Significance
Lipoprotein biosynthesis is crucial for Gram-negative bacterial viability and involves the activities of three essential integral membrane proteins embedded in the inner membrane (Lgt, LspA, and Lnt). These enzymes function sequentially to produce mature triacylated lipoproteins, many of which are then transported to the outer membrane. Lnt is responsible for catalyzing the addition of palmitate to the N terminus of diacylated apolipoproteins. Despite a number of studies that have biochemically characterized Escherichia coli Lnt, the structural basis for substrate engagement and catalysis remains unclear. Here we present the crystal structures of wild-type E. coli Lnt and a C387S active-site mutant. These structures provide insights into the molecular mechanisms of apolipoprotein N-acylation by Lnt and shed further light on the mechanism of lipoprotein biosynthesis by these essential bacterial enzymes.
Keywords: lipoprotein biosynthesis, apolipoprotein N-acyltransferase, crystal structure, enzyme mechanism
Abstract
Gram-negative bacteria express a diverse array of lipoproteins that are essential for various aspects of cell growth and virulence, including nutrient uptake, signal transduction, adhesion, conjugation, sporulation, and outer membrane protein folding. Lipoprotein maturation requires the sequential activity of three enzymes that are embedded in the cytoplasmic membrane. First, phosphatidylglycerol:prolipoprotein diacylglyceryl transferase (Lgt) recognizes a conserved lipobox motif within the prolipoprotein signal sequence and catalyzes the addition of diacylglycerol to an invariant cysteine. The signal sequence is then cleaved by signal peptidase II (LspA) to give an N-terminal S-diacylglyceryl cysteine. Finally, apolipoprotein N-acyltransferase (Lnt) catalyzes the transfer of the sn-1-acyl chain of phosphatidylethanolamine to this N-terminal cysteine, generating a mature, triacylated lipoprotein. Although structural studies of Lgt and LspA have yielded significant mechanistic insights into this essential biosynthetic pathway, the structure of Lnt has remained elusive. Here, we present crystal structures of wild-type and an active-site mutant of Escherichia coli Lnt. The structures reveal a monomeric eight-transmembrane helix fold that supports a periplasmic carbon–nitrogen hydrolase domain containing a Cys–Glu–Lys catalytic triad. Two lipids are bound at the active site in the structures, and we propose a putative phosphate recognition site where a chloride ion is coordinated near the active site. Based on these structures and complementary cell-based, biochemical, and molecular dynamics approaches, we propose a mechanism for substrate engagement and catalysis by E. coli Lnt.
The essential outer membrane (OM) of Gram-negative bacteria has a distinct composition, containing a diverse set of phospholipids, lipopolysaccharides, OM proteins, and lipoproteins (1). Escherichia coli expresses at least 90 lipoproteins that play critical roles in multiple cellular processes ranging from OM biogenesis to cell division and virulence (2). A common feature of these proteins is an N-terminal N-acyl, S-diacylglyceryl cysteine, which ensures their proper localization (3, 4). Before triacylation, preprolipoproteins are translated in the cytoplasm and transferred across the inner bacterial membrane via the Sec or Tat pathway machineries (5). These nascent preprolipoproteins have an N-terminal hydrophobic signal peptide that contains a conserved lipobox motif with the consensus sequence [LVI]-3[ASTVI][GAS]C+1, which directs them to the lipoprotein biosynthetic pathway (6–8). The maturation of lipoproteins is then catalyzed by the consecutive action of three essential integral membrane proteins (Fig. 1A). In the first step of this pathway, phosphatidylglycerol:prolipoprotein diacylglyceryl transferase (Lgt) catalyzes the covalent attachment of the sn-1,2-diacylglyceryl group from phosphatidylglycerol to the sulfydryl group of the invariant Cys+1 residue in the lipobox motif (9). This S-diacylglyceryl–modified protein is then recognized by a lipoprotein signal peptidase (LspA), which cleaves the hydrophobic signal peptide, liberating the α-amino group of the S-diacylglyceryl cysteine (10–12). Finally, apolipoprotein N-acyltransferase (Lnt) catalyzes the transfer of the sn-1-acyl chain of preferentially phosphatidylethanolamine (PE) to the free α-amino group of this cysteine, resulting in a mature, triacylated lipoprotein (13, 14). This final N-acylation step is essential for engaging the Localization of Lipoproteins (Lol) machinery that is responsible for transporting lipoproteins to the inner leaflet of the OM (15). Lgt and LspA homologs can be found in both Gram-negative and Gram-positive bacteria, whereas Lnt is only present in Gram-negative bacteria and certain GC-rich Gram-positive bacteria (16–19).
Fig. 1.
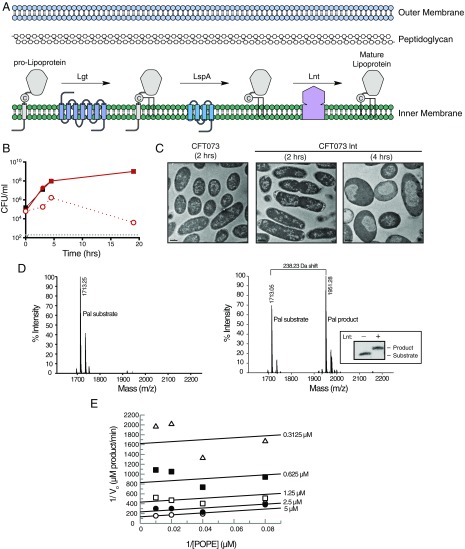
Lnt is required for in vitro and in vivo growth and is enzymatically active in vitro. (A) Schematic of the lipoprotein biosynthetic pathway in Gram-negative bacteria. (B) Lack of in vitro growth of CFT073 lnt mutant after depletion of Lnt. WT CFT073 (black) and CFT073 lnt (red) were incubated in 2% arabinose (filled symbols) or 0.2% glucose (open symbols), and cfus were enumerated at various times posttreatment. The gray dashed line indicates the limit of detection for the assay. These data are representative of two independent experiments. (C) Transmission electron microscopy of WT CFT073 and CFT073 lnt mutant after treatment for 2 or 4 h with 0.2% glucose. Bars represent 0.5 μm. (D) The Pal substrate peptideshort (Pam2Cys-SSNKNGGK-Biotin, molecular mass = 1,713 Da) and product peptideshort (Pam3Cys-SSNKNGGK-Biotin, molecular mass = 1,951 Da), as detected by SAMDI mass spectrometry. A total of 50 nM recombinant Lnt was incubated with 10 μM Pal peptide and 100 μM POPE. Reactions were either prequenched (Left) or allowed to react for 21.5 h at room temperature (Right). Inset shows a Western blot of a reaction that has been allowed to go to completion. Substrate and product peaks were monitored to determine the peptide fraction conversion [AUCproduct/(AUCproduct + AUCsubstrate)] and product concentration [fraction conversion × (peptidetotal)]. (E) SAMDI mass spectrometry was used to monitor wild-type Lnt activity over 180 min while simultaneously varying the concentrations of POPE from 12.5–100 μM and Pal peptideshort from 0.3125–5 μM. Global fitting of the initial velocities with a two-substrate ping-pong model was used to calculate the intrinsic Km(POPE), Km(Pal peptide), and kcat (55 μM, 14 μM, and 0.0093 s−1, respectively). Lineweaver–Burk double reciprocal plot was generated from the global fitting results, and the data represent velocities determined from average of duplicate samples.
The atomic structures of both Lgt and LspA have recently been elucidated, providing insight into the first two steps in the lipoprotein biosynthetic pathway (20, 21). The crystal structure of Lnt, however, has not been determined. In the absence of structural information, considerable biochemical efforts have focused on determining the general topology and essential residues of Lnt. Lnt consists of a large periplasmic carbon–nitrogen hydrolase domain that has been predicted to be anchored in the cytoplasmic membrane by at least six transmembrane helices (22). Carbon–nitrogen hydrolases comprise a large superfamily of enzymes involved in diverse activities including nitrilase, amidase, carbamylase, and N-acyltransferase catalysis. These enzymes are often multimeric and adopt a characteristic α–β–β–α sandwich fold that contains a canonical Lys–Cys–Glu catalytic triad (23). The Lnt active site is contained within its carbon–nitrogen hydrolase domain, with a catalytic triad comprised of E267, K335, and C387, each of which is essential for Lnt activity (24). A general two-step ping-pong mechanism for N-acylation by Lnt has been proposed in which a thioester acyl-enzyme intermediate is first formed at the catalytic cysteine through nucleophilic attack on the carbonyl of the sn-1-glycerophospholipid of PE, resulting in S-palmitoylated Lnt. In the second step, the acyl-enzyme intermediate is resolved by nucleophilic attack by the α-amino group of an apolipoprotein, resulting in a mature triacylated lipoprotein (24–26). Lnt has been shown to exist primarily in the acylated form in vivo, priming it for the more efficient second step of the N-acylation reaction (25). Other residues have been reported to be essential for the activity of Lnt, however their function remains unclear due to a lack of an atomic structure (24, 25, 27).
Despite considerable biochemical insights, the molecular mechanisms that underlie lipid binding and apolipoprotein N-acylation by Lnt remain unknown. Here we present the high-resolution crystal structures of wild type and a C387S mutant of E. coli Lnt. These structures reveal an eight-transmembrane helix fold and shed light on the role of conserved residues in the two-step reaction mechanism. We use a variety of approaches to confirm the mechanism of lipid binding and acyl-enzyme intermediate formation, postulating a mechanism for subsequent engagement of diacylated apolipoprotein substrates and N-acylation.
Results
In Vitro and in Vivo Characterization of E. coli Lnt.
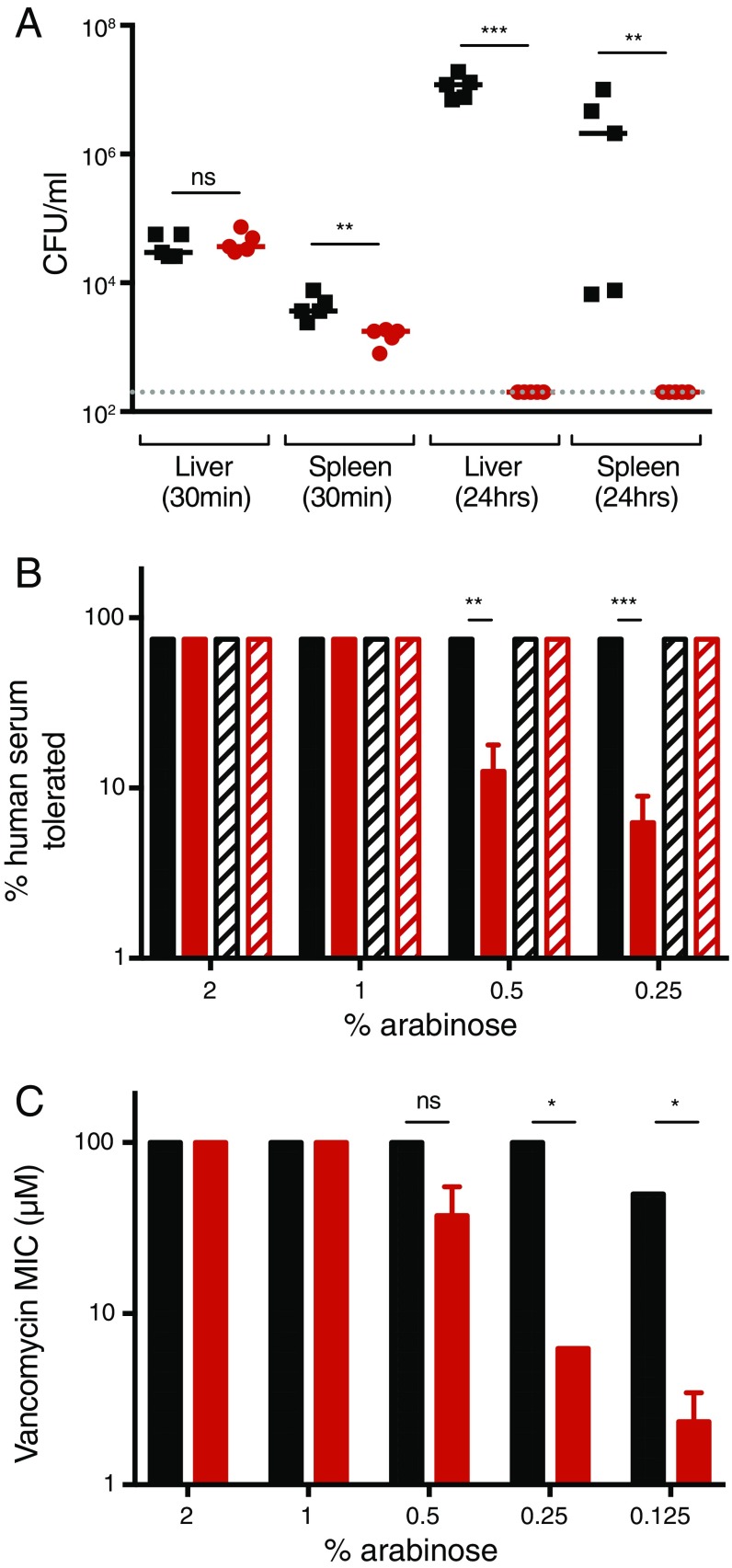
To determine the role of Lnt in clinical isolates, we generated an lnt mutant in the uropathogenic E. coli strain CFT073 (CFT073 lnt) (SI Materials and Methods and Table S1 for bacterial strains and plasmids) that allowed Lnt protein levels to be induced or repressed (28). Briefly, in the presence of arabinose, Lnt is expressed under the control of the PBAD promoter of the araBAD operon, whereas expression is repressed in the presence of glucose, a negative regulator of this operon. We then confirmed that Lnt is essential for growth in vitro, as CFT073 lnt did not grow when Lnt transcription is repressed (Fig. 1B). Further, Lnt was shown to be essential for virulence in mice, as no viable CFT073 lnt colony-forming units (cfus) were detected in the liver and spleen 24 h after i.v. infection compared with wild-type CFT073 (Fig. S1A). On a cellular level, depletion of Lnt in the CFT073 lnt mutant initially led to swelling of the periplasmic space at the bacterial poles (CFT073 lnt, 2 h), as measured by transmission electron microscopy (Fig. 1C). At later times, we noted cellular swelling and loss of density in the cytoplasm surrounding the nucleoid. Depletion of Lnt to levels that still support bacterial growth in vitro resulted in increased sensitivity to human serum killing (Fig. S1B) as well as to vancomycin (Fig. S1C), a Gram-positive antibiotic that is inactive against E. coli due to its impermeable OM. These data suggest that Lnt is essential for bacterial viability in vitro and in vivo and that depletion of Lnt results in increased OM permeability.
Table S1.
Bacterial strains and plasmids
| Strain | Description/use | Source |
| E. coli | ||
| CFT073 | Bacteremia isolate, wild-type (O6:K2:H1) | 28 |
| TOP10 | pWQ601, general cloning strain | Invitrogen |
| CFT073 lnt | CFT073 Δlnt::kan | This study |
| Plasmids | ||
| pKD4 | Kanamycin resistance (KanR) cassette flanked by FRT (FLP recognition target) sites, oriRγ | 42 |
| pKD46 | Expresses the phage λ Red recombinase, AmpR, temperature sensitive, oriRγ | 42 |
| pCP20 | Thermal induction of FLP recombinase expression, AmpR, temperature sensitive | 43 |
| pLMG18tet | IPTG-inducible lacI promoter, low copy plasmid, TetR, p15a origin | This study |
| pET52 | T7lac promoter, AmpR, high copy plasmid, f1 origin | Novagen |
Fig. S1.
E. coli Lnt is required for in vivo infection. (A) I.v. infection of neutropenic A/J mice with WT CFT073 (black) or CFT073 lnt (red) cells. At 30 min and 24 h postinfection, bacterial burdens in the liver and spleen were enumerated. These data are representative from two independent experiments. Overall P value for the ANOVA is P < 0.0001. Pairwise comparisons were analyzed using regular unpaired t test (spleen 30 min, **P = 0.0058; liver 24 h, ***P < 0.001; spleen 24 h, **P = 0.0032) and are denoted in the graphs. The gray dashed line indicates the limit of detection for the assay. (B) Increased sensitivity to serum killing after depletion of Lnt. WT CFT073 (black) and CFT073 lnt (red) were treated with decreasing concentrations of arabinose, which still allowed for CFT073 lnt in vitro growth, and incubated overnight with various concentrations of normal human serum (filled bars) or heat-inactivated human serum (hatched bars). Percent human serum that still allowed for bacterial growth overnight was graphed. These data are representative from two independent experiments performed in triplicate. Pairwise comparisons were analyzed using regular unpaired t test (**P = 0.0013, ***P < 0.001) and are denoted in the graphs. (C) Increased sensitivity to vancomycin after depletion of Lnt. WT CFT073 (black) and CFT073 lnt (red) were treated with decreasing concentrations of arabinose and incubated overnight with various concentrations of vancomycin, and MIC values were calculated the following day. These data are representative from two independent experiments. Pairwise comparisons were analyzed using regular unpaired t test (*P = 0.0121) and are denoted in the graphs. ns, nonsignificant.
In addition to these cell-based approaches to examining Lnt activity, we assessed the in vitro biochemical activity of recombinant E. coli Lnt using self-assembled monolayer desorption/ionization (SAMDI) mass spectrometry to measure accumulation of a modified peptide product (29). Following biochemical reactions, biotinylated, diacylated peptide substrate and product were captured on a SAMDI surface, and all other reaction constituents were washed away before analysis by mass spectrometry. Enzymatic activity was monitored by ratiometrically assessing the fraction conversion of the substrate to product. In the absence of Lnt, only the substrate peptide was observed. Both substrate and product were observed following an incubation with recombinantly purified Lnt at room temperature (Fig. 1D). Reactions containing wild-type Lnt were linear with regard to time and enzyme concentration in the presence of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) with both a short and long version (two versions were used due to limited availability of either peptide) of diacylated Peptidoglycan-associated lipoprotein (Pal) peptide substrates (Fig. S2). Similar to previous studies, these data suggested that wild-type Lnt functions in two steps via a ping-pong mechanism (Fig. 1E) (26). The kinetics of wild-type Lnt reacting with PE in this assay are in line with results reported previously, with a kcat/Km value of 674·M−1·s−1 (26).
Fig. S2.
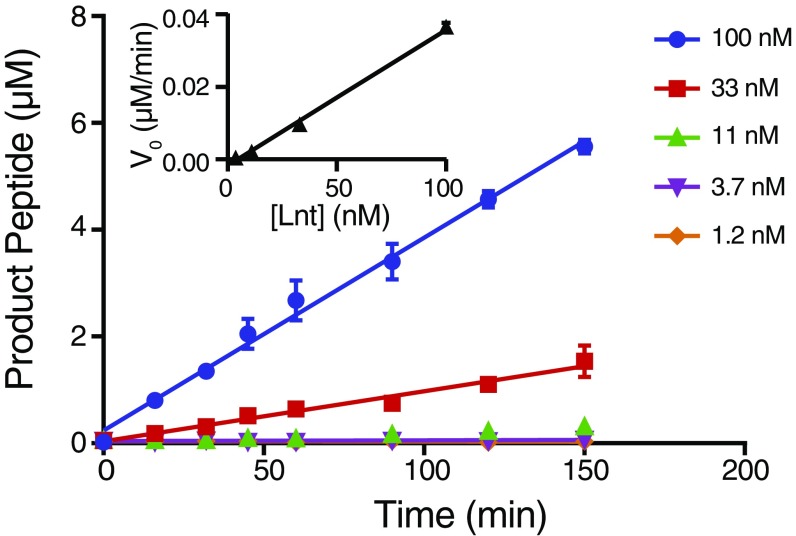
Recombinant E. coli Lnt is biochemically active. An in vitro activity assay using 1.2–100 nM purified E. coli wild-type Lnt, 11 µM Pal peptidelong, and 100 µM E. coli PE. SAMDI mass spectrometry was used to monitor the peptidelong substrate and product peaks. Error bars represent SEM, n = 3. (Inset) The activity of purified E. coli wild-type Lnt follows a linear increase in reaction velocity with increasing enzyme concentration.
Overall Structure of Wild-Type E. coli Lnt.
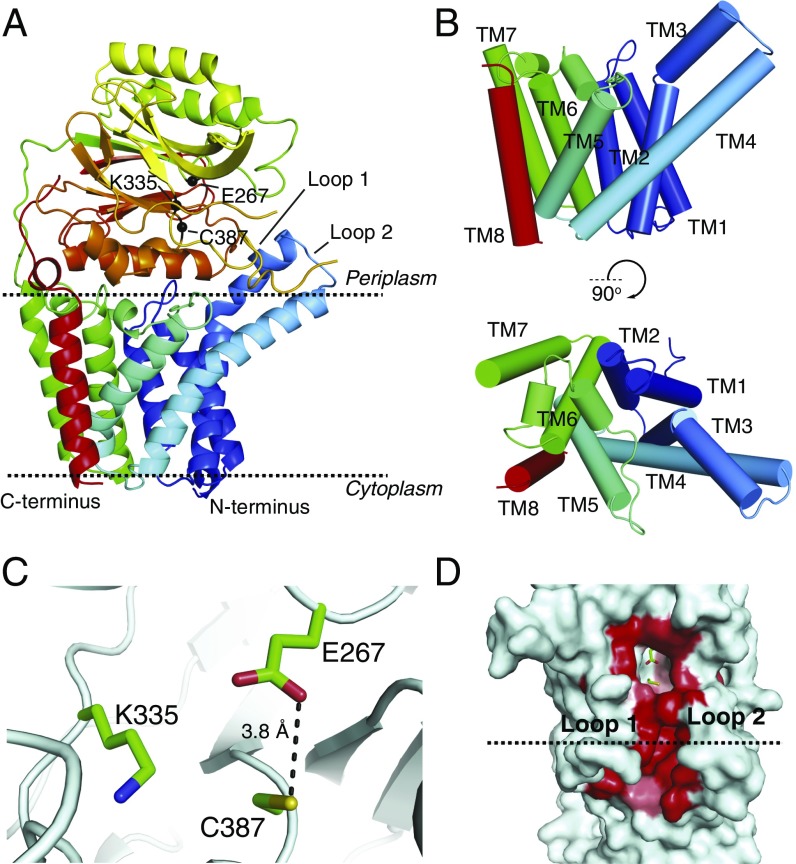
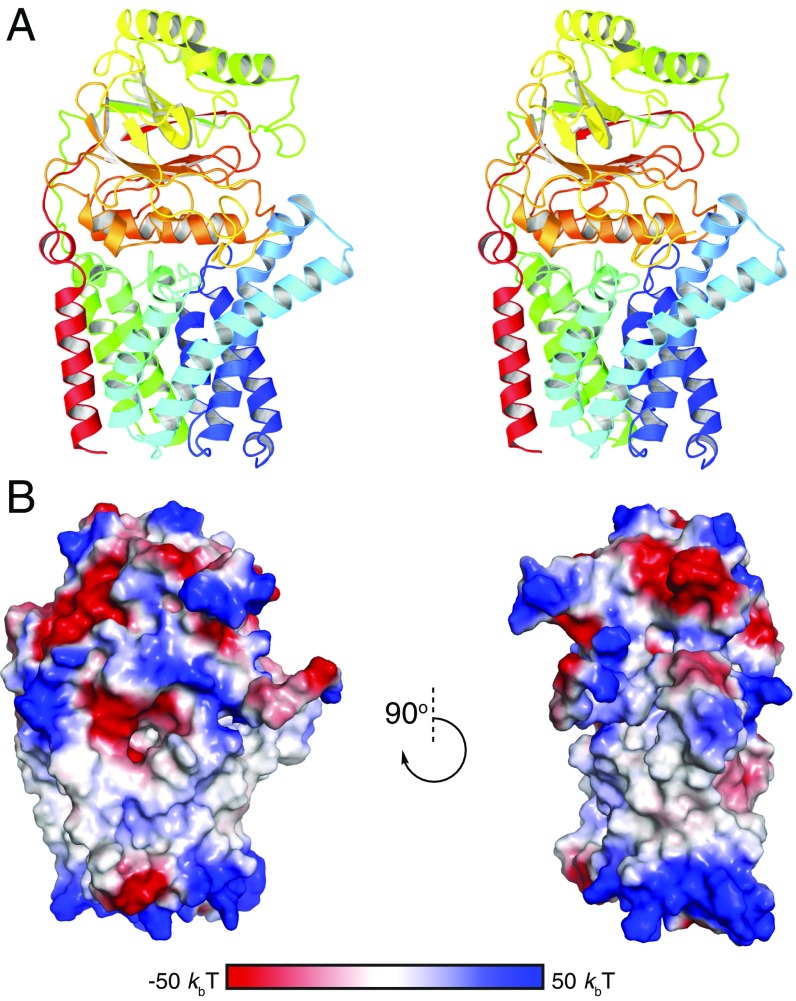
To gain mechanistic insights into lipoprotein N-acylation by E. coli Lnt, we crystallized a wild-type, selenomethionine (SeMet)-labeled Lnt by vapor diffusion and also obtained crystals of native Lnt using in meso methods. We determined the crystal structure of the SeMet-labeled Lnt at a resolution of 3.29 Å by single-wavelength anomalous dispersion (SAD) phasing. This structure was then used to phase the higher resolution (2.52 Å) native wild-type dataset (Fig. 2A and Fig. S3). Data collection and refinement statistics are shown in Table 1. Lnt crystallized as a monomer and is comprised of eight transmembrane helices (TMs 1–8; Fig. 2B) with both the N and C termini located in the cytoplasm. TM3 contains a pronounced kink at G71 such that the C-terminal portion of the helix extends away from the rest of the transmembrane helical bundle at an ∼70° angle. Side chain interactions contribute to similar angles through the membrane for TM4 and TM5, which are slightly splayed near the outer leaflet of the inner membrane to form a groove leading up into the periplasmic domain of the enzyme. TMs 5 and 6 comprise a portion of the protein that was previously presumed to form two partially membrane-embedded helices (27). The large loop between these helices (W141–V169) is primarily embedded in the membrane except for a short, amphipathic helix (K161–G168) that lies against the periplasmic side of the membrane before TM6. These helices and the intervening loop form a small, aqueous cavity that appears to allow solvent to penetrate into the membrane.
Fig. 2.
Structure of wild-type Lnt. (A) Wild-type E. coli Lnt crystal structure with the predicted boundaries of the cytoplasmic membrane indicated by dashed lines. The protein is colored in a rainbow scheme, with the N terminus in blue and the C terminus in red. The Cα positions for E267, C387, and K335 are shown as black spheres. (B) Cylinder representations of the wild-type Lnt transmembrane helices viewed in-plane with the membrane and rotated looking down from the periplasmic side of the protein. The protein is colored in a rainbow scheme, with the N terminus in blue and the C terminus in red. (C) Wild-type E. coli Lnt active site. Catalytic residues are shown in green as stick representations. (D) Surface representation of wild-type Lnt showing the hydrophobic lipid-binding groove. Active-site residues are shown in green in stick representations. Residues lining the hydrophobic groove are colored in a white-to-red scale based on degree of hydrophobicity, with the most hydrophobic residues shown in red (40).
Fig. S3.
Structure of E. coli Lnt. (A) Stereoview of the wild-type E. coli Lnt crystal structure. The protein is colored in a rainbow scheme, with the N terminus in blue and the C terminus in red. (B) Surface electrostatic potential of E. coli Lnt. Positively charged regions of the molecular surface are shown in blue. Negatively charged regions are shown in red.
Table 1.
Crystallographic data and refinement statistics
| Data collection | Lnt wild type, SeMet | Lnt wild type, native | Lnt C387S |
| Beamline | ALS 5.0.2 | APS 22ID | APS 22ID |
| Wavelength | 0.97957 | 0.9792 | 0.9792 |
| Resolution rangea | 43.82–3.29 (3.41–3.29) | 45.16–2.52 (2.61–2.52) | 42.04–2.14 (2.21–2.14) |
| Space group | P 3221 | P 212121 | P 1211 |
| Unit cell | 153.54, 153.54, 89.51 | 87.76, 158.03, 44.76 | 52.67, 72.59, 75.61 |
| 90, 90, 120 | 90, 90, 90 | 90, 101.73, 90 | |
| Total reflections | 36,544 (3,700) | 43,030 (4,220) | 60,043 (6,032) |
| Unique reflections | 18,294 (1,184) | 21,533 (1,937) | 30,212 (3,039) |
| Multiplicity | 2.0 (2.0) | 2.0 (2.0) | 2.0 (2.0) |
| Completeness, % | 93.08 (63.76) | 98.03 (91.71) | 97.36 (98.99) |
| Mean I/sigma I | 15.27 (2.34) | 10.42 (2.06) | 9.79 (2.01) |
| Wilson B factor | 55.38 | 32.82 | 27.61 |
| R-merge | 0.06087 (0.3954) | 0.06562 (0.3566) | 0.06904 (0.4642) |
| CC1/2 | 0.999 (0.751) | 0.995 (0.773) | 0.974 (0.715) |
| Refinement | |||
| Reflections used in refinement | 21,380 (1,937) | 30,207 (3,039) | |
| Reflections used for R-free | 1,099 (83) | 1,461 (153) | |
| R-work | 0.2265 (0.2729) | 0.2068 (0.2927) | |
| R-free | 0.2674 (0.3364) | 0.2502 (0.2976) | |
| Number of nonhydrogen atoms | 4,143 | 4,247 | |
| Macromolecules | 3,878 | 3,851 | |
| Ligands | 119 | 138 | |
| Solvent | 146 | 258 | |
| Protein residues | 494 | 490 | |
| rmsd bonds (Å) | 0.01 | 0.003 | |
| rmsd angles (o) | 1.14 | 0.69 | |
| Ramachandran favored, % | 94.69 | 96.71 | |
| Ramachandran allowed, % | 4.69 | 3.09 | |
| Ramachandran outliers, % | 0.61 | 0.21 | |
| Rotamer outliers, % | 1.71 | 1.72 | |
| Clashscore | 11.07 | 5.67 | |
| Average B factor | 34.28 | 33.91 | |
| Macromolecules | 33.91 | 32.45 | |
| Ligands | 44.05 | 57.45 | |
| Solvent | 36.05 | 43.05 |
Numbers in parentheses represent the high-resolution shell.
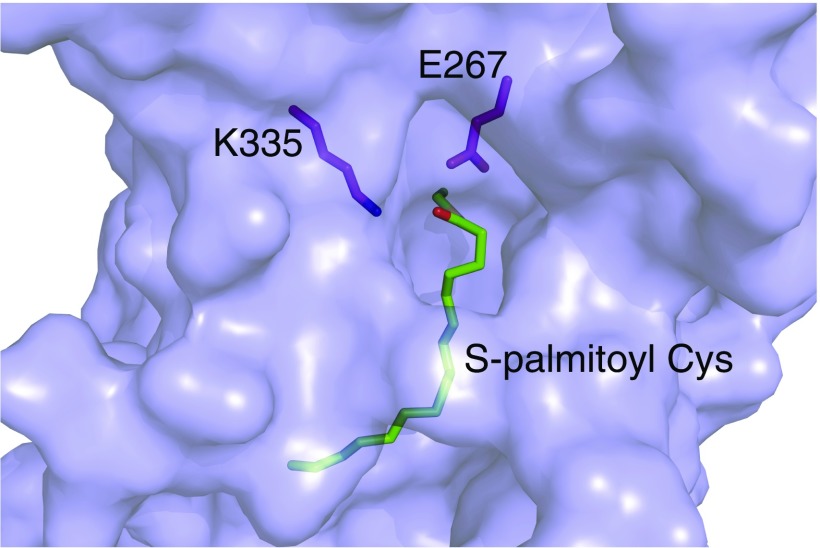
Lnt has a large periplasmic carbon–nitrogen hydrolase domain with a characteristic α–β–β–α sandwich fold between TMs 7 and 8, as expected from sequence homology analyses. The two helices near the N terminus of this domain are located fully in the periplasm, whereas the helices C-terminal to the carbon–nitrogen hydrolase beta sheets pack against the membrane. Within this domain, the catalytic residues K335, E267, and C387 exist as a preformed active site, with E267 properly positioned to act as the general base that will abstract a hydrogen from C387 and initiate the first step of catalysis (Fig. 2C). Interestingly, these residues are located in a cavity that is raised above the outer leaflet of the inner membrane, with the Cα of C387 located ∼10 Å above the predicted membrane interface. A small channel formed by highly conserved hydrophobic residues leads from the active site toward the cytoplasmic membrane (Fig. 2D). This hydrophobic cleft is enclosed on one side by a loop comprised of residues V339–V345 (loop 1) and on the other side by another loop (S78–G87; loop 2) directly preceding TM4. Both loops have high crystallographic B factors, suggesting that they are dynamic relative to the rest of the molecule. F82 is poorly ordered and extends across the hydrophobic groove.
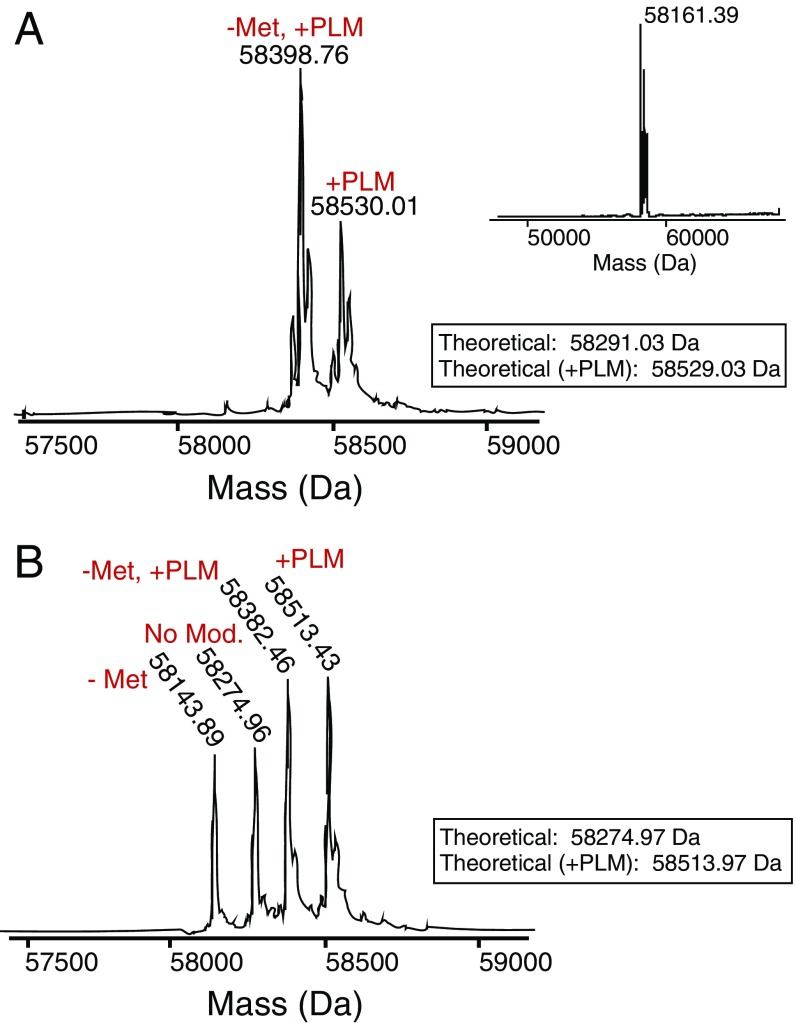
A number of lipids are bound to Lnt in the wild-type structure. Interestingly, although intact mass spectrometry analysis indicated that a large portion of the purified protein was covalently bound to palmitate (Fig. S4A), no electron density was observed that was consistent with a covalent modification of the catalytic cysteine. This lack of covalent density is likely due to hydrolysis of the lipid during crystallization, as we noted an ∼238-Da mass shift in the predominant peak following a prolonged incubation in crystallization buffer (Fig. S4A, Inset). Despite this lack of a covalent modification, the structures of wild-type Lnt showed Fo–Fc density likely corresponding to two aliphatic lipid tails leading from the active-site cavity down the hydrophobic cleft mentioned above. The presence of this density provides evidence that this hydrophobic groove represents the binding site for substrate lipids on Lnt. Due to low occupancy of lipid and poor electron density, we were unable to unambiguously identify the lipids bound in the active site of the crystal structures, and they have been built as monoolein molecules that are derived from the lipid phase of the LCP matrix.
Fig. S4.
In vitro palmitoylation of E. coli Lnt. (A) Intact MS spectrum of recombinant wild-type E. coli Lnt. “–Met” denotes a subspecies of protein that lost its N-terminal methionine during purification. Inset shows the intact MS spectrum of recombinant wild-type E. coli Lnt following an incubation in crystallization buffer with the predominant mass corresponding to an ∼238 Da shift. (B) Intact MS spectrum of recombinant C387S mutant of E. coli Lnt. “–Met” denotes a subspecies of protein that lost its N-terminal methionine during purification.
Structure of a Lipid-Bound Lnt C387S Mutant.
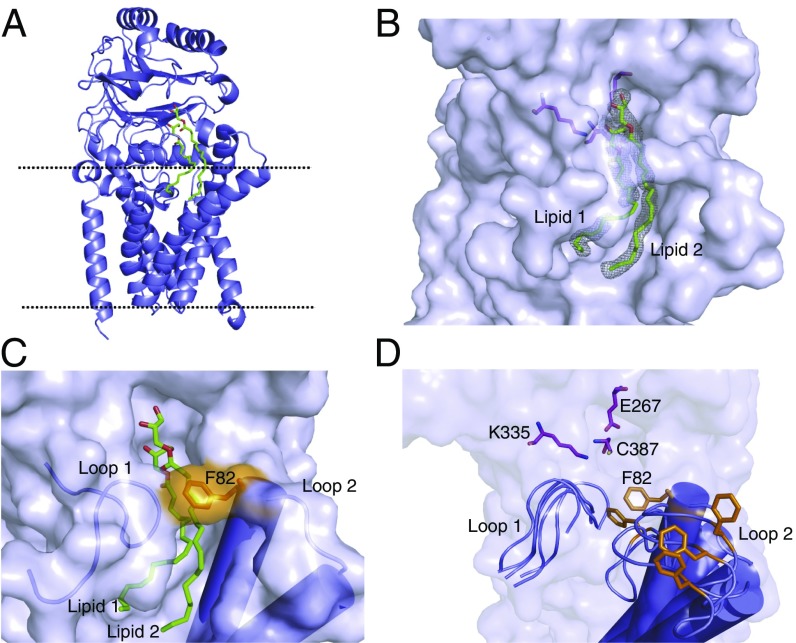
Previous studies have demonstrated that mutating the active-site cysteine of Lnt to a serine leads to the formation of an acyl-enzyme intermediate that is resistant to reducing agents (25). Such mutants are also unable to efficiently resolve the acyl enzyme intermediate in the second step of N-acylation, likely owing to the substantially higher pKa of the serine side chain, which makes it a poor leaving group compared with the cysteine in the wild-type enzyme. In an attempt to obtain a structure of Lnt that is stably and covalently bound to palmitate, we purified a C387S mutant and solved its structure in the presence of POPE to 2.14 Å resolution (Fig. 3A). In terms of overall architecture, this structure is highly similar to the wild-type Lnt structure, with an overall average Cα rmsd of 0.268 Å.
Fig. 3.
Structure of the Lnt C387S mutant. (A) Lnt C387S mutant crystal structure with the predicted boundaries of the cytoplasmic membrane indicated by dashed lines. Two bound molecules of monoolein are shown as green sticks. (B) Surface representation of the Lnt lipid-binding site. Two bound molecules of monoolein are shown as green sticks. The gray mesh represents a feature-enhanced electron density map contoured at 1 σ (41). Active-site residue side chains are shown as magenta sticks. (C) Representation of the Lnt lipid-binding groove “gate.” Two monoolein molecules are shown as green sticks. The F82 side chain is shown as orange sticks. (D) Overlay of several lipid-binding groove “gate” poses from a molecular dynamics simulation of an apo version of wild-type Lnt in a membrane environment. Active-site residues are shown as green sticks. Loops 1 and 2 are shown in a cartoon representation with cylindrical helices. The conformational flexibility of the F82 loop is highlighted by showing different poses of the residue as orange sticks.
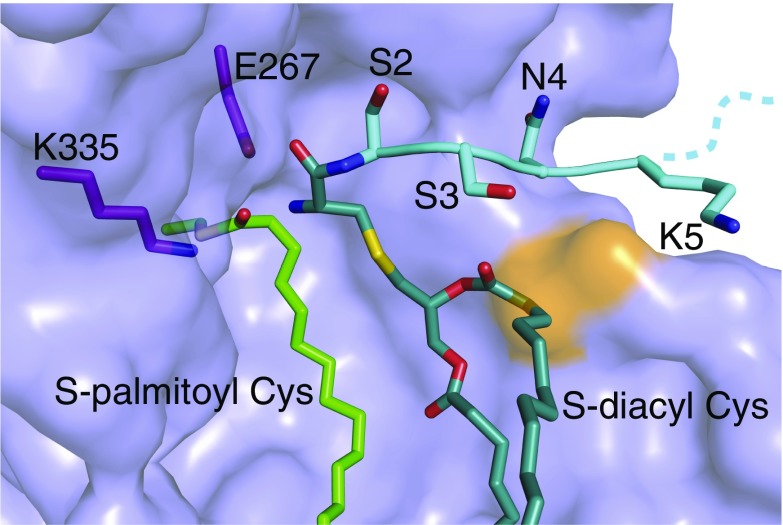
We were surprised to find that no discernable electron density corresponded to a covalently bound lipid in the C387S mutant structure. This lack of covalent modification was again in contrast to mass spectrometry analysis performed before protein crystallization (Fig. S4B). However, the crystal structure revealed the presence of two monooelin lipids (lipid 1 and lipid 2) bound at two sites (site 1 and site 2, respectively) within the hydrophobic channel that connects the active site to the membrane (Fig. 3B). Both lipid acyl chains are partially embedded in the membrane. The structure suggests that lipid 1 binds in the site that would normally be occupied by the acyl chain of the S-palmitoyl cysteine (based on its proximity to C387), representing the acyl donor. Lipid 2 binds further from the C387, likely in the site that would normally contain the lyso-PE byproduct of the first half of the reaction.
The poorly ordered loop following TM3 in the wild-type Lnt structure (loop 2) is well ordered in the C387S structure, with F82 positioned across the lipid-binding groove (Fig. 3C). This positioning has the result that both bound lipids are completely encircled by protein, indicating that Lnt may bind lipids via an induced-fit mechanism before nucleophilic attack on the sn-1-acyl chain by C387. To probe the possibility of this induced-fit mechanism further, we performed a 10-ns molecular dynamics simulation using the crystal structure of wild-type apo Lnt embedded in a POPE lipid bilayer. Indeed, this simulation shows F82 and the rest of loop 2 to be highly flexible with regard to the rest of the protein, leading us to propose that this residue may serve as a gate for lipid binding and release (Fig. 3D and Movie S1).
Lnt Active Site and a Putative Phosphate-Binding Site.
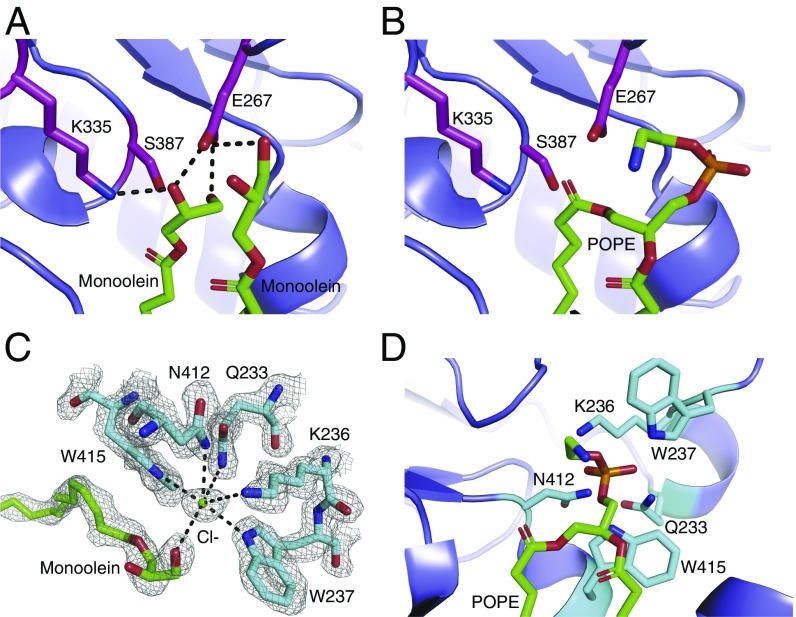
The active-site geometry in our lipid-bound C387S mutant structure supports a mechanism wherein E267 acts as a general base, abstracting a hydrogen to activate C387 for nucleophilic attack on the carbonyl group of the phospholipid sn-1-acyl chain. K335 is positioned to stabilize the oxyanion of the tetrahedral intermediate formed in both steps of the N-acylation reaction (Fig. 4 A and B). Using our structure to model a full POPE substrate places the carbonyl oxygen of the sn-1-acyl chain within hydrogen-bonding distance of K335, an interaction that would likely strengthen the electrophilic nature of the carbonyl carbon before nucleophilic attack by the catalytic cysteine. Nucleophilic attack would then form a thioester acyl-enzyme intermediate, which we have modeled in Fig. S5.
Fig. 4.
Substrate lipid recognition and catalysis by Lnt. (A) The active site of the Lnt C387S mutant. Catalytic triad residues are shown as magenta sticks. Two molecules of monoolein are shown as green sticks. Black dashed lines denote hydrogen bonds. (B) Model of POPE bound in the Lnt C387S mutant active site. Catalytic residues are shown as magenta sticks, and POPE is shown as green sticks. (C) Putative Lnt phosphate-binding site with a chloride ion bound. A monoolein molecule is shown as green sticks. Lnt residues that are coordinating the chloride ion are shown as cyan sticks. The gray mesh represents a feature-enhance electron density map contoured at 1 σ. (D) Model of POPE bound to the Lnt C387S mutant active site with the phosphoryl group positioned in the putative phosphate-binding site. POPE is shown as green sticks, and binding-site residues are shown in cyan stick representation.
Fig. S5.
Structural model of the E. coli Lnt acyl-enzyme intermediate. S-palmitoyl cysteine is shown in green stick representation, and other active-site residues are shown in magenta stick representation.
In our crystal structures, a chloride ion is bound in a cavity surrounded by the side chains of W237, W415, N412, Q233, and K236, each with coordination distances of ∼3.1–3.6 Å (Fig. 4C). Although it is possible that this ion-binding site is simply a crystallization artifact, we are intrigued that the chloride ion binds where we expect the negatively charged phosphate portion of the POPE headgroup to bind to Lnt based on our model of this substrate bound to Lnt (Fig. 4D). Additionally, it is well-established that W237 is essential for Lnt activity (24, 25, 27), although the reason behind this requirement has been elusive. Based on our structure, we therefore propose this structural element to be a potential phosphate-recognition site.
Critical Residues for Lnt Activity and E. coli Growth.
To probe the importance of key active-site, lipid-binding groove, and phosphate-binding site residues of Lnt, we examined the ability of mutations in these regions to functionally complement E. coli in the absence of wild-type Lnt expression. In addition to these cell-based assays, a subset of mutants were recombinantly expressed and purified for in vitro biochemical activity assays using SAMDI mass spectrometry.
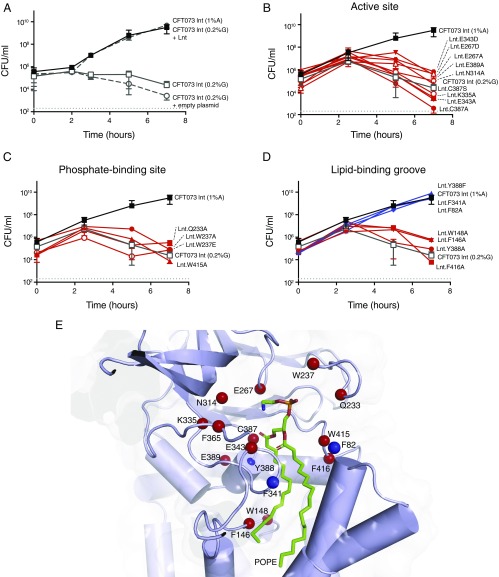
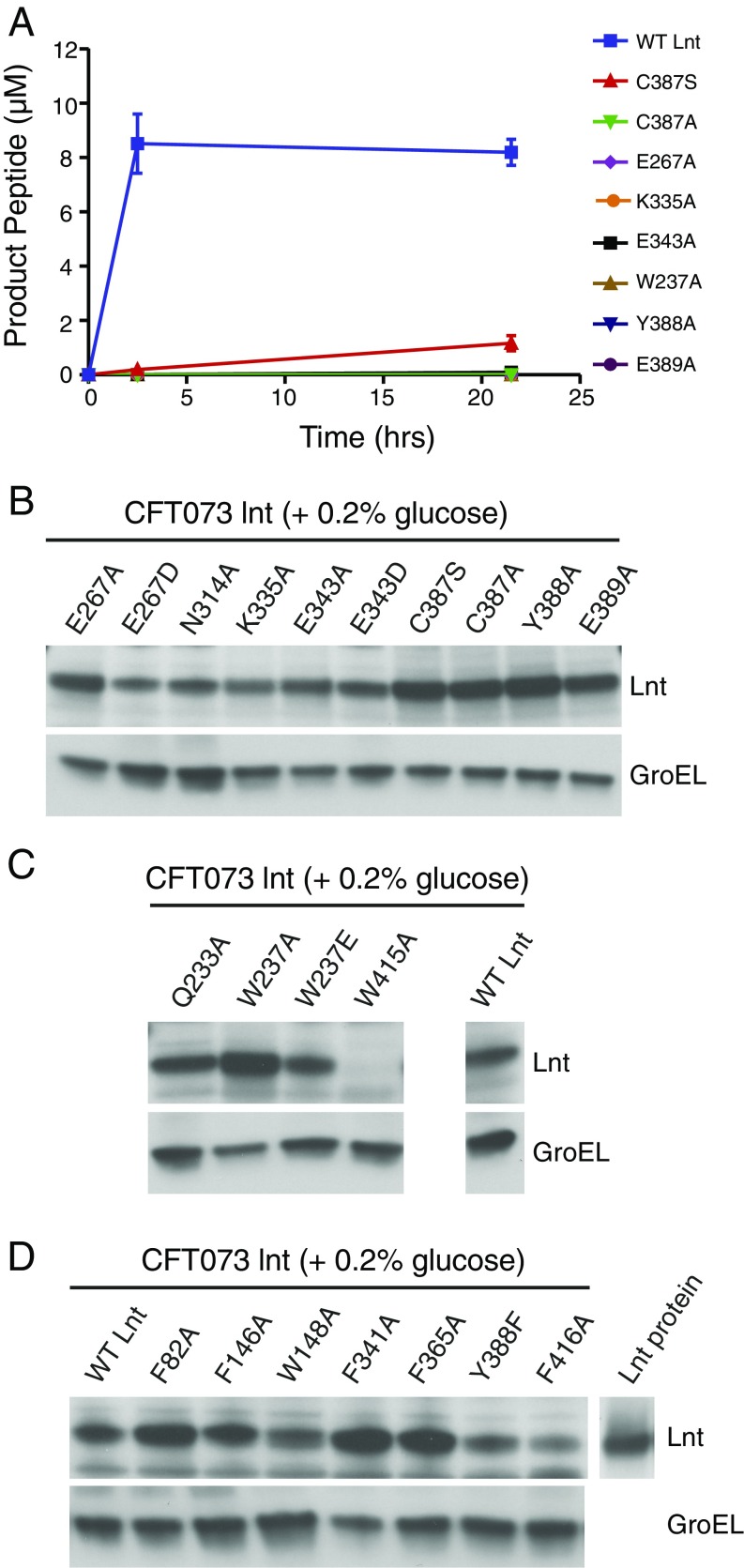
As expected, no growth of CFT073 lnt cells was detected in the absence of wild-type Lnt expression. This growth defect was rescued by a plasmid expressing wild-type Lnt but not by the empty plasmid (Fig. 5A). The first panel of mutants we tested included residues at or near the active site that have previously been demonstrated to be essential for Lnt function in E. coli (24, 25, 27): namely, C387S, C387A, K335A, E267A, E267D, E343A, E343D, and E389A. Although previous studies have not examined N314, we also included a N314A mutant in our panel, given the proximity of this residue to the active site. This mutant was unable to rescue growth of the CFT073 lnt mutant (Fig. 5B) and likely plays an important role in stabilizing the active site due to the fact that it hydrogen bonds with both E267 and E343. In accordance with previous studies, alanine mutations for all of the other residues listed above also did not rescue growth of the CFT073 lnt mutant or exhibit significant biochemical activity (Fig. 5B and Fig. S6A). All of these mutant proteins were, however, expressed in the CFT073 lnt strain (Fig. S6B). Although the C387S mutant protein showed modest in vitro activity after an overnight reaction, this was not sufficient to rescue bacterial growth of the Lnt-depleted CFT073 lnt mutant (Fig. 5B and Fig. S6A). Even the more conservative E267D and E343D mutations did not rescue CFT073 lnt growth in vitro, underscoring the importance of a properly positioned general base at position 267. In the absence of structural information, the specific role of E343 was previously unknown. In our structures, this residue hydrogen bonds with K335, likely ensuring its proper positioning for stabilizing the transition state of catalysis. Additionally, it is possible that E343 plays the dual role of substrate binding given that it makes a second hydrogen bond with the monoolein molecule that binds in site 2 in our C387S structure (this lipid represents the sn-2-acyl chain of PE). It is unclear if this hydrogen bond would also occur on a native lipid substrate or if it is simply an artifact of the crystallization system. E389 potentially serves two roles. This residue appears to stabilize the active-site conformation through a network of hydrogen bonds with the peptide backbone and a hydrogen bond with the side-chain of Y333. Beyond these interactions, this residue also likely creates a steric block that further stabilizes the positioning of K335.
Fig. 5.
Critical residues for Lnt function in E. coli. (A) Rescue of CFT073 lnt mutant growth in vitro with wild-type Lnt but not with empty plasmid. CFT073 lnt mutant was grown under Lnt depleting conditions (media containing glucose; open gray squares) containing either an empty plasmid (open gray circles) or a plasmid expressing wild-type Lnt (filled gray squares). As a control, CFT073 lnt mutant was grown under Lnt inducing conditions (media containing arabinose; filled black squares). (B) Active-site residues, (C) phosphate-binding residues, and (D) lipid-binding groove residues critical for E. coli growth. CFT073 lnt mutants lacking wild-type Lnt expression were complemented with plasmids expressing various Lnt mutants. Lnt mutants that rescue growth of the Lnt-depleted CFT073 lnt cells are shown in blue, whereas those that could not rescue growth are shown in red. The gray dashed line indicates the limit of detection for the assay. (E) Map of Lnt mutational analysis onto a model of the Lnt crystal structure with POPE bound. Mutants that were unable to rescue wild-type growth of CFT073 lnt in glucose-containing media are shown with a red sphere at the Cα position. Mutants that showed growth similar to wild type in glucose-containing media are denoted with a blue sphere at the Cα position.
Fig. S6.
Analysis of Lnt mutants. (A) Biochemical analysis of E. coli Lnt mutants. Lnt activity measurements for 500 nM wild-type and mutant enzymes at 0, 2.5, and 21.5 h in the presence of 10 μM Pal peptideshort and 100 μM POPE substrates at ambient temperature as determined by SAMDI mass spectrometry. Error bars represent SEM, n = 3. (B) Western blot analysis of Lnt expression levels in Lnt-depleted CFT073 lnt mutant (grown in 0.2% glucose) complemented with wild-type Lnt (WT Lnt) or mutants in the active-site residues. Recombinant Lnt protein was loaded to confirm the Lnt-specific band. GroEL expression was assessed as a loading control. (C) Western blot analysis of Lnt mutants of phosphate-binding residues. (D) Western blot analysis of Lnt mutants of lipid-binding groove residues.
We also sought to test the importance of the putative phosphate-binding residues that coordinate a chloride ion in our C387S mutant structure (Fig. 5C). W237 has already been shown to be critical for Lnt activity (24, 25, 27). Indeed, the W237A mutant enzyme was completely inactive biochemically, and both this mutant and a W237E mutant failed to rescue growth of the Lnt-depleted CFT073 lnt mutant despite showing protein expression in this strain (Fig. 5C and Fig. S6 A and C). The other putative phosphate-binding residues (Q233A and W415A) also failed to rescue growth of the CFT073 lnt mutant (Fig. 5C). It should be noted that in addition to participating in ion coordination, W415 also contacts the aliphatic chain of lipid 2. Interestingly, expression of the W415A Lnt protein was undetectable in CFT073 lnt cells, as detected by Western blot (Fig. S6C). These findings underscore the importance of the Lnt ion-coordinating residues, which are distant from the active site, in Lnt function, bolstering the hypothesis that they may be involved in phosphate recognition, although we cannot rule out the possibility that this site is in fact an ion coordination element that is important for overall Lnt protein stabilization.
Finally, we probed the importance of several residues located along the Lnt lipid-binding groove. Lnt-inducible knockout strains harboring F82A, F341A, and Y388F mutants all grew to levels similar to wild type, whereas F146A, W148A, F365, Y388A, and F416A mutants all could not rescue growth of the CFT073 lnt mutant despite protein being expressed (Fig. 5D and Fig. S6D). We were somewhat surprised to see that the F82 and F341 mutants exhibited normal growth, given that they are involved in enclosing lipid substrates in the lipid-binding groove of Lnt. One possible explanation for this normal growth could be that the affinity of the site for POPE is still high enough to maintain sufficient N-acylation for normal bacterial cell viability. Alternatively, it is possible that the removal of this gating mechanism may have actually increased the inherent substrate turnover, yielding a more efficient enzyme. Y388 contributes to the hydrophobic nature of the lipid-binding groove and likely makes Van der Waals interactions with what would be the acyl tail of the acyl enzyme intermediate. This residue is also located near the active site and makes backbone hydrogen-bonding interactions, but these interactions are not likely to be important for catalysis given that a Y388F mutant showed growth similar to wild type. Overall, our data confirm that the lipid-binding groove observed in our structures is important for Lnt function biochemically and in bacterial cells. The results of our mutagenesis assays are mapped onto the Lnt structure in Fig. 5E.
SI Materials and Methods
Bacterial Strains and Mutant Construction.
Bacterial strains and plasmids used are listed in Table S1. E. coli strain CFT073 was purchased from ATCC (28). E. coli cells were grown in LB medium (0.5% yeast extract, 1% tryptone, 0.5% NaCl) at 37 °C. Where indicated, kanamycin was added to culture media at a 50 μg/mL final concentration. Gene disruption in CFT073 was performed as previously described (42). The primers used to generate the CFT073 mutants are listed in Table S2. Plasmids pKD46 for the l Red recombinase, pKD4 for the integration construction, and pCP20 (43) for the FLP recombinase were used in this study.
Table S2.
Primers used in this study
| Primer | Sequence, 5′ to 3′ |
| Generation of CFT073 lnt | |
| CFT073 lnt.F | CACCCCAGCCGAAGCTGGATGAATAAAACCGAAACTGGATAGATAACTACGTGTAGGCTGGAGCTGCTTC |
| CFT073 lnt.R | CCACAACTCAATGAGAAAGTATGTTTTGTTCATGCCGGATACATCAGGCACACATATGAATATCCTCCTTAGTTCCTATTC |
| Site-directed mutagenesis for pLMG18tet complementation | |
| pLMG18tet.Lnt-his.F | GGAATTCGAGCTCGGTACCATGGCTTTTGCCTCATTAATTGAAC |
| pLMG18tet.Lnt-his.R | TGCAGGTCGACTCTAGATCAATGATGATGATGATGATGATGTTTACGTGCTCGCAGACTC |
| pLMG18tet.LntC23A-his.F | TTCGGTGCCTGCGGAACGCTG |
| pLMG18tet.LntC23A-his.R | CAGCGTTCCGCAGGCACCGAA |
| pLMG18tet.LntC62A-his.F | ATTGGCTTTGCCTGGGGATTTG |
| pLMG18tet.LntC62A-his.R | CAAATCCCCAGGCAAAGCCAAT |
| pLMG18tet.LntI74A-his.F | GGTATTAACGCGGTCTATGTC |
| pLMG18tet.LntI74A-his.R | GACATAGACCGCGTTAATACC |
| pLMG18tet.LntI79A-his.F | CTATGTCAGCGCCGCGACCTTTG |
| pLMG18tet.LntI79A-his.R | CAAAGGTCGCGGCGCTGACATAG |
| pLMG18tet.LntF82A-his.F | CATCGCGACCGCTGGCGGAATG |
| pLMG18tet.LntF82A-his.R | CATTCCGCCAGCGGTCGCGATG |
| pLMG18tet.LntF82A-his.F | CATCGCGACCGCTGGCGGAATG |
| pLMG18tet.LntF82A-his.R | CATTCCGCCAGCGGTCGCGATG |
| pLMG18tet.LntF146A-his.F | CTAACAGGCGCCCCGTGGTTAC |
| pLMG18tet.LntF146A-his.R | GTAACCACGGGGCGCCTGTTAG |
| pLMG18tet.LntW148A-his.F | CAGGCTTCCCGGCGTTACAGTTC |
| pLMG18tet.LntW148A-his.R | GAACTGTAACGCCGGGAAGCCTG |
| pLMG18tet.LntK220A-his.F | CAACCGGAAGCAACTATTCAG |
| pLMG18tet.LntK220A-his.R | CTGAATAGTTGCTTCCGGTTG |
| pLMG18tet.LntQ233A-his.F | GATATTCCGGCATCGCTGAAATG |
| pLMG18tet.LntQ233A-his.R | CATTTCAGCGATGCCGGAATATC |
| pLMG18tet.LntK236A-his.F | CGCAATCGCTGGCATGGGACGG |
| pLMG18tet.LntK236A-his.R | CCGTCCCATGCCAGCGATTGCG |
| pLMG18tet.LntW237A-his.F | CCGCAATCGCTGAAAGCGGACGGAGACCAGC |
| pLMG18tet.LntW237A-his.R | GCTGGTCTCCGTCCGCTTTCAGCGATTGCGG |
| pLMG18tet.LntW237E-his.F | CGCTGAAAGAGGACGGAGACC |
| pLMG18tet.LntW237E-his.R | GGTCTCCGTCCTCTTTCAGCG |
| pLMG18tet.LntW267A-his.F | CGTTGATTATCTGGCCGGCGTCGGCGATAACCGATC |
| pLMG18tet.LntW267A-his.R | GATCGGTTATCGCCGACGCCGGCCAGATAATCAACG |
| pLMG18tet.LntE267D-his.F | CTGGCCGGACTCGGCGAT |
| pLMG18tet.LntE267D-his.R | ATCGCCGAGTCCGGCCAG |
| pLMG18tet.LntN314A-his.F | CGATACCTACGCCACCATCATC |
| pLMG18tet.LntN314A-his.R | GATGATGGTGGGCTAGGTATCG |
| pLMG18tet.LntW335A-his.F | GCCGATCGCTATAACGCAAACCATCTGGTGCCG |
| pLMG18tet.LntW335A-his.R | CGGCACCAGATGGTTTGCGTTATAGCGATCGGC |
| pLMG18tet.LntF314A-his.F | CTGGTGCCGGCTGGCGAG |
| pLMG18tet.LntF341A-his.R | CTCGCCAGCCGGCACCAG |
| pLMG18tet.LntW343A-his.F | CTGGTGCCGTTTGGCGCGTTTGTCCCACTGG |
| pLMG18tet.LntW343A-his.R | CCAGTGGGACAAACGCGCCAAACGGCACCAG |
| pLMG18tet.LntE343D-his.F | GCCGTTTGGCGACTTTGTCCC |
| pLMG18tet.LntE343D-his.R | GGGACAAAGTCGCCAAACGGC |
| pLMG18tet.LntF365A-his.F | ATGTCTTCAGCCAGCCGCGG |
| pLMG18tet.LntF365A-his.R | CCGCGGCTGGCTGAAGACAT |
| pLMG18tet.LntW387A-his.F | CTTACTGCGGCCATTGCCTACGAAATCATTCTCGGCG |
| pLMG18tet.LntW387A-his.R | CGCCGAGAATGATTTCGTAGGCAATGGCCGCAGTAAG |
| pLMG18tet.LntW387S-his.F | CTTACTGCGGCCATTTCTTACGAAATCATTCTCGGCG |
| pLMG18tet.LntW387S-his.R | CGCCGAGAATGATTTCGTAAGAAATGGCCGCAGTAAG |
| pLMG18tet.LntY388A-his.F | CTTACTGCGGCCATTTGCGCCGAAATCATTCTCGGCG |
| pLMG18tet.LntY388A-his.R | CGCCGAGAATGATTTCGGCGCAAATGGCCGCAGTAAG |
| pLMG18tet.LntY388F-his.F | CCATTTGCTTCGAAATCATTCT |
| pLMG18tet.LntY388F-his.R | AGAATGATTTCGAAGCAAATGG |
| pLMG18tet.LntY389A-his.F | CTGCGGCCATTTGCTACGCAATCATTCTCGGCGAGC |
| pLMG18tet.LntY389A-his.R | GCTCGCCGAGAATGATTGCGTAGCAAATGGCCGCAG |
| pLMG18tet.LntW415A-his.F | AACGATGCGGCGTTTGGTAAG |
| pLMG18tet.LntW415A-his.R | CTTACCAAACGCCGCATCGTT |
| pLMG18tet.LntF416A-his.F | CGATGCGTGGGCTGGTAAGTC |
| pLMG18tet.LntF416A-his.R | GACTTACCAGCCCACGCATCG |
| Site-directed mutagenesis for recombinant Lnt protein expression | |
| pET.LntW237A-his.F | GATATTCCGCAGTCTCTGAAAGCGACGAAGGCCAGCTGCTGAACACGC |
| pET.LntW237A-his.R | CGTGTTCAGCAGCTGGCCTTCGTCGCTTTCAGAGACTGCGGAATATC |
| pET.LntW267A-his.F | GCAGCCTGATTATCTGGCCGGCGGCGCTATTACCGATCTGG |
| pET.LntW267A-his.R | CCAGATCGGTAATAGCGCCGCCGGCCAGATAATCAGGCTGC |
| pET.LntW335A-his.F | GCGCGGATCGCTATAACGCGAATCATCTGGTCCCGTTTGGTG |
| pET.LntW335A-his.R | CACCAAACGGGACCAGATGATTCGCGTTATAGCGATCCGCGC |
| pET.LntW343A-his.F | TCATCTGGTCCCGTTTGGTGCGTTTGTGCCGCTGGAATCTATTC |
| pET.LntW343A-his.R | GAATAGATTCCAGCGGCACAAACGCACCAAACGGGACCAGATGA |
| pET.LntW387A-his.F | TTGAACTGACCGCGGCGATTGCGTATGAAATTATCCTGGGCGAAC |
| pET.LntW387A-his.R | GTTCGCCCAGGATAATTTCATACGCAATCGCCGCGGTCAGTTCAA |
| pET.LntW387S-his.F | TTGAACTGACCGCGGCGATTTCGTATGAAATTATCCTGGGCGAAC |
| pET.LntW387S-his.R | GTTCGCCCAGGATAATTTCATACGAAATCGCCGCGGTCAGTTCAA |
| pET.LntY388A-his.F | GAACTGACCGCGGCGATTTGCGCGGAAATTATCCTGGGCGAACAAGTG |
| pET.LntY388A-his.R | CACTTGTTCGCCCAGGATAATTTCCGCGCAAATCGCCGCGGTCAGTTC |
| pET.LntY389A-his.F | CTGACCGCGGCGATTTGCTATGCGATTATCCTGGGCGAACAAGTGCG |
| pET.LntY389A-his.R | CGCACTTGTTCGCCCAGGATAATCGCATAGCAAATCGCCGCGGTCAG |
Anti-Lnt Antibody.
For recombinant protein expression, a DNA fragment encoding Lnt was cloned into a modified pAcGP67A vector downstream of the polyhedron promoter with an N-terminal 8×His-tag. Recombinant baculovirus was generated using the Baculogold system (BD Biosciences) following standard protocols, and Trichoplusia ni cells were infected for protein production and harvested 48 h postinfection. Cells expressing Lnt were resuspended in lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 10% glycerol, 1 mM TCEP, 20 mM imidazole) with complete EDTA-free protease inhibitor tablets (Roche) added and stirred for 1 h at 4 °C. Following three passages through an LV1 microfluidizer (Microfluidics), the lysate was centrifuged at 40,000 rpm for 1 h at 4 °C and supernatant passed through a 0.45-nm filter. Clarified supernatant was passed over a Ni-NTA resin and washed with 45 CVs of buffer A + 0.1% Triton X-114 and then subsequently washed with another 50 CVs of buffer A (20 mM Tris, pH 7.5, 300 mM NaCl, 10% glycerol, 20 mM imidazole, 0.5 mM TCEP). Protein was eluted with buffer B (20 mM Tris, pH 7.5, 300 mM NaCl, 10% glycerol, 250 mM imidazole, 0.5 mM TCEP) and then passed over a Superdex 200 16/60 column in buffer C (50 mM Tris, pH 7.5, 150 mM NaCl, 10% glycerol, 0.5 mM TCEP). The peak fraction was pooled and diluted with buffer D (20 mM Mes, pH 6.0, 10% glycerol, 0.5 mM TCEP) to an NaCl concentration of 100 mM and loaded onto a HiTrap SP column. Following a column wash, the protein was eluted with a gradient of buffer E (20 mM Mes, pH 6.0, 1 M NaCl, 10% glycerol, 0.5 mM TCEP). Finally, the Lnt peak fraction was collected, concentrated, and desalted into buffer C. Two rabbits were each immunized with 500 μg Lnt in Complete Freund’s Adjuvant (via multiple s.c. and intradermal site injections) followed up by four biweekly boosts with 250 μg Lnt in Incomplete Freund’s Adjuvant, after which production bleeds were obtained and Lnt affinity-purified polyclonal antibody was generated.
SDS/PAGE and Western Immunoblotting.
Lnt substrate peptide (Pam2Cys-SSNKNASNDGSEGMLGAGTGMDK-Biotin) was custom synthesized at AnaSpec. Total in vitro Lnt reactions (performed as described in the main text) were separated by tricine SDS/PAGE (4% stacking, 10% spacer, and 16% resolving gels). Western blot analysis was performed using the iBlot 2 Dry Blotting System (ThermoFisher Scientific). Nitrocellulose membranes were blocked in blocking buffer (PBS containing 5% BSA and 0.05% Tween-20) for 30 min and then incubated overnight at 4 °C with an anti-biotin antibody (Invitrogen) followed by an anti-mouse secondary antibody (Licor IFDye). Detection was performed using Licor Odyssey software. For detection of Lnt expression in bacterial cells, total lysates expressing wild-type or mutant Lnt proteins were resuspended in lysis buffer (4% SDS, 10 mM Tris·HCl, pH 7.5, 10 mM EDTA) and separated by SDS/PAGE (10–20% resolving gels). Western blot analysis was performed using the iBlot 2 Dry Blotting System (ThermoFisher Scientific). PVDF membranes were blocked in blocking buffer (PBS containing 5% BSA and 0.05% Tween-20) for 30 min and then incubated overnight at 4 °C with one of the following antibodies: anti-His monoclonal antibody (Cell Signaling Technology; 1:1,000 final dilution), rabbit anti-Lnt polyclonal antibody (1:10,000 final dilution), and rabbit anti-GroEL (1:10,000 final dilution). The membranes were washed with PBS + 0.05% Tween-20 for 1 h and then incubated with secondary antibodies: donkey anti-rabbit conjugated to horseradish peroxidase (ThermoFisher Scientific) at a 1:10,000 final dilution, goat anti-rabbit conjugated to alkaline phosphatase (Cedarlane Laboratories) at a 1:5,000 final dilution, and goat anti-mouse conjugated to alkaline phosphatase (Jackson ImmunoResearch) at a 1:3,333 final dilution.
Statistical Analyses.
All statistical analyses were performed using Prism software (GraphPad). All graphs represent the mean ± the SEM. Unless stated otherwise, P values for all data were determined using regular unpaired t test (*P < 0.05, **P < 0.01, and ***P < 0.001). The data were tested for being parametric, and statistical analyses were performed on log-transformed data. P values for mouse lethality studies were determined using log-rank (Mantel–Cox) test and adjusted for multiple testing using Bonferroni correction.
Discussion
We have determined the crystal structures of both wild-type and a C387S mutant of E. coli Lnt with two lipids bound in the active site. These structures provide a detailed view of lipid substrate binding and the catalytic mechanism, which is supported by our mutational analysis. One question that remains unanswered is how a lipid or diacylated substrate peptide is extracted from the lipid bilayer and transported ∼10 Å into the periplasmic catalytic site of Lnt. Our molecular dynamics simulations suggest that the helices and loops that make up the walls of the lipid-binding groove are quite mobile. Given that this is a highly dynamic region of the protein, it is reasonable to assume that Brownian motion and appropriate gating by the protein may be the primary driver of lipid movement into the Lnt active site.
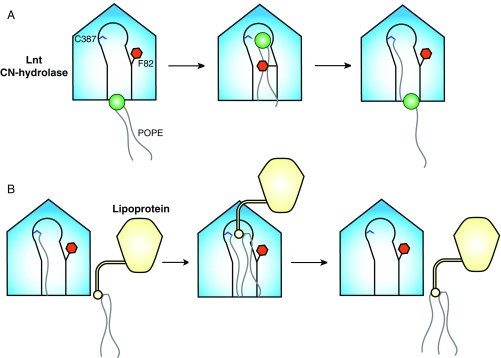
The Lnt crystal structures allow us to suggest a mechanism by which the protein regulates the entry of substrates to the active-site cavity and the exit of products (Fig. 6). We propose that in the crystal structure of the C387S mutant, lipid 2 occupies the site that would normally contain either the lyso-PE product of the first half of the reaction or one of the substrate peptide acyl groups that will help direct the α-amino group of the lipoprotein to the thioester carbonyl of the acyl-enzyme intermediate for nucleophilic attack. Based on our studies, initial substrate binding likely involves an induced-fit mechanism wherein loop 1 and loop 2 open to allow the PE substrate entry to the active site. Upon substrate binding, the loops close and the F82 and F341 side chains clamp down on the acyl chains of the lipid, restricting their motion and facilitating the nucleophilic attack of the catalytic C387 on the carbonyl of the PE sn-1-acyl chain. Upon formation of the acyl-enzyme intermediate, the Phe clamp is capable of opening and allowing the noncovalent product to exit site 2 (Fig. 6A). This would then enable entry of one of the acyl chains of the diacylated protein substrate into site 2. Based on modeling this interaction into the C387S mutant crystal structure (Fig. S7), it is possible that the Phe clamp would remain open during this step to accommodate the presence of a third acyl chain. Binding would likely be mediated by the diacylglyceryl moiety of the substrate peptide rather than the peptide itself, given that E. coli lipoproteins have diverse sequences downstream of the conserved cysteine and are all substrates of Lnt. It is possible, however, that Lnt makes hydrogen-bonding interactions with the main chain of the substrate peptide. Binding would position the α-amino group of the lipoprotein substrate for nucleophilic attack on the carbonyl of the S-palmitoyl cysteine intermediate. The mature, triacylated lipoprotein product would then be released, and the now vacant lipid-binding groove would be available to bind another PE substrate (Fig. 6B).
Fig. 6.
Model of lipoprotein N-acylation by E. coli Lnt. (A) Formation of an acyl-enzyme intermediate. Loop 2 of apo Lnt is highly flexible, with the gatekeeper F82 in the open position some portion of the time. A molecule of POPE first binds to the lipid-binding groove, which causes F82 to close, placing the sn-1-acyl chain carbonyl in position for nucleophilic attack by C387. This reaction forms a S-palmitoyl cysteine-modified form of Lnt. The F82 gate then opens, allowing the lyso-PE byproduct to exit. (B) Lipoprotein N-acylation. The F82 gate is mobile in the acyl-enzyme intermediate form of Lnt. This allows entrance of one acyl chain of an S-diacylated lipoprotein substrate. Entry of the S-diacylglyceryl cysteine into the vacant lipid channel positions the α-amino group of the lipoprotein N-terminal cysteine for nucleophilic attack on the carbonyl of the S-palmitoyl cysteine of Lnt. This mature, triacylated lipoprotein then exits for translocation to the OM by the Lol machinery.
Fig. S7.
Structural model of Pal substrate peptide engagement by the E. coli Lnt acyl-enzyme. S-palmitoyl cysteine is shown in green stick representation, and other active-site residues are shown in magenta stick representation. A diacylated Pal substrate peptide is shown in cyan stick representation. Loop 1 has been hidden for clarity. F82 is shown in an orange surface representation and has been truncated to accommodate the diacylated substrate peptide. The need to truncate this residue suggests that a conformational change in loop 2 would likely be required for peptide engagement.
Overall, our structures provide a more complete picture of the lipoprotein biosynthetic pathway in Gram-negative bacteria. One question that remains to be answered is how lipoproteins are transferred from one enzyme to the next during this biosynthetic process. It is unclear if this is achieved by simple 2D diffusion through the membrane or if these proteins may exist as a higher order lipoprotein biosynthetic complex to facilitate more efficient lipidation. Further studies into this possibility will certainly deepen our understanding of this important biochemical pathway in Gram-negative bacteria.
Materials and Methods
Lnt Expression and Purification.
E. coli Lnt constructs containing a noncleavable C-terminal 6×His tag were expressed recombinantly in E. coli BL21 (DE3) cells by 64 h autoinduction at 16 °C. Cells were harvested by centrifugation at 4,668 × g for 20 min at 4 °C, resuspended in lysis buffer (20 mM Tris, pH 8.0, 500 mM NaCl, 10% glycerol, 1× complete protease inhibitor mixture; Roche), and lysed by three passages through a microfluidizer at 10,000 psi. Cell debris was removed by centrifugation at 18,000 g for 15 min at 4 °C. The membrane fraction was then isolated by ultracentrifugation at 125,000 g for 1 h at 4 °C. The membrane pellet was resuspended in buffer A (20 mM Tris, pH 8.0, 300 mM NaCl, 5 mM imidazole, 10% glycerol, 1% DDM) supplemented with 1× complete protease inhibitor mixture and agitated overnight at 4 °C using a magnetic stirrer. Insoluble material was separated by ultracentrifugation at 125,000 g for 1 h at 4 °C. The supernatant was then incubated with cobalt affinity resin for 1 h at 4 °C on a nutator. The resin was added to a gravity flow column and washed with five column volumes (CVs) of buffer A followed by five CVs of buffer B (20 mM Tris, pH 8.0, 300 mM NaCl, 5 mM imidazole, 10% glycerol, 0.02% DDM). Bound protein was eluted with buffer B containing 250 mM imidazole. Pure fractions were combined and applied to a Superdex 200 16/60 column that had been pre-equilibrated with buffer C (20 mM Tris, pH 8.0, 250 mM NaCl, 1 mM TCEP, 5% glycerol, 0.02% DDM). Peak fractions containing Lnt were combined and concentrated to 25–40 mg/mL.
Crystallization.
Lnt at 40 mg/mL was reconstituted into the lipidic cubic phase (LCP) by mixing with monoolein at a 2:3 (wt/vol) protein-to-lipid ratio using two coupled syringes. A Mosquito LCP robot (TTP Labtech) was used to dispense 50 nL boluses of protein-laden mesophase overlayed with 800 nL precipitant in 96-well glass sandwich plates. Crystals with P212121 symmetry were obtained at 20 °C using a precipitant solution containing 50 mM ADA, pH 6.5, and 24% PEG 400. Crystals were harvested using MiTeGen cryoloops and flash-cooled in liquid nitrogen without added cryoprotectant.
The C387S mutant of Lnt was also crystallized in LCP and grew in a condition containing 0.1 M MES, pH 5.7–6.3, 27% PEG 500 DME, 0.1 M sodium chloride, 0.1 M magnesium chloride, and 0.01 M copper(II) chloride dihydrate as the precipitant. The LCP host lipid contained a monoolein mixture containing 0.25 mol% POPE. These crystals grew at 20 °C, had P21 symmetry, and were harvested as above.
For phasing efforts, aliquots of SeMet-labeled Lnt at 25 mg/mL were adjusted to 10 mg/mL with buffer C. Twenty-five microliter aliquots of SeMet-labeled Lnt were then supplemented with 300 μg E. coli polar lipid extract (Avanti Polar Lipids) and incubated overnight at 4 °C on a nutator. Aggregates were removed from the lipidated sample by centrifugation at 20,000 g for 90 min at 4 °C. Crystallization was carried out at 18 °C using the hanging drop vapor diffusion method combined with streak seeding using a seed stock containing native Lnt crystals obtained by the same method. Crystals with P3221 symmetry were obtained in a precipitant solution containing 50 mM sodium acetate, pH 5.0, 50 mM magnesium acetate, and 28–36% PEG 200. Crystals were cryoprotected in reservoir solution containing 40% PEG 200 and flash-cooled in liquid nitrogen.
Structure Determination.
Initial phases of wild-type Lnt were obtained by SAD from data collected using beamline 5.0.2 at the Advanced Light Source (ALS) on SeMet derivative crystals. The heavy atom substructure of 12 selenium atoms was determined by the Hybrid Substructure Search submodule in the Phenix package (30), and initial phases were obtained using Autosolve (31). Automatic chain tracing using Autobuild (32) yielded an interpretable electron density map, and this was used as an initial guide for building an intermediate model. The intermediate model was used as a search model in Phaser (33) to phase a native dataset collected on crystals grown using LCP at beamline 22-ID of the Advanced Photon Source (APS). Iterative rounds of refinement and manual rebuilding using PHENIX (34) and Coot (35) resulted in an almost complete chain of Lnt. The wild-type model was used to phase the Lnt C387S datasets.
Molecular Dynamics Simulations.
Molecular dynamics simulations of wild-type Lnt embedded in a POPE lipid bilayer was performed using Desmond Molecular Dynamics submodule from the Schrödinger suite of programs (36). Briefly, the crystal structure of wild-type apo Lnt was prepared by building missing loops using Prime (37), bond orders assigned, and energy minimized with hydrogen bond optimization within Maestro (Schrödinger). The minimized protein was then placed in a simulation box that satisfied the periodic boundary conditions, embedded in a POPE membrane bilayer, and solvent and counterions were added to neutralize the system. The position of the membrane was adjusted based on the Orientation of Proteins in Membranes database (38). The membrane-embedded protein system was relaxed before running the 10-ns molecular dynamics simulation. The trajectory was analyzed using the tools in Desmond and exported to PyMOL to generate the movie of the molecular dynamics simulation.
In Vitro Growth and Serum-Killing Assays.
For in vitro growth curves, overnight cultures of WT CFT073 and CFT073 lnt grown in Luria–Bertani (LB) medium containing 2% arabinose were back-diluted 1:100, grown to midexponential phase (OD600 = 0.6), and then diluted 1:100 in media containing either 2% arabinose or 0.2% glucose to initiate growth curves. For complementation studies in the CFT073 lnt mutant, Lnt mutants were generated using the QuikChange Lightning Site-Directed Mutagenesis kit (Agilent Technologies) as per the manufacturer recommendations. Primers used to generate the mutant Lnt-expressing plasmids are shown in Table S2. Wild-type or mutant lnt coding regions were cloned into the pLMG18tet plasmid. The cfus were counted by plating on LB + kanamycin (50 μg/mL) agar. Serum sensitivity assays were carried out using normal human sera (nHS) pooled from six donors, at 50% serum concentration. A Checkerboard microdilution minimal inhibition concentration (MIC) assay was performed to determine if decreasing Lnt expression (by decreasing arabinose concentrations in the growth medium) from the pBAD plasmid increases sensitivity to vancomycin or human serum killing. WT CFT073 or CFT073 lnt colonies were picked from an overnight LB plate, and MIC assays were performed according to Clinical and Laboratory Standards Institute (CLSI) guidelines.
Biochemical Assays.
The enzymatic activity of wild-type and mutant E. coli Lnt was quantified using SAMDI mass spectrometry (SAMDI Tech, Inc.) by monitoring the ratio of substrate (diacylated) and product (triacylated) peptides and calculating the fraction conversion as AUCtriacylated/(AUCtriacylated + AUCdiacylated). The amount of product peptide formed was determined by multiplying the fraction conversion by the total peptide concentration. Two peptide substrates were synthesized based on the Pal lipoprotein, Pal peptideshort (Pam2Cys-SSNKNGGK-Biotin; CPC Scientific, Inc.) and Pal peptidelong (Pam2Cys-SSNKNASNDGSEGMLGAGTGMDK-Biotin; AnaSpec, Inc.). Reactions were carried out at room temperature in Lnt assay buffer [50 mM Tris, pH 7.5, 150 mM NaCl, 0.05% DDM (Anatrace); 0.05% bovine skin gelatin (Sigma); and 1 mM TCEP in a 384-well polypropylene plate (Greiner)]. Two lipid substrates were used, POPE (Avanti Polar Lipids) formulated as POPE/DDM mixed micelles, and E. coli l-α-PE in chloroform (Avanti Polar Lipids). Enzymatic activity was monitored in the presence of PE or POPE and peptideshort or peptidelong. Reactions were quenched by the addition of a final concentration of 0.5% formic acid. The kinetic mechanism of wild-type Lnt was assessed by simultaneously varying the concentration of the peptideshort and POPE substrates and monitoring enzyme activity over 3 h. Intrinsic Km and kcat values for each substrate were determined by global fitting the initial, linear enzymatic rates as a function of concentration of both substrates according to two substrate ping-pong (ν = Vmax[A][B]/(KB[A]+KA[B]+[A][B])) or ternary mechanisms (ν = Vmax[A][B]/(KA´KB+ KB[A]+KA[B]+[A][B])), where A is the POPE substrate and B is the peptideshort substrate. Reaction rates were determined using Prism software (GraphPad Software), and global fitting was done using Grafit software (Erithacus Software, Ltd.). Global fitting was also used to distinguish between these two general mechanisms. Lineweaver–Burk plots of the reciprocal initial velocity (1/V0) versus the reciprocal of the POPE concentration (1/[A]) at each fixed peptide substrate concentration were generated for visualization purposes using the global fitting results.
Transmission Electron Microscopy.
Bacteria were grown to midexponential phase, washed with PBS, and incubated in modified Karnovsky’s fixative (0.1 M sodium cacodylate, pH 7.2, 2% paraformaldehyde, and 2.5% glutaraldehyde). Samples were then postfixed in 1% aqueous osmium tetroxide (EM Sciences) for 1 h followed by overnight incubation in 0.5% uranyl acetate. The samples were then dehydrated through an ascending series of ethanol followed by propylene oxide and embedded in Eponate 12 (Ted Pella, Inc.). Ultrathin sections (80 nm) were cut with an Ultracut microtome (Leica), stained with 0.2% lead citrate, and examined in a JEOL JEM-1400 transmission electron microscope at 80 kV. Digital images were captured with a GATAN Ultrascan 1000 CCD camera.
Mouse Infection Model.
Overnight bacterial cultures of WT CFT073 and CFT073 lnt grown in medium containing 2% arabinose were back diluted 1:100 in M9 media and grown to an OD600 of 0.8–1 at 37 °C. Cells were harvested, washed once with PBS, and resuspended in PBS containing 10% glycerol. Cells were frozen in aliquots, and thawed aliquots were measured for cfus before mouse infections. Virulence of WT CFT073 and CFT073 lnt was measured using the neutropenic E. coli infection model (39). Briefly, 7-wk-old A/J mice (Jackson Laboratory) were rendered neutropenic by peritoneal injection of two doses of cyclophosphamide (150 mg/kg on day –4 and 100 mg/kg on day –1). On day 0, mice were infected with 5 × 105 cfu mid-exponential phase bacteria diluted in PBS by i.v. injection through the tail vein. At 30 min and 24 h post infection, bacterial burdens in the liver and spleen were determined by serial dilutions of tissue homogenates on LB plates.
Supplementary Material
Acknowledgments
We thank members of the Genentech Structural Biology and Infectious Diseases departments for their valuable feedback on this research. Jim Kiefer collected the SAD data. Yiming Xu and SAMDI Technology provided assistance with assay development and data analysis. Michael Laird and Luis Masip helped in the generation of the pLMG18 IPTG-inducible plasmid. We also thank the members of the Biomolecular Resource Group for providing the necessary reagents to enable this study. This research used the resources of the Advanced Light Source, which is a Department of Energy (DOE) Office of Science User Facility under contract no. DE-AC02-05CH11231 and the resources of the Advanced Photon Source, a DOE Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. We are grateful for the support of the beamline staff on beamlines ALS 5.0.2 and APS 22ID.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5VRG and 5VRH).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707813114/-/DCSupplemental.
References
- 1.Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2:1–16. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Narita S-I, Matsuyama S-I, Tokuda H. Lipoprotein trafficking in Escherichia coli. Arch Microbiol. 2004;182:1–6. doi: 10.1007/s00203-004-0682-4. [DOI] [PubMed] [Google Scholar]
- 3.Braun V, Rehn K. Chemical characterization, spatial distribution and function of a lipoprotein (murein‐lipoprotein) of the E. coli cell wall. FEBS J. 1969;10:426–438. doi: 10.1111/j.1432-1033.1969.tb00707.x. [DOI] [PubMed] [Google Scholar]
- 4.Hantke K, Braun V. Covalent binding of lipid to protein. Diglyceride and amide-linked fatty acid at the N-terminal end of the murein-lipoprotein of the Escherichia coli outer membrane. Eur J Biochem. 1973;34:284–296. doi: 10.1111/j.1432-1033.1973.tb02757.x. [DOI] [PubMed] [Google Scholar]
- 5.Natale P, Brüser T, Driessen AJM. Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane—Distinct translocases and mechanisms. Biochim Biophys Acta. 2008;1778:1735–1756. doi: 10.1016/j.bbamem.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 6.Heijne von G The structure of signal peptides from bacterial lipoproteins. Protein Eng Des Sel. 1989;2:531–534. doi: 10.1093/protein/2.7.531. [DOI] [PubMed] [Google Scholar]
- 7.Juncker AS, et al. Prediction of lipoprotein signal peptides in Gram‐negative bacteria. Protein Sci. 2003;12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inouye S, Wang S, Sekizawa J, Halegoua S, Inouye M. Amino acid sequence for the peptide extension on the prolipoprotein of the Escherichia coli outer membrane. Proc Natl Acad Sci USA. 1977;74:1004–1008. doi: 10.1073/pnas.74.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sankaran K, Wu HC. Lipid modification of bacterial prolipoprotein. Transfer of diacylglyceryl moiety from phosphatidylglycerol. J Biol Chem. 1994;269:19701–19706. [PubMed] [Google Scholar]
- 10.Hussain M, Ichihara S, Mizushima S. Mechanism of signal peptide cleavage in the biosynthesis of the major lipoprotein of the Escherichia coli outer membrane. J Biol Chem. 1982;257:5177–5182. [PubMed] [Google Scholar]
- 11.Tokunaga M, Tokunaga H, Wu HC. Post-translational modification and processing of Escherichia coli prolipoprotein in vitro. Proc Natl Acad Sci USA. 1982;79:2255–2259. doi: 10.1073/pnas.79.7.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tokunaga M, Loranger JM, Wolfe PB, Wu HC. Prolipoprotein signal peptidase in Escherichia coli is distinct from the M13 procoat protein signal peptidase. J Biol Chem. 1982;257:9922–9925. [PubMed] [Google Scholar]
- 13.Jackowski S, Rock CO. Transfer of fatty acids from the 1-position of phosphatidylethanolamine to the major outer membrane lipoprotein of Escherichia coli. J Biol Chem. 1986;261:11328–11333. [PubMed] [Google Scholar]
- 14.Gupta SD, Wu HC. Identification and subcellular localization of apolipoprotein N-acyltransferase in Escherichia coli. FEMS Microbiol Lett. 1991;78:37–41. doi: 10.1016/0378-1097(91)90251-5. [DOI] [PubMed] [Google Scholar]
- 15.Fukuda A, et al. Aminoacylation of the N-terminal cysteine is essential for Lol-dependent release of lipoproteins from membranes but does not depend on lipoprotein sorting signals. J Biol Chem. 2002;277:43512–43518. doi: 10.1074/jbc.M206816200. [DOI] [PubMed] [Google Scholar]
- 16.Tschumi A, et al. Identification of apolipoprotein N-acyltransferase (Lnt) in mycobacteria. J Biol Chem. 2009;284:27146–27156. doi: 10.1074/jbc.M109.022715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Widdick DA, et al. Dissecting the complete lipoprotein biogenesis pathway in Streptomyces scabies. Mol Microbiol. 2011;80:1395–1412. doi: 10.1111/j.1365-2958.2011.07656.x. [DOI] [PubMed] [Google Scholar]
- 18.Mohiman N, et al. The ppm operon is essential for acylation and glycosylation of lipoproteins in corynebacterium glutamicum. PLoS One. 2012;7:1–14. doi: 10.1371/journal.pone.0046225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brülle JK, Tschumi A, Sander P. Lipoproteins of slow-growing mycobacteria carry three fatty acids and are N-acylated by apolipoprotein N-acyltransferase BCG_2070c. BMC Microbiol. 2013;13:1–15. doi: 10.1186/1471-2180-13-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mao G, et al. Crystal structure of E. coli lipoprotein diacylglyceryl transferase. Nat Commun. 2015;7:1–12. doi: 10.1038/ncomms10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogeley L, et al. Structural basis of lipoprotein signal peptidase II action and inhibition by the antibiotic globomycin. Science. 2016;351:876–880. doi: 10.1126/science.aad3747. [DOI] [PubMed] [Google Scholar]
- 22.Robichon C, Vidal-Ingigliardi D, Pugsley AP. Depletion of apolipoprotein N-acyltransferase causes mislocalization of outer membrane lipoproteins in Escherichia coli. J Biol Chem. 2005;280:974–983. doi: 10.1074/jbc.M411059200. [DOI] [PubMed] [Google Scholar]
- 23.Pace HC, Brenner C. The nitrilase superfamily: Classification, structure and function. Genome Biol. 2001;2:1–9. doi: 10.1186/gb-2001-2-1-reviews0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vidal-Ingigliardi D, Lewenza S, Buddelmeijer N. Identification of essential residues in apolipoprotein N-acyl transferase, a member of the CN hydrolase family. J Bacteriol. 2007;189:4456–4464. doi: 10.1128/JB.00099-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buddelmeijer N, Young R. The essential Escherichia coli apolipoprotein N-acyltransferase (Lnt) exists as an extracytoplasmic thioester acyl-enzyme intermediate. Biochemistry. 2010;49:341–346. doi: 10.1021/bi9020346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hillmann F, Argentini M, Buddelmeijer N. Kinetics and phospholipid specificity of apolipoprotein N-acyltransferase. J Biol Chem. 2011;286:27936–27946. doi: 10.1074/jbc.M111.243519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gélis-Jeanvoine S, Lory S, Oberto J, Buddelmeijer N. Residues located on membrane‐embedded flexible loops are essential for the second step of the apolipoprotein N‐acyltransferase reaction. Mol Microbiol. 2015;95:692–705. doi: 10.1111/mmi.12897. [DOI] [PubMed] [Google Scholar]
- 28.Mobley HL, et al. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: Role of hemolysin in some strains. Infect Immun. 1990;58:1281–1289. doi: 10.1128/iai.58.5.1281-1289.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su J, Mrksich M. Using mass spectrometry to characterize self-assembled monolayers presenting peptides, proteins, and carbohydrates. Angew Chem Int Ed Engl. 2002;41:4715–4718. doi: 10.1002/anie.200290026. [DOI] [PubMed] [Google Scholar]
- 30.Grosse-Kunstleve RW, Adams PD. Substructure search procedures for macromolecular structures. Acta Crystallogr D Biol Crystallogr. 2003;59:1966–1973. doi: 10.1107/s0907444903018043. [DOI] [PubMed] [Google Scholar]
- 31.Terwilliger TC, et al. Decision-making in structure solution using Bayesian estimates of map quality: The PHENIX AutoSol wizard. Acta Crystallogr D Biol Crystallogr. 2009;65:582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terwilliger TC. Automated side-chain model building and sequence assignment by template matching. Acta Crystallogr D Biol Crystallogr. 2003;59:45–49. doi: 10.1107/S0907444902018048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Shivakumar D, et al. Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. J Chem Theory Comput. 2010;6:1509–1519. doi: 10.1021/ct900587b. [DOI] [PubMed] [Google Scholar]
- 37.Jacobson MP, et al. A hierarchical approach to all-atom protein loop prediction. Proteins. 2004;55:351–367. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- 38.Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: Orientations of proteins in membranes database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 39.Cross AS, Siegel G, Byrne WR, Trautmann M, Finbloom DS. Intravenous immune globulin impairs anti-bacterial defences of a cyclophosphamide-treated host. Clin Exp Immunol. 1989;76:159–164. [PMC free article] [PubMed] [Google Scholar]
- 40.Eisenberg D, Schwarz E, Komaromy M, Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 41.Afonine PV, et al. FEM: Feature-enhanced map. Acta Crystallogr D Biol Crystallogr. 2015;71:646–666. doi: 10.1107/S1399004714028132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmF cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.