Summary
Vertebrate centromeres are epigenetically defined by nucleosomes containing the histone H3 variant, CENP-A. CENP-A nucleosome assembly requires the three-protein Mis18 complex (Mis18α, Mis18β, and M18BP1) that recruits the CENP-A chaperone HJURP to centromeres, but how the Mis18 complex recognizes centromeric chromatin is unknown. Using Xenopus egg extract, we show that direct, cell cycle-regulated binding of M18BP1 to CENP-A nucleosomes recruits the Mis18 complex to interphase centromeres to promote new CENP-A nucleosome assembly. We demonstrate that Xenopus M18BP1 binds CENP-A nucleosomes using a motif that is widely conserved except in mammals. The M18BP1 motif resembles a CENP-A nucleosome binding motif in CENP-C, and we show that CENP-C competes with M18BP1 for CENP-A nucleosome binding at centromeres. We show that both CENP-C and M18BP1 recruit HJURP to centromeres for new CENP-A assembly. This study defines cellular mechanisms for recruiting CENP-A assembly factors to existing CENP-A nucleosomes for the epigenetic inheritance of centromeres.
Introduction
The centromere provides the assembly site for the mitotic kinetochore, enabling chromosome-microtubule interaction and chromosome segregation (Foley and Kapoor, 2013). Functional centromeres are specified epigenetically by the presence of nucleosomes containing the histone H3 variant, centromere protein A (CENP-A) (Fukagawa and Earnshaw, 2014; Guse et al., 2011; Mendiburo et al., 2011; Stoler et al., 1995; Yoda et al., 2000). Failure to assemble CENP-A at centromeres results in chromosome missegregation (Fachinetti et al., 2013), and CENP-A misincorporation into non-centromeric regions can promote neocentromere formation and chromosome instability (Mendiburo et al., 2011; Scott and Sullivan, 2014). Faithful genome transmission during cell division therefore requires CENP-A chromatin to be properly maintained.
During DNA replication, CENP-A nucleosomes are equally distributed to each new DNA strand (Jansen et al., 2007). To replenish centromeric chromatin following replicative dilution, existing CENP-A nucleosomes seed new CENP-A assembly during G1 (Westhorpe and Straight, 2015). HJURP, complexed with nucleophosmin1 (Npm1), CENP-A, and histone H4, acts as a chaperone complex to provide new CENP-A for incorporation into chromatin (Dunleavy et al., 2009; Foltz et al., 2009). The Mis18 complex (Mis18α, Mis18β, and M18BP1/KNL2), identified for its role in CENP-A assembly (Fujita et al., 2007; Hayashi et al., 2004; Maddox et al., 2007), is required for targeting HJURP to CENP-A chromatin (Barnhart et al., 2011; Moree et al., 2011; Nardi et al., 2016; Pidoux et al., 2009; Wang et al., 2014; Williams et al., 2009), but its molecular functions are still unknown. In addition to HJURP and the Mis18 complex, CENP-A nucleosome assembly requires components of the constitutive centromere associated network (CCAN), a 17-protein complex recruited by CENP-A nucleosomes throughout the cell cycle (Carroll et al., 2010; Carroll et al., 2009; Hori et al., 2013; Moree et al., 2011; Okada et al., 2006; Shono et al., 2015). Two CCAN proteins, CENP-C and CENP-N, are the only two factors known to directly and selectively interact with CENP-A nucleosomes (Carroll et al., 2010; Carroll et al., 2009; Kato et al., 2013). CENP-C, in particular, has been implicated in CENP-A assembly through its interactions with the Mis18 complex (Dambacher et al., 2012; Moree et al., 2011; Nardi et al., 2016; Stellfox et al., 2016), though CENP-C is not required for Mis18 complex recruitment during interphase in Xenopus, C. elegans, or chicken (Maddox et al., 2007; Moree et al., 2011; Perpelescu et al., 2015). Although selective co-purification of M18BP1 (Maddox et al., 2007) and HJURP (Foltz et al., 2009) with centromeric chromatin suggests tight association with CENP-A nucleosomes, no assembly factor has been shown to directly interact with the underlying centromeric chromatin. Thus, how CENP-A nucleosome assembly is spatially and temporally regulated to restrict new CENP-A nucleosome assembly to the pre-existing centromere at the right stage of the cell cycle remains a key unanswered question.
Here, we discover that Xenopus M18BP1 interacts directly with CENP-A nucleosomes specifically during interphase. We find that M18BP1 binds CENP-A nucleosomes via a conserved ‘CENP-C motif’ as suggested by recent bioinformatics and genetic analyses in zebrafish and Arabidopsis (Kral, 2015; Sandmann et al., 2017). M18BP1 chromatin recognition requires nucleosomes containing both the CENP-A Targeting Domain (CATD) and the CENP-A C-terminus (CAC), analogous to CENP-C. M18BP1 targeting to the centromere is negatively regulated during mitosis through post-translational modification of M18BP1. Using the native sperm centromere as a template for new CENP-A assembly, we find that CENP-C limits M18BP1 recruitment by direct competition. Using a combination of in vitro CENP-A assembly on reconstituted chromatin arrays, artificial tethering of LacI-M18BP1 to LacO-containing arrays, and mutants of M18BP1 that do not bind CENP-A, we show that M18BP1 binding to CENP-A nucleosomes is necessary for new CENP-A incorporation. Furthermore, both the Mis18 complex and CENP-C recruit HJURP to centromeres by directly binding both CENP-A nucleosomes and HJURP. Together, we propose that direct recognition of CENP-A nucleosomes by M18BP1 and CENP-C provides a mechanism to spatially restrict HJURP localization and CENP-A nucleosome assembly to centromeric chromatin.
Results
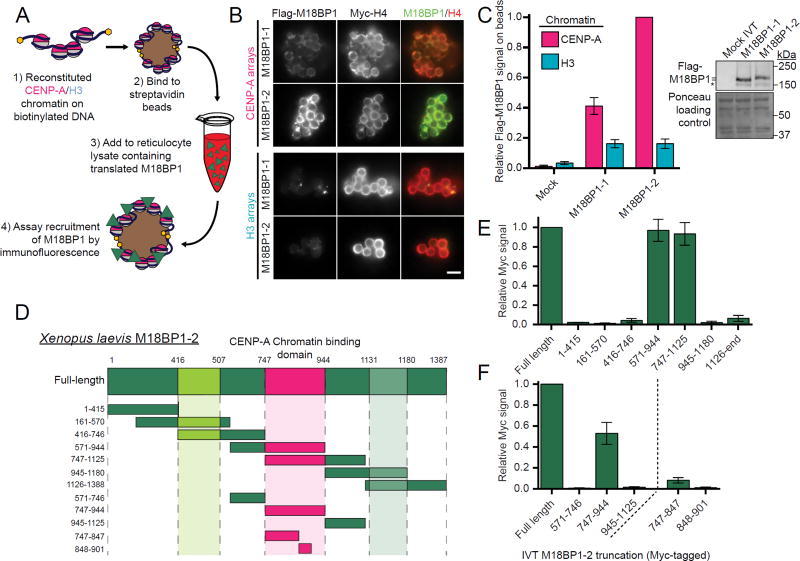
Xenopus M18BP1 selectively binds CENP-A nucleosomes
In Xenopus, M18BP1 does not require CENP-C to bind to interphase centromeres or reconstituted CENP-A chromatin (Moree et al., 2011; Westhorpe et al., 2015). This appears conserved not only in C. elegans and chicken (Maddox et al., 2007; Perpelescu et al., 2015), but also in humans, as auxin-induced degradation of endogenous CENP-C in G1 human cells does not prevent M18BP1 localization to centromeres (Figure S1A-C). As CENP-C is required for recruiting the rest of the CCAN (Carroll et al., 2010; Guse et al., 2011; Weir et al., 2016), we tested the possibility that M18BP1 binds chromatin directly by challenging immobilized, reconstituted CENP-A and H3 chromatin (Guse et al., 2012) with M18BP1 translated in rabbit reticulocyte lysate (Figure 1A). Xenopus laevis is allotetraploid and, as with many genes in its genome, has two isoforms of M18BP1: M18BP1-1 and M18BP1-2 which share 74% sequence identity (Moree et al., 2011; Session et al., 2016). When we tested the binding of each isoform of Xenopus M18BP1 to chromatin we found that M18BP1-1 was 2.5-fold and M18BP1-2 was 5-fold enriched on CENP-A chromatin relative to H3 chromatin (Figure 1B,C). Together, this indicates that M18BP1 binds CENP-A nucleosomes, in addition to its proposed DNA binding activity (Maddox et al., 2007).
Figure 1. Xenopus M18BP1 directly binds CENP-A chromatin.
A) Schematic of in vitro chromatin binding assay. Reconstituted CENP-A or H3 chromatin, immobilized on streptavidin-coated beads, was incubated in rabbit reticulocyte lysate containing M18BP1. Following chromatin isolation M18BP1 binding was assayed by immunofluorescence.
B) Both isoforms of Xenopus M18BP1 bind selectively to CENP-A chromatin. Representative immunofluorescence images of M18BP1 binding to CENP-A chromatin (top) and H3 chromatin (bottom), both labeled with Myc-tagged histone H4. The M18BP1 isoform is indicated to the left; immunolocalized protein is indicated above. Scale bar, 5 µm.
C) (Left) Quantification of B. Values are normalized to M18BP1-2 signal on CENP-A beads. (Right) Representative Western blot of samples from B. Top portion (anti-Flag) shows similar translation of M18BP1 isoforms; bottom portion (Ponceau stained) is a loading control.
D) Schematic representation of truncations of M18BP1- 2 used to define the CENP-A chromatin binding domain (magenta) in E-F.
E-F) M18BP1-2 binds CENP-A nucleosomes via amino acids 747-944. M18BP1-2 truncations spanning the indicated amino acids were assayed for CENP-A chromatin binding by the method described in A. Values are normalized to full-length M18BP1-2 signal.
All graphs show the mean ± SEM of at least three experiments.
See also Figure S1-S3
To better understand how M18BP1 recognizes CENP-A chromatin we mapped its nucleosome-binding domain using a series of overlapping M18BP1-2 truncations (Figure 1D). Two truncations, collectively spanning amino acids 571-1125, showed CENP-A nucleosome binding similar to full-length M18BP1-2 (Figure 1E). Amino acids 747-944, the region common between these truncations, bound CENP-A nucleosomes at 53 ± 11% of the full-length M18BP1-2, whereas amino acids 571-746 and 945-1125 did not bind nucleosomes (Figure 1F). Similar results were obtained for M18BP1-1 (Figure S1D). The region between 747-944 is predicted to be largely unstructured, and was prone to aggregation during purification, thus the decreased binding of 747-944 versus full-length M18BP1 may reflect protein aggregation rather than truncation of additional binding residues. CENP-A nucleosome recognition was not dependent on M18BP1’s SANT DNA-binding domain or SANTA protein interaction domain (Figure 1D-F) (Zhang et al., 2006). Taken together, Xenopus laevis M18BP1 binds CENP-A nucleosomes directly via a binding site within amino acids 747-944.
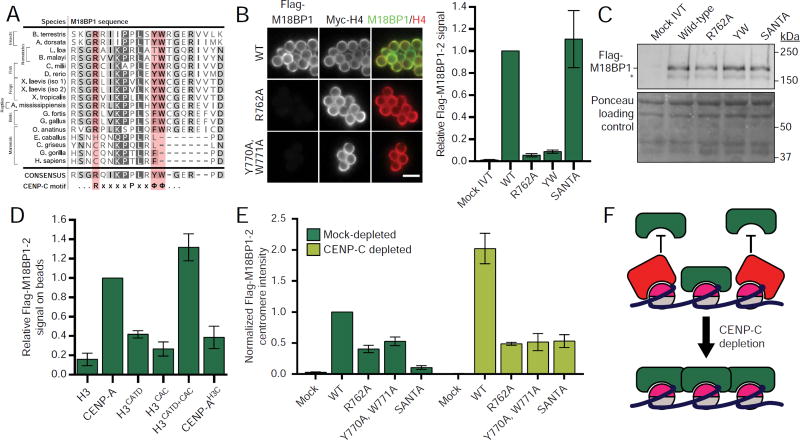
M18BP1-2 engages CENP-A nucleosomes using a ‘CENP-C motif’
To test the hypothesis that direct binding of CENP-A nucleosomes recruits M18BP1 to centromeres, we sought to identify M18BP1 mutants that could not recognize CENP-A nucleosomes. The ‘CENP-C motif’ within CENP-C acts as a CENP-A nucleosome binding domain (Carroll et al., 2010; Kato et al., 2013). A recent bioinformatic analysis of CENP-C and M18BP1 identified a similar motif within M18BP1 proteins of fish, frogs and birds (Kral, 2015). Furthermore, a recent study identified a CENP-C motif-like sequence within Arabidopsis thaliana M18BP1/KNL2 that, when mutated, disrupts KNL2 targeting to centromeres (Sandmann et al., 2017). Alignment of diverse M18BP1 orthologues revealed conservation of a putative CENP-C motif within the Xenopus laevis M18BP1 nucleosome binding region (Figure 2A) consistent with the idea that M18BP1 may engage CENP-A nucleosomes via a CENP-C-like mechanism (Kral, 2015; Sandmann et al., 2017).
Figure 2. Characterization of the CENP-C motif within M18BP1-2.
A) M18BP1 contains a CENP-C motif conserved through most of Eukarya. Alignment of M18BP1 sequences among select eukaryotes. Red columns highlight conserved positively-charged and hydrophobic residues previously shown to be required for CENP-C binding to CENP-A nucleosomes(Carroll et al., 2010; Kato et al., 2013). Grey shading indicates conserved amino acids in alignment of ~300 M18BP1 homologs in eukaryotes.
B) Mutation of the M18BP1-2 CENP-C motif abolishes nucleosome binding. (Left) Representative images of full-length M18BP1-2 binding to CENP-A chromatin. The M18BP1-2 mutant is indicated to the left; immunolocalized protein is indicated above. Scale bar, 5 µm. (Right) Quantification of Flag-M18BP1-2 signal on chromatin. Values are normalized to wild-type M18BP1-2 signal.
C) Representative Western blot of samples in B. Top portion (anti-Flag) shows similar translation of all Flag-M18BP1-2 mutants; bottom portion (Ponceau stained) is a loading control.
D) Nucleosome binding by M18BP1-2 requires both the CATD and the CAC. Quantification of M18BP1-2 binding to chromatin reconstituted with the CENP-A/H3 chimeras indicated. Representative images are shown in Figure S1I. Values are normalized to Flag-M18BP1-2 signal on CENP-A beads.
E) Nucleosome binding is required for M18BP1-2 localization to interphase centromeres. Quantification of Flag-M18BP1-2 signal at sperm centromeres in mock-depleted or CENP-C-depleted interphase extract. All extracts have also been depleted of M18BP1. Representative images are shown in Figure S1J. Values are normalized to wild-type M18BP1-2 centromere signal in mock-depleted extract.
All graphs show the mean ± SEM of at least three experiments.
F) Schematic of proposed competition between M18BP1 and CENP-C for CENP-A nucleosome binding. CENP-C binding to CENP-A nucleosomes prevents M18BP1 binding and limits its centromere localization. Depletion of CENP-C results in increased M18BP1 localization (E).
See also Figure S1.
The CENP-C motif contains two aromatic residues that form hydrophobic contacts with the CENP-A C-terminus (CAC) and an arginine that forms electrostatic contacts with the H2A/H2B acidic patch (Kato et al., 2013) (Figure 2A). Mutating the analogous residues in M18BP1-2 (Y770A, W771A or R762A, respectively) abolished binding to CENP-A nucleosomes in vitro (Figure 2B). Mutating the SANTA domain (L418A, W421A, F491A, F495A, W499A) based on previous analysis of this domain (Zhang et al., 2006) had no effect (Figure 2B).
In interphase, CENP-C requires the presence of both the CATD and CAC regions of the CENP-A nucleosome to localize to CENP-A chromatin (Westhorpe et al., 2015). Using chromatin containing CENP-A/H3 chimeric histones we tested if M18BP1 requires the same domains of the CENP-A nucleosome for its interaction with chromatin. We found that, similar to CENP-C, M18BP1 binding to CENP-A chromatin required both the CATD and CAC (Figure 2D, Figure S1H,I). Together, these data demonstrate that M18BP1 binds CENP-A chromatin in a manner similar to CENP-C.
To determine whether CENP-A nucleosome binding is required for M18BP1 localization to centromeres, we assessed the localization of wild-type or nucleosome-binding mutant M18BP1 to sperm centromeres in interphase egg extract that had been depleted of endogenous M18BP1. Relative to wild-type centromere localization, M18BP1-2R762A showed a 60 ± 6% reduction and M18BP1-2Y770A, W771A (M18BP1-2YW) a 47 ± 7% reduction (Figure 2E, Figure S1J), showing that M18BP1 binding to the CENP-A nucleosome is important for M18BP1 localization to interphase centromeres. Mutations in the nucleosome binding domain of M18BP1-1 also showed a defect in localizing to centromeres in interphase (Figure S1E,F). Residual interphase M18BP1 centromere localization suggests that additional mechanisms act to recruit M18BP1 to sperm chromatin centromeres during interphase (Shono et al., 2015). Consistent with this idea, SANTA domain mutations that do not affect CENP-A nucleosome binding in vitro (Figure 2B) reduced M18BP1 localization to centromeres by 90 ± 2% (Figure 2E, S1J).
Previously, we have shown that CENP-C depletion causes an increase in M18BP1 localization to sperm centromeres in interphase (Moree et al., 2011). Consistent with this, wild-type and M18BP1SANTA localization increased 2- and 5.6-fold, respectively, in response to CENP-C depletion (Figure 2E). In contrast, the localization of M18BP1-2R762A and M18BP1-2YW was unaffected by CENP-C depletion (Figure 2E). These data demonstrate that increased CENP-A nucleosome binding by M18BP1 accounts for the increase of M18BP1 at centromeres in the absence of CENP-C. Conversely, M18BP1 depletion does not result in increased CENP-C localization (Moree et al., 2011). Together, our data imply that CENP-C occupies potential M18BP1 binding sites, limiting the amount of M18BP1 bound to CENP-A nucleosomes (Figure 2F). Thus, while CENP-C is required for CENP-A assembly (Carroll et al., 2010; Moree et al., 2011; Westhorpe et al., 2015), it may also regulate CENP-A assembly by competing with M18BP1 for CENP-A nucleosome binding.
Regulation of M18BP1 binding to CENP-A nucleosomes
Although M18BP1/KNL2 localizes to centromeres throughout the cell cycle in C. elegans, Arabidopsis, and Xenopus (Lermontova et al., 2013; Maddox et al., 2007; Moree et al., 2011), in humans M18BP1 is only enriched at centromeres during G1 (Fujita et al., 2007; Maddox et al., 2007; Silva et al., 2012). Furthermore, in Xenopus M18BP1 localization transitions from a CENP-C-dependent mechanism in metaphase to a CENP-C-independent mechanism in interphase (Moree 2011), suggesting that M18BP1 binding to CENP-A nucleosomes is cell cycle regulated.
We tested the cell cycle dependence of M18BP1 binding to CENP-A nucleosomes by measuring binding of in vitro translated M18BP1-2 to chromatin-coated beads in metaphase or interphase extract. While M18BP1-2 bound to CENP-A chromatin in interphase, binding in metaphase was completely inhibited (Figure S2A). Importantly, depletion of CENP-C did not permit M18BP1-2 binding in metaphase, suggesting that inhibition of M18BP1-2 binding in metaphase is not due to competition with CENP-C. Unlike on sperm chromatin, interphase M18BP1-2 binding to reconstituted chromatin arrays does not increase following CENP-C depletion (Westhorpe et al., 2015) – possibly because CENP-A nucleosomes are in excess and the concentration of CENP-C in extract is therefore too low to compete with M18BP1-2.
We sought to understand whether modification of chromatin or of M18BP1 inhibited CENP-A nucleosome binding in metaphase. In vitro, purified recombinant Flag-M18BP1-2747-944 recapitulated CENP-A nucleosome binding and the Y770A, W771A mutations abolished binding (Figure S2B,C). In extract, M18BP1-2747-944 bound CENP-A chromatin specifically in interphase (Figure S2C,D), recapitulating the regulation of full-length M18BP1-2 (Figure S2A). To test whether an inhibitory modification occurs on chromatin to prevent M18BP1 binding, we preincubated CENP-A chromatin in extract, recovered the chromatin, and assayed the binding of unmodified M18BP1-2747-944 protein in vitro (Figure S2E). We observed no difference in M18BP1-2747-944 binding between chromatin preincubated in metaphase extract, interphase extract and no extract, suggesting that modification of chromatin does not regulate M18BP1 binding to nucleosomes (Figure S2E). This is consistent with the fact that nucleosome modifications would likely affect both CENP-C and M18BP1 recruitment, given that CENP-C and M18BP1 bind the same CENP-A nucleosome epitope via a common motif.
In humans, Cdk phosphorylation of M18BP1 prevents binding to Mis18α and Mis18β (McKinley and Cheeseman, 2014; Pan et al., 2017; Spiller et al., 2017) and prevents premature localization of M18BP1 to centromeres in mitosis (Silva et al., 2012; Stankovic et al., 2016). To test if Cdk phosphorylation of M18BP1-2747-944 might regulate nucleosome binding we incubated MBP-M18BP1-2747-944 in metaphase egg extract, recovered the MBP fusion, and performed mass spectrometry to identify post-translational modifications of MBP-M18BP1-2747-944 (Figure S3B). MBP-M18BP1-2747-944 binding to chromatin is inhibited by incubation in metaphase extract (Figure S3A) and we identified six phosphorylated Cdk sites with greater than 50% modification stoichiometry (Figure S3C). However, simultaneously mutating all 6 sites to phosphomimetic glutamate (Cdk-E) or to non-phosphorylatable alanine (Cdk-A) in full-length M18BP1-2, did not affect nucleosome binding in vitro (Figure S3D,E). Thus, although inhibition of M18BP1 centromere localization in metaphase is likely by post-translation modification of M18BP1, it does not appear mediated by direct Cdk phosphorylation of the M18BP1-2 nucleosome binding domain.
M18BP1 CENP-A nucleosome binding is required for CENP-A assembly
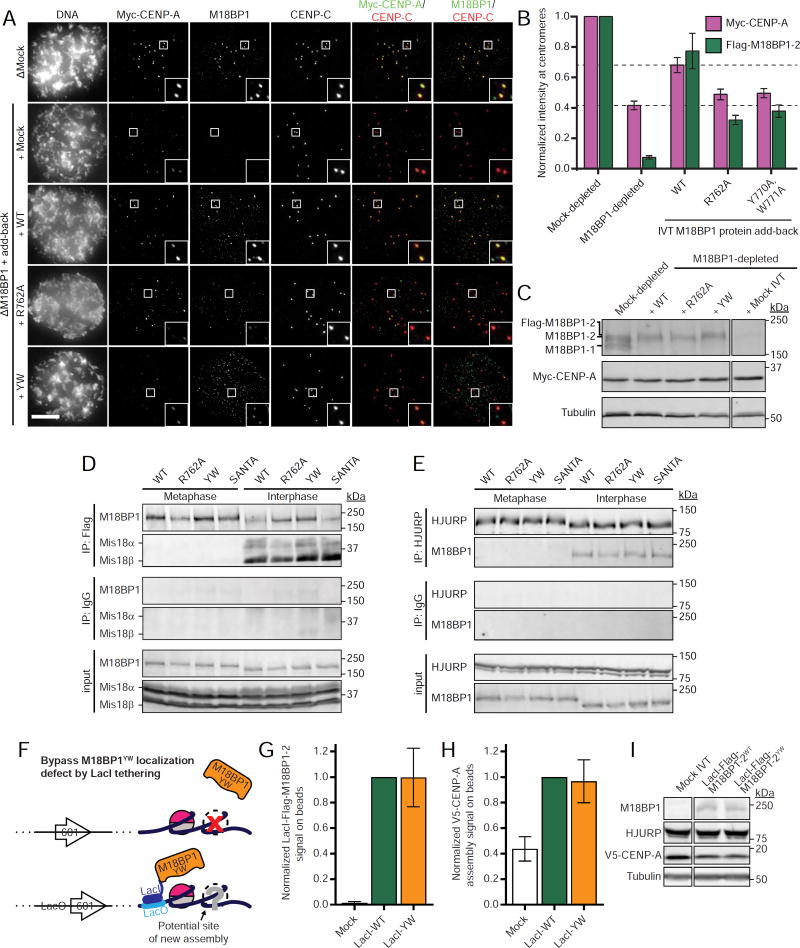
To test the importance of M18BP1 binding to CENP-A nucleosomes in the process of new CENP-A nucleosome assembly, we depleted endogenous M18BP1 from Xenopus egg extracts, replaced it with nucleosome binding mutants in M18BP1-2, and assayed the assembly of exogenous Myc-CENP-A at sperm centromeres (Moree et al., 2011). Consistent with previous results (Moree et al., 2011), depletion of M18BP1 reduced M18BP1 centromere levels by >90%, and caused a 58 ± 3% decrease in the amount of Myc-CENP-A assembled (Figure 3A,B, lower dotted line). Addition of wild-type M18BP1-2 protein rescued M18BP1 centromere localization and partially rescued Myc-CENP-A assembly (Figure 3A,B, upper dotted line). In contrast, the level of CENP-A assembly in extracts complemented with M18BP1-2R764A and M18BP1-2YW was comparable to that observed in M18BP1-depleted extract despite their partial localization (Figure 3A-C). These data suggest that M18BP1 must bind CENP-A nucleosomes to promote new CENP-A assembly.
Figure 3. M18BP1 binding to CENP-A nucleosomes is required for CENP-A assembly.
A) M18BP1-2 that cannot bind to CENP-A nucleosomes do not rescue new CENP-A assembly at sperm centromeres. Representative images of sperm nuclei incubated in M18BP1-depleted interphase Xenopus egg extracts complemented with the M18BP1-2 protein indicated at left. Extracts are supplemented with RNA encoding Myc-CENP-A to track new CENP-A assembly and in vitro translated HJURP. The immunolocalized protein is indicated above. Scale bar, 10 µm. Insets magnified 3X.
B) Quantification of A. Values are normalized to the centromere signals in mock-depleted extract. Dashed lines indicate the Myc-CENP-A assembly signal observed upon M18BP1 depletion (bottom) and Flag-M18BP1-2WT add-back (top) as points of reference for mutant rescue.
C) Representative Western blot of CENP-A assembly reactions in A probed with anti-M18BP1 (top), anti-Myc (middle), and anti-tubulin as a loading control (bottom). Efficient M18BP1 depletion is indicated by comparing lanes 1 and 5. Add-back of wild-type or mutant M18BP1-2 is near endogenous levels.
D) Interphase Mis18 complex formation is unaffected by M18BP1 nucleosome binding mutations. Extract depleted of endogenous M18BP1 was supplemented with Myc-Mis18α, Myc-Mis18β, and Flag-M18BP1-2. M18BP1-2 species added to each reaction and cell cycle state of the extract is indicated at the top. Co-immunoprecipitation of Myc-Mis18α/β was assessed by anti-Myc immunoblot following Flag precipitation. Mock precipitations using whole mouse IgG serve as a negative control.
E) HJURP association with M18BP1 is unaffected by CENP-C motif mutations. Extract depleted of endogenous M18BP1 was supplemented with Flag-M18BP1-2. M18BP1-2 species added to each reaction and cell cycle state of the extract are indicated at the top. Coimmunoprecipitation of Flag-M18BP1-2 was assessed by anti-Flag immunoblot following HJURP precipitation. Mock precipitations using whole rabbit IgG serve as a negative control.
F) Schematic of LacI/LacO tethering experiment in G-I. To force localization of mutant M18BP1 to chromatin despite defective nucleosome binding, M18BP1 was fused to the DNA-binding protein LacI. The cognate LacO DNA sequence was inserted into the repeat unit of the nucleosome array DNA such that there is one LacO sequence per 601 nucleosome positioning sequence.
G) Fusion of LacI to Flag-M18BP1-2YW rescues localization to CENP-A chromatin. Quantification of Flag signal on LacO chromatin-coated beads tethered with the indicated LacI-Flag-M18BP1 species.
H) Rescue of M18BP1-2YW localization by artificial tethering rescues CENP-A assembly. Quantification of V5-CENP-A on beads from (G).
All graphs show the mean ± SEM of five experiments.
I) Representative Western blot of samples from G-H. Immunolocalized protein is indicated at the top. M18BP1 immunoblot indicates equivalent levels of LacI-Flag-M18BP1-2 proteins in V5-CENP-A assembly reactions.
See also Figure S4.
We considered the possibility that M18BP1-2R762A and M18BP1-2YW were defective in CENP-A assembly due to an inability to interact with other CENP-A assembly factors. We therefore assayed formation of the Mis18 complex (Fujita et al., 2007). We observed interphase-specific association between Xenopus Mis18α, Mis18β, and M18BP1-2 by immunoprecipitation (Figure 3D) as previously demonstrated in humans (McKinley and Cheeseman, 2014). Wild-type M18BP1-2, M18BP1-2R762A and M18BP1-2YW all bound Mis18α and Mis18β, suggesting that defects in CENP-A assembly do not result from defective Mis18 complex assembly (Figure 3D). Moreover, we also observed no difference in wild-type M18BP1-2, M18BP1-2R762A and M18BP1-2YW binding to HJURP in interphase (Figure 4E). These data suggest that interaction of M18BP1 with proteins required for CENP-A assembly is not affected by M18BP1 nucleosome binding mutations.
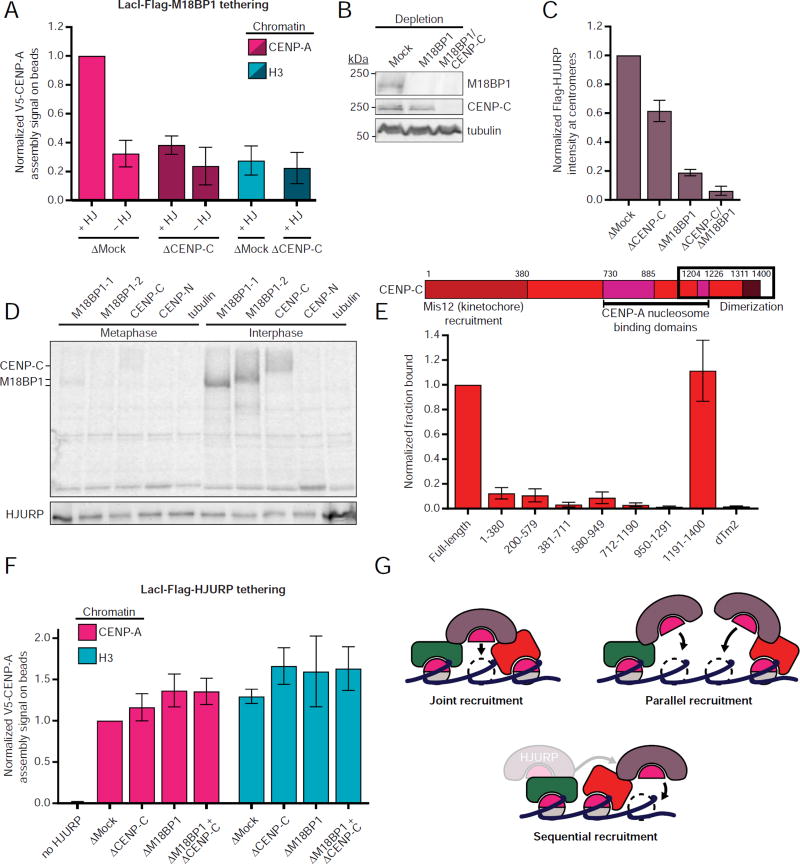
Figure 4. The Mis18 complex and CENP-C coordinate HJURP recruitment to CENP-A chromatin.
A) M18BP1 localization is not sufficient to promote new CENP-A assembly on H3 chromatin or in the absence of CENP-C. Quantification of V5-CENP-A assembly on LacO chromatin-coated beads in M18BP1-depleted extracts complemented with LacI-Flag-M18BP1-2. Extracts were additionally depleted of CENP-C or mock-depleted to assess the role of CENP-C. Depletion of CENP-C or tethering to H3 rather than CENP-A chromatin reduced V5-CENP-A assembly to background levels, indicated by signal equivalent to that observed when HJURP was omitted from the assembly reaction (-HJ). Plot shows mean V5-CENP-A signal ± SEM of at least three experiments.
B) Representative Western blot of egg extract samples used for A. Depletion condition is indicated above; immunoblotted species is indicated at right. Efficient immunodepletion of M18BP1 and CENP-C are indicated by comparing lanes 2 and 3 to lane 1.
C) M18BP1 and CENP-C make distinct contributions to Flag-HJURP localization at interphase sperm centromeres. Flag-HJURP translated in rabbit reticulocyte lysate was added to interphase egg extract depleted of CENP-C, M18BP1 or both. Plot shows mean centromeric Flag signal ± SEM of three experiments.
D) HJURP associates with CENP-C specifically in interphase. Metaphase or interphase extract was supplemented with rabbit reticulocyte lysate containing putative HJURP binding partners (indicated above) translated in the presence of 35S-methionine. Co-immunoprecipitation with endogenous HJURP was detected by autoradiography. Equivalent precipitation of HJURP in all reactions is indicated by anti-HJURP immunoblot below.
E) HJURP directly interacts with the CENP-C C-terminus. Full-length CENP-C, CENP-C truncations, or Drosophila tropomyosin 2 (dTm2) as a negative control were translated in rabbit reticulocyte lysate in the presence of 35S-methionine and then mixed with in vitro translated Flag-HJURP. Co-immunoprecipitation with Flag-HJURP was detected by autoradiography. (Top) Schematic of CENP-C with conserved domains indicated by colored bars and the region corresponding to amino acids 1191-1400 boxed. (Bottom) Plot shows fraction of input bound for each species normalized to full-length CENP-C ± SD from two experiments. Representative autoradiograph in Figure S4C.
F) The functions of M18BP1 and CENP-C in CENP-A assembly are effectively bypassed by direct tethering of HJURP to CENP-A or H3 chromatin. Quantification of V5-CENP-A assembly on LacO chromatin-coated beads in extract (depletion condition indicated below) supplemented with LacI-Flag-HJURP. Plot shows mean V5 signal on beads ± SEM of at least three experiments. Corresponding LacI-Flag-HJURP localization quantification and retention of V5-CENP-A signal after 1 mM IPTG treatment shown in Figure S4D-F.
G) Possible models for coordinated HJURP recruitment by M18BP1 and CENP-C.
See also Figure S4.
We next considered that defective CENP-A assembly by mutants with disrupted nucleosome binding may be due primarily to the reduction of centromeric M18BP1. If so, then forced localization of mutant M18BP1 to centromeres would rescue CENP-A assembly. We forced the localization of M18BP1-2YW by artificially tethering LacI-Flag-M18BP1-2YW to reconstituted CENP-A chromatin containing LacO in M18BP1-depleted extract (Figure 3F). We observed LacO-LacI specific recruitment to these arrays, as LacI-Flag protein bound to LacO-containing chromatin but did not bind LacO-negative chromatin (Figure S4A). Likewise, fusion of LacI to Flag-M18BP1-2YW rescued its recruitment to CENP-A chromatin only when the LacO sequence was within the underlying DNA (Figure S4A). When LacI-Flag-M18BP1-2YW was tethered directly to chromatin, both localization (Figure 3G) and CENP-A assembly (Figure 3H) were restored to near wild-type levels. These data demonstrate that CENP-A nucleosome binding by M18BP1 is required to target M18BP1 to centromeres for new CENP-A assembly.
HJURP localizes to CENP-A chromatin by directly binding the Mis18 complex and CENP-C
The Mis18 complex directly binds HJURP via Mis18β (Stellfox et al., 2016; Wang et al., 2014) and HJURP localization is sufficient to promote local CENP-A nucleosome assembly (Barnhart et al., 2011). We hypothesized that tethering M18BP1 to chromatin might be sufficient to promote CENP-A assembly on CENP-A or H3 chromatin. CENP-A assembly in frog egg extracts requires the addition of the HJURP chaperone. Consistent with its function in localizing HJURP to centromeres, tethered LacI-Flag-M18BP1-2 promoted CENP-A assembly on CENP-A chromatin only when HJURP was added to the extract (Figure 4A). However, tethering LacI-Flag-M18BP1-2 to H3 chromatin was not sufficient to promote CENP-A assembly, even in the presence of added HJURP (Figure 4A).
These data indicate one or more factors, in addition to M18BP1, are required to promote CENP-A assembly. Consistent with this, depletion of CENP-C prevented CENP-A assembly even when M18BP1 was tethered to CENP-A chromatin (Figure 4A). To understand how M18BP1 and CENP-C promote CENP-A assembly in interphase, we measured the localization of Flag-HJURP at sperm centromeres after removal of M18BP1, CENP-C or both. Despite an increase in centromeric M18BP1 levels after CENP-C depletion (Figure 2E), Flag-HJURP localization decreased by 38 ± 7% (Figure 4B,C). M18BP1 depletion caused an 81 ± 4% defect in Flag-HJURP localization, and co-depletion of M18BP1 and CENP-C reduced centromeric HJURP levels by 94±6% (Figure 4C). Together, these data indicate that both CENP-C and M18BP1 are required for localizing HJURP to centromeric chromatin. Because the effects of CENP-C depletion and M18BP1 depletion on HJURP centromere localization are additive and both are required for CENP-A assembly, M18BP1 and CENP-C appear to coordinate CENP-A assembly.
In addition to the established interaction between the Mis18 complex and HJURP, we hypothesized that CENP-C might recruit HJURP to centromeres by a direct interaction. We tested whether CENP-C associates with HJURP in Xenopus by immunoprecipitation from metaphase and interphase extracts. We radiolabeled M18BP1-1, M18BP1-2, CENP-C and CENP-N by in vitro translation in the presence of 35S-methionine and added the labeled protein to metaphase or interphase egg extract. Following immunoprecipitation of endogenous HJURP, we detected interphase-specific association of both M18BP1 isoforms and CENP-C by autoradiography (Figure 4D). Coexpression of CENP-C or CENP-C truncations with Flag-HJURP in rabbit reticulocyte lysates demonstrated a direct interaction between HJURP and CENP-C that occurs through the C-terminal domain of CENP-C (Figure 4E, S4C), consistent with a recent report in human cells (Tachiwana et al., 2015). Based upon the interaction between CENP-C and HJURP and the function for CENP-C in localizing HJURP to interphase centromeres, we propose that CENP-C directly recruits HJURP to promote new CENP-A assembly (Figure 4G).
We tested whether we could bypass the need for CENP-C and/or M18BP1 in CENP-A assembly by directly tethering LacI-Flag-HJURP to CENP-A or H3 chromatin and measuring V5-CENP-A assembly in egg extracts depleted of CENP-C and/or M18BP1. In the absence of HJURP tethering, CENP-C and/or M18BP1 depletion caused defects in CENP-A assembly (Figure 3B) (Moree et al., 2011; Westhorpe et al., 2015). However, tethering LacI-Flag-HJURP supported CENP-A assembly on CENP-A or H3 chromatin in the absence of CENP-C, M18BP1 or both (Figure 4F, S4E-F). Importantly, the V5-CENP-A signal persisted after treatment with 1 mM IPTG, which releases LacI from LacO, demonstrating that V5-CENP-A is not merely tethered to chromatin by LacI-Flag-HJURP (Figure S4D,E). Thus, during the process of CENP-A assembly, M18BP1 and CENP-C both function to localize HJURP to centromeres.
Discussion
CENP-A nucleosome binding by M18BP1 has now been demonstrated in frog (this work) and in accompanying work in chicken (Hori et al., 2017). We and others have observed that the M18BP1 CENP-C motif is conserved in insect, nematode, bird, reptile, frog, fish, and plant M18BP1 orthologues (Figure 2A) (Kral, 2015; Sandmann et al., 2017). With the exception of platypus (O. anatinus), the key nucleosome binding residues of the CENP-C motif are mutated in all mammalian orthologues we examined. Indeed, human M18BP1 does not show selective binding to reconstituted CENP-A chromatin (Figure S1G). We therefore propose that the mode of M18BP1 centromere recruitment has diverged specifically in the mammalian clade. Mis18α/β may govern Mis18 complex recruitment in mammals (Stellfox et al., 2016). In most other eukaryotes with an M18BP1 orthologue, M18BP1 interaction with CENP-A nucleosomes is likely to play a dominant role in the epigenetic determination of new CENP-A assembly.
We show that, in Xenopus, M18BP1 must bind CENP-A nucleosomes to support new CENP-A assembly. Disrupting the interaction between M18BP1 and the CENP-A nucleosome by mutating M18BP1 impairs CENP-A assembly, but assembly can be restored by artificially targeting mutant M18BP1 to CENP-A chromatin. M18BP1 shares a CENP-A nucleosome recognition mechanism with CENP-C and both are required to recruit HJURP to chromatin in interphase through direct HJURP binding. In addition, we show that the functions of M18BP1 and CENP-C in CENP-A assembly can be bypassed by directly tethering HJURP to CENP-A chromatin. This suggests a model in which M18BP1 and CENP-C coordinately recruit HJURP and soluble CENP-A to ensure that new CENP-A assembly is restricted to sites near existing CENP-A nucleosomes (Figure 4G).
It is unclear why both M18BP1 and CENP-C are required to bring functional HJURP to centromeres. Because M18BP1 tethering does not bypass the requirement for CENP-C, we disfavor a model in which M18BP1 and CENP-C provide redundant, parallel pathways for HJURP recruitment and CENP-A nucleosome assembly (Figure 4G). Instead, M18BP1 and CENP-C may coordinate HJURP recruitment to adjacent nucleosomes or two faces of the same CENP-A nucleosome (Figure 4G, ‘Joint recruitment’). One alternative model is that CENP-C and M18BP1 recruit HJURP sequentially (Figure 4G). A two-step ‘licensing’ and assembly reaction was recently proposed for the Mis18 complex (Nardi et al., 2016), one way to control the directionality of the assembly reaction and ensure it occurs only once would be to separate the recruitment of nascent CENP-A to centromeres from the transfer of CENP-A into a new nucleosome (Figure 4G).
Although, M18BP1 and CENP-C appear to possess a redundant function (HJURP recruitment), their precise roles in CENP-A assembly must be unique for both factors to be required. Depletion of either M18BP1 or CENP-C causes defects in CENP-A assembly, yet artificially tethering HJURP to chromatin can bypass the requirement for both proteins in CENP-A assembly. This phenomenon, where the presence of only CENP-C or only M18BP1 cannot support assembly, may represent a quality control mechanism for the CENP-A assembly process to allow new CENP-A nucleosome formation only proximal to existing CENP-A nucleosomes. A model in which HJURP is transferred from CENP-C to M18BP1 (or vice versa) may help to explain how loss of either protein causes a defect in CENP-A assembly yet forcing HJURP localization can bypass both. Thus, a major goal for the field will be to understand what the respective activities of the Mis18 complex and CENP-C are in CENP-A assembly and how they are coordinated to promote faithful transmission of centromeric chromatin across generations.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Aaron Straight (astraigh@stanford.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mature Xenopus laevis females (Nasco, LM00535MX) were housed and maintained in the Stanford Aquatic Facility staffed by the Veterinary Service Center. For ovulation, frogs were primed 2–14 days before ovulation by subcutaneous injection of 50 U pregnant mare serum gonadotropin (PMSG; Sigma) at the dorsal lymph sac, then induced 18 hours before ovulation with 500 U human chorionic gonadotropin (hCG; Chorulon). During ovulation, frogs were housed individually in 2 L MMR buffer (6 mM Na-HEPES, pH 7.8, 0.1 mM EDTA, 100 mM NaCl, 2 mM KCl, 1 mM MgCl2, and 2 mM CaCl2) at 17°C. Animal work was carried out in accordance with the guidelines of the Stanford University Administrative Panel on Laboratory Animal Care (APLAC).
Human DLD1 cell line (male, pseudo-diploid, colorectal adenocarcinoma) endogenously tagged at both CENP-C alleles with auxin-inducible degron (AID)-EYFP and stably expressing the F-box protein osTIR1 was a gift from Daniele Fachinetti (Institut Curie, Paris) (Fachinetti et al., 2015). Production of CENP-C-AID cells stably expressing mRuby-Flag-M18BP1 is described below. Cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum, penicillin/streptomycin, and 2g/L sodium bicarbonate and maintained in a 37°C/5% CO2 incubator.
METHOD DETAILS
Protein purification
Histones were purified as described previously (Guse et al., 2012; Westhorpe et al., 2015). Briefly, Histone H2A, H2B, H3, H3CAC and H4 were expressed from a pET3a vector in BL21 (DE3) Codon Plus RIPL Escherichia coli (Agilent Technologies, 230280), grown in 2xYT medium (20 g/liter tryptone, 10 g/liter yeast extract, and 5 g/liter NaCl), and induced for 3 hours with 0.25 mM IPTG at an OD600 of 0.6. Bacteria were harvested and resuspended in lysis buffer (20 mM potassium phosphate, pH 6.8, 1M NaCl, 5 mM 2-mercaptoethanol, 1 mM PMSF, 1 mM benzamidine, 0.05% NP-40, and 0.2 mg/ml lysozyme), homogenized using an EmulsiFlex-C5 (Avestin, Inc.) followed by 1 × 30 second sonication. The lysate was centrifuged (18,000 × g, 20 min, 4°C) and the insoluble pellet was washed in lysis buffer and resuspended in unfolding buffer (7 M guanidine-HCl, 20 mM Tris-HCl, pH 7.5, and 10 mM DTT). After another round of centrifugation, the supernatant from this resuspension was dialyzed into urea buffer (6 M deionized urea, 200 mM NaCl, 10 mM Tris-HCl, pH 8, 1 mM EDTA, 5 mM 2-mercaptoethanol, and 0.1 mM PMSF). Histones were isolated by chromatography through HiTrap Q and HiTrap S columns in series. Histones were eluted from the HiTrap S column with urea buffer containing 1 M NaCl, dialyzed into water and lyophilized. To reconstitute H2A/B dimer or H3/H4 tetramer, histones were mixed and resuspended in unfolding buffer followed by dialysis into 2 M NaCl, 10 mM Tris-HCl, pH 7.6, 1 mM EDTA, and 5 mM 2-mercaptoethanol. H2A/H2B dimers and H3/H4 tetramers were then purified by size-exclusion chromatography.
CENP-A-Myc-H4 tetramer, and CENP-A/H3-Myc-H4 chimeric tetramers containing the CATD were expressed as soluble tetramers as described(Guse et al., 2012) using the pST39 vector(Tan, 2001). The initial supernatant, isolated as described above, was run through hyroxyapatite (HA) (type II 20 µM HA; Bio-Rad Laboratories) pre-equilibrated with 20 mM potassium phosphate, pH 6.8. After washing with 6 column volumes (20 mM potassium phosphate, pH 6.8, 1 M NaCl, and 5 mM 2-mercaptoethanol) tetramer was eluted with HA buffer containing 3.5 M NaCl. The elution was dialyzed into 10 mM Tris-HCl, pH 7.4, 0.75 M NaCl, 10 mM 2-mercaptoethanol, and 0.5 mM EDTA. The dialyzed protein was passed through a 1 ml HiTrap SP FastFlow column, washed, and tetramer was eluted over a 20–column volume gradient into dialysis buffer containing 2 M NaCl. Fractions containing concentrated tetramer were pooled, aliquoted, frozen in liquid nitrogen, and stored at −80°C.
MBP-Prescission-M18BP1-2747-944 and MBP-Prescission-Flag-M18BP1-2747-944-His (wild-type and YW mutant) were expressed from modified pMal-c2x and pMal-c5x-His plasmids, respectively, (NEB) in BL21 (DE3) Codon Plus RIPL E. coli. Two 2L cultures were grown in 2xYT medium to OD600 of 0.6 at 37°C and then induced with 0.5 mM IPTG for 4 hours at 21°C. Bacteria were lysed in MBP lysis buffer (10 mM sodium phosphate pH 7.4, 500 mM NaCl, 2.7 mM KCl, 1 mM EDTA, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM benzamidine hydrochloride, 10 ug/mL LPC [leupeptin, pepstatin A, chymostatin], 0.2 mg/mL lysozyme) by three passes through an EmulsiFlex-C5 (Avestin, Inc.) and then clarified by centrifugation at 40,000 rpm in a Type 45ti rotor (Beckman) for 1.5 hours at 4°C. Clarified lysates were flowed over 2 mL amylose resin (NEB) equilibrated with MBP lysis buffer to capture MBP-tagged protein. Resin was subsequently washed with 30 column volumes lysis buffer. MBP-Prescission-M18BP1-2747-944 was then washed with 10 column volumes MBP wash buffer (20 mM Na-HEPES pH 7.7, 150 mM NaCl, 5 mM 2-mercaptoethanol) and eluted with MBP wash buffer containing 10 mM maltose. This protein was dialyzed against protein storage buffer (20 mM Na-HEPES, pH 7.7, 150 mM NaCl, 5 mM 2-mercaptoethanol, 10% glycerol), concentrated to 1 mL by ultracentrifugation (Amicon Ultra 15 MWCO 30k; Millipore), and stored in 50 uL aliquots at −80°C. MBP-Prescission-Flag-M18BP1-2747-944-His bound to amylose resin was washed with 10 column volumes of His binding buffer (20 mM Tris-HCl, pH 7.4, 500 mM NaCl, 5 mM 2-mercaptoethanol, 10 mM imidazole). Flag-M18BP1-2747-944-His was eluted by incubating the resin in 2 column volumes of His binding buffer containing 160U Prescission Protease overnight at 4°C with rotation. Eluted material was bound to NiNTA agarose (QIAGEN) equilibrated with His binding buffer and then washed with an additional 30 column volumes. Flag-M18BP1-2747-944-His was eluted with His binding buffer containing 300 mM imidazole, dialyzed against protein storage buffer, concentrated to 1 mL, and stored at −80°C as above.
Chromatin bead reconstitution
Biotinylated 19 × 601 array DNA was purified as previously described(Guse et al., 2012). In brief, puC18 containing 19 repeats of the ‘601’ nucleosome positioning sequence (Lowary and Widom, 1998) was purified with a Gigaprep kit (QIAGEN) from SURE2 bacteria grown in Luria Broth media. The plasmid was restriction digested with EcoRI, XbaI, DraI and HindIII, and the positioning array purified by polyethylene glycol (PEG) precipitation using 0.5% incremental increases in PEG concentration, each with a 10 minute, 5,000 × g centrifugation. PEG precipitations with 19 × 601 DNA were dialyzed into TE buffer (10 mM Tris, pH 8 and 0.5 mM EDTA) and concentrated by ethanol precipitation. The EcoRI overhangs were filled with biotin-14-dATP (Invitrogen), dCTP, α-thiο-dGTP, and α-thio-dTTP (ChemCyte) using Klenow fragment 3’-5’ exo- (New England Biolabs). Nucleosome array DNA containing LacO was grown in DH5α cells at 30°C instead of SURE2 cells. The repeating unit of the 12 × LacO positive array is AGCTTGGATCTCTGGAGAATCCCGGTGCCGAGGCCGCTCAATTGGTCGTAGCAAGCT CTAGCACCGCTTAAACGCACGTACGCGCTGTCCCCCGCGTTTTAACCGCCAAGGGGA TTACTCCCTAGTCTCCAGGCACGTGTCAGATATATACATCCTGTCCTCGAGCAATTGT GAGCGGCTCACAATTCGTCCCTATCAGTGATAGAGACTAGTTTCGTTGGAAACGGGA CTGA, with the nucleosome positioning ‘601’ site underlined and the LacO site italicized. pCS2 plasmid containing 12 × LacO positive array (ASP 3317) was digested with BamHI, HaeII and DraI and the array was isolated by PEG precipitation and biotinylated as above for the19 × 601 array.
Reconstituted chromatin containing recombinant nucleosomes was prepared at 2 µM nucleosome concentration by salt dialysis as previously described(Guse et al., 2012). DNA and histones were pooled in pre-dialysis buffer (2 M NaCl, 10 mM Tris pH 7.5, 0.25 mM EDTA) and dialyzed over 36 hours into array dialysis buffer (2.5 mM NaCl, 10 mM Tris pH 7.5, 0.25 mM EDTA). H2A/H2B dimers were added at 2.2 × tetramer ratio, and 1.8 tetramers per nucleosome positioning sequence. Reconstituted chromatin was assessed by SyBr Gold (Life Technologies) staining of a 5% acrylamide native PAGE gel after chromatin digestion with AvaI.
Chromatin was bound to streptavidin-coated M-280 Dynabeads (Invitrogen, 11205D) prewashed in bead buffer (50 mM Tris pH 7.4, 75 mM NaCl, 0.25 mM EDTA, 0.05% Triton X-100 and 2.5% polyvinyl alcohol). Chromatin was bound at 2.6 fmol arrays per microgram of bead for 30–60 minutes at room temperature with constant shaking. After washing in bead buffer, beads were added to either Xenopus egg extract, rabbit reticulocyte lysate with in vitro translated protein (below), or a solution of purified protein.
In vitro translation and chromatin binding assays
In vitro translated (IVT) proteins were generated in rabbit reticulocyte lysate using the SP6 TNT Quick-Coupled Transcription/Translation kit (Promega, L2080) according to the manufacturer’s instructions.
For chromatin binding assays, lysates containing in vitro translated protein were diluted with 5x CSF-XBT (50 mM K-HEPES, pH 7.7, 500 mM KCl, 250 mM sucrose, 10 mM MgCl2, 0.5 mM CaCl2, 25 mM EGTA, 0.25% Triton X-100) to a final concentration of 1x CSF-XBT. Protein was bound to reconstituted CENP-A or H3 chromatin by incubating 10 uL diluted IVT protein with 0.25 uL chromatin-coated beads for 60 minutes at 21°C. Beads were isolated by magnetization and washed three times with CSF-XBT. In Figures 1–4, beads were fixed for 5 minutes at 21°C in CSF-XBT containing 2% formaldehyde, washed into antibody dilution buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl with 0.1% Triton X-100, and 2% bovine serum albumin), and settled on poly-L-lysine-coated coverslips for 30–60 minutes before processing for immunofluorescence microscopy. In Figures S2–3, beads were fixed in antibody dilution buffer containing 2% formaldehyde and settled on poly-L-lysine-coated coverslips for 30 minutes. In Figure S2C,E, recombinant Flag-M18BP1-2747-944 was diluted to the indicated concentrations in 1x CSF-XBT and incubated with 0.25 uL chromatin beads.
Xenopus egg extracts and immunodepletions
CSF-arrested (metaphase) Xenopus egg extracts were prepared as described previously (Desai et al., 1999; Guse et al., 2012). In brief, eggs were washed with MMR buffer (6 mM Na-HEPES, pH 7.8, 0.1 mM EDTA, 100 mM NaCl, 2 mM KCl, 1 mM MgCl2, and 2 mM CaCl2) and then dejellied in MMR + 2% L-cysteine. Dejellied eggs were washed in CSF-XB buffer (10 mM K-HEPES, pH 7.7, 100 mM KCl, 50 mM sucrose, 2 mM MgCl2, 0.1 mM CaCl2, 5 mM EGTA), followed by CSF-XB containing 10 µg/ml LPC [leupeptin/pepstatin A/chymostatin]. Eggs were packed in a 13 × 51-mm polyallomer tube (Beckman Coulter, 326819) by a low-speed spin in a table top clinical centrifuge for 45 s. After removal of buffer, packed eggs were centrifuged in a SW55Ti rotor (Beckman Coulter) for 15 min at 10,000 rpm. The soluble cytoplasm was removed by side puncture with a 18G needle and supplemented with 50 mM sucrose, 10 µg/ml LPC, 10 µg/ml cytochalasin D, and energy mix (7.5 mM creatine phosphate, 1 mM ATP, and 1 mM MgCl2).
Immunodepletions of CENP-C and M18BP1 from Xenopus extract was performed essentially as described (Moree et al., 2011). Protein A Dynabeads (Invitrogen, 10001D) were washed in TBSTx (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% Triton X-100) or CSF-XBT and then coupled to affinity purified antibody for 30–60 minutes at 4°C. For 100 uL extract, 2–5 ug anti-xCENP-C antibody (rabbit, raised and purified against xCENP-C207-296 (Milks et al., 2009)), or 4–5 ug anti-xM18BP1 (rabbit, raised against GST-xM18BP1-2 amino acids 161-415 and purified against xM18BP1-1161-375 (Moree et al., 2011)) were used. An equivalent amount of whole rabbit IgG (Jackson ImmunoResearch Laboratories, Inc.) was used for mock depletions. Antibody-coupled beads were washed three times in TBSTx or CSF-XBT and then resuspended in extract. Depletions were rotated at 4°C for 1 hour. Beads were removed from extract by 2–3 5-minute magnetizations.
CENP-A assembly assays
CENP-A assembly assays on sperm chromatin or reconstituted chromatin were performed as previously described (Moree et al., 2011; Westhorpe et al., 2015). RNAs used to express protein in Xenopus egg extract were prepared with the SP6 mMessage mMachine kit (Life Technologies, AM1340). NotI linearized pCS2+ plasmids were used in the transcription reaction following manufacturer’s instructions, using double the amount of DNA. RNA was purified using RNeasy mini columns (QIAGEN). RNA was typically concentrated to at least 1 ug/uL using a SpeedVac (Savant) to avoid excessive dilution of the extract.
For sperm assembly assays, 30 uL assembly reactions contained 1.5 uL xHJURP IVT protein, 0.75 uL Flag-M18BP1-2 IVT protein, and 25 ng/uL Myc-xCENP-A RNA. CSF-arrested, M18BP1-depleted extract (prepared as above) was supplemented with IVT protein and RNA and incubated at 16–18°C for 30 minutes to permit RNA translation. Cycloheximide was then added to 0.1 mg/mL to terminate translation. Reactions were then released to interphase by addition of 750 uM CaCl2 and supplemented with demembranated sperm chromatin to 3,000/uL. After incubation for 75 minutes at 16–18°C, assembly reactions were diluted in 1 mL dilution buffer (BRB-80 [80 mM K-PIPES, pH 6.8, 1 mM MgCl2, 1 mM EGTA], 0.5% Triton X-100, 30% glycerol) containing 150 mM KCl and incubated on ice for 5 minutes. These samples were then fixed by addition of 1 mL dilution buffer containing 4% formaldehyde and spun through cushions of 40% glycerol in BRB-80 onto poly-L-lysine-coated coverslips to be processed for immunofluorescence. Quantification of CENP-A loading (centromeric Myc-CENP-A signal) was performed as described below.
For assembly assays using reconstituted chromatin, V5-xCENP-A and xHJURP RNA were added to CSF-arrested extract (prepared as above) at 20 and 40 ng/µl, respectively, and translated for 30 minutes at 16–20°C. After the translation period, 0.1 mg/ml cycloheximide and 750 uM CaCl2 was added to the extract. Reactions were added to washed chromatin-coated beads to a final bead concentration of 25 µg beads/100 µl extract. For experiments comparing CENP-A assembly across chromatin templates, large translation reactions were prepared and aliquoted to each type of chromatin-coated beads. After incubation for 75 minutes at 16–20°C, chromatin arrays were washed three times in CSF-XBT, fixed for five minutes in CSF-XBT containing 2% formaldehyde, mounted onto poly-L-lysine-coated coverslips, and allowed to settle for 30–60 minutes prior to immunofluorescence. Quantification of CENP-A loading (V5-xCENP-A signal on beads) was performed as described below.
For experiments looking at V5-CENP-A retention after IPTG treatment, large reactions were split into two at the end of the 75 minute CENP-A assembly period, and half was treated with 1 mM IPTG for 10 minutes at 16–18°C. These samples were washed three times with CSF-XBT containing 1 mM IPTG and then processed for immunofluorescence as above.
M18BP1-mRuby-Flag human cell line
CENP-C-AID-EYFP/osTIR1/TRex DLD1 cells harboring an integrated FRT site were a gift from Daniele Fachinetti (Institut Curie, Paris). Cells were co-transfected with pcDNA5/FRT/TO containing the full-length human M18BP1 cDNA with mRuby2 and 3xFlag fused to the C-terminus and with pOG44 encoding Flp recombinase in a mass ratio of 1:9. Transfections were carried out with FugeneHD (Promega, E2311) at a ratio of 6 µL FugeneHD:1 µg DNA according to the manufacturer’s instructions. 48 h after transfection, culture medium was replaced and supplemented with 100 µg/mL hygromycin to select for stable M18BP1-mRuby-Flag integrants. Hygromycin resistant colonies were obtained after 1 month of selection and pooled for localization experiments.
Degradation of endogenous CENP-C was induced by supplementation of the culture medium with 1 mM indole-3-acetic acid (Fisher, 50-213-368) for 24 h.
To visualize CENP-C degradation by immunoblotting, cells were collected by trypsinization, washed once in PBS, and then lysed by sonication in denaturing lysis buffer (20 mM Tris-HCl pH 7.4, 15 mM NaCl, 10 mM EDTA, 0.5% Igepal, 0.1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate, 10 µg/mL LPC, 1 mM PMSF). Lysate concentrations were measured by Bradford Assay (BioRad) and normalized across samples by dilution with additional denaturing lysis buffer. After addition of sample buffer, immunoblotting was performed as described below.
For localization experiments, cells were grown on acid washed coverglass. Cells were fixed with 2% formaldehyde in PBS at 37°C for five minutes and then permeablized with 0.5% Triton X-100 in PBS for 5 minutes at room temperature. Coverslips were then processed for immunofluorescence.
Immunofluorescence preparation
Coverslips were first blocked in antibody dilution buffer (AbDil; 20 mM Tris-HCl, pH 7.4, 150 mM NaCl with 0.1% Triton X-100, and 2% bovine serum albumin) for 30 minutes. Coverslips were exposed to primary antibody diluted in AbDil for 15–30 minutes, washed three times with AbDil, and then exposed to AlexaFluor-conjugated secondary antibodies (Life Technologies; Jackson ImmunoResearch Laboratories, Inc.). When required, coverslips were blocked with 1 mg/mL whole rabbit or whole mouse IgG in AbDil followed by staining with directly-conjugated primary antibodies. Sperm nuclei and human cell coverslips were stained with 10 ug/mL Hoechst 33258 in AbDil for DNA. Coverslips were mounted in 90% glycerol, 10 mM Tris-HCl, pH 8.8, 0.5% p-phenylenediamine, sealed to a slide with clear nail polish and stored at −20°C.
Primary antibodies used were: 2 µg/mL anti-Flag (Sigma, F7425 [rabbit] and F1804 [mouse]), 0.25-0.5 µg/mL mouse anti-Myc (EMD Millipore, 4A6 [mouse]), 1 µg/mL rabbit anti-Myc (EMD Millipore, 06-549 [rabbit]), 1:400 mouse anti-V5 (Thermo Fisher, R960-25), 1µg/mL rabbit-anti-xCENP-A, 1µg/mL rabbit-anti-xCENP-C(Milks et al., 2009), 1.5 µg/ml rabbit-anti-xM18BP1, 1 µg/mL rabbit-anti-hCENP-A (raised and purified against GST-hCENP-A1-42), and 1:100 human anti-centromere serum (Antibodies, Inc., 15-234-0001). Secondary antibodies used were AlexaFluor 488-, 568-, and 647-conjugated goat anti-mouse or anti-rabbit (Life Technologies), and AlexaFluor 647-conjugated donkey anti-rabbit (Jackson ImmunoResearch Laboratories, Inc.). All secondaries were used at 1–2 µg/mL.
Image acquisition and processing
Imaging was performed on an IX70 Olympus microscope with a DeltaVision system (Applied Precision) a Sedat quad-pass filter set (Semrock) and monochromatic solid-state illuminators, controlled via softWoRx 4.1.0 software (Applied Precision). Images of sperm nuclei were acquired using a 60× 1.4 NA Plan Apochromat oil immersion lens (Olympus). Images of chromatin-coated beads and human cells were acquired using a 100× 1.4 NA oil immersion objective (Olympus). Images were acquired with a charge-coupled device camera (CoolSNAP HQ; Photometrics) and digitized to 12 bits. Z-sections were taken at 0.2-um intervals (for sperm nuclei and human cells) or 0.4-µm intervals (for beads). Displayed images of sperm nuclei are maximum intensity projections of deconvolved z-stacks; images of human cells are maximum intensity projections.
Image analysis of Xenopus sperm and human cell centromeres (Moree et al., 2011) or chromatin-coated beads (Westhorpe et al., 2015) was performed using custom software as previously described. For sperm, at least 200 centromeres among at least 15 nuclei were counted in each condition in each experiment. For human cells, at least 400 centromeres among at least 12 pairs of G1 nuclei were quantified per condition per experiment. For reconstituted chromatin, ~50–100 beads were quantified per condition per experiment.
Immmunoblotting
Immunoblots were performed as previously described (Westhorpe et al., 2015). Briefly, samples were resolved by SDS-PAGE and transferred onto polyvinylidene fluoride membrane (Bio-Rad Laboratories). For immunoblotting histones (Figure 3C, 3I), samples were transferred in CAPS transfer buffer (10 mM 3- (cyclohexylamino)-1-propanesulfonic acid, pH 11.3, 0.1% SDS, 20% methanol). All other samples were transferred in Tris-glycine transfer buffer (20 mM Tris-HCl, 200 mM glycine). Samples containing 0.75-1.5 uL IVT protein, 1.5 uL Xenopus egg extract, or 50 µg sonicated whole cell lysate were loaded per lane.
Membranes were blocked in 4% milk in PBSTx (10 mM sodium phosphate pH 7.4, 150 mM NaCl, 2.7 mM KCl, 0.2% Tween 20), probed with primary antibody, and detected using AlexaFluor 647-conjugated goat-anti-rabbit or goat-anti-mouse antibodies (Jackson ImmunoResearch Laboratories, Inc.). Detection was performed with a fluorescence imager (VersaDoc; Bio-Rad Laboratories, Inc.).
Primary antibodies used were: 1 ug/mL mouse-anti-Flag, 1 ug/mL mouse-anti-Myc, 1:2500 mouse anti-V5, 5 ug/mL rabbit-anti-xM18BP1, 2 ug/mL rabbit-anti-xHJURP (raised against GST-xHJURP42-194 and purified against His-xHJURP42-194), and 0.2 ug/mL mouse-anti-tubulin (Dm1α; Sigma). All secondary antibodies were used at 0.4 ug/mL.
Immunoprecipitation
To assay association of M18BP1-2 with Mis18α and Mis18β 50 uL reactions containing 10 uL Flag-M18BP1-2, 5 uL Myc-Mis18α, and 5 uL Mis18β IVT proteins in CSF-arrested or interphase M18BP1-depleted extract were added to 10 uL Protein A Dynabeads (Invitrogen) coupled to 2.5 ug mouse anti-Flag antibody (Sigma). To assay association of M18BP-2 with HJURP, 50 uL reactions containing 10 uL Flag-M18BP1-2 in CSF-arrested or interphase M18BP1-depleted extract were added to 10 uL Protein A Dynabeads (Invitrogen) coupled to 2.5 ug rabbit anti-xHJURP antibody. Following rotation for 1 hour at 4°C, beads were collected by magnetization and washed four times in TBSTx. Immunoprecipitates were eluted in sample buffer (50 mM Tris, pH 6.8, 15 mM EDTA, 1 M β-mercaptoethanol, 3.3% SDS, 10% glycerol, and 1 µg/ml Bromophenol blue), resolved by SDS-PAGE, and immunoblotted as described above.
To assay association of endogenous HJURP with CENP-C, CENP-C and other putative binding partners were translated in rabbit reticulocyte lysate as above except that 4 µCi/µL L-[35S]-methionine (PerkinElmer, NEG709A500UC) was used in place of the unlabeled methionine provided with the Promega kit. 50 µL reactions containing 5µL radiolabeled protein in metaphase-arrested or interphase egg extract were added to 10 uL Protein A Dynabeads (Invitrogen) coupled to 2.5 ug rabbit anti-xHJURP antibody. After washing beads and eluting as above, bound material was detected by autoradiography.
To assay direct binding of HJURP and CENP-C, unlabeled Flag-HJURP and radiolabeled full-length Myc-CENP-C or Myc-CENP-C truncations were translated in vitro as above. 50 uL binding reactions containing 10 uL Flag-HJURP and 10 uL radiolabeled protein in IVT binding buffer (50 mM Tris-HCl, pH 8.0, 50 mM NaCl, 0.25 mM EDTA, 0.05% Triton X-100, and 2% BSA) were incubated at 4°C for 1 h. Bound material was recovered by subsequent incubation of binding reactions with 10 µL Protein A Dynabeads (Invitrogen) coupled to 2.5 µg mouse anti-Flag at 4°C for 1 h. Beads were then washed three times with TBSTx and bound material eluted with sample buffer. Samples were resolved by SDS-PAGE and bound material visualized by autoradiography.
For identification of post-translational modifications of MBP-Prescission-xM18BP1747-944, protein was added to 200 ul Xenopus extract at 500 nM and incubated for 60 minutes at 20°C. Reactions were then diluted 2-fold in CSF-XBT, and MBP-tagged protein was reisolated by incubation at 4°C for 1 hour with 10 ug rabbit-anti-MBP antibody (generated by Cocalico Biologicals and purified from rabbit serum ) coupled to 40 ul Protein A Dynabeads (Invitrogen). After extensive washing in CSF-XBT, beads were incubated in sample buffer.
Mass spectrometry
Isolated MBP-Prescission-xM18BP1747-944 protein separated by SDS-PAGE in 10% acrylamide, stained with Coomassie R-250 and the protein band excised. Mass spectrometry was performed by the Taplin Mass-Spectrometry facility. Excised gel bands were cut into approximately 1 mm3 pieces, reduced with 1 mM dithiothreitol (DTT) for 30 minutes at 60°C and then alkylated with 5mM iodoacetamide for 15 minutes in the dark at room temperature. Gel pieces were trypsin digested in the gel, washed and dehydrated with acetonitrile for 10 min followed by removal of acetonitrile. Gel pieces were dried in a SpeedVac and rehydrated with 50 mM ammonium bicarbonate solution containing 12.5 ng/µl modified sequencing-grade trypsin (Promega, Madison, WI) at 4°C. Samples were then incubated at 37°C overnight. Peptides were later extracted by removing the ammonium bicarbonate solution, washing once with a solution containing 50% acetonitrile and 1% formic acid, and the extracts were dried in a SpeedVac (~1 hr). The samples were then stored at 4°C until analysis. For analysis, samples were reconstituted in 5–10 µl of HPLC solvent A (2.5% acetonitrile, 0.1% formic acid). A nano-scale reverse-phase HPLC capillary column was created by packing 2.6 µm C18 spherical silica beads into a fused silica capillary (100 µm inner diameter × ~30 cm length) with a flame-drawn tip. After equilibrating the column. each sample was loaded via a Famos auto sampler (LC Packings, San Francisco CA). A gradient was formed and peptides were eluted with increasing concentrations of solvent B (97.5% acetonitrile, 0.1% formic acid).
As each peptide was eluted they were subjected to electrospray ionization before entering into an LTQ Orbitrap Velos Pro ion-trap mass spectrometer (Thermo Fisher Scientific, San Jose, CA). Eluting peptides were detected, isolated, and fragmented to produce a tandem mass spectrum of specific fragment ions for each peptide. Peptide sequences (and hence protein identity) were determined by matching protein or translated nucleotide databases with the acquired fragmentation pattern by the software program, Sequest (ThermoFinnigan, San Jose, CA). The modification of 79.9663 mass units to serine, threonine, and tyrosine, and 42.0106 mass units to lysine, was included in the database searches to determine phospho and acetylpeptides, respectively. Phosphorylation/Acetylation assignments were determined by the Ascore algorithm. All databases include a reversed version of all the sequences and the data was filtered to between a one and two percent peptide false discovery rate. We estimated the prevalence of each modification by summing the peak max intensity of unique peptides including each modified residue and dividing by the sum of the max intensities of all unique peptides containing that residue, using a custom Matlab script (https://gitlab.com/straightlab/mass_spec).
Protein alignment
Protein sequences homologous to the nucleosome binding domain of Xenopus M18BP1-2 were identified using NCBI PSI-BLAST with an input query of amino acids 747-944. Multiple sequence alignments were performed with MAFFT (version 7.305b)(Katoh et al., 2002) using default parameters.
Quantification and Statistical Analysis
Image analysis of Xenopus sperm and human cell centromeres was performed using custom software as previously described (Moree et al., 2011). To identify centromere regions, images were normalized by median filtering the image and then dividing the original image by the filtered intensity value. Centromere masks were then generated by thresholding the channel with the indicated centromere marker (eg, CENP-C, ACA, etc.) followed by size filtering to remove regions larger or smaller than centromeres. Masks were then manually inspected to remove any remaining errant regions. Mean pixel intensity per centromere region was then measured for the channels of interest from maximum intensity projections. Reported values represent the average of this quantity across all centromere regions per condition, normalized to the indicated positive control. For sperm, at least 200 centromeres among at least 15 nuclei were counted in each condition in each experiment. For human cells, at least 400 centromeres among at least 12 pairs of G1 nuclei were quantified per condition per experiment.
Image analysis of chromatin-coated beads was performed using custom software as previously described (Westhorpe et al., 2015). Briefly, bead masks were generated by thresholding a single plane of a multi-z-section image representing the middle of a bead cluster and segmenting on the bead autofluorescence in the 605-nm emission channel. Regions larger than the known size of a bead or smaller than one-third of a bead were discarded. Mean pixel intensity per bead region was then measured for the channels of interest from maximum intensity projections. To correct for variation in the amount of chromatin coating each bead, these values were normalized by the Myc-H4 signal per bead. Reported values represent the average of this ratio across all beads per condition, normalized to the indicated positive control. ~50–100 beads were quantified per condition per experiment.
In Figure 4E, the fraction of CENP-C truncation bound to HJURP was quantified using ImageJ (National Institutes of Health). Briefly, a rectangular region of fixed size was drawn around each band in the ‘input’ and ‘bound’ lanes and the mean integrated density measured. To estimate the local background signal, the mean integrated density was also measured for a region of the same size above the bands, and this value was subtracted from the values measured for each band. The ‘bound’ signal was divided by the ‘input’ (adjusted for the amount of the sample loaded per lane) to obtain the fraction bound.
To estimate the prevalence of M18BP1 post-translational modifications identified by mass spectrometry, we summed the peak max intensity of unique peptides that included each modified residue and divided by the sum of the max intensities of all unique peptides containing that residue, using a custom Matlab script (https://gitlab.com/straightlab/mass_spec).
Supplementary Material
Highlights.
Xenopus M18BP1 directly binds CENP-A nucleosomes via a conserved ‘CENP-C’ motif
CENP-C competitively limits M18BP1 binding to CENP-A nucleosomes
CENP-A chromatin binding is required for M18BP1 recruitment and new CENP-A assembly
CENP-C directly binds HJURP, recruits HJURP to centromeres with the Mis18 complex
Acknowledgments
We thank the members of the Straight Laboratory for support and feedback on the project. We thank Tatsuo Fukagawa and colleagues for sharing their data prior to publication. B.T.F was supported by NIH T32 GM007276 and an NSF Graduate Research Fellowship. This work was supported by NIH R01 GM074728 to A.F.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
B.T.F. and F.G.W. performed the experiments in the manuscript. C.L. performed analysis of the mass spectrometry data. B.T.F., F.G.W., and A.F.S designed the experiments and wrote the manuscript.
References
- Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol. 2011;194:229–243. doi: 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Milks KJ, Straight AF. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol. 2010;189:1143–1155. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Silva MC, Godek KM, Jansen LE, Straight AF. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat Cell Biol. 2009;11:896–902. doi: 10.1038/ncb1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambacher S, Deng W, Hahn M, Sadic D, Frohlich J, Nuber A, Hoischen C, Diekmann S, Leonhardt H, Schotta G. CENP-C facilitates the recruitment of M18BP1 to centromeric chromatin. Nucleus. 2012;3:101–110. doi: 10.4161/nucl.18955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Murray A, Mitchison TJ, Walczak CE. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods in cell biology. 1999;61:385–412. doi: 10.1016/s0091-679x(08)61991-3. [DOI] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- Fachinetti D, Diego Folco H, Nechemia-Arbely Y, Valente LP, Nguyen K, Wong AJ, Zhu Q, Holland AJ, Desai A, Jansen LE, et al. A two-step mechanism for epigenetic specification of centromere identity and function. Nat Cell Biol. 2013;15:1056–1066. doi: 10.1038/ncb2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachinetti D, Han JS, McMahon MA, Ly P, Abdullah A, Wong AJ, Cleveland DW. DNA Sequence-Specific Binding of CENP-B Enhances the Fidelity of Human Centromere Function. Developmental cell. 2015;33:314–327. doi: 10.1016/j.devcel.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nature reviews Molecular cell biology. 2013;14:25–37. doi: 10.1038/nrm3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR, 3rd, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Developmental cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Fukagawa T, Earnshaw WC. The centromere: chromatin foundation for the kinetochore machinery. Developmental cell. 2014;30:496–508. doi: 10.1016/j.devcel.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guse A, Carroll CW, Moree B, Fuller CJ, Straight AF. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 2011;477:354–358. doi: 10.1038/nature10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guse A, Fuller CJ, Straight AF. A cell-free system for functional centromere and kinetochore assembly. Nature protocols. 2012;7:1847–1869. doi: 10.1038/nprot.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Hori T, Shang WH, Hara M, Ariyoshi M, Arimura Y, Fujita R, Kurumizaka H, Fukagawa T. Association of M18BP1/KNL2 with CENP-A nucleosome is essential for centromere formation in non-mammalian vertebrates. Developmental cell. 2017 doi: 10.1016/j.devcel.2017.06.019. [DOI] [PubMed] [Google Scholar]
- Hori T, Shang WH, Takeuchi K, Fukagawa T. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J Cell Biol. 2013;200:45–60. doi: 10.1083/jcb.201210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Jiang J, Zhou BR, Rozendaal M, Feng H, Ghirlando R, Xiao TS, Straight AF, Bai Y. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science. 2013;340:1110–1113. doi: 10.1126/science.1235532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kral L. Possible identification of CENP-C in fish and the presence of the CENP-C motif in M18BP1 of vertebrates. F1000Res. 2015;4:474. doi: 10.12688/f1000research.6823.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lermontova I, Kuhlmann M, Friedel S, Rutten T, Heckmann S, Sandmann M, Demidov D, Schubert V, Schubert I. Arabidopsis kinetochore null2 is an upstream component for centromeric histone H3 variant cenH3 deposition at centromeres. Plant Cell. 2013;25:3389–3404. doi: 10.1105/tpc.113.114736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. Journal of molecular biology. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- Maddox PS, Hyndman F, Monen J, Oegema K, Desai A. Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J Cell Biol. 2007;176:757–763. doi: 10.1083/jcb.200701065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley KL, Cheeseman IM. Polo-like kinase 1 licenses CENP-A deposition at centromeres. Cell. 2014;158:397–411. doi: 10.1016/j.cell.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiburo MJ, Padeken J, Fulop S, Schepers A, Heun P. Drosophila CENH3 is sufficient for centromere formation. Science. 2011;334:686–690. doi: 10.1126/science.1206880. [DOI] [PubMed] [Google Scholar]
- Milks KJ, Moree B, Straight AF. Dissection of CENP-C-directed centromere and kinetochore assembly. Molecular biology of the cell. 2009;20:4246–4255. doi: 10.1091/mbc.E09-05-0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moree B, Meyer CB, Fuller CJ, Straight AF. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J Cell Biol. 2011;194:855–871. doi: 10.1083/jcb.201106079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardi Isaac K, Zasadzińska E, Stellfox Madison E, Knippler Christina M, Foltz Daniel R. Licensing of Centromeric Chromatin Assembly through the Mis18α -Mis18β Heterotetramer. Mol Cell. 2016;61:774–787. doi: 10.1016/j.molcel.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR, 3rd, Desai A, Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- Pan D, Klare K, Petrovic A, Take A, Walstein K, Singh P, Rondelet A, Bird AW, Musacchio A. CDK-regulated dimerization of M18BP1 on a Mis18 hexamer is necessary for CENP-A loading. Elife. 2017;6 doi: 10.7554/eLife.23352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perpelescu M, Hori T, Toyoda A, Misu S, Monma N, Ikeo K, Obuse C, Fujiyama A, Fukagawa T. HJURP is involved in the expansion of centromeric chromatin. Mol Biol Cell. 2015;26:2742–2754. doi: 10.1091/mbc.E15-02-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux AL, Choi ES, Abbott JK, Liu X, Kagansky A, Castillo AG, Hamilton GL, Richardson W, Rappsilber J, He X, et al. Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol Cell. 2009;33:299–311. doi: 10.1016/j.molcel.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann M, Talbert P, Demidov D, Kuhlmann M, Rutten T, Conrad U, Lermontova I. Targeting of Arabidopsis KNL2 to Centromeres Depends on the Conserved CENPC-k Motif in Its C Terminus. Plant Cell. 2017;29:144–155. doi: 10.1105/tpc.16.00720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KC, Sullivan BA. Neocentromeres: a place for everything and everything in its place. Trends Genet. 2014;30:66–74. doi: 10.1016/j.tig.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Session AM, Uno Y, Kwon T, Chapman JA, Toyoda A, Takahashi S, Fukui A, Hikosaka A, Suzuki A, Kondo M, et al. Genome evolution in the allotetraploid frog Xenopus laevis. Nature. 2016;538:336–343. doi: 10.1038/nature19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shono N, Ohzeki J, Otake K, Martins NM, Nagase T, Kimura H, Larionov V, Earnshaw WC, Masumoto H. CENP-C and CENP-I are key connecting factors for kinetochore and CENP-A assembly. J Cell Sci. 2015;128:4572–4587. doi: 10.1242/jcs.180786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MC, Bodor DL, Stellfox ME, Martins NM, Hochegger H, Foltz DR, Jansen LE. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Developmental cell. 2012;22:52–63. doi: 10.1016/j.devcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Spiller F, Medina-Pritchard B, Abad MA, Wear MA, Molina O, Earnshaw WC, Jeyaprakash AA. Molecular basis for Cdk1-regulated timing of Mis18 complex assembly and CENP-A deposition. EMBO Rep. 2017;18:894–905. doi: 10.15252/embr.201643564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankovic A, Guo LY, Mata JF, Bodor DL, Cao XJ, Bailey AO, Shabanowitz J, Hunt DF, Garcia BA, Black BE, et al. A Dual Inhibitory Mechanism Sufficient to Maintain Cell-Cycle-Restricted CENP-A Assembly. Mol Cell. 2016;65:231–246. doi: 10.1016/j.molcel.2016.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellfox Madison E, Nardi Isaac K, Knippler Christina M, Foltz Daniel R. Differential Binding Partners of the Mis18α/β YIPPEE Domains Regulate Mis18 Complex Recruitment to Centromeres. Cell Reports. 2016;15:2127–2135. doi: 10.1016/j.celrep.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes & Development. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- Tachiwana H, Muller S, Blumer J, Klare K, Musacchio A, Almouzni G. HJURP involvement in de novo CenH3(CENP-A) and CENP-C recruitment. Cell Rep. 2015;11:22–32. doi: 10.1016/j.celrep.2015.03.013. [DOI] [PubMed] [Google Scholar]
- Tan S. A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli. Protein expression and purification. 2001;21:224–234. doi: 10.1006/prep.2000.1363. [DOI] [PubMed] [Google Scholar]
- Wang J, Liu X, Dou Z, Chen L, Jiang H, Fu C, Fu G, Liu D, Zhang J, Zhu T, et al. Mitotic regulator Mis18beta interacts with and specifies the centromeric assembly of molecular chaperone holliday junction recognition protein (HJURP) The Journal of biological chemistry. 2014;289:8326–8336. doi: 10.1074/jbc.M113.529958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir JR, Faesen AC, Klare K, Petrovic A, Basilico F, Fischbock J, Pentakota S, Keller J, Pesenti ME, Pan D, et al. Insights from biochemical reconstitution into the architecture of human kinetochores. Nature. 2016;537:249–253. doi: 10.1038/nature19333. [DOI] [PubMed] [Google Scholar]
- Westhorpe FG, Fuller CJ, Straight AF. A cell-free CENP-A assembly system defines the chromatin requirements for centromere maintenance. J Cell Biol. 2015;209:789–801. doi: 10.1083/jcb.201503132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhorpe FG, Straight AF. The centromere: epigenetic control of chromosome segregation during mitosis. Cold Spring Harbor perspectives in biology. 2015;7:a015818. doi: 10.1101/cshperspect.a015818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Hayashi T, Yanagida M, Russell P. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell. 2009;33:287–298. doi: 10.1016/j.molcel.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda K, Ando S, Morishita S, Houmura K, Hashimoto K, Takeyasu K, Okazaki T. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. P Natl Acad Sci USA. 2000;97:7266–7271. doi: 10.1073/pnas.130189697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Martyniuk CJ, Trudeau VL. SANTA domain: a novel conserved protein module in Eukaryota with potential involvement in chromatin regulation. Bioinformatics. 2006;22:2459–2462. doi: 10.1093/bioinformatics/btl414. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.