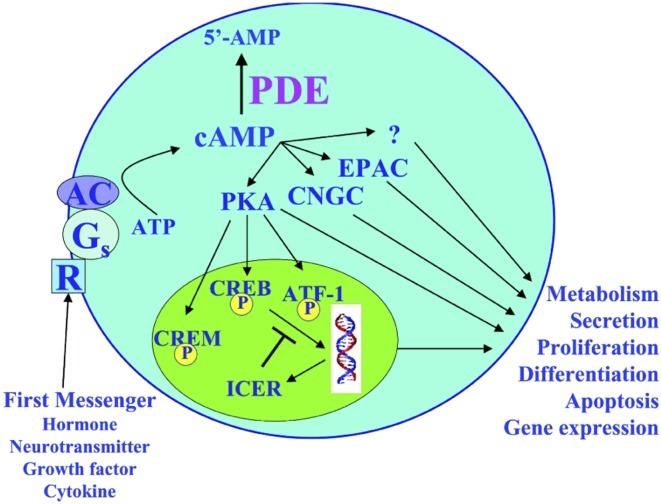
Sixty years ago, Sutherland and Rall identified cAMP as the first second messenger involved in cell–cell communication (reviewed in ref. 1). For many years, Sutherland had been interested in understanding how hormones, such as catecholamines, released by one cell type could alter the characteristics of a target cell because hormones of this type were charged, and therefore could not enter the cell. Sutherland termed compounds, such as hormones and neurotransmitters, released by one cell type that were capable of altering the properties of target cells in cell–cell communication, as “first messengers.” He reasoned that these first messengers, upon binding to the external surface of the target cell, might stimulate the synthesis of a “second messenger” on the inner surface of the target cell’s membrane, which would travel into the target cell’s interior and redirect the machinery of that cell (Fig. 1). With the discovery of cAMP, Sutherland and Rall proved this theory and thus the field of signal transduction was born. Ten years later it was discovered that the actions of cAMP resulted from its binding to and activating effector proteins, termed cAMP-dependent protein kinases, or PKAs (reviewed in ref. 1). PKAs are serine/threonine kinases that phosphorylate a wide range of protein substrates, almost all of which contain the consensus amino acid sequence R-R/K-X-S/T-Y, where X represents any small amino acid and Y represents a large hydrophobic amino acid. Phosphorylation of a target enzyme by PKA can lead to either activation or inhibition of enzymatic activity, depending on the particular target enzyme. In addition to this posttranslational modification of enzymatic activities, cAMP can regulate the transcriptional expression of a wide range of proteins through the PKA phosphorylation and regulation of transcription factors (Fig. 1).
Fig. 1.
Role of PDEs in regulation of signal transduction. In the model of the second messenger concept originally put forth by Sutherland and Rall (14), first messengers—such as hormones, neurotransmitters, cytokines, and growth factors—upon interacting with receptors on the cell surface, generate the production of a second messenger, such as cAMP, which then redirects the machinery of the cell, affecting many physiological processes. PDEs, by controlling the steady-state levels and temporal and spatial components of cAMP, are central to controlling and regulating this signal transduction. ATF-1, activating transcription factor-1; CREB, cAMP-response element binding protein; CREM, cAMP-response element modulator; Gs, stimulatory guanine nucleotide-binding protein; ICER, inducible cAMP early repressor protein; R, G protein-coupled metabotropic receptor. Reproduced with permission from ref. 4.
Thirty years after the discovery of PKA as an effector of cAMP, another cAMP effector, exchange protein activated by cAMP (EPAC), was discovered as a Rap1 guanine-nucleotide exchange factor directly activated by cAMP (2), and a third class of effectors, cyclic nucleotide-gated channels (CNGC), was also found to exist (3). Several reports have provided evidence for actions of cAMP independent of these three effectors, suggesting that additional effectors of cAMP action may yet be uncovered (Fig. 1) (reviewed in ref. 4). The level of cAMP within cells is controlled by its rate of synthesis from ATP by adenylyl cyclases (ACs), its rate of degradation to 5′-AMP by cyclic nucleotide phosphodiesterases (PDEs), and to some extent by extrusion out of the cell (Fig. 1) (5, 6). In the late 1970s, Brunton, Hayes, and Mayer observed a differential effect of two agonists of adenylyl cyclase, isoproterenol (ISO) and prostaglandin E1 (PGE1), on cardiac myocytes. Whereas both agonists increased cAMP levels, and both activated a soluble fraction of PKA, only ISO activated a particulate PKA, and only ISO produced the responses of activation of phosphorylase and enhanced contractile activity. PGE1, by contrast, only activated a soluble PKA and caused no other measurable response. Based on this observation, these investigators proposed that different signals lead to cAMP accumulation and the activation of PKAs in different subcellular compartments, with the consequent phosphorylation of specific, rather than all, substrates of PKA (reviewed in ref. 7). Decades later, the development of FRET reporter sensors, based on cAMP binding domains on EPAC and the regulatory subunit of PKAs, enabled visualization of cAMP microdomains following cell stimulation, providing clear evidence for signal compartmentalization, and with this technique in hand, work by a multitude of laboratories indicated that PDEs are critically important in regulating the spatial and temporal dynamics of cAMP signaling and the creation of these cAMP microdomains within the cell (8–13).
Although Sutherland and Rall, when they first reported on the discovery of cAMP, also described a caffeine-sensitive enzymatic activity in tissue extracts capable of hydrolyzing cAMP to 5′-AMP, which they thought was a PDE, little more was known about it at the time (14). Since then, studies have revealed a remarkable complexity of the PDE system, showing it to represent a superfamily of enzymes encoded by 21 different genes, grouped into 11 gene families, based on sequence similarity, mode of regulation, and preference for cAMP or cGMP as substrate. With the existence of multiple transcription initiation sites, as well as alternatively spliced forms of many of these genes, more than 100 different forms of PDE have been identified and cloned to date, and many of these PDE forms are localized to different cells and different subcellular compartments as part of complexes or signalosomes composed of scaffolding proteins, such as A-kinase anchoring proteins, cAMP effectors (PKA, EPAC, CNGC), ACs, and distinct PDEs, thus achieving targeted cAMP degradation and the creation of localized intracellular cAMP gradients, and allowing the control of specific cellular functions by specific PDE isoforms during cellular signaling (15). Although the use of subcellularly targeted FRET reporter sensors have helped to confirm the role of PDEs in regulating the temporal and spatial control of cAMP during signal transduction, this approach only has limited ability to link specific PDEs to regulation of downstream effector molecules and biological functions. In PNAS, Beltejar et al. (16) use mass spectrometry coupled with the use of specific isozyme-selective PDE inhibitors to characterize, for the first time, the phosphoproteomes of functional pools of cAMP regulated by specific PDEs, to delineate which PDEs control phosphorylation of which proteins, leading to regulation of different responses by different PDEs.
Using CD3/CD28-stimulated Jurkat T leukemic cells, Beltejar et al. (16) coupled mass spectrometry phosphoproteomic analyses with treatment using selective inhibitors of PDEs 1, 3, 4, 7, and 8, in the presence and absence of low physiological concentrations of PGE2, to characterize the PDE-regulated phosphoproteome of these cells. To determine which sets of PDE inhibitors to use in these studies, the authors first treated Jurkat cells with individual isozyme-selective PDE inhibitors or various combinations of them, and measured resulting changes in cAMP levels. Surprisingly treatment with individual PDE inhibitors alone did not cause a significant increase in total cAMP, either in the presence or absence of low PGE2, and it required two or more PDE inhibitors to obtain an increase in cAMP. Based on this approach, Beltejar et al. chose the inhibitor combinations that seemed most likely to influence the greatest number of PDE-regulated compartments for follow-up by phosphoproteomic analysis. Specifically, this approach included two combinations of PDE-selective inhibitors, PDE3 and -4 inhibitors as one combination, and PDE1, -7, and -8 inhibitors as a second combination. Beltejar et al. also used a combination of a PDE8 inhibitor with the nonselective inhibitor isobutylmethylxanthine (IBMX), to determine changes occurring in response to inhibiting all known cAMP-hydrolyzing PDEs, because IBMX inhibits all cAMP-hydrolyzing PDE families, with the exception of PDE8. In total, through these analyses, Beltejar et al. identified 13,589 phosphopeptides and 3,241 proteins, of which 618 phosphoproteins distributed among 461 unique proteins were significantly regulated by the PDE treatments. Consistent with the cAMP assays, no phosphosites were significantly regulated by individual PDE inhibitor treatments alone, and it required inhibitors of two or more PDEs to see such a change. A portion of the total phosphosites regulated by total PDE inhibition
As Beltejar et al. point out, a big advantage of the phosphoproteome analysis they used is the unbiased identification of regulated phosphosites and the incredible sheer number of sites identified.
were uniquely regulated by the PDE3 and -4 inhibitor combination (40 sites in the absence of PGE2 and 65 in the presence), and another portion was uniquely regulated by the PDE1, -7, and -8 inhibitor combination (35 in the absence of PGE2 and 122 in the presence), with only a single phosphosite regulated by both inhibitor combinations in the presence of PGE2. This striking nonoverlap between the phosphosites regulated by these two inhibitor combinations strongly suggests that the pools of cAMP regulated by these two inhibitor combinations are functionally distinct from each other.
Further analysis was done to predict which kinases might be most likely to phosphorylate the PDE inhibitor-dependent sites. This was done by analyzing a peptide sequence of four amino acids flanking the regulated phosphosites with a NetPhorest program, a web based tool for kinase prediction. Results showed that the majority of the phosphosites increased by inhibition of PDE3 and -4 are primarily phosphorylated by PKA, as these sites contain the classic PKA consensus sequence. In contrast, the majority of phosphosites regulated by inhibition of PDEs 1, 7, and 8 did not appear to be phosphorylated by PKA, as they did not contain the PKA consensus sequence; rather, they were mostly predicted to be phosphorylated by a diversity of other kinases, with casein kinase 2 (CK2) being the most prominent. This sharp contrast in the kinases predicted to phosphorylate the phosphosites regulated by the PDE3 and -4 inhibitor combination from those regulated by the PDE1, -7, and -8 inhibitor combination further supports the concept of different functional pools of cAMP regulated by different PDE isoforms, and suggests that the cAMP pool regulated by the PDE1, -7, and -8 inhibitors may be regulating its targets indirectly by activating PKA or EPAC upstream of CK2 and the other non-PKA kinases predicted to phosphorylate these regulated target phosphosites.
Beltejar et al. (16) carried out several other important bioinformatics analyses. First, by searching a database of phosphosites annotated for predicted regulatory function, the authors were able to identify which phosphosites were most likely to be biologically relevant. In so doing, Beltejar et al. found 50 potential regulatory sites in the PDE3 and -4 inhibitor treatment group and 30 in the PDE1, -7, and 8 inhibitor treatment group, for which a regulatory role had not yet been determined, making them prime candidates for further follow-up. Beltejar et al. also used STRING analysis to suggest which biological processes or pathways might be regulated in each PDE inhibitor treatment group. This analysis revealed clusters of interacting proteins associated with particular cell pathways, which differed between the PDE3 and -4 and the PDE1, -7, and -8 inhibitor treatment groups. Finally, Beltejar et al. conducted Gene Ontology analysis, which grouped 90 of 133 genes regulated by the PDE1, -7, and -8 inhibitors into 17 functional clusters, and 20 of 74 genes regulated by the PDE3 and -4 inhibitors into 6 functional clusters, representing different biological functions. These results further indicate that different combinations of PDE inhibitors likely subserve different functional pools of cAMP, and that different functional pools in turn regulate the different functions as identified by the Gene Ontology analysis.
This study by Beltejar et al. (16) has important clinical implications. Because so many fundamental physiological processes are regulated by cAMP signaling, PDE inhibitors are under intense development for a whole host of disorders and are seeing increasing approval for clinical use. PDE5 inhibitors (sildenafil and others) have seen widespread use for treatment of erectile dysfunction and pulmonary hypertension; PDE3 inhibitors for treatment of intermittent claudication (cilostazol) and congestive heart failure (milrinone); and PDE4 inhibitors for chronic obstructive pulmonary disease (roflumilast), plaque psoriasis and psoriatic arthritis (apremilast), and atopic dermatitis (crisaborole). As Beltejar et al. point out, a big advantage of the phosphoproteome analysis they used is the unbiased identification of regulated phosphosites and the incredible sheer number of sites identified. As the authors show, this can lead to the discovery of many new relevant targets in the pathways of cAMP signaling and this approach of coupling inhibition of selective PDE isozymes with phosphoproteomic analysis should also provide a means of preclinical screening to determine which PDEs should be inhibited to maximize a therapeutic effect or minimize an unwanted side effect, thus enhancing and streamlining the development of PDE inhibitors for a wide range of disorders, which is sure to come in the near future.
Acknowledgments
I thank Lea’s Foundation for Leukemia Research, Inc. and the Smart Family Foundation for their continued support of my work.
Footnotes
The author declares no conflict of interest.
See companion article on page E6240.
References
- 1.Beavo JA, Brunton LL. Cyclic nucleotide research—Still expanding after half a century. Nat Rev Mol Cell Biol. 2002;3:710–718. doi: 10.1038/nrm911. [DOI] [PubMed] [Google Scholar]
- 2.de Rooij J, et al. Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature. 1998;396:474–477. doi: 10.1038/24884. [DOI] [PubMed] [Google Scholar]
- 3.Yau KW. Cyclic nucleotide-gated channels: An expanding new family of ion channels. Proc Natl Acad Sci USA. 1994;91:3481–3483. doi: 10.1073/pnas.91.9.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lerner A, Epstein PM. Cyclic nucleotide phosphodiesterases as targets for treatment of haematological malignancies. Biochem J. 2006;393:21–41. doi: 10.1042/BJ20051368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Houslay MD, Milligan G. Tailoring cAMP-signalling responses through isoform multiplicity. Trends Biochem Sci. 1997;22:217–224. doi: 10.1016/s0968-0004(97)01050-5. [DOI] [PubMed] [Google Scholar]
- 6.Barber R, Butcher RW. The egress of cyclic AMP from metazoan cells. Adv Cyclic Nuclotide Res. 1983;15:119–138. [Google Scholar]
- 7.Steinberg SF, Brunton LL. Compartmentation of G protein-coupled signaling pathways in cardiac myocytes. Annu Rev Pharmacol Toxicol. 2001;41:751–773. doi: 10.1146/annurev.pharmtox.41.1.751. [DOI] [PubMed] [Google Scholar]
- 8.Baillie GS. Compartmentalized signalling: Spatial regulation of cAMP by the action of compartmentalized phosphodiesterases. FEBS J. 2009;276:1790–1799. doi: 10.1111/j.1742-4658.2009.06926.x. [DOI] [PubMed] [Google Scholar]
- 9.Houslay MD. Underpinning compartmentalised cAMP signalling through targeted cAMP breakdown. Trends Biochem Sci. 2010;35:91–100. doi: 10.1016/j.tibs.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Fischmeister R, et al. Compartmentation of cyclic nucleotide signaling in the heart: The role of cyclic nucleotide phosphodiesterases. Circ Res. 2006;99:816–828. doi: 10.1161/01.RES.0000246118.98832.04. [DOI] [PubMed] [Google Scholar]
- 11.Conti M, Mika D, Richter W. Cyclic AMP compartments and signaling specificity: Role of cyclic nucleotide phosphodiesterases. J Gen Physiol. 2014;143:29–38. doi: 10.1085/jgp.201311083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Otero C, et al. Temporal and spatial regulation of cAMP signaling in disease: Role of cyclic nucleotide phosphodiesterases. Fundam Clin Pharmacol. 2014;28:593–607. doi: 10.1111/fcp.12080. [DOI] [PubMed] [Google Scholar]
- 13.Brescia M, Zaccolo M. Modulation of compartmentalised cyclic nucleotide signalling via local inhibition of phosphodiesterase activity. Int J Mol Sci. 2016;17:E1672. doi: 10.3390/ijms17101672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sutherland EW, Rall TW. Fractionation and characterization of a cyclic adenine ribonucleotide formed by tissue particles. J Biol Chem. 1958;232:1077–1091. [PubMed] [Google Scholar]
- 15.Maurice DH, et al. Advances in targeting cyclic nucleotide phosphodiesterases. Nat Rev Drug Discov. 2014;13:290–314. doi: 10.1038/nrd4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beltejar M-CG, Lau H-T, Golkowski MG, Ong S-E, Beavo JA. Analyses of PDE-regulated phosphoproteomes reveal unique and specific cAMP-signaling modules in T cells. Proc Natl Acad Sci USA. 2017;114:E6240–E6249. doi: 10.1073/pnas.1703939114. [DOI] [PMC free article] [PubMed] [Google Scholar]