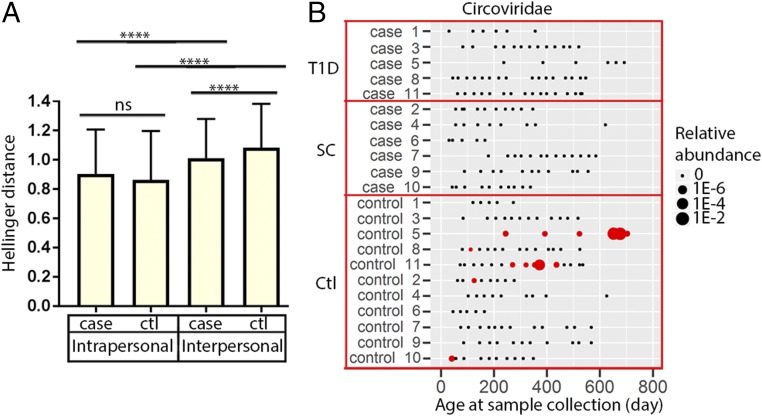
We measured virome β diversity using the Hellinger distance metric on the RPKM matrix (34). Distances were computed between fecal viromes sampled from a given individual over time (intrapersonal variation). Additional comparisons were made between individuals (interpersonal variation). The greatest similarity between fecal viromes was within an individual over time, as reported previously (34). The intrapersonal variation for both controls and cases was significantly (adjusted P < 0.0001) lower than the interpersonal variation within these two groups (Fig. 2A), and there was no significant difference in this measure between the two groups. However, the interpersonal variation between cases was significantly lower than that of controls (adjusted P < 0.0001), indicating that the virome of cases as a group was less diverse than that of the controls as a group (Fig. 2A).
Fig. 2.
Hellinger distance-based β-diversity measurements of viromes and quantification of Circoviridae viral sequences in fecal samples. (A) Mean ± SD values for pairwise β-diversity measurements are shown for within-individual (intrapersonal) or between individual (interpersonal) distances. Differences between groups were considered statistically significant if P < 0.05 using nonparametric Kruskal–Wallis one-way ANOVA with Dunn’s multiple comparisons test. ns, not significant; ****P ≤ 0.0001. ctl, control. (B) The presence and relative abundance of Circoviridae sequences in controls and cases. SC, seroconverters, subjects who exhibited autoantibodies without progression to T1D. T1D, subjects who seroconverted and developed T1D.
Changes in the Eukaryotic Virome Suggest Protective Effects Against Autoimmunity.
We next analyzed the relationships between sequences from specific types of viruses and the development of autoimmunity. We first compared the relative abundance of each eukaryotic virus taxon between controls and cases using repeated measures ANOVA with permutation test (Table 2). We found a higher relative abundance of sequences associated with Circoviridae in control subjects compared with cases (P = 0.026) (Table 2 and Fig. 2B). When we compared the relative abundance of these sequences in control subjects with 6 seroconverter subjects and 5 T1D subjects as separate subgroups, the differences failed to reach significance (Table 2), perhaps owing to small sample size. Importantly, Circoviridae-related sequences were detected only in the control subjects (5 of 11; 45.5%) but not cases (0 of 11; P = 0.035, Fisher’s exact test) (Fig. 2B). However, not all control subjects exhibited Circoviridae-related sequences, and so the association with the absence of development of autoimmunity was not absolute. Circoviridae-related sequences were detected in multiple samples from two subjects who appeared chronically infected. Both the repeated measures ANOVA with permutation test, which accounts for the dependency of samples within a subject, and Fisher’s exact test by subject detected a significant association between Circoviridae-related sequences and controls.
Table 2.
Comparison of relative abundance of viral sequences between controls and cases
| Virus taxonomy | Control vs. case* (P value) | Control, seroconverter vs. T1D* (P value) |
| Circoviridae | 0.026 | NS (0.167) |
| Anelloviridae | NS (0.509) | NS (0.749) |
| Picobirnaviridae | NS (0.062) | NS (0.191) |
| Picornaviridae | NS (0.811) | NS (0.669) |
| Enterovirus | NS (0.259) | NA |
| Kobuvirus | NS (0.321) | NA |
| Parechovirus | NS (0.250) | NA |
| Parvoviridae | NS (0.619) | NS (0.392) |
| Reoviridae | NS (1.000) | NS (0.739) |
NA, not analyzed; NS, not significant.
Repeated measures ANOVA with permutation test.
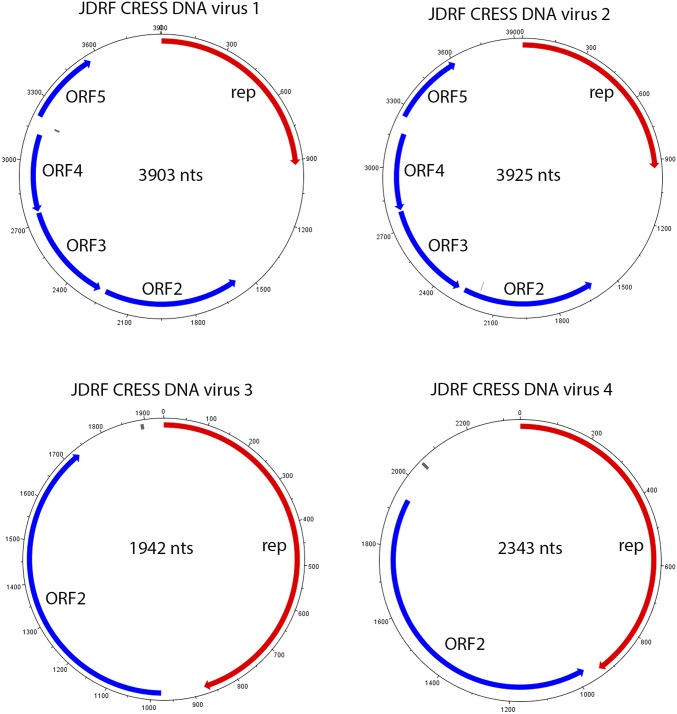
To further define the Circoviridae-related sequences in control subjects, we performed multiple displacement amplification (MDA) of the fecal samples in which these sequences were detected, followed by next-generation sequencing. Sequence assembly revealed six full-length circular genomes. Four of these genomes encoded a replication-associated protein (Rep) and are referred as rep-encoding single-stranded DNA virus (CRESS DNA virus 1–4) according to the current nomenclature (30) (Dataset S2). The other two genomes did not encode an apparent Rep protein. Without the ability to culture these viruses and resequence, we were concerned that they might be an artifact from MDA and did not analyze them further. Overlapping PCR amplification and Sanger sequencing confirmed the full-length circular genomes of CRESS DNA viruses 1–4. These genomes shared 86.8–92.3% nucleotide identity. The novel CRESS DNA viruses were most closely related to porcine circo-like viruses 21 and 22 (CRESS DNA virus 1 and 2 shared 37–44% amino acid sequence identity), bat circovirus (CRESS DNA virus 3 shared 36% amino acid identity), and Farfantepenaeus duorarum circovirus (CRESS DNA virus 4 shared 34% amino acid sequence identity) (Dataset S2). Further analysis revealed three different genome structures for the CRESS DNA viruses. Viruses 1 and 2 had genomes of ∼3.9 kb and encoded 5 ORFs (Fig. S1). Both the genome size and the number as well as the organization of encoded ORFs were similar to those of the viruses of recently proposed Kirkoviridae (35); however, in contrast to viruses in this proposed family, ORF5 was in the same orientation as the Rep-encoding ORF in CRESS DNA viruses 1 and 2. Virus 3 had a genome size (1.9 kb) similar to the Circoviridae; however, the two ORFs encoded by this virus were in the same direction (Fig. S1) instead of in opposite directions as observed for the Circoviridae (31). Virus 4 was similar to circoviruses (30, 31) in terms of both genome size (2.3 kb) and genome structure (Fig. S1). Overall, the low level of similarity to even the most closely related circoviruses strongly suggests that these viruses are novel.
Fig. S1.
Genome structure of CRESS DNA viruses. Full-length genomes of CRESS DNA virus 1, 2, 3, and 4. rep, replication-associated protein.
We next compared the abundance of each of the CRESS DNA viruses between cases and controls using a contig-based method. Using repeated measures ANOVA with permutation test, we detected a significantly higher abundance of CRESS DNA virus 1 in control subjects compared with cases (P = 0.008).
Eukaryotic Viruses Not Associated with Risk of Autoimmunity.
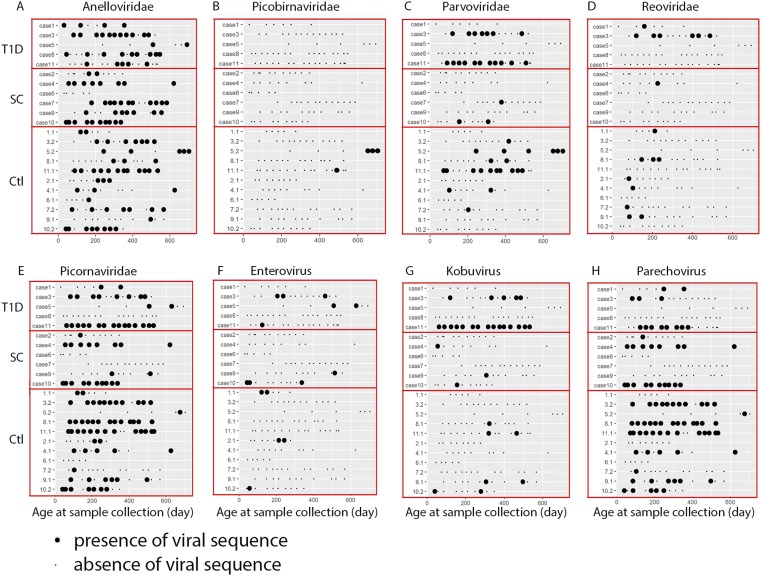
Studies of eukaryotic viruses in T1D have suggested predisposing and disease-triggering effects. Such effects have been suggested for viruses in the Enterovirus and Parechovirus genera of the Picornaviridae (36–38), cytomegalovirus (39), mumps virus (40), parvovirus (41), rubella (42, 43), and rotavirus (44). Although we did not observe any sequences originating from cytomegalovirus, rubella virus, or mumps virus, we detected a high prevalence of Picornaviridae sequences, including sequences of enteroviruses, kobuviruses, and parechoviruses (Fig. S2 E, F, G, and H). We assembled eight nearly full-length genomic sequences of human parechoviruses and one virus most closely related to human coxsackievirus A4 (Dataset S2). At the family level, Picornaviridae sequences were detected in the majority of subjects, and there was no statistically significant difference in the abundance or prevalence of Picornaviridae sequences between cases and controls (Table 2 and Fig. S2E). At the genus level, no significant differences in virus abundance or prevalence were detected between cases and controls for enteroviruses, kobuviruses, and parechoviruses (Table 2 and Fig. S2 F, G, and H).
Fig. S2.
Eukaryotic virus sequences in fecal samples. The presence of eukaryotic virus sequences in controls and cases. (A) Anelloviridae. (B) Picobirnaviridae. (C) Parvoviridae. (D) Reoviridae. (E) Picornaviridae. (F) Enterovirus. (G) Kobuvirus. (H) Parechovirus. T1D, T1D cases; SC, seroconverters; Ctl, controls.
Parvovirus B19 in the Erythroparvovirus genus is reportedly associated with human T1D (41). We did not observe any sequences from parvovirus B19 or the Erythroparvovirus genus. Instead, we detected human bocavirus 2, human bocavirus 3, and porcine bocavirus in multiple subjects and at different time points (Fig. S2C). Three nearly full-length genomes were obtained from case 3 (human bocavirus 3), case 11 (human bocavirus 2), and control 11 (human bocavirus 3) (Dataset S2). The contigs obtained from case 3 and control 11 were 99.8% identical over 5,261 nt at the nucleotide level. No significant difference in relative abundance or prevalence was detected between cases and controls (Table 2). Rotavirus has been identified as a candidate trigger or exacerbating factor for T1D (44, 45). We detected rotavirus in multiple subjects, including three cases and six controls (Fig. S2D), but found no difference in relative abundance or prevalence between cases and controls (Table 2). Anelloviridae sequences were present in nearly every subject (Fig. S2A). There were no statistically significant differences in the relative abundance and prevalence of Anelloviridae sequences between cases and controls (Table 2).
Changes in Bacteriophages Associated with Development of Auto-immunity.
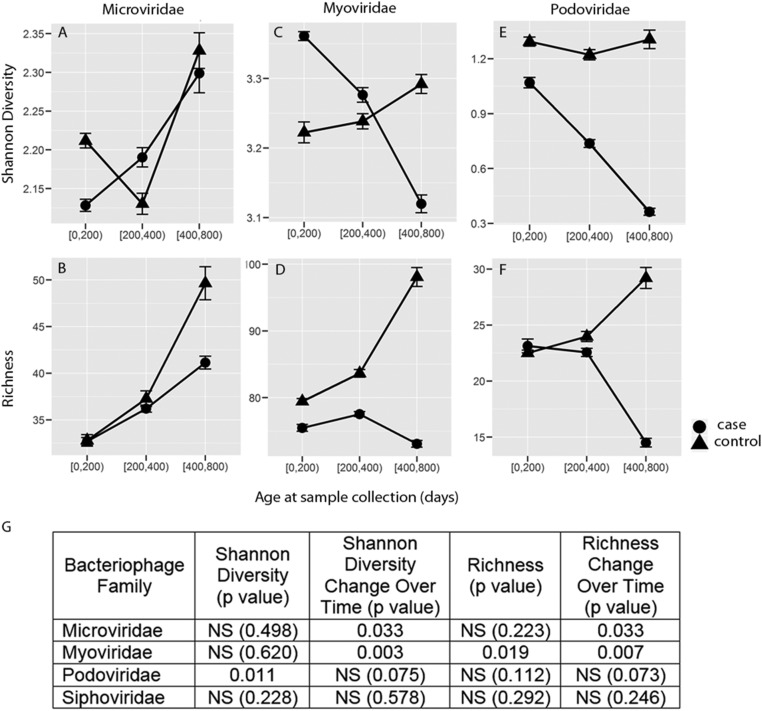
We observed no differences between cases and controls in the relative abundance for Microviridae, Myoviridae, Podoviridae, and Siphoviridae using read-based analysis. However, populations of bacteriophages differed significantly between cases and controls by a number of measures. Controls exhibited significantly higher Shannon diversity for the Podoviridae than cases (P = 0.011; Fig. 3 E and G). In addition, the change of Shannon diversity over time differed between controls and cases, especially for Myoviridae (P = 0.003; Fig. 3 C and G), but also in Microviridae (P = 0.033; Fig. 3 A and G) and Podoviridae (P = 0.075; Fig. 3 E and G).
Fig. 3.
Alterations in viral community composition in subjects who developed autoimmunity. The Shannon diversity (Top) and richness (Bottom) of Microviridae (A and B), Myoviridae (C and D), and Podoviridae (E and F) were compared between cases (n = 11) and controls (n = 11). Siphoviridae were not significantly different between cases and controls and thus are not shown. Viral abundance was measured by the RPKM abundance matrix. Differences between groups were considered statistically significant at P ≤ 0.05 using repeated measures ANOVA with permutation test. (G) Comparison of Shannon diversity and richness of bacteriophages between controls and cases. NS, not significant.
Controls exhibited higher richness than cases for the Myoviridae (P = 0.019; Fig. 3 D and G). Controls and cases had similar levels of richness at younger ages, but the differences between cases and controls increased with increasing age. Interestingly, in controls, bacteriophage richness increased consistently over time, and controls generally exhibited higher richness than cases for all three bacteriophage families for all age groups. Importantly, the changes in richness over time differed between controls and cases, especially for Myoviridae (P = 0.007; Fig. 3 D and G), but also in Microviridae (P = 0.033; Fig. 3 B and G) and Podoviridae (P = 0.073; Fig. 3 F and G).
Overall, these data indicate that bacteriophage populations differed between cases and controls. Interestingly, these differences were observed even though analysis of bacterial populations in the same samples failed to reveal a bacterial signature associated with seroconversion (11).
Age-Discriminatory Viral Contigs.
To identify bacteriophage contigs associated with autoimmunity, we first identified age-discriminative contigs to remove them as a confounding factor. Age affects virome composition during the first 3 y of life (34, 46). Consistent with this, we observed a significant correlation between age and the relative abundance of specific bacteriophages and Shannon diversity, as well as richness, using both reads-based methods and contig-based methods (Table S1).
Table S1.
Spearman correlation between age and the relative abundance, richness, and Shannon diversity of bacteriophages
| Reads-based analysis | Contig-based analysis | ||||
| Bacteriophage family | Relative abundance | Richness | Shannon diversity | Richness | Shannon diversity |
| Microviridae | 0.001 | 2.88e-07 | 3.18e-07 | 0.036 | 0.007 |
| Myoviridae | 0.016 | NS (0.892) | NS (0.062) | NS (0.493) | 0.016 |
| Podoviridae | 0.016 | NS (0.224) | NS (0.471) | NS (0.399) | 0.011 |
| Siphoviridae | 1.00e-04 | NS (0.667) | 2.09e-12 | 2.14e-05 | NS (0.891) |
NS, not significant.
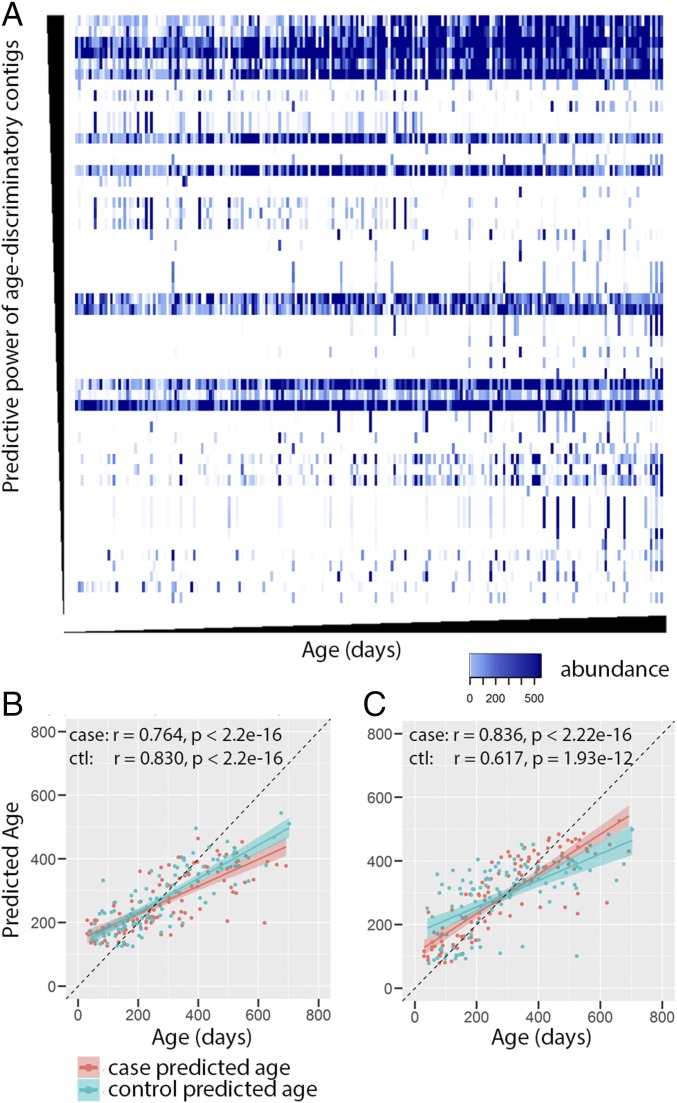
To identify viral contigs whose presence correlated with age, we trained a Random Forests regression model for predicting subject age using contig abundances from the control group. This identified contigs with high feature importance, indicating their importance for accurate prediction of age. Fig. 4A shows a heat map of the 55 age-discriminatory contigs identified using this approach. When used to train a new Random Forests model, these 55 age-discriminatory contigs explained 65.04% of the observed variance in age, compared with 54.98% when using the full set of contigs. An out-of-bag (OOB) estimate of errors, which iteratively uses a training set of the data to predict the behavior of remaining data, provides a rigorous estimate of the generalization error in Random Forests models (47). The correlation coefficient of the Pearson correlation between OOB-predicted age with the chronological age of control subjects was 0.830 (P < 2.2e-16; Fig. 4B), indicating that the 55 age-discriminatory contigs accurately predicted the age of controls. The resulting Random Forests model also successfully predicted the age of cases (r = 0.764, P < 2.2e-16; Fig. 4B), suggesting that cases experience similar developmental virome changes as controls. The prediction accuracy was slightly worse compared with the training data, however (Fig. 4B). Although this difference may be explained by the training data bias, it also suggests the virome of cases diverged from those presented during normal development within these age-discriminatory contigs.
Fig. 4.
Age-discriminatory contigs. (A) Heat map of the abundance of 55 age-discriminatory contigs from controls. Each row represents a contig, and contigs are arranged according to decreasing Random Forests feature importance. Each column represents a sample, and samples are arranged according to increasing chronological age at sample collection. (B) Comparison of OOB-predicted age of controls (green) and predicted age of cases (red) using the age-discriminatory contigs identified from controls. (C) Comparison of predicted age of controls (green) and OOB-predicted age of cases (red) using the age-discriminatory contigs identified from cases.
We then performed the reciprocal analysis using contig abundance information from cases. Sixty-five age-discriminatory contigs were identified in cases. Thirteen of these contigs were shared with the age-discriminative contigs identified in the controls. When used to train a new Random Forests model, these 65 age-discriminatory contigs explained 69.15% of the observed variance in age of cases, compared with 66.51% when using the full set of contigs. As observed for controls, the model accurately predicted the age of cases and controls, with only a small difference in the predictions of cases and controls (Fig. 4C). Here the Pearson correlation coefficient between the predicted age (OOB-predicted age used for case subjects) with the chronological age was 0.836 (P < 2.2e-16) for cases and 0.617 (P = 1.93e-12) for controls (Fig. 4C).
Nonetheless, even though the age-discriminative contigs identified from the training data had the same predictive power for the training data (r = 0.830 for controls compared with r = 0.836 for cases; Fig. 4 B and C), they performed differently for the testing data. The age of controls predicted using age-discriminative contigs identified from cases (r = 0.617, P = 1.93e-12; Fig. 4C) were less accurately predicted than the age of cases predicted using age-discriminative contigs identified from controls (r = 0.764, P < 2.22e-16; Fig. 4B). This suggests that some of the bacteriophage signatures associated with age change in cases may be disease-related rather than age-associated. Therefore, only the 55 contigs identified in controls were considered age-discriminatory contigs and were subtracted from total contigs for the downstream analysis of establishing disease-discriminatory contigs.
Disease-Discriminatory Viral Contigs.
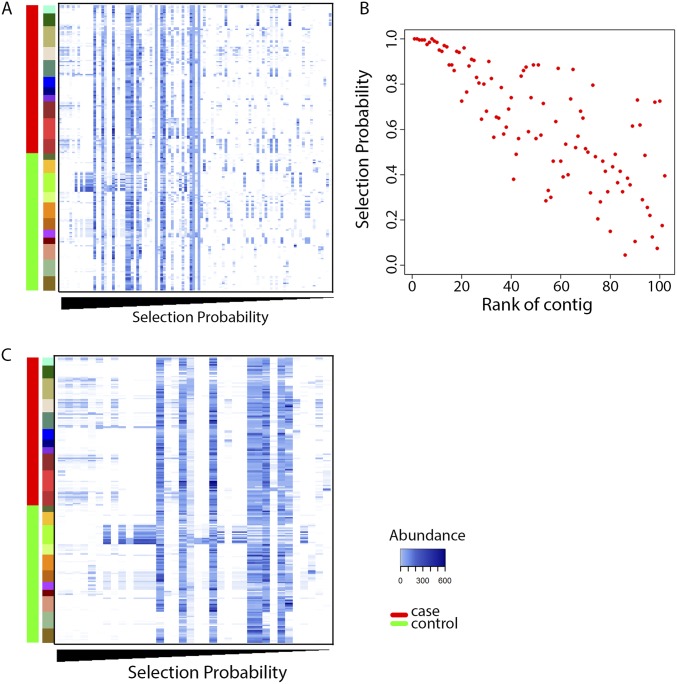
After removing potentially confounding age-discriminatory contigs, we used Random Forests to identify contigs that can distinguish the viromes of cases and controls from within the remaining 3,618 viral contigs. We identified these disease-discriminative contigs using a method that iteratively fits Random Forests models with each iteration building a new forest after discarding contigs with the smallest feature importance score (48). This analysis identified 102 contigs as disease-discriminative (Fig. S3A). We measured the frequency of a contig being selected as the discriminative contig to quantify sampling variability and confidence in discriminative-feature identification (49). Fig. S3B shows the selection probability against the feature importance rank of the top 102 contigs. The best contig was selected as discriminatory 100% of the time, whereas 16 additional contigs were selected in >90% of the bootstrap samples (Fig. S3B). We defined those contigs with a selection probability of ≥80% as the most discriminative contigs for disease status. This identified 36 contigs that were most discriminative for future disease status (Fig. 5), and these were ranked by the selection probability in the bootstrap samples (Dataset S3).
Fig. S3.
Identification of disease-discriminatory viral contigs. (A) Heatmap of the abundance in the controls and cases for the 102 disease-discriminatory contigs. Each column represents a disease-discriminatory contig. Contigs were ordered by decreasing selection probability. (B) Selection probability of the top 102 contigs ranked by feature importance. (C) Heat map of abundance distribution in cases and controls for the 36 most discriminatory contigs for disease status. Each column represents a disease-discriminatory contig. Samples from each subject were clustered together and each color in the second column represented a subject.
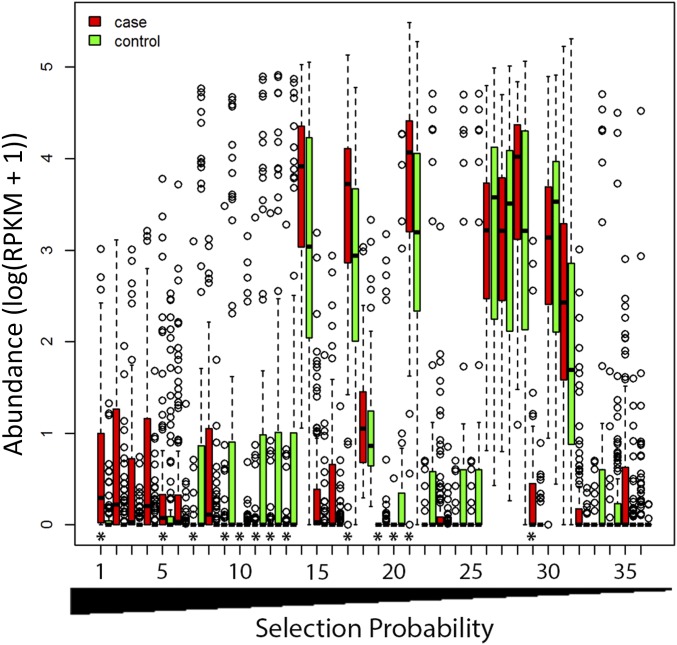
Fig. 5.
Disease-discriminatory viral contigs. Abundances of the 36 most frequently selected contigs. Differences between groups were considered statistically significant at P ≤ 0.05 using repeated measures ANOVA with permutation test. *Indicates that the abundance was significantly different between cases and controls (contig 1 only) or the change over time was significantly different between cases and controls. The x-axis labels the contig number.
We next compared the abundance of these 36 contigs via an independent approach using repeated measures ANOVA with permutation test. The abundance of contig 1 differed significantly between cases and controls. Furthermore, the direction of change in average contig abundance over time differed between cases and controls for 12 contigs (Dataset S3 and Fig. 5). Contig 1, a 65,411-bp complete circular genome, was always selected as a disease-discriminative contig in the bootstrap samples (Dataset S3). Cases had significantly higher abundance of this contig than controls using repeated measures ANOVA with permutation test (P = 0.025; Fig. 5). This contig shared the highest nucleotide sequence similarity to the genome of bacteria B. dorei CL03T12C01 (5,769 of 6,464, 89%; e-value = 0). At the amino acid level, specific ORFs in this contig shared significant sequence similarity with several bacteriophage proteins: phage tail length tape measure protein of B. dorei (805 of 806, 99%; e-value = 0), phage portal protein of Bacteroides vulgatus (490 of 497, 99%; e-value = 0), gp64 of Mycobacterium phage GUmbie (441 of 836, 53%; e-value = 0), as well as the integrase (413 of 413, 100%; e-value = 0) and transposase (404 of 404, 100%; e-value = 0) of multiple bacteriophage proteins of the prophage of Bacteroides.
Contig 5, a 71,649-bp complete circular genome, was selected as disease-discriminative in 98% of the bootstrap samples (Dataset S3). Contig 5 was more abundant in cases than in controls, and the change over time differed significantly between cases and controls (P = 0.018; Fig. 5). This contig shared the greatest nucleotide sequence similarity to the genome of bacteria B. dorei CL03T12C01 (20,546 of 20,823, 99%; e-value = 0). At the amino acid level, specific ORFs in this contig shared significant sequence similarity with several bacteriophage proteins: phage tail length tape measure protein of Bacteroides pyogenes DSM 20611 = JCM 6294 (804 of 1,182, 68%; e-value = 0), phage portal protein of Bacteroides sp. 2_2_4, Bacteroides ovatus and multiple other species (485 of 485, 100%; e-value = 0), phage/plasmid primase of various Bacteroides strains (545 of 545, 100%; e-value = 0), as well as the phage terminase large subunit of Bacteroides sp. 9_1_42FAA (541 of 544, 99%; e-value = 0), integrase (379 of 379, 100%; e-value = 0), and transposase (404 of 404, 100%; e-value = 0) of multiple bacteriophage proteins of prophages of Bacteroides. Even though both contig 1 and contig 5 shared sequence similarity to prophages of Bacteroides, they had very limited sequence similarity to one another (209 of 310, 67% at the nucleotide level and 94 of 155, 61% at the amino acid level). The abundances of contig 1 and contig 5 were significantly correlated (2.50e-13), but the correlation was weak (Pearson correlation coefficient r = 0.467) with some samples containing tens- to hundreds-fold more of one contig than the other, suggesting that contig 1 and contig 5 may represent phages of different bacteria.
Interestingly, we detected one bacteriophage contig (contig 10) that was absent in cases. With a circular complete genome of 97,014 nt and a selection probabilty of 0.945 as disease-discriminative, contig 10 is distantly related to crAssphage, with nucleotide sequence similarity in a 201-bp region. At the amino acid level, several ORFs encoded by contig 10 share 25–34% sequence identity with different proteins of crAssphage.
Altered Relationships Between Bacteria and Bacteriophages in the Cases.
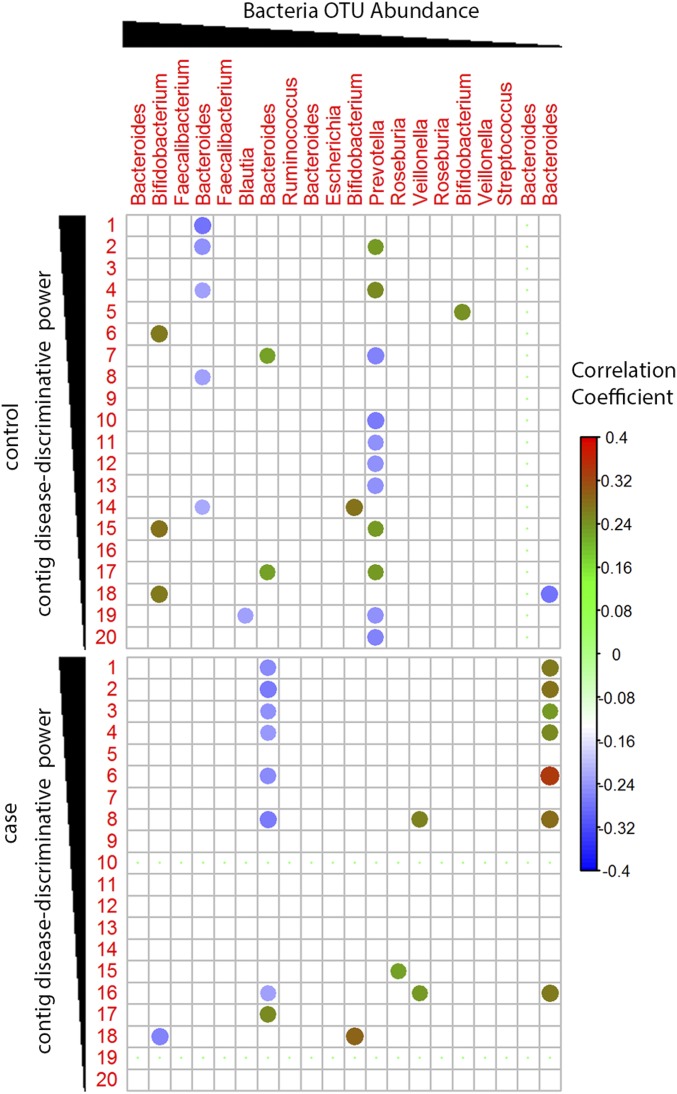
After identifying certain bacteriophage sequences as disease-discriminative, we next characterized the relationships between viruses and bacterial taxa present in the samples. We calculated the Spearman correlation between the disease-discriminatory contigs and the bacterial OTUs from previously published data for the same samples (11). Significantly correlated OTUs were selected if the false discovery rate (FDR)-corrected P was ≤0.1. This analysis detected 27 significant associations between 15 disease-discriminatory contigs and nine unique OTUs from four different families (Dataset S4). The abundance of contig 1 was positively associated with B. ovatus in the Bacteroidaceae family and OTUs of Bifidobacterium genus in the Bifidobacteriaceae family (Dataset S4). Contig 5 was inversely correlated with OTUs of the Roseburia genus in the Lachnospiraceae family and positively associated with Bacteroides fragilis and B. ovatus in the Bacteroidaceae family. Significant correlations of disease-discriminative contigs with different bacteria OTUs, including both positive and negative correlations, were found in controls and cases (Fig. 6), suggesting a distinct virome-bacterial microbiome relationship in controls versus cases even before seroconversion occurred.
Fig. 6.
Statistical relationship between abundance of bacterial OTUs and disease-discriminative viral contigs. Spearman correlation plots of the relative abundances of the top 20 disease-discriminatory contigs and top 20 most abundant bacterial OTUs. Statistical significance was determined for all pairwise correlations. The FDR-corrected P value ≤ 0.1. Color intensity is proportional to correlation coefficient.
Discussion
Here we report a comprehensive analysis of the intestinal eukaryotic and bacteriophage virome in children at risk for the development of T1D, an important human autoimmune disease. This analysis revealed many correlations between the intestinal virome and human autoimmune disease risk. Although the cohort was relatively small (11 cases and 11 controls), the longitudinal collection of samples and the total number of samples analyzed (220) provided sufficient statistical power to detect significant changes in the virome related to autoimmunity. For eukaryotic viruses, we detected a higher prevalence and abundance of Circoviridae-related sequences in controls compared with cases. We frequently detected anellovirus, enterovirus, parechovirus, parvovirus, and reovirus sequences, but these were not statistically associated with autoimmune disease. For bacteriophages, we detected significantly higher virome diversity in controls using multiple approaches. Furthermore, we identified disease-discriminatory bacteriophage contigs whose presence correlated with the abundance of specific bacteria taxa, such as Bacteroides and Bifidobacterium. Taken together, our data reveal substantial and complex relationships between the intestinal virome and the risk of a major human autoimmune disease.
Cases and Controls Share the Same Developmental Change in the Virome.
There is great variation in the gut virome among individuals (28, 34, 46, 50). We found the greatest similarity in fecal viromes within an individual over time as reported previously (28, 34, 46, 50). Intrapersonal variation for both control subjects as a group and cases as a group was significantly lower than the interpersonal variation within these two groups (Fig. 2A). In addition, our findings suggest the presence of a set of viral signatures that were remarkably consistent across individuals and defined the normal development of the virome. The application of Random Forests regression established a set of age-discriminative contigs that were highly predictive of the developmental changes in the intestinal virome. These data also suggest that time-dependent changes in these viruses were similar across biologically unrelated individuals and were related to the developmental stage of the subject.
Changes in the Virome in Subjects at Risk for T1D.
Previous studies found no association between viruses and T1D in stool samples collected 3, 6, and 9 mo before the date of the first islet autoantibody-positive result (20, 21). In the present study, we detected a significantly lower abundance of Circoviridae-related sequences and low virome diversity in cases. The differences between these studies could be related to differences in sampling method, sequencing technique, or viral sequence detection methods (27), or to the smaller cohort of the previous study (96 samples vs. our 220 samples) (20, 21). We found reduced statistical significance when we divided the case subjects into two subgroups, with and without progression to T1D. The fact that the previous study analyzed only a random selection of 100,000 raw sequence reads from each sample also could help explain the discrepant results, given that we analyzed a significantly larger number of sequences from each subject.
Viruses in the Circoviridae family have small circular genomes. This group of viruses includes the circoviruses. Porcine circovirus type 2 has been linked to postweaning multisystemic wasting disease in pigs, and disease onset and symptom severity are influenced by intrinsic factors, such as immune system status and genetic predisposition (51). No circoviruses have yet been linked to human diseases. Our observation of greater abundance and prevalence of Circoviridae-related sequences in controls suggests that infection with the virus offers benefits to the host, as observed in animal models for other viruses (22–24) and might offer protection from the development of autoimmune diabetes, as has been shown in animal studies for other types of viruses (25, 26). However, the sample size in terms of number of subjects is small in this cohort, and these sequences were not detected in every control subject. Caution should be taken in interpreting these results, and future studies including studies in a validation cohort are important for validating this significant observation.
Multiple lines of evidence suggest that over time, the developmental trajectory of viromes differed between children who developed autoimmunity and those who did not. First, the dynamic change in Shannon diversity over time was different between controls and cases for Microviridae, Myoviridae, and Podoviridae. Second, changes in richness over time differed between controls and cases for Microviridae, Myoviridae, and Podoviridae. Third, Random Forests analysis of age-discriminative contigs suggested differences between cases and controls. This is consistent with a model in which cases acquire disease-associated contigs in addition to the normal changes in bacteriophages that occur as children age.
Because the virome changes reported here occurred before seroconversion, our data raise the possibility that virome change could trigger seroconversion. The detection of disease-discriminative contigs provides targets for future mechanistic studies to distinguish between causal and incidental association relationships among bacteriophages and the development of autoimmunity in subjects at risk for T1D.
Relationship Between Bacteriophage Sequences and Bacterial Dysbiosis in Children at Risk for T1D.
No overt changes in the bacterial microbiome community structure before seroconversion have been reported in previous studies, including a study using the same cohort and samples studied here (11, 52). In contrast, pronounced alterations of the intestinal bacterial microbiome occurring after seroconversion and preceding overt T1D have been reported (7, 11, 53). However, an increase of a single species of bacteria, B. dorei, in cases before seroconversion was observed in another study (8). Bacteria in the Bacteroides genus, particularly the B. fragilis group, which includes B. fragilis, B. ovatus, and B. vulgatus, were found to enhance bacterial translocation (54). Bacteroides lipopolysaccharide (LPS) is structurally distinct from Escherichia coli LPS and inhibits innate immune signaling and endotoxin tolerance, suggesting that it may preclude immune development in early infancy (9). Notably, some of the disease-discriminative bacteriophage contigs detected were related to prophages in Bacteroides. It is interesting to speculate that changes in lytic and lysogenic cycles before seroconversion might contribute to the observed virome changes without substantially altering the bacterial microbiome composition. Furthermore, it is possible that changes in the bacterial microbiome observed later in the course of the development of autoimmune disease might be secondary to these earlier changes in bacteriophages.
In the present study, 27 significant associations between disease-discriminate contigs and bacteria OTUs were detected, and the significantly associated bacteria OTUs were clustered in four genera: Bifidobacterium (Bifidobacteriaceae), Bacteroides (Bacteroidaceae), Roseburia (Lachnospiraceae), and Veillonella (Veillonellaceae). Most of the disease-discriminatory contigs were significantly correlated with bacterial OTUs belonging to the Bacteroides genus (B. ovatus, B. vulgates, and B. fragilis; Dataset S4). These taxa were found to be increased in cases in multiple studies (6). In addition, multiple disease-discriminatory contigs were significantly correlated with bacterial OTUs that belonged to the Bifidobacterium, whereas decreases in the relative abundance of Lachnospiraceae and Veillonellaceae were detected in cases after seroconversion in a bacteria microbiome study using the same cohort (11). Bifidobacterium was significantly decreased in children who developed autoimmunity but had not progressed to T1D (7). The levels of Bifidobacterium have been related to improved glucose metabolism, insulin resistance, and low-grade inflammation (55, 56).
Given the relationship between bacteriophages and bacterial hosts, it is conceivable that the changes in bacteriophages before seroconversion modulate bacterial abundance and eventually lead to overt bacteria dysbiosis. Because single nucleotide changes in the bacteriophage genome could change the tropism of the bacteriophage (57), the possible molecular basis for these associations between bacterial OTUs and the disease-discriminatory contigs is unclear; however, it is interesting that our virome study and previous bacteria microbiome study detected signals from the same genera. Importantly, based on our findings, the possibility exists that these changes are regulated by, or the effects are countered by, CRESS DNA viruses. This suggests the hypothesis that the balance between protective and disease causing changes in the virome may modulate human autoimmune disease in genetically susceptible individuals. This would be consistent with the finding that viruses can interact with host disease risk genes to modulate diseases in mice (22, 58, 59). Taken together, these findings in a single cohort suggest a model in which the risk for human autoimmunity (in this case T1D) is related to a combination of protective vs. disease risk-conferring effects of the virome.
Methods
Details of cohort recruitment, sample acquisition, and information collection are provided elsewhere (11) and SI Methods. The study was approved by the local Ethics Committees in Helsinki (228/13/03/03/2008) and in Tartu (172/T-15; 20.08.2008). Informed consent was previously obtained at sample collection for the DIABIMMUNE study. Procedures for sample collection, VLP preparation, shotgun sequencing of VLP-derived DNA, assembly of viral contigs, viral genome confirmation, cross-contig comparisons, calculations of viral α- and β-diversity, Random Forests analysis, bacterial 16S rRNA sequencing data analysis, and statistical analyses are described in detail in SI Methods. Sequences of disease-discriminative contigs are provided as Dataset S5.
SI Methods
Study Cohort.
The children were recruited to the study cohort from Espoo, Finland (n = 18) and Tartu, Estonia (n = 4) between September 2008 and August 2010 as part of the international DIABIMMUNE Study (www.diabimmune.org/). Inclusion criteria included positive cord blood testing for HLA DR-DQ alleles conferring risk of T1D. The participating children were monitored prospectively for infections, use of drugs, (antibiotics in particular), and other life events. Data on breastfeeding and introduction of complementary foods was registered continuously. Serum samples were collected from all infants during visits to the clinic at the following time points: 0 (cord blood), 3, 6, 12, 18, 24, and 36 mo. The following four diabetes-associated autoantibodies were analyzed from each serum sample with specific radiobinding assays: insulin autoantibodies (IAA), glutamic acid decarboxylase antibodies (GADA), islet antigen-2 antibodies (IA-2A), and zinc transporter 8 antibodies (ZnT8A) as described previously (60). Islet cell antibodies (ICA) were analyzed with immunofluoresence in those subjects who tested positive for at least one of the biochemical autoantibodies. The cutoff values were based on the 99th percentile in nondiabetic children and were 2.80 relative units (RU) for IAA, 5.36 RU for GADA, 0.77 RU for IA-2A, and 0.61 RU for ZnT8A, The detection limit in the ICA assay was 2.5 Juvenile Diabetes Foundation units. The parents were asked to collect stool samples from their child once a month starting from the age of 1 mo up to age 36 mo. The stool samples were frozen immediately after sampling and stored in the home freezer until they were brought to the study center for storage at −70 °C. Subject metadata are available in Dataset S1.
Virome Sequencing.
For each sample, 100–200 mg of frozen feces was resuspended in a 6× volume of SM buffer and centrifuged (61). The fecal supernatant was passed through a 0.45-μm-pore-size membrane, and the filtrates were treated with lysozyme and DNase at 37 °C for 1 h. Total RNA and DNA was extracted on a COBAS AmpliPrep instrument (Roche) according to the manufacturer’s recommendations. To evaluate samples for both RNA and DNA viruses, the total nucleic acids were randomly amplified as described previously (61, 62) using barcoded primers consisting of a base-balanced 16-nt–specific sequence upstream of a random 15-mer and used for NEBNext library construction (New England BioLabs). Libraries were multiplexed (12 samples per flow cell) on an Illumina MiSeq instrument at the Washington University Center for Genome Sciences using the paired-end 2 × 250 protocol.
For MDA, total nucleic acid was amplified with Phi29 polymerase (GenomiPhi V2 Kit; GE Healthcare) according to the manufacturer’s instructions and used for Nextera DNA library construction (Illumina). Libraries were multiplexed and sequenced as described above.
Identification and Analysis of Viral-Like Sequences.
VirusSeeker Virome (27) was used to detect sequences sharing nucleotide and protein level sequence similarity to known viruses. VirusSeeker was designed to remove potentially ambiguous or false-positive viral sequences and identify only sequences unambiguously classified as viral sequences. In brief, sequences are adapter-trimmed, joined if read 1 and read 2 of paired-end reads overlap, quality controlled, and dereplicated. Potential unique viral sequences were queried against the NCBI nt/nr databases, and only sequences matching exclusively to viral sequences were retained for further analysis. All sequences aligning to viruses were further classified into viral families based on the NCBI taxonomic identity of the best hit.
Absolute read counts were normalized by dividing individual taxon sequence counts by the total number of deduplicated sequence count in a sample (referred to as “relative abundance”). Richness and diversity were calculated using the diversityresult function of the vegan package. Heatmaps were generated using the gplots R package. Virus presence/absence graphs were generated using the ggplot2 R package.
Assembly, Confirmation, and Annotation of Viral Genomes.
VirusSeeker-Discovery (27) was used to assemble sequences into contigs and to detect contigs sharing nucleotide and protein level sequence similarity to known viruses. In brief, sequences were adapter-trimmed, quality controlled, and dereplicated. Host sequences were filtered by aligning sequences to human reference genomes. For each sample, CD-HIT (63) was used to cluster sequences at 95% identity with 95% overlap of length. The longest sequence from each cluster and the top three sequences were taken as representatives and used for de novo assembly using Newbler v2.8 (454 Life Sciences) and Phrap (www.phrap.org/) in a two-step assembly. Potential unique viral contigs and reads were sequentially queried against the NCBI NT/NR databases, and only sequences matching exclusively to viral sequences were retained for further analysis. All sequences aligning to viruses were further classified into viral families based on the NCBI taxonomic identity of the best hit. The resulting contigs from each sample were pooled and deduplicated using CD-HIT to cluster sequences at 95% sequence identity with 95% overlap. A total of 10,715 contigs ≥1,000 bp in length and 6,550 unique contigs were obtained after deduplication.
Six full length circovirus genomes were obtained by sequence assembly from original sequencing data, as well as sequencing data generated using the MDA amplification method. Overlapping PCR amplification and Sanger sequencing were used to confirm the full-length circular genome of all six viruses. ORFs were predicted and annotated using Artemis (64).
Cross-Contig Comparison.
To analyze viral abundance and diversity within and between individuals, we mapped all high-quality deduplicated sequences from each sample against the assembled contigs using FR-HIT (33) at 90% identity cutoff. The crAssphage (32) genome and the six full-length circovirus-like virus genomes were added to the 6,550 unique contigs, producing the 6,557 final contigs used for the analysis. A RPKM matrix was generated and used for analysis of α and β diversity.
Viral α and β Diversity.
For α diversity analysis, the sequence counts mapping to each contig per sample was normalized to 400,000 sequences per sample based on the average number of deduplicated sequences per sample. Observed species and the Shannon diversity index were calculated for each sample using BiodiversityR R package. To calculate α diversity metrics associated with Microviridae, Myoviridae, Podoviridae, and Siphoviridae only the rows corresponding to contigs assigned to the given taxonomy were selected from the matrix. β diversity analyses were performed using the RPKM abundance matrix; the Hellinger distance matrices were calculated, and GraphPad Prism was used to evaluate within-virome and between-virome sample distances.
Random Forests Analysis.
For age-discriminative contig analysis, Random Forests was used for regression models on the quantitative variable “Age” as implemented in randomForest 4.6–12 R package. For disease-discriminative contig analysis, Random Forests was used as a classifier for the discrete variable “Case_Control status”. A total of 200 bootstraps with 2,000 random trees for each bootstrap dataset were generated. The OOB error rate, feature importance, and prediction from the 200 bootstrap dataset were averaged for the results presented. Feature importance score for a given contig was defined as the mean decrease in accuracy over all classes when the contig was removed from a random tree. The VarSelRF R package (65) was used for feature selection for discrete variables (“Case_Control status”) and the Boruta R package was used for feature selection for quantitative variables (“Age”). Both packages select the minimum number of contigs capable of obtaining the same or better classification accuracy, thereby minimizing the error. In the 200 bootstrap iterations, each with 2,000 trees, the frequency of a contig being selected as discriminatory was calculated (referred to as “selection probability” herein), and the contigs were ranked by the selection probability. The higher the selection probability for a contig, the greater our confidence in that contig’s discriminative properties. We defined those contigs with a selection probability ≥80% as the most discriminative contigs for disease status.
16S rRNA Gene Profiling.
16S rRNA gene sequencing and data have been described previously (11). The Dada2 (66) pipeline was used to generate sequence variants and their sample-wise abundances after removing substitution and chimera errors. The GreenGenes 13.8 release clustered at 97% identity (67) was used for taxonomic classification.
Statistical Analysis.
The aovp function of repeated measures ANOVA with permutation test in the lmPerm R package was used to compare relative abundances between cases and controls. Associations between groups using number of subjects were determined using 2 × 2 contingency tables, and significance was determined with Fisher’s exact test. Spearman rank correlations were calculated, and significance was tested using the rcorr functions in the Hmisc R package. Correlation plot was drawn using the corrplot R package. The p.adjust function in the stats R package was used for FDR P value correction. Univariate and multivariate statistical tests were performed in GraphPad Prism version 6. A P value ≤0.05 was considered significant, except for the FDR-adjusted P value where ≤0.1, was considered significant.
The CRESS DNA virus genomic sequences have been deposited in the NCBI’s GenBank database (accession nos. MF118166–MF118169). Raw data have been deposited in the Sequence Read Archive (accession no. PRJNA387903).
Supplementary Material
Acknowledgments
We thank Brian Koebbe and Eric Martin from the High-Throughput Computing Facility at the Center for Genome Sciences and Systems Biology for providing high-throughput computational resources and support; Chuck Goss at the Division of Biostatistics, Washington University School of Medicine for statistics consultation (supported by UL1 TR000448 for an Institute of Clinical and Translational Sciences subsidized activity); Jessica Hoisington-Lopez from the DNA Sequencing Innovation Lab at the Center for Genome Sciences and Systems Biology for her sequencing expertise; and Dr. Scott A. Handley for his critical comments. This work was supported by Juvenile Diabetes Research Foundation Grant 2-SRA-2015-305-Q-R and National Institutes of Health Grants R01 DK101354, R24 OD019793, and R01 AI111918. T.V., A.D.K., and R.J.X. are supported by National Institutes of Health Grants DK43351, DK92405, and the Juvenile Diabetes Research Foundation. The DIABIMMUNE study was supported by the European Union Seventh Framework Programme (Grant 202063). M.K. was supported by the Academy of Finland (Centre of Excellence in Molecular Systems Immunology and Physiology Research, Grant 250114, 2012-17).
Footnotes
Conflict of interest statement: M.K. serves as an advisor to Vactech Oyj and Provention Bio, Inc.
Data deposition: The CRESS DNA virus genomic sequences have been deposited in the GenBank database (accession nos. MF118166–MF118169). Raw data have been deposited in the Sequence Read Archive (accession no. PRJNA387903).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706359114/-/DCSupplemental.
References
- 1.Stappenbeck TS, Virgin HW. Accounting for reciprocal host-microbiome interactions in experimental science. Nature. 2016;534:191–199. doi: 10.1038/nature18285. [DOI] [PubMed] [Google Scholar]
- 2.Monaco CL, et al. Altered virome and bacterial microbiome in human immunodeficiency virus-associated acquired immunodeficiency syndrome. Cell Host Microbe. 2016;19:311–322. doi: 10.1016/j.chom.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Handley SA, et al. SIV infection-mediated changes in gastrointestinal bacterial microbiome and virome are associated with immunodeficiency and prevented by vaccination. Cell Host Microbe. 2016;19:323–335. doi: 10.1016/j.chom.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Norman JM, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160:447–460. doi: 10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Needell JC, Zipris D. The role of the intestinal microbiome in type 1 diabetes pathogenesis. Curr Diab Rep. 2016;16:89. doi: 10.1007/s11892-016-0781-z. [DOI] [PubMed] [Google Scholar]
- 6.Knip M, Siljander H. The role of the intestinal microbiota in type 1 diabetes mellitus. Nat Rev Endocrinol. 2016;12:154–167. doi: 10.1038/nrendo.2015.218. [DOI] [PubMed] [Google Scholar]
- 7.de Goffau MC, et al. Fecal microbiota composition differs between children with β-cell autoimmunity and those without. Diabetes. 2013;62:1238–1244. doi: 10.2337/db12-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis-Richardson AG, et al. Bacteroides dorei dominates gut microbiome prior to autoimmunity in Finnish children at high risk for type 1 diabetes. Front Microbiol. 2014;5:678. doi: 10.3389/fmicb.2014.00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vatanen T, et al. DIABIMMUNE Study Group Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. 2016;165:842–853. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown CT, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One. 2011;6:e25792. doi: 10.1371/journal.pone.0025792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kostic AD, et al. DIABIMMUNE Study Group The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe. 2015;17:260–273. doi: 10.1016/j.chom.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foxman EF, Iwasaki A. Genome-virome interactions: Examining the role of common viral infections in complex disease. Nat Rev Microbiol. 2011;9:254–264. doi: 10.1038/nrmicro2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Craighead JE, McLane MF. Diabetes mellitus: Induction in mice by encephalomyocarditis virus. Science. 1968;162:913–914. doi: 10.1126/science.162.3856.913. [DOI] [PubMed] [Google Scholar]
- 14.Coleman TJ, Gamble DR, Taylor KW. Diabetes in mice after Coxsackie B 4 virus infection. BMJ. 1973;3:25–27. doi: 10.1136/bmj.3.5870.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christen U, et al. A viral epitope that mimics a self antigen can accelerate but not initiate autoimmune diabetes. J Clin Invest. 2004;114:1290–1298. doi: 10.1172/JCI22557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oldstone MB, Nerenberg M, Southern P, Price J, Lewicki H. Virus infection triggers insulin-dependent diabetes mellitus in a transgenic model: Role of anti-self (virus) immune response. Cell. 1991;65:319–331. doi: 10.1016/0092-8674(91)90165-u. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez-Calvo T, Sabouri S, Anquetil F, von Herrath MG. The viral paradigm in type 1 diabetes: Who are the main suspects? Autoimmun Rev. 2016;15:964–969. doi: 10.1016/j.autrev.2016.07.019. [DOI] [PubMed] [Google Scholar]
- 18.Filippi CM, von Herrath MG. Viral trigger for type 1 diabetes: Pros and cons. Diabetes. 2008;57:2863–2871. doi: 10.2337/db07-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coppieters KT, Boettler T, von Herrath M. Virus infections in type 1 diabetes. Cold Spring Harb Perspect Med. 2012;2:a007682. doi: 10.1101/cshperspect.a007682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kramná L, et al. Gut virome sequencing in children with early islet autoimmunity. Diabetes Care. 2015;38:930–933. doi: 10.2337/dc14-2490. [DOI] [PubMed] [Google Scholar]
- 21.Cinek O, et al. Imbalance of bacteriome profiles within the Finnish Diabetes Prediction and Prevention study: Parallel use of 16S profiling and virome sequencing in stool samples from children with islet autoimmunity and matched controls. Pediatr Diabetes. November 17, 2016 doi: 10.1111/pedi.12468. [DOI] [PubMed] [Google Scholar]
- 22.Barton ES, et al. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature. 2007;447:326–329. doi: 10.1038/nature05762. [DOI] [PubMed] [Google Scholar]
- 23.Virgin HW. The virome in mammalian physiology and disease. Cell. 2014;157:142–150. doi: 10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kernbauer E, Ding Y, Cadwell K. An enteric virus can replace the beneficial function of commensal bacteria. Nature. 2014;516:94–98. doi: 10.1038/nature13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tracy S, et al. Toward testing the hypothesis that group B coxsackieviruses (CVB) trigger insulin-dependent diabetes: Inoculating nonobese diabetic mice with CVB markedly lowers diabetes incidence. J Virol. 2002;76:12097–12111. doi: 10.1128/JVI.76.23.12097-12111.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hermitte L, et al. Paradoxical lessening of autoimmune processes in non-obese diabetic mice after infection with the diabetogenic variant of encephalomyocarditis virus. Eur J Immunol. 1990;20:1297–1303. doi: 10.1002/eji.1830200615. [DOI] [PubMed] [Google Scholar]
- 27.Zhao G, et al. VirusSeeker, a computational pipeline for virus discovery and virome composition analysis. Virology. 2017;503:21–30. doi: 10.1016/j.virol.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reyes A, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mokili JL, Rohwer F, Dutilh BE. Metagenomics and future perspectives in virus discovery. Curr Opin Virol. 2012;2:63–77. doi: 10.1016/j.coviro.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosario K, et al. Revisiting the taxonomy of the family Circoviridae: Establishment of the genus Cyclovirus and removal of the genus Gyrovirus. Arch Virol. 2017;162:1447–1463. doi: 10.1007/s00705-017-3247-y. [DOI] [PubMed] [Google Scholar]
- 31.Rosario K, Duffy S, Breitbart M. A field guide to eukaryotic circular single-stranded DNA viruses: Insights gained from metagenomics. Arch Virol. 2012;157:1851–1871. doi: 10.1007/s00705-012-1391-y. [DOI] [PubMed] [Google Scholar]
- 32.Dutilh BE, et al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat Commun. 2014;5:4498. doi: 10.1038/ncomms5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niu B, Zhu Z, Fu L, Wu S, Li W. FR-HIT, a very fast program to recruit metagenomic reads to homologous reference genomes. Bioinformatics. 2011;27:1704–1705. doi: 10.1093/bioinformatics/btr252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reyes A, et al. Gut DNA viromes of Malawian twins discordant for severe acute malnutrition. Proc Natl Acad Sci USA. 2015;112:11941–11946. doi: 10.1073/pnas.1514285112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li L, et al. Exploring the virome of diseased horses. J Gen Virol. 2015;96:2721–2733. doi: 10.1099/vir.0.000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeung WC, Rawlinson WD, Craig ME. Enterovirus infection and type 1 diabetes mellitus: Systematic review and meta-analysis of observational molecular studies. BMJ. 2011;342:d35. doi: 10.1136/bmj.d35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nilsson AL, et al. Serological evaluation of possible exposure to Ljungan virus and related parechovirus in autoimmune (type 1) diabetes in children. J Med Virol. 2015;87:1130–1140. doi: 10.1002/jmv.24127. [DOI] [PubMed] [Google Scholar]
- 38.Kolehmainen P, et al. Human parechovirus and the risk of type 1 diabetes. J Med Virol. 2013;85:1619–1623. doi: 10.1002/jmv.23659. [DOI] [PubMed] [Google Scholar]
- 39.Pak CY, Eun HM, McArthur RG, Yoon JW. Association of cytomegalovirus infection with autoimmune type 1 diabetes. Lancet. 1988;2:1–4. doi: 10.1016/s0140-6736(88)92941-8. [DOI] [PubMed] [Google Scholar]
- 40.Hyöty H, et al. Mumps infections in the etiology of type 1 (insulin-dependent) diabetes. Diabetes Res. 1988;9:111–116. [PubMed] [Google Scholar]
- 41.Kasuga A, Harada R, Saruta T. Insulin-dependent diabetes mellitus associated with parvovirus B19 infection. Ann Intern Med. 1996;125:700–701. doi: 10.7326/0003-4819-125-8-199610150-00030. [DOI] [PubMed] [Google Scholar]
- 42.Gale EA. Congenital rubella: Citation virus or viral cause of type 1 diabetes? Diabetologia. 2008;51:1559–1566. doi: 10.1007/s00125-008-1099-4. [DOI] [PubMed] [Google Scholar]
- 43.Burgess MA, Forrest JM. Congenital rubella and diabetes mellitus. Diabetologia. 2009;52:369–370, author reply 373. doi: 10.1007/s00125-008-1227-1. [DOI] [PubMed] [Google Scholar]
- 44.Honeyman MC, et al. Association between rotavirus infection and pancreatic islet autoimmunity in children at risk of developing type 1 diabetes. Diabetes. 2000;49:1319–1324. doi: 10.2337/diabetes.49.8.1319. [DOI] [PubMed] [Google Scholar]
- 45.Honeyman MC, Stone NL, Harrison LC. T-cell epitopes in type 1 diabetes autoantigen tyrosine phosphatase IA-2: Potential for mimicry with rotavirus and other environmental agents. Mol Med. 1998;4:231–239. [PMC free article] [PubMed] [Google Scholar]
- 46.Lim ES, et al. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat Med. 2015;21:1228–1234. doi: 10.1038/nm.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 48.Díaz-Uriarte R, Alvarez de Andrés S. Gene selection and classification of microarray data using random forest. BMC Bioinformatics. 2006;7:3. doi: 10.1186/1471-2105-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pepe MS, Longton G, Anderson GL, Schummer M. Selecting differentially expressed genes from microarray experiments. Biometrics. 2003;59:133–142. doi: 10.1111/1541-0420.00016. [DOI] [PubMed] [Google Scholar]
- 50.Minot S, et al. The human gut virome: Inter-individual variation and dynamic response to diet. Genome Res. 2011;21:1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finsterbusch T, Mankertz A. Porcine circoviruses—small but powerful. Virus Res. 2009;143:177–183. doi: 10.1016/j.virusres.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 52.Endesfelder D, et al. Compromised gut microbiota networks in children with anti-islet cell autoimmunity. Diabetes. 2014;63:2006–2014. doi: 10.2337/db13-1676. [DOI] [PubMed] [Google Scholar]
- 53.Giongo A, et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 2011;5:82–91. doi: 10.1038/ismej.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Romond MB, et al. Does the intestinal bifidobacterial colonisation affect bacterial translocation? Anaerobe. 2008;14:43–48. doi: 10.1016/j.anaerobe.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 55.Philippe D, et al. Bifidobacterium lactis attenuates onset of inflammation in a murine model of colitis. World J Gastroenterol. 2011;17:459–469. doi: 10.3748/wjg.v17.i4.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cani PD, et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut. 2009;58:1091–1103. doi: 10.1136/gut.2008.165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu M, et al. Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science. 2002;295:2091–2094. doi: 10.1126/science.1067467. [DOI] [PubMed] [Google Scholar]
- 58.MacDuff DA, et al. Phenotypic complementation of genetic immunodeficiency by chronic herpesvirus infection. eLife. 2015;4:4. doi: 10.7554/eLife.04494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cadwell K, et al. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Knip M, et al. Finnish TRIGR Study Group Dietary intervention in infancy and later signs of beta-cell autoimmunity. N Engl J Med. 2010;363:1900–1908. doi: 10.1056/NEJMoa1004809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Finkbeiner SR, et al. Human stool contains a previously unrecognized diversity of novel astroviruses. Virol J. 2009;6:161. doi: 10.1186/1743-422X-6-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang D, et al. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 2003;1:E2. doi: 10.1371/journal.pbio.0000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li W, Godzik A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 64.Rutherford K, et al. Artemis: Sequence visualization and annotation. Bio4nformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 65.Diaz-Uriarte R. GeneSrF and varSelRF: A web-based tool and R package for gene selection and classification using random forest. BMC Bioinformatics. 2007;8:328. doi: 10.1186/1471-2105-8-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Callahan BJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McDonald D, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.