Significance
We found that tRNA-derived small RNAs (tsRNAs) are dysregulated in many cancers and that their expression is modulated during cancer development and staging. Indeed, activation of oncogenes and inactivation of tumor suppressors lead to a dysregulation of specific tsRNAs, and tsRNA-KO cells display a specific change in gene-expression profile. Thus tsRNAs could be key effectors in cancer-related pathways. These results indicate active crosstalk between tsRNAs and oncogenes and suggest that tsRNAs could be useful markers for diagnosis or targets for therapy. Additionally, ts-46 and ts-47 affect cell growth in lung cancer cell lines, further confirming the involvement of tsRNAs in cancer pathogenesis.
Keywords: tsRNA, tRF, tDR, ncRNA, tRNA fragments
Abstract
Small, noncoding RNAs are short untranslated RNA molecules, some of which have been associated with cancer development. Recently we showed that a class of small RNAs generated during the maturation process of tRNAs (tRNA-derived small RNAs, hereafter “tsRNAs”) is dysregulated in cancer. Specifically, we uncovered tsRNA signatures in chronic lymphocytic leukemia and lung cancer and demonstrated that the ts-4521/3676 cluster (now called “ts-101” and “ts-53,” respectively), ts-46, and ts-47 are down-regulated in these malignancies. Furthermore, we showed that tsRNAs are similar to Piwi-interacting RNAs (piRNAs) and demonstrated that ts-101 and ts-53 can associate with PiwiL2, a protein involved in the silencing of transposons. In this study, we extended our investigation on tsRNA signatures to samples collected from patients with colon, breast, or ovarian cancer and cell lines harboring specific oncogenic mutations and representing different stages of cancer progression. We detected tsRNA signatures in all patient samples and determined that tsRNA expression is altered upon oncogene activation and during cancer staging. In addition, we generated a knocked-out cell model for ts-101 and ts-46 in HEK-293 cells and found significant differences in gene-expression patterns, with activation of genes involved in cell survival and down-regulation of genes involved in apoptosis and chromatin structure. Finally, we overexpressed ts-46 and ts-47 in two lung cancer cell lines and performed a clonogenic assay to examine their role in cell proliferation. We observed a strong inhibition of colony formation in cells overexpressing these tsRNAs compared with untreated cells, confirming that tsRNAs affect cell growth and survival.
In the last decade, small noncoding RNAs (ncRNAs), including miRNAs and Piwi-interacting RNAs (piRNAs) (1–3), have been associated with cancer onset, progression, and drug response. Recently, a class of small ncRNAs was reported to derive from tRNA precursor or mature sequences, and their connection with cancer is currently under investigation (4). In eukaryotic cells, tRNAs are transcribed by RNA polymerase III, with transcription terminating after a stretch of four or more Ts located 10–60 nt downstream of the 3′ end of the tRNA mature sequence (5, 6). Pre-tRNAs and mature tRNAs undergo extensive modifications before and after exportation to the cytoplasm (7) resulting in the production of three types of tRNA-derived ncRNAs: tRNA-derived small RNAs (tsRNAs) (4), tRNA halves (tiRNAs) (8), and tRNA-derived fragments (tRFs or tDRs) (9, 10). tsRNAs are generated in the nucleus as a consequence of the pre-tRNA 3′ end cleavage (4), whereas tiRNAs are generated from mature tRNAs by cytoplasmic angiogenin activated in response to stress (6, 11). The biogenesis of tRFs is currently under investigation, but a Dicer-dependent cleavage of mature tRNAs in the cytoplasm has been proposed as a possible mechanism of tRF production (12–14). Considering that tsRNAs do not cover any of the consensus sequences present within the tRNA itself, they mostly are unique sequences (4). Therefore we focused on these molecules, which we defined as single-stranded small RNAs, 16–48 nt long, ending with a stretch of four Ts (4). When tsRNAs accumulate in the nucleus, they can be exported, suggesting that tsRNAs could regulate gene expression at different levels (15). Indeed, we previously showed that tsRNAs can interact with both Ago and Piwi proteins, potentially affecting the regulation of gene expression at a pretranscriptional level (by interacting with the epigenetic machinery while in the nucleus, similar to piRNAs) and at a posttranscriptional level (by 3′ UTR targeting after exportation to the cytoplasm, similar to miRNAs) (4, 12, 16).
In 2009, Lee et al. (9) showed that expression of ts-36, which the authors called “tRF-1001,” correlates with cell proliferation. In 2010, Haussecker et al. (12) reported that the same tsRNA (called “cand45” in that report) associates with Ago proteins, and in 2013, Maute et al. (17) showed that tRNA fragments can function like miRNAs in B-cell lymphomas. In 2016, we demonstrated that ts-53 and ts-101 (previously designated “miR-3676” and “miR-4521” and here renamed “ts-53” and “ts-101,” respectively) interact not only with Ago proteins but also with PiwiL2 protein and are down-regulated in chronic lymphocytic leukemia (CLL) and lung cancer (16). We showed that ts-53 targets the 3′ UTR of TCL1, a key oncogene in the development of aggressive CLL, and that its down-regulation in leukemic cells is inversely correlated with TCL1 expression. Additionally, we described two mutations at the ts-53 locus in CLL patients and found several mutations located mainly in the genomic region of ts-101 in lung cancer samples (4). By using a custom tsRNA microarray chip, we determined the presence of tsRNA signatures in CLL and lung cancer. Ts-46 and ts-47 were the most down-regulated tsRNAs in these malignancies (4, 16). Here we describe the results of our most recent experiments aimed at clarifying the role of tsRNAs in cancer and finding tsRNA signatures in different malignancies.
Results
tsRNA Signatures in Cancers.
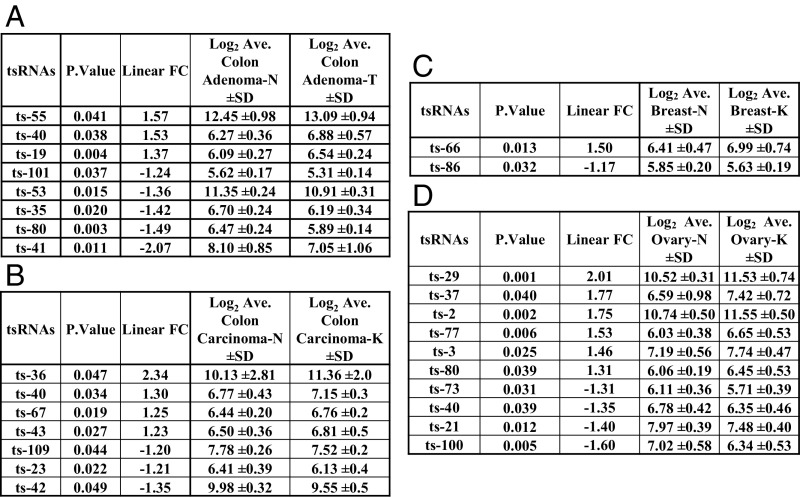
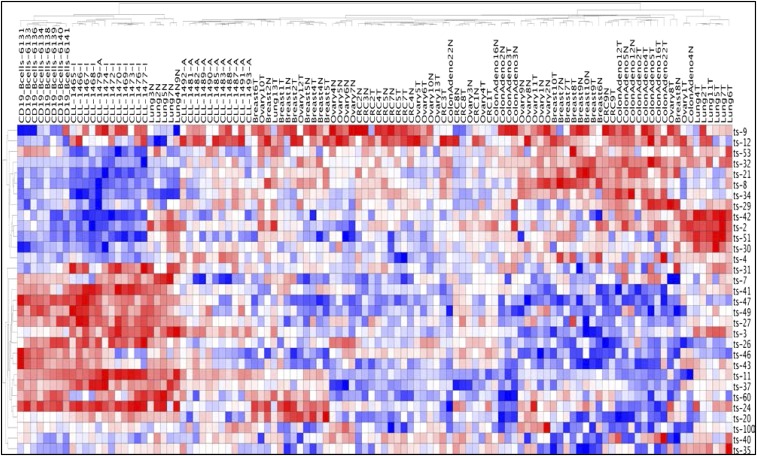
We studied tsRNA regulation/signatures in cancer by hybridizing total RNA samples from patients to our custom tsRNA microarray chip (4). Previously, we found a signature of 17 tsRNAs differentially expressed in CLL and a signature of six tsRNAs in lung cancer. Specifically, we identified ts-46 and ts-47 as tsRNAs that were strongly down-regulated in both malignancies (4). Thus, we examined the expression profile of tsRNAs in other cancer samples. We profiled 14 paired samples from seven patients with colon adenoma and 16 paired samples from eight patients with colon adenocarcinoma cancer. We found a signature of eight tsRNAs characterizing adenomas and a signature of seven tsRNAs for adenocarcinomas (Fig. 1 A and B and Fig. S1 A and B). Ts-53 and ts-101 were down-regulated in adenomas but not in adenocarcinomas, suggesting that they may have a role in the initial phases of transformation. Ts-40 was up-regulated in both comparisons; thus this tsRNA could be an oncogenic tsRNA in colon cancer development. Ts-36, which was previously correlated with cell proliferation (9), shows a twofold increase in its expression level in carcinomas but not in adenomas, indicating that this tsRNA could have a role in the final stages of the malignant transformation of colon cells. Then we studied samples from breast cancer and ovarian cancer patients. In breast cancer, we found only two tsRNAs significantly dysregulated: ts-66 and ts-86. Ts-66 was up-regulated in cancer, whereas ts-86 was down-regulated (Fig. 1C and Fig. S1C). In ovarian cancer we found a signature of 10 tsRNAs. Among these, ts-29 was overexpressed more than twofold in cancer samples as compared with normal controls (Fig. 1D and Fig. S1D). To identify common sets of tsRNAs potentially playing an oncogenic or tumor-suppressor role in multiple malignancies, we examined the signatures from all the profiled types of cancer, including our previous results from CLL and lung cancer (4, 16). Our analysis revealed that five tsRNAs are down-regulated in both CLL and lung cancer (ts-46, ts-47, ts-49, ts-53, and ts-101), whereas ts-4 is up-regulated in these two malignancies. Ts-53 and ts-101 are also down-regulated in colon adenomas. Ts-3 is up-regulated in CLL and ovarian cancer. Importantly, a signature of 31 tsRNAs can discriminate among different cancers and tissue types (Fig. 2).
Fig. 1.
tsRNA signatures in colon adenomas and adenocarcinomas, breast invasive ductal carcinoma, and ovary cancers. (A) Paired comparison between samples of colon adenomas and respective surrounding normal tissues from seven patients. We found a signature of eight tsRNAs. (B) Paired comparison between samples of colon carcinomas and respective surrounding normal tissues from eight patients. We found a signature of seven tsRNAs. (C) Paired comparison between breast cancer samples and respective surrounding normal tissues from nine patients. We found two dysregulate tsRNAs. (D) Unpaired comparison between ovary cancer samples from nine patients and normal ovary samples from ten different patients. We found a signature of 10 tsRNAs. All data were analyzed by applying normexp negative background correction, quantile data normalization, and moderated t-statistics for differential expression analysis. All tsRNAs considered are significantly differentially expressed (P < 0.05).
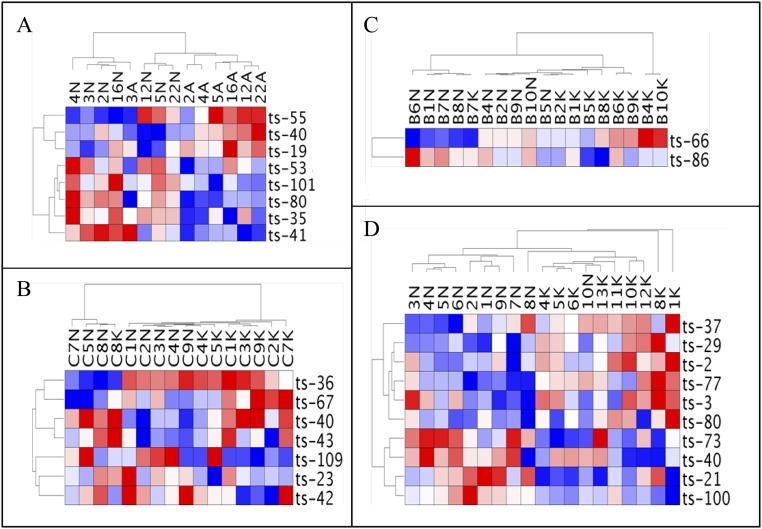
Fig. S1.
(A) Heat map of the tsRNA signature generated with data from paired samples from colon adenoma patients. (B) Heat map of the tsRNA signature generated with data from paired samples from colon adenocarcinoma patients. (C) Heat map of the tsRNA signature generated with data from paired samples from breast cancer patients. (D) Heat map of the tsRNA signature generated with data from samples from ovary cancer patients and normal ovary tissues from controls.
Fig. 2.
tsRNA tissues signatures. tsRNA heat map across tissue samples from patients with CLL, lung, colon, ovary, or breast cancer. All values are in log2 scale; red depicts overexpression, and blue represents underexpression.
tsRNAs Expression Is Modulated by Oncogenes.
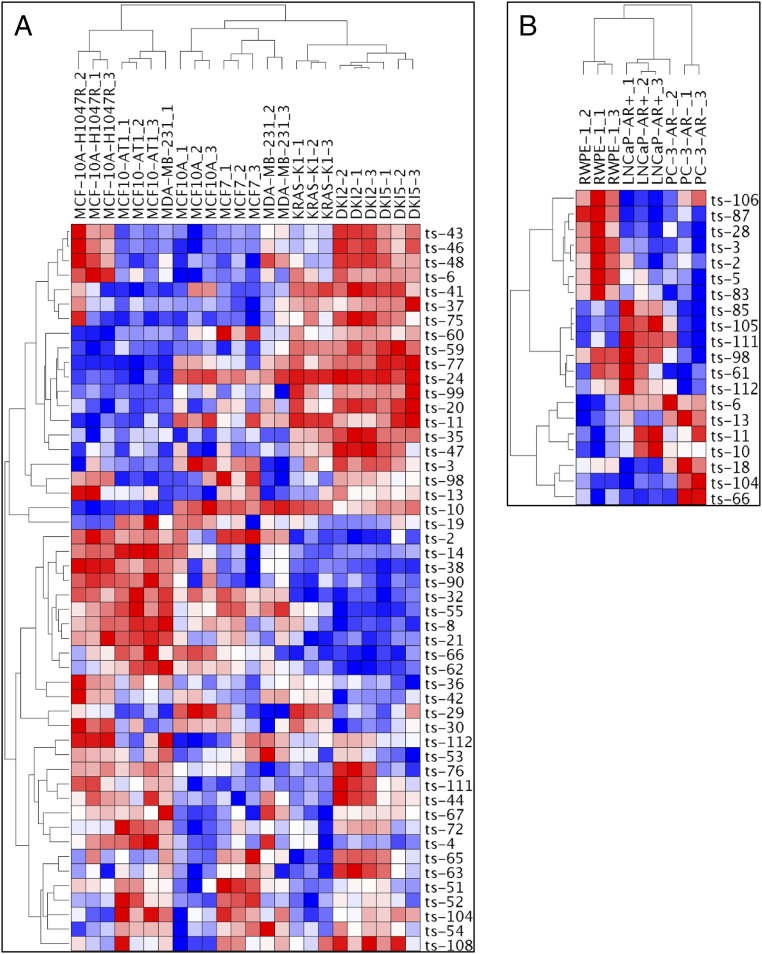
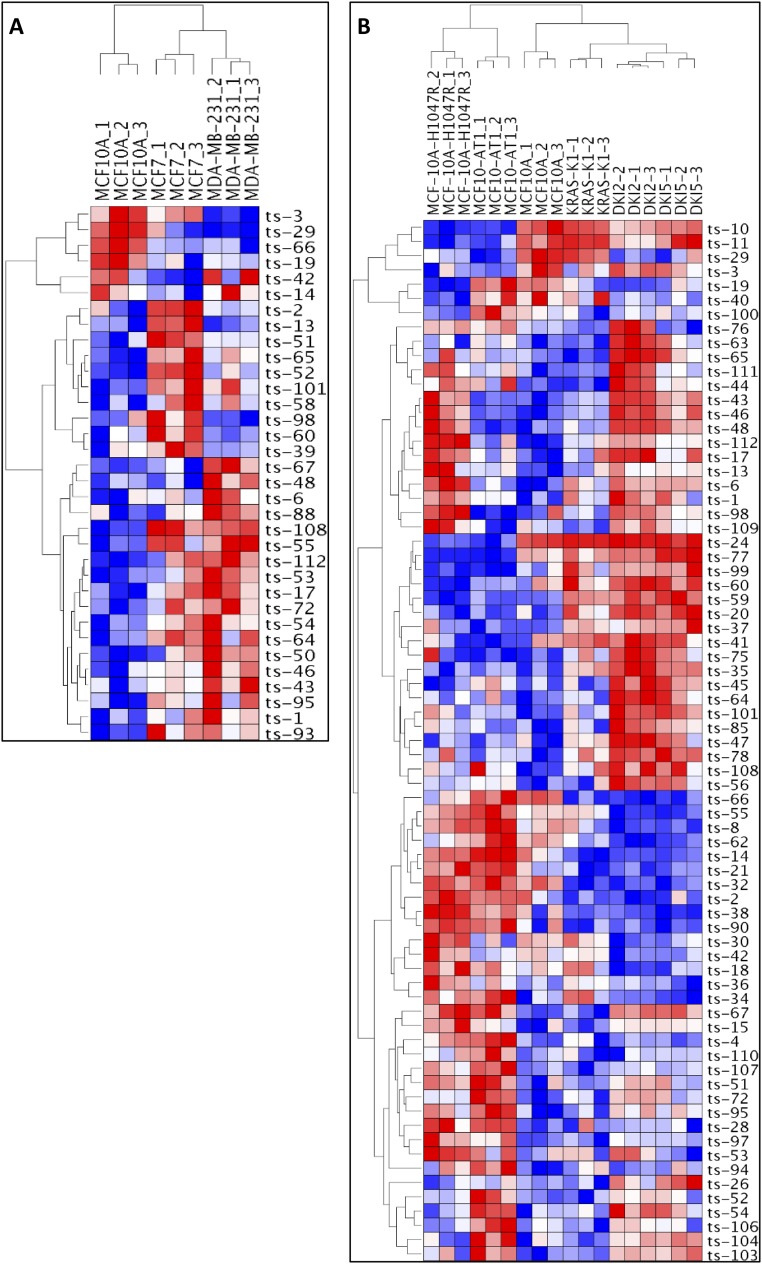
By using our custom tsRNA microarray chip, we investigated tsRNA expression patterns in human lymphocytes with and without activation of the MYC oncogene and found a signature of 15 tsRNAs; among these ts-47 was the most down-regulated by MYC activation (Table S1). Given these results, we examined the effect of other oncogenes on tsRNA expression. Because the tsRNA signature in breast cancer showed only two dysregulated tsRNAs, we hypothesized that cell lines derived from breast tissue would provide a good model to study the effect of specific oncogenes on tsRNA expression. Thus we used cell lines derived from MCF10A (normal breast epithelial cells) carrying activating mutations of the HRAS, KRAS, and PIK3CA genes and two cell lines from different stages of breast cancer: MCF7 cells, representing an early-stage, noninvasive, luminal type of breast cancer and displaying hormone sensitivity through the expression of estrogen receptor (ER+), and MDA-MB-231 cells, representing a later stage of breast cancer from a metastatic, invasive, triple-negative basal breast cancer not responsive to hormonal therapy and poorly responsive to chemotherapy (Table S2). When performing an unsupervised analysis to compare all these cell lines together, we detected a signature of 50 tsRNAs able to cluster into eight groups following a hierarchical order: PIK3CA-mutated cells and HRAS-transformed cells clustered together in the first subtree; normal cells, early-stage breast cancer, and late-stage breast cancer cells clustered in a second subtree; and KRAS-mutated cells and cell lines harboring both KRAS and PIK3CA mutations clustered in a third subtree (Fig. 3A). This comparison shows that the activation of KRAS significantly affects tsRNA expression profiles in breast cells. In particular, nine tsRNAs (ts-2, ts-14, ts-38, ts-90, ts-32, ts-8, ts21, ts-66, and ts-62) show a remarkable down-regulation in all cell lines with KRAS mutations (KRAS-KI, DKI2, and DKI5), whereas ts-55, ts-42, ts-29, and ts-30 are down-regulated in cell lines with mutations on both KRAS and PIK3CA (DKI2 and DKI5). Surprisingly, ts-46 is up-regulated in all cell lines with a PIK3CA mutation (MCF10A-H1047R, DKI2, and DKI5), and ts-47 is up-regulated in all cell lines with a KRAS mutation (KRAS-KI, DKI2, and DKI5). HRAS and PIK3CA mutated cells (MCF10A-H1047R and MCF10-AT1) are clustered together and show a profile very different from that of normal, KRAS, and KRAS+PIK3CA cells. For example ts-24, ts-11, and ts-10 are strongly down-regulated only in HRAS and in PIK3CA mutated cells. We then performed two additional unsupervised analyses by separating the cell lines according to their origin (Fig. S2): one comparison among all MCF10A-derived cell lines and another between the two breast cancer cell lines and normal breast cell lines. We confirmed that the HRAS and the PIK3CA cells cluster together. Indeed, eight tsRNAs (ts-10, ts-11, ts-29, ts-3, ts-24, ts-77, ts-99, and ts-60) are down-regulated in both the MCF10A-H1047R and MCF10-AT1 cell lines. Among these, only ts-29 is also down-regulated in KRAS/PIK3CA cells (DKI2 and DKI5). KRAS mutated cells cluster in another group, and all tsRNAs found down-regulated in the first comparison were confirmed. Thus, activation of KRAS strongly affected the tsRNA profile, and cells carrying any activating mutation of KRAS, alone or in combination with PIK3CA, show a tsRNA expression pattern significantly different from that of cell lines harboring activation of PIK3CA alone or HRAS (Fig. S2B). These results suggest that tsRNAs could be key effectors in the pathways regulated by these oncogenes. When comparing normal cells with cancer cells, we found a signature of 34 tsRNAs able to distinguish the two groups. This comparison revealed that ts-3 is strongly down-regulated in aggressive late-stage breast cancer, whereas ts-29 expression decreases progressively from the early stage to the late stage. Ts-66 is down-regulated in both cancer cell lines. Ts-108 and ts-55 are up-regulated in cancer cell lines, but ts-67, ts-48, and ts-6 are up-regulated only in the late-stage cell line. Surprisingly, ts-101, ts-53, and ts-46 are also up-regulated in cancer cell lines. In addition, we studied the expression of tsRNAs in cell lines derived from prostate cancer at different stages: LNCaP cells, derived from early-stage p53 wild-type lymph node metastasis, are positive for androgen receptor signaling (AR+) and thus are androgen responsive; PC-3 cells, derived from a late-stage p53-defective bone metastasis, are AR− and thus are nonresponsive to androgen. When the normal prostate epithelium cell line RWPE-1 was compared with LNCaP and with PC-3 cells, we found a signature of 20 dysregulated tsRNAs (Fig. 3B). Among these, ts-18, ts-104, and ts-66 are up-regulated in the p53-impaired cell line compared with the normal or the early-stage p53 WT cells.
Fig. 3.
Heatmaps from breast and prostate cell lines. (A) Unsupervised clustering of all cell lines from breast. (B) Unsupervised clustering of normal vs. cancer prostate cell lines. All values are in log2 scale; red depicts overexpression, and blue represents underexpression.
Fig. S2.
(A) Unsupervised clustering of normal vs. cancer breast cell lines. (B) Unsupervised clustering of breast cell lines derived from MCF10A. All values are in log2 scale; red depicts overexpression, and blue represents underexpression.
Gene and miRNA Expression Are Dysregulated in tsRNA-KO Cell Models.
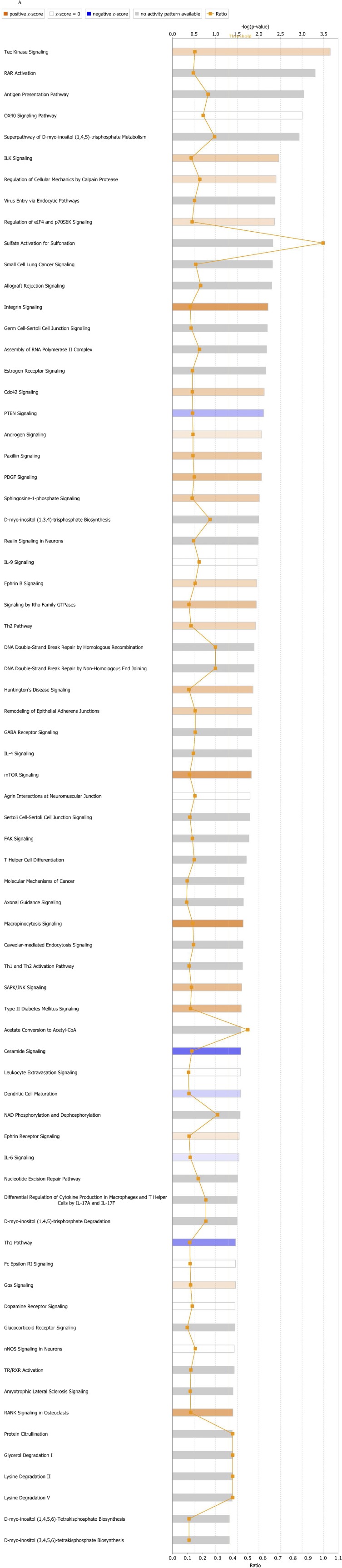
To understand better whether tsRNAs have a role in regulating gene expression, we generated ts-101 and ts-46 KO stable cell lines from HEK293 cells by using CRISPR technology. Affymetrix gene-expression profiling was performed using RNA from two HEK293_KO-ts46 clones, two HEK293_KO-ts101 clones, and two WT controls transfected with the empty vector. We found 270 genes differentially expressed in the ts-46 clones (211 coding genes plus 59 noncoding genes) and 216 genes differentially expressed in the ts-101 clones (170 coding genes plus 46 noncoding genes) compared with the WT controls (Fig. S3 and Table S3). Among up-regulated genes in the ts-46 and ts-101 KO clones we found miR-222, IRS4, MAP3K19, PLK4, MED28, EPS8L2, CDH3, GINS1, SMARCA5, and PKP3. Among down-regulated genes we found TAF9B, METTL23, FILIP1, OCLN, BMP2, TAX1BP1, RASGRF2, MIR613, OGN, SMC1A, and all the components of the H3A1 histone subfamily (HIST1H3A, HIST1H3B, HIST1H3C, HIST1H3D, HIST1H3E, HIST1H3F, HIST1H3G, HIST1H3H, HIST1H3I, and HIST1H3J). As expected mir-4521 (corresponding to ts-101) is strongly down-regulated in ts-101 clones. The down-regulation of all members of the H3A1 histone subfamily encoding for one of the components of the nucleosome core suggests that tsRNAs can interfere with chromatin configuration and thus with the epigenetic control of gene expression. Additionally, we performed a functional enrichment study by using the Ingenuity Pathway Analysis (IPA) software to evaluate whether cancer-related pathways are altered in ts-46 and ts-101 KO cells. Ts-46 and ts-101 KO cells showed a significant activation of networks associated with cell proliferation and inhibition of networks associated with apoptosis. Indeed, in ts-46 KO cells several pathways related to cell transformation and cancer development are over-activated, including ILK signaling (18), integrin signaling (19), PDGF signaling (20, 21), sphingosine-1-phosphate (S1P) signaling (22), and mTOR signaling (23–26). In addition, we observed an inhibition of the tumor suppressor PTEN signaling (27–29) and ceramide signaling (30). In ts-101 KO cells, two pathways related to cell transformation and cancer development are over-activated, namely, S1P signaling (22) and glutamate receptor signaling (31). Nonetheless, no pathways related to cell growth inhibition were found to be impeded (Fig. S4).
Fig. S3.
Gene-expression profile. We generated ts-46 (A) and ts-101 (B) KO stable clones from HEK-293 cells by using CRISPR technology. RNA from two ts-46 KO clones, two ts-101 KO clones, and two samples transfected with the empty pCAS9 vector were used for Affymetrix gene-expression profiles. By setting a linear 1.5-fold change, we found 270 genes differentially expressed in the ts-46 clones and 216 genes differentially expressed in the ts-101 clones compared with the WT. Some examples of up-regulated genes in ts-46 and ts-101 clones are MAP3K19, miR-222, and MED28. As expected, mir-4521 is strongly down-regulated in ts-101 clones. Among down-regulated genes we found FILIP1, OCLN, and HIST1H3-A, -B, -C, -D, -E, -F, -G, -H, -I, -J, supporting our hypothesis that tsRNAs can interfere with chromatin structure. All values are in log2 scale; red depicts overexpression, and blue represents underexpression.
Fig. S4.
Ingenuity Pathway Analyses of canonical pathways in gene-expression profiles from ts-46 KO (A) and ts-101 KO (B) cells.
Evidence of the Tumor Suppression Function of ts-46 and ts-47: An Inhibiting Effect on Colony Formation in Lung Cancer Cell Lines.
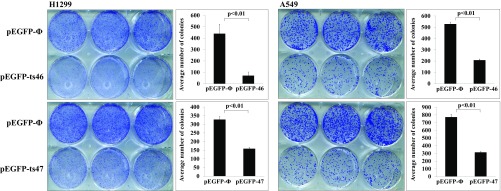
We previously carried out a clonogenic formation experiment to determine if ts-53 can function as tumor suppressor in lung cancer cell lines A549 and H1299. We indeed observed that exogenous expression of ts-53 resulted in a decrease in colony formation, indicating that ts-53 can function as a tumor suppressor. Considering that ts-46 and ts-47 are also down-regulated in both CLL and lung cancer, we studied their effect on the colony-formation ability of lung cancer cell lines. We overexpressed ts-46 and ts-47 in the lung cancer cell lines H1299 and A549 and performed a colony assay by seeding the same number of transfected cells for each treatment and negative controls. After 10 d for A549 and 15 d for H1299, we counted the number of colonies developed from each condition and observed a significant decrease in the colony-formation ability of the cells overexpressing ts-46 or ts-47 (Fig. 4).
Fig. 4.
Colony assay on lung cancer cell lines transfected with ts-46 or ts-47. Colony assay performed on H1299 (Left) and A549 (Right) cells. (Upper) The top wells were transfected with the empty vector, and the lower wells were transfected with the vector expressing ts-46. (Lower) The top wells were transfected with the empty vector, and the lower wells were transfected with the vector expressing ts-47. A graphic representation of the colony count is provided beside each plate.
Discussion
While studying the miR-4521/3676 cluster in CLL, we found that these two miRNAs are tsRNAs, which we now call ts-101 and ts-53, respectively (16). We showed that ts-53 and ts-101 are deleted in 17p- CLL and that ts-53 targets TCL1 (16) in an miRNA fashion. Later, we found that ts-46 and ts-47 are down-regulated in CLL and lung cancer and showed that tsRNAs can act like piRNAs by interacting with Piwi proteins. Thus, tsRNAs can interfere with the epigenetic regulation of genes (4). The data presented here combined with these previous reports indicate that tsRNAs play a key role in the onset and progression of several types of cancers. Indeed, by analyzing the data from CLL, lung, colon, breast, and ovarian cancer samples, we found a cancer-specific signature of 31 tsRNAs able to discriminate these cancers (Fig. 2). We previously described CLL and lung cancer signatures (4, 16). In this study we report that tsRNA signatures also can be identified in adenomas and carcinomas of the colon, breast invasive ductal carcinoma, and ovarian cancers (Fig. 1). Indeed, we found that tsRNA expression differs in normal and tumor tissues, although these differences could reflect discrepancies related to the transformed phenotypes or cell-type compositions among the samples. To minimize this possibility, we used paired samples from colon and breast cancer patients, with the samples of normal tissue harvested from the normal tissue surrounding the cancer samples. However, for the comparisons of ovary samples, it was not possible to obtain a specimen of normal tissue from the same patient. Thus, ovary normal tissues representative of the different origins of all cancer sample histotypes were collected from cervical cancer patients undergoing radical hysterectomy. The analysis of colon samples shows eight tsRNAs dysregulated in adenomas and seven in carcinomas. Only two tsRNAs, ts-66 and ts-86, were found to be dysregulated in breast cancer. Interestingly, when the tsRNA expression profile of breast cell lines was compared with that of prostate cell lines, ts-66 was always dysregulated: It was down-regulated in breast cancer cell lines but was up-regulated in the prostate AR− late-stage cell line compared with AR+ early-stage and normal cells. Because tRNA halves and piRNAs were recently shown to be involved in hormone response in breast and prostate cancer (32, 33), it is possible that tsRNAs also may be implicated in the response to sex hormones in hormone-related cancers. Last, we found a signature of 10 tsRNAs dysregulated in ovarian cancers.
To verify that tsRNA expression can be dysregulated by activation of specific oncogenes, we studied the effect of MYC activation in lymphocytes and found that ts-47, which is lost in CLL and lung cancer, is strongly down-regulated by MYC activation, indicating that MYC may be involved in cancer by turning off an epigenetic silencer belonging to the tsRNA gene family (Table S1). We then studied the effects of other oncogenes on tsRNAs by using well-established breast-derived cell lines carrying specific mutations that activate key oncogenes such as HRAS, KRAS, and PIK3CA and two cancer cell lines. The results of these experiments suggest that tsRNAs could be key effectors in pathways regulated by these oncogenes and that these molecules could have important roles in the cell transformation process and in cancer development/progression. Additionally, the oncogene-driven dysregulation of tsRNA could be involved in feedback loops, as previously shown for miRNAs (34). Indeed, when profiling the gene-expression patterns of ts-46 and ts-101 KO cells, we found that pathways such as PTEN and ceramide signaling are inhibited. PTEN is a major negative regulator of the PI3-kinase pathway, and recent studies indicate that ceramide signaling induces apoptosis by reducing the activity of p42/44-MAPK and Akt (35, 36). Pathways involved in cell growth and cancer development are up-regulated in ts-101 and ts-46 KO cells (Fig. S4). In both KO cell types the S1P and glutamate receptor signaling pathways are significantly overactivated, and production of S1P promotes tumor growth, resistance to apoptosis, and metastasis. Interestingly, ceramide and S1P counter-regulate the phosphorylation of Bax and Bad to control apoptosis and cell survival (37); thus a simultaneous activation of S1P and inhibition of the ceramide signal can significantly damage the cell growth control system. Therefore, ts-46 could control cell growth by interfering with the regulation of the S1P/ceramide pathways (22). Additionally, ts-46 KOs show an increase of (i) ILK signaling, associated with tumor growth and metastasis (18), (ii) integrin signaling, associated with cell survival (19, 38), and (iii) PDGF and mTOR signaling, well-known cancer-related pathways. Furthermore, in ts-101 KO cells, genes related to the chromatin structure are down-regulated. We previously showed that ts-53 can interact with Piwil2, a protein involved in DNA methylation (4); thus these results support our hypothesis that tsRNAs could be involved in the epigenetic control of gene expression. In light of a very recent study reporting that tRF-5030c [named td-piR(glu) in this study] interacts with Piwi proteins and with DNA- and histone-methyltransferases affecting methylation of genes and chromatin condensation (39), it is possible that tsRNAs could also have a role in DNA and histone methylation, affecting chromatin functionality and structure.
Last, we show that ts-46 and ts-47 have an inhibiting effect on the ability of lung cancer cells to form colonies, as previously observed for ts-53. This effect suggests that the deficiency of expression of these tsRNAs can favor cell proliferation and cancer onset/progression (Fig. 4). We performed this experiment with two cell lines differing in p53 expression and KRAS mutation status and obtained the same inhibitory effect in both cases. These tsRNAs therefore could counterbalance the oncogenic activity of KRAS mutation in lung cancer and positively affect the p53 pathway in cells in which this tumor suppressor is impeded.
All these findings indicate that tsRNAs have a key role in cancer onset and progression and suggest that tsRNAs can be studied as a class that may have oncogenic or tumor-suppressor functions in cancer.
Methods
Tissue Samples.
This study was carried out in accordance with a protocol approved by the Institutional Review Board of The Ohio State University. Cancer samples and normal counterparts were obtained from patients enrolled in a clinical study of the role of ncRNAs in solid tumors at the Institute Regina Elena and University La Sapienza, Rome; patients’ written informed consent was obtained in accordance with the Declaration of Helsinki. Samples were grouped as follows: seven tumor samples and seven samples of normal surrounding tissue from colorectal adenomas patients; eight cancer samples and eight samples of normal surrounding tissue from colorectal adenocarcinoma patients; nine cancer samples and nine samples of normal surrounding tissue from breast cancer patients with invasive ductal carcinomas; nine cancer samples from nine ovarian cancer patients; and ten samples of normal ovary tissue from ten cervical cancer patients undergoing radical hysterectomy. Histology reports are showed in Table S4.
RNA was extracted using the standard TRIzol method (Invitrogen), and RNA quality was assayed using an Agilent 2100 Bioanalyzer.
Cell Cultures.
Cell lines A549 (p53 WT, KRAS bearing the G12S activating mutation), H1299 (p53 null and KRAS WT), and HEK293 were cultured in RPMI (Sigma-Aldrich) supplemented with 10% FBS and 100 µg/L gentamicin at 37 °C.
P493-6 cells carrying a conditional, tetracycline-regulated MYC (40, 41) were grown in RPMI medium 1640 supplemented with 10% FCS, 100 U/mL penicillin, 100 µg/mL streptomycin, and 2 mM l-glutamine (Life Technologies). For repression of MYC, 0.1 µg/mL tetracycline was added to the culture medium.
MCF10A cells (nontransformed human breast epithelial cells) were grown in DMEM:F12 (HyClone-SH30271), 5% (vol/vol) horse serum (Gibco no. 16050, lot no. 1075876), 10 µg/mL human insulin (Sigma I-1882), 20 ng/mL recombinant hEGF (PeproTech AF-100-15), 100 ng/mL cholera toxin (Sigma C-8052), 0.5 µg/mL hydrocortisone (Sigma H-0888) penicillin/streptomycin (Life Technologies), and glutamine (Life Technologies) (hereafter referred to as “supplemented DMEM:F12 medium”). The isogenic model system of cell lines derived from MCF10A were cultured as follows: premalignant MCF10AT1 cells (with a constitutively active HRAS) and KRAS-KI cells (carrying a heterozygous KRAS G12V gain-of-function mutation) were maintained in supplemented DMEM:F12. MCF10A-H1047R cells (carrying a heterozygous PIK3CA H1047R activating mutation) and DKI2 cells and DKI5 cells [both derived from KRAS-KI cells and carrying an additional gain-of-function mutation for PIK3CA exon 9 (E545K) or PIK3CA exon 20 (H1047R), respectively] were grown in supplemented DMEM:F12 medium without EGF. The cell lines derived from different stages of breast cancer MCF7 cells (harboring an E545K activating mutation in the PIK3CA gene) and MDA-MB-231 cells (harboring a G13D gain-of-function mutation in the KRAS gene) were cultured in DMEM:F12 medium (HyClone-SH30271) plus 10% (vol/vol) FBS (Atlanta Biologicals), penicillin/streptomycin (Gibco), and glutamine (Gibco).
RWPE-1 cells derived from normal prostate epithelium were cultured in Keratinocyte Serum-Free Medium (KSFM; Gibco) supplemented with 12.5 mg/L bovine pituitary extract and 1.25 µg/L EGF. LNCaP prostate cancer cells were grown in RPMI 1640 medium (Sigma) containing penicillin (100 units/mL), streptomycin (100 μg/mL), and 10% FBS. PC-3 prostate cancer cells were grown in T-medium [80% DMEM (4.5 g/L glucose), 20% F12K (Gibco), 3 g/L NaHCO3 (Sigma), 13.6 pg/mL triiodothyronine (Sigma), 5 μg/mL transferrin (Sigma), 0.25 μg/mL biotin (Sigma), and 25 μg/mL adenine (Sigma)]. At the time of use, 5 μg/mL insulin (Sigma), 5% FBS (Atlanta), 100 units/mL penicillin, and 100 μg/mL streptomycin (1%) (Gibco) were added (Table S2).
tsRNA Nomenclature.
All names and sequences of tsRNAs in this study can be found in Table S5.
SI Methods
Plasmid Construction.
pEGFP-53 and pEGFP-101 were previously described. pEGFP-46 and pEGFP-47 were generated by cloning the genomic region containing the ts-46 and the ts-47 sequences (chr15:45,493,267–45,493,549 and chr15:89,878,214–89,878,488, respectively) upstream of the EGFP gene of the pEGFP-N1 vector (Clontech).
For pCAS9-101-P1 and pCAS9-101-P2 and for pCAS9-46-P1 and pCAS9-46-P2, the guide RNA (gRNA) were designed using the benchling software (https://benchling.com) and were cloned into the pSpCas9(BB)-2A-GFP (Addgene plasmid ID: 48138). The gRNA sequences are the following:
Ts-101_P1_TS: 5′-CACCGCAGGACTTCCTTAGCCGACG-3′
Ts-101_P1_BS: 5′- AAACCGTCGGCTAAGGAAGTCCTGC-3′
Ts-101_P2_TS: 5′- CACCGAAACTGAATGGAGTGACTGG-3′
Ts-101_P2_BS: 5′-AAACCCAGTCACTCCATTCAGTTTC-3′
Ts-46_P1_TS: 5′- CACCGGTTTCCACAATGCCGTGACT-3′
Ts-46_P1_BS: 5′- AAACAGTCACGGCATTGTGGAAACC-3′
Ts-46_P2_TS: 5′-CACCGTAGCTTCCCAGAAGAATGTG-3′
Ts-46_P2_BS: 5′- AAACCACATTCTTCTGGGAAGCTAC-3′
The sequence of the gRNA-top is located immediately upstream of a proto-spacer adjacent motif (PAM) sequence (NGG), because Cas9 nuclease cuts 3 nt upstream of a PAM site, and the overhang sequences CACC (sense oligo) and AAAC (antisense oligo) are complementary to overhangs generated by BbsI digestion. The bottom and top strands of each flanking guide were aligned, 5′ phosphorylated, and cloned into the BbsI cloning site of the pSpCas9(BB)-2A-GFP vector following standard protocols.
Transfections, FACS Analysis and Sorting, Clonogenic Assay, and KO Clone Selection.
For the clonogenic assay, 10 μg of pEGFP-46, pEGFP-47, or pEGFP empty vector were transfected into the A549 and H1299 lung cancer cell lines by using the Lipofectamine 2000 standard protocol (Invitrogen). Forty-eight hours after transfection, cells were GFP-sorted and seeded in triplicate in six-well plates. Cell sorting for each transfection (A549-ts46, A549-ts47, A549-EV, H1299-ts46, H1299-ts47, H1299-EV) was performed on a FACSAria III cell sorter (BD Biosciences) in sterile conditions. For A549-derived cells, 5,000 sorted cells were seeded. For H1299-derived cells, 10,000 sorted cells were seeded. Ten days later, A549 cells were fixed and stained with crystal violet using the standard procedure, and colonies were counted. For H1299 cells the staining was performed 15 d after sorting.
To generate the CRISPR-KO cell line, pCAS9-46-P1 and pCAS9-46-P2, or pCAS9-101-P1 and pCAS9-101-P2, or pCAS9 empty vector were cotransfected into HEK293 cells by using the Lipofectamine 2000 standard protocol (Invitrogen). Transfected cells were GFP-sorted after 48 h. Cell sorting was performed on a FACSAria III cell sorter in sterile conditions for each transfection (HEK293_KO-ts46, HEK293_KO-ts101, HEK293_KO-control), and sorted cells were seeded in an appropriate vessel. After 3 d, a limiting dilution was performed by seeding serial dilutions of cells in 96-well plates to obtain single-cell clonal populations. Single-cell clones were harvested 2–3 wk later.
Sequencing, Real-Time RT-PCR, and Array Hybridization.
HEK293-KO-CRISPR clones were screened by PCR sequencing and real-time PCR to confirm the deletion of the genomic region containing the targeted tsRNA in both alleles and the absence of the targeted tsRNA expression. Sequencing was performed on PCR products obtained from DNA extracted using the Blood and Tissue DNA Extraction Kit (Qiagen). Primers for PCR and sequencing are the following:
ts-46_F: 5′-GCTTAGGAAAGCTGGAATGG-3′
ts-46_R: 5′-TGTGCACACTTCATTGTCCG-3′
ts-46_ F sequencing: 5′-ATGGCCGCCATCTAGACGAG-3′
ts-46_R sequencing: 5′-ATAATTTCAGCCCTGGCCAG-3′
ts-53_F: 5′-AGTTCTAGACCTCTCAGCAG-3′
ts-53_R: 5′-GACAGTACTGGCAAGTACAG-3′
ts-53_F sequencing: 5′-ATGGAAGAAGTCGGTCTCTG-3′
ts-53_R sequencing: 5′- GACTGGTGTAACTGCAGGTC-3′
To amplify the ts-46 sequence, we used the Advantage 2 Polymerase Mix (Clontech), and to amplify the ts-101 sequence we used the CG-rich high-fidelity Advantage 2 Polymerase Master Mix (Clontech). PCR products were cleaned up using ExoSAP-IT (USB) and sequenced.
RNAs were extracted with TRIzol standard methods (Invitrogen). Expression of ts-46 was assayed by real-time RT-PCR using a customized Taqman miRNA assay (hsa-ts-46-pr3 assay ID: CSBJW1T; batch ID: w1511782051000, catalog no. 4440418). Expression of ts-101 was assayed using a Taqman inventoried miRNA assay (hsa-miR-4521 assay ID: 465004_mat, catalog no. 4427975). The procedure was executed following the manufacturer’s protocol, and results were normalized using RNU6B (assay ID 001093, catalog no. 4427975).
Total RNAs from tissue samples (2.5 μg) and from cell lines (500 ng) were extracted with TRIzol and were hybridized on our custom tsRNA microarray chips or on Affymetrix chips for tsRNA or gene-expression profiles, respectively, by following standard protocols. In particular, for the custom tsRNA microarray chips we used a Tecan HS4800 hybridization station, and images were analyzed by using GenePix Pro Software (Molecular Devices). For gene expression, we used GeneChip Human Transcriptome Array 2.0 (HTA 2.0; Affymetrix).
Computational Analysis: tsRNA Microarray Analysis.
Custom tsRNA microarray data were analyzed by applying normexp negative background correction, quantile data normalization, and moderated t-statistics for differential expression analysis calculated using the Linear Models for Microarray Data (LIMMA) package from the Bioconductor R project (42). P values were adjusted for multiple testing using the Benjamini–Hochberg (43) method to control the false-discovery rate. All transcripts differentially expressed with a P value <0.05 were considered statistically significant.
Affymetrix Analysis.
Gene-expression profiling was performed by GeneChip Human Transcriptome Array 2.0 (HTA 2.0; Affymetrix). Gene-level normalization was carried out using Affymetrix Expression Console software, and differential expression (DE) analysis was performed using Affymetrix Transcriptome Analysis Console (TAC) 3.0 Software. The one-way between-subject ANOVA algorithm was used on normalized data to calculate the statistical significance of pairwise comparisons, considering all those transcripts differentially expressed with a P value <0.05 as statistically significant.
Enrichment Analysis.
Functional enrichment analysis was performed on significant differentially expressed targets (P < 0.05) resulting from both ts-46 and ts-101 knockdowns by using the Ingenuity Pathway Analysis (IPA) software. Settings used included experimentally observed data for human species.
Generation of Heat Maps.
Custom tsRNA microarray data were analyzed in multiple groups by applying normexp negative background correction, quantile data normalization, and moderated F statistics for differential expression analysis calculated using the LIMMA package from the Bioconductor R project (44). P values were adjusted for multiple testing using the Benjamini–Hochberg (43) method to control the false-discovery rate. All transcripts differentially expressed with a P value <0.05 were considered statistically significant. Distinct sets of tsRNAs differentially expressed with P values under this significance threshold were considered for hierarchical clustering using the HierarchicalClustering module provided by version 3.9.9 of the GenePattern suite (https://genepattern.broadinstitute.org/gp/pages/login.jsf), by using pairwise average-linkage with the Euclidean distance for the samples (columns of the heat maps), and Spearman’s rank correlation for the individual tsRNA expression profiles across all samples (rows of the heat maps). Heat map images then were generated via the HierarchicalClustering Image module provided by GenePattern (42).
Supplementary Material
Acknowledgments
This work was supported by NIH Grants R35 CA197706 (to C.M.C.), R35 CA197582 (to P.K.V.), U01 CA196383 (to G.S.S.), and P01 CA082834 (to G.S.S.); Department of Defense CDRMP PCRP Postdoctoral Fellowship W81XWH-14-1-0468 (to N.H.F.); and by the Office of the Assistant Secretary of Defense for Health Affairs, through the Breast Cancer Research Program under Award W81XWH-15-1-0033 (to P.K.V.). Opinions, interpretations, conclusions and recommendations are those of the author and are not necessarily endorsed by the National Institutes of Health or the Department of Defense.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706908114/-/DCSupplemental.
References
- 1.Müller S, et al. Next-generation sequencing reveals novel differentially regulated mRNAs, lncRNAs, miRNAs, sdRNAs and a piRNA in pancreatic cancer. Mol Cancer. 2015;14:94. doi: 10.1186/s12943-015-0358-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calin GA, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 4.Pekarsky Y, et al. Dysregulation of a family of short noncoding RNAs, tsRNAs, in human cancer. Proc Natl Acad Sci USA. 2016;113:5071–5076. doi: 10.1073/pnas.1604266113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arimbasseri AG, Rijal K, Maraia RJ. Transcription termination by the eukaryotic RNA polymerase III. Biochim Biophys Acta. 2013;1829:318–330. doi: 10.1016/j.bbagrm.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maraia RJ, Lamichhane TN. 3′ processing of eukaryotic precursor tRNAs. Wiley Interdiscip Rev RNA. 2011;2:362–375. doi: 10.1002/wrna.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohira T, Suzuki T. Precursors of tRNAs are stabilized by methylguanosine cap structures. Nat Chem Biol. 2016;12:648–655. doi: 10.1038/nchembio.2117. [DOI] [PubMed] [Google Scholar]
- 8.Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selitsky SR, Sethupathy P. tDRmapper: Challenges and solutions to mapping, naming, and quantifying tRNA-derived RNAs from human small RNA-sequencing data. BMC Bioinformatics. 2015;16:354. doi: 10.1186/s12859-015-0800-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emara MM, et al. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem. 2010;285:10959–10968. doi: 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haussecker D, et al. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson P, Ivanov P. tRNA fragments in human health and disease. FEBS Lett. 2014;588:4297–4304. doi: 10.1016/j.febslet.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cole C, et al. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao JY, et al. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PLoS One. 2010;5:e10563. doi: 10.1371/journal.pone.0010563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balatti V, et al. TCL1 targeting miR-3676 is codeleted with tumor protein p53 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2015;112:2169–2174. doi: 10.1073/pnas.1500010112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maute RL, et al. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci USA. 2013;110:1404–1409. doi: 10.1073/pnas.1206761110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sawai H, et al. Integrin-linked kinase activity is associated with interleukin-1 alpha-induced progressive behavior of pancreatic cancer and poor patient survival. Oncogene. 2006;25:3237–3246. doi: 10.1038/sj.onc.1209356. [DOI] [PubMed] [Google Scholar]
- 19.Playford MP, Schaller MD. The interplay between Src and integrins in normal and tumor biology. Oncogene. 2004;23:7928–7946. doi: 10.1038/sj.onc.1208080. [DOI] [PubMed] [Google Scholar]
- 20.Heldin CH. Targeting the PDGF signaling pathway in tumor treatment. Cell Commun Signal. 2013;11:97. doi: 10.1186/1478-811X-11-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu G, et al. FoxM1 promotes breast tumorigenesis by activating PDGF-A and forming a positive feedback loop with the PDGF/AKT signaling pathway. Oncotarget. 2015;6:11281–11294. doi: 10.18632/oncotarget.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pyne NJ, Pyne S. Sphingosine 1-phosphate and cancer. Nat Rev Cancer. 2010;10:489–503. doi: 10.1038/nrc2875. [DOI] [PubMed] [Google Scholar]
- 23.Pópulo H, Lopes JM, Soares P. The mTOR signalling pathway in human cancer. Int J Mol Sci. 2012;13:1886–1918. doi: 10.3390/ijms13021886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guerrero-Zotano A, Mayer IA, Arteaga CL. PI3K/AKT/mTOR: Role in breast cancer progression, drug resistance, and treatment. Cancer Metastasis Rev. 2016;35:515–524. doi: 10.1007/s10555-016-9637-x. [DOI] [PubMed] [Google Scholar]
- 25.Henry RE, et al. Acquired savolitinib resistance in non-small cell lung cancer arises via multiple mechanisms that converge on MET-independent mTOR and MYC activation. Oncotarget. 2016;7:57651–57670. doi: 10.18632/oncotarget.10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zining J, Lu X, Caiyun H, Yuan Y. Genetic polymorphisms of mTOR and cancer risk: A systematic review and updated meta-analysis. Oncotarget. 2016;7:57464–57480. doi: 10.18632/oncotarget.10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chalhoub N, Baker SJ. PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol. 2009;4:127–150. doi: 10.1146/annurev.pathol.4.110807.092311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He E, Pan F, Li G, Li J. Fractionated ionizing radiation promotes epithelial-mesenchymal transition in human esophageal cancer cells through PTEN deficiency-mediated Akt activation. PLoS One. 2015;10:e0126149. doi: 10.1371/journal.pone.0126149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao D, et al. Synthetic essentiality of chromatin remodelling factor CHD1 in PTEN-deficient cancer. Nature. 2017;542:484–488. doi: 10.1038/nature21357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bieberich E. Ceramide signaling in cancer and stem cells. Future Lipidol. 2008;3:273–300. doi: 10.2217/17460875.3.3.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willard SS, Koochekpour S. Glutamate signaling in benign and malignant disorders: Current status, future perspectives, and therapeutic implications. Int J Biol Sci. 2013;9:728–742. doi: 10.7150/ijbs.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Honda S, et al. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc Natl Acad Sci USA. 2015;112:E3816–E3825. doi: 10.1073/pnas.1510077112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Honda S, Kirino Y. SHOT-RNAs: A novel class of tRNA-derived functional RNAs expressed in hormone-dependent cancers. Mol Cell Oncol. 2015;3:e1079672. doi: 10.1080/23723556.2015.1079672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fabbri M, et al. Association of a microRNA/TP53 feedback circuitry with pathogenesis and outcome of B-cell chronic lymphocytic leukemia. JAMA. 2011;305:59–67. doi: 10.1001/jama.2010.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fox TE, et al. Ceramide recruits and activates protein kinase C zeta (PKC zeta) within structured membrane microdomains. J Biol Chem. 2007;282:12450–12457. doi: 10.1074/jbc.M700082200. [DOI] [PubMed] [Google Scholar]
- 36.Stoica BA, Movsesyan VA, Lea PM., 4th Faden AI. Ceramide-induced neuronal apoptosis is associated with dephosphorylation of Akt, BAD, FKHR, GSK-3beta, and induction of the mitochondrial-dependent intrinsic caspase pathway. Mol Cell Neurosci. 2003;22:365–382. doi: 10.1016/s1044-7431(02)00028-3. [DOI] [PubMed] [Google Scholar]
- 37.Jarvis WD, et al. Coordinate regulation of stress- and mitogen-activated protein kinases in the apoptotic actions of ceramide and sphingosine. Mol Pharmacol. 1997;52:935–947. doi: 10.1124/mol.52.6.935. [DOI] [PubMed] [Google Scholar]
- 38.Giancotti FG, Ruoslahti E. Integrin signaling. Science. 1999;285:1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- 39.Zhang X, et al. IL-4 Inhibits the biogenesis of an epigenetically suppressive PIWI-interacting RNA to upregulate CD1a molecules on monocytes/dendritic cells. J Immunol. 2016;196:1591–1603. doi: 10.4049/jimmunol.1500805. [DOI] [PubMed] [Google Scholar]
- 40.Pajic A, et al. Cell cycle activation by c-myc in a burkitt lymphoma model cell line. Int J Cancer. 2000;87:787–793. doi: 10.1002/1097-0215(20000915)87:6<787::aid-ijc4>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 41.Schuhmacher M, et al. Control of cell growth by c-Myc in the absence of cell division. Curr Biol. 1999;9:1255–1258. doi: 10.1016/s0960-9822(99)80507-7. [DOI] [PubMed] [Google Scholar]
- 42.Reich M, et al. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 43.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Methodol. 1995;57:289–300. [Google Scholar]
- 44.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.