Abstract
Phenotypes are rarely consistent across genetic backgrounds and environments, but instead vary in many ways depending on allelic variants, unlinked genes, epigenetic factors, and environmental exposures. In the extreme, individuals carrying the same causal DNA sequence variant but on different backgrounds can be classified as having distinct conditions. Similarly, some individuals that carry disease alleles are nevertheless healthy despite affected family members in the same environment. These genetic background effects often result from the action of so-called “modifier genes” that modulate the phenotypic manifestation of target genes in an epistatic manner. While complicating the prospects for gene discovery and the feasibility of mechanistic studies, such effects are opportunities to gain a deeper understanding of gene interaction networks that provide organismal form and function as well as resilience to perturbation. Here, we review the principles of modifier genetics and assess progress in studies of modifier genes and their targets in both simple and complex traits. We propose that modifier effects emerge from gene interaction networks whose structure and function vary with genetic background and argue that these effects can be exploited as safe and effective ways to prevent, stabilize, and reverse disease and dysfunction.
Main Text
Introduction
Functional annotation of the genome has progressed at a remarkable pace since the Human Genome Project was completed in 2003, aided tremendously by continual advancements in high-throughput DNA-sequencing technologies and the computational capacity to analyze genome-scale datasets. With these resources, many contributing genes have been identified for specific diseases and phenotypes. To date, causative genes have been identified for more than 3,300 of the ∼4,900 Mendelian disorders reported in humans.1, 2 In parallel, systematic efforts to assign molecular function and phenotypic outcome to every gene in the genome via targeted genetic engineering and high-throughput phenotyping infrastructures have been undertaken in a variety of model organisms,3, 4, 5, 6, 7, 8, 9 and similar strategies to characterize the impact of naturally occurring loss-of-function mutations in humans are being pursued.10 But, as has been realized for a century, genes do not act in isolation.11, 12, 13, 14 The ultimate phenotypic manifestation of most genetic variants depends on interactions with several additional genetic elements, usually in the context of functional networks.15, 16, 17 Effects of these modifier genes can result from direct interaction with the target gene product, mechanistic contribution to the same biological process, and functional compensation through alternative pathways.
Evidence for modifier genes is extensive, both in humans and model organisms (Table 1).18, 19, 20 A comprehensive review found only one allele at one gene that failed to show genetic background effects.20 In this exceptional case—albinism—failure to produce melanin due to complete tyrosinase deficiency leaves no opportunities to modulate protein activity, whereas pigmentation in hypomorphic mutants is readily modified. Modifier effects can be subtle (e.g., slightly decreased latency for onset of disease) or profound (e.g., complete suppression of disease in genetically predisposed individuals). The clearest examples involve phenotypic variation among individuals harboring an allelic variant or mutation known to produce a specific Mendelian (i.e., single-gene) trait. In humans, congenital abnormalities can differ among family members who inherit the same disease-causing mutation. In laboratory mice, identical mutations often produce distinct phenotypes across different inbred strain backgrounds. These types of observations demonstrate the importance of modifier genes in determining phenotypic outcomes. They also highlight conceptual and experimental challenges; disease alleles are harder to identify when their effects depend on modifier genes and genetic background. Such heterogeneity reduces statistical power and creates ambiguity in defining relevant phenotypes. Despite these challenges, modifier effects provide opportunities to more deeply understand functional gene interaction networks and the processes they regulate. Modifier genes also represent promising targets for intervening in disease initiation and progression.
Table 1.
Examples of Modifier Genes and Their Functional Effects
|
TARGET |
MODIFIER |
||||||
|---|---|---|---|---|---|---|---|
| Gene(s) | Nature of Allele(s) | Condition | Gene(s) | Nature of Allele(s) | Effect Type | Modifier Effect | Refs |
| Examples in Humans | |||||||
| PRPF31 | LOF mutation | retinitis pigmentosa | CNOT3 | polymorphism | penetrance | reduced incidence | 27, 28 |
| KCNQ1 | dominant-negative mutation | long QT syndrome | NOS1AP | polymorphism | expressivity | increased QT interval, risk of cardiac arrest, and sudden death | 32 |
| HFE | LOF mutation | hereditary hemochromatosis | HAMP | LOF mutation | dominance | heterozygote susceptibility | 34 |
| BBS1/9/10 | LOF mutation | Bardet-Biedl syndrome | MKS1 | LOF mutation | pleiotropy | novel seizure phenotype | 38 |
| SMAD6 | LOF mutation | non-syndromic craniosynostosis | BMP2 | polymorphism | penetrance | increased incidence | 63 |
| SMN1 | LOF mutation | spinal muscular atrophy | PLS3 | polymorphism | penetrance | prevention of disease | 64 |
| CFTR | LOF mutation | cystic fibrosis | MUC4/20, SLC9A3, HLA-DRA, EHF/APIP, AGTR2/SLC6A14 | polymorphism | expressivity | altered severity of lung disease | 65 |
| CFTR | LOF mutation | cystic fibrosis | DCTN4 | polymorphism | expressivity | altered time to initial lung infection | 66 |
| MYH7 | LOF mutation | hypertrophic cardiomyopathy | MYBPH | polymorphism | expressivity | altered severity of cardiac hypertrophy | 67 |
| HBB | LOF mutation | sickle cell disease | BCL11A, HBS1L-MYB | polymorphism | expressivity | increased levels of fetal hemoglobin, decreased rate of pain crisis | 96 |
| Examples in Mice | |||||||
| Grhl3 | spontaneous LOF mutation (curly tail) | spina bifida | Lmnb1 | polymorphism | penetrance | reduced incidence | 30 |
| Pkd1 | engineered LOF mutation | polycystic kidney disease | Nedd9 | engineered LOF mutation | expressivity | increased kidney size and cyst number | 33 |
| Atp2b2 | spontaneous LOF mutation (deaf waddler) | deafness | Cdh23 | polymorphism | dominance | heterozygote susceptibility | 36 |
| Bcl2 | engineered LOF mutation | overactive apoptosis | Bid | engineered LOF mutation | pleiotropy | improved lymphocyte count and survival with no effect on hypopigmentation or kidney disease | 40 |
| 129/Sv background | inbred strain genetic background | testicular germ cell tumors | Kitl, Dnd1, Stra8, Eif2s2, Apobed1, A1cf, Ago2, Trp53 | engineered LOF mutation | penetrance | altered incidence | 45, 46, 47, 48, 49 |
| Apoe | engineered LOF mutation | atherosclerosis | Raet1e | polymorphism | expressivity | decreased aortic root lesion area | 69, 70, 71 |
| Mpl | engineered LOF mutation | thrombocytopenia | Myb | ENU-induced hypomorphic mutation | expressivity | increased platelet count | 72 |
| Mecp2 | engineered LOF mutation | Rett syndrome | Sqle | ENU-induced LOF mutation | pleiotropy | increased motor function and lifespan with no effect on malocclusion or inflammation | 73 |
| Nr2e3 | spontaneous LOF mutation | enhanced S cone syndrome | Nr1d1 | polymorphism | penetrance | reduced incidence | 92, 93 |
| Gars | ENU-induced novel function mutation | Charcot-Marie Tooth disease type 2D | Nrp1 | engineered LOF mutation | expressivity | increased loss of motor function | 75 |
Abbreviations are as follows: LOF, loss-of-function; ENU, ethylnitrosourea.
As our ability to assign function to specific genes and alleles expands, so too does our understanding that the phenotypes they produce depend on other genes. The challenge is to define the genetic basis of modifier effects, their mechanisms of action, and the gene interaction networks in which they exert their influence. Detecting modifier effects is primarily a matter of surveying phenotypes within a genetically diverse cohort that shares a functional genetic variant and similar environment. Subsequent identification and investigation of underlying modifier genes provide opportunities for increased functional understanding of individual genes, the networks in which they act, the phenotypes they affect, and the systems properties that buffer organisms from environmental exposures and genetic dysfunctions. Here, we review the principles of modifier genetics, strategies to identify modifier genes, and progress characterizing their influence and mechanisms. We discuss modifier effects in the context of gene interaction networks and provide examples of how these effects may be exploited therapeutically to improve human health. We emphasize modifier genes in mice and humans but recognize that great progress is being made with gene interaction networks in yeast,21 plants,22 worms,23 flies,24 and other species.25 Together, these studies characterize the genetic and phenotypic architecture of simple and complex traits, and they suggest ways that modifiers might be used to prevent, stabilize, and reverse disease and dysfunction.
Modifier Effects and Functional Significance
Principles of Modifier Genetics
“Modifier gene” describes a locus at which DNA sequence variation alters phenotypes normally associated with independent “target genes.” Interactions between specific alleles of modifier and target genes is the defining feature of modifier genetics. Unlike conventional genetic variants (QTLs) that because of analytical limitations are surveyed for additive effects, modifiers act non-additively with target genes and often do not affect phenotypes in an obvious way in their absence (Figure 1). Both target alleles and their modifiers can be spontaneous, induced, or engineered mutations, or they can be naturally occurring allelic variants. In some cases, modifier variants cause loss of gene function, highlighting the sensitivity of biological systems to modifier dosage. In general, the pervasive nature of modifier effects across all types of biological traits means that phenotypes are rarely an intrinsic property of target alleles, but instead result from integration of their activities with environmental context and genetic background.12, 13, 14
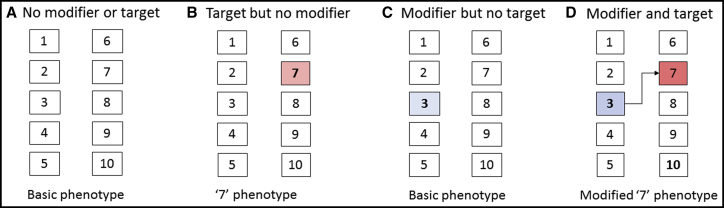
Figure 1.
Targets and Modifiers in a Hypothetical 10-Gene Genome
Blue and red boxes highlight allelic variants that have potential modifier or target effects, respectively, on a phenotype of interest.
(A) No modifier or target alleles affecting the baseline phenotype.
(B) A gene 7 variant changes the basic phenotype but the genetic background does not also carry a variant with modifier activity.
(C) The potential of a gene 3 variant to modify the phenotype cannot be realized because the genome does not carry an appropriate gene 7 allele.
(D) Only where both modifier and target alleles occur together is the hypothetical phenotype altered. The arrow shows the functional relation between the modifier (gene 3) and its target (gene 7).
Types of Target Genes
We focus on two types of target genes: classic single-gene targets and those embedded in multigenic traits. In the latter case, the number of targets is ambiguous. All that is certain is that the modifier gene alters the phenotypic effects of at least one and perhaps several genes underlying the multigenic trait. Unfortunately, we currently lack sufficient understanding of either network biology or genetic architectures to resolve this question, despite the obvious implications for controlling outcomes for common conditions.
Types of Modifier Effects
In general, modifier effects can be classified into four major categories based on the phenotypic aspect they influence: penetrance, expressivity, dominance, and pleiotropy (Figure 2). In the following section, we describe these classes of modifier effects and provide examples of mouse and human studies that have identified specific modifier genes for each. With this background, we then discuss strategies to discover modifiers experimentally.
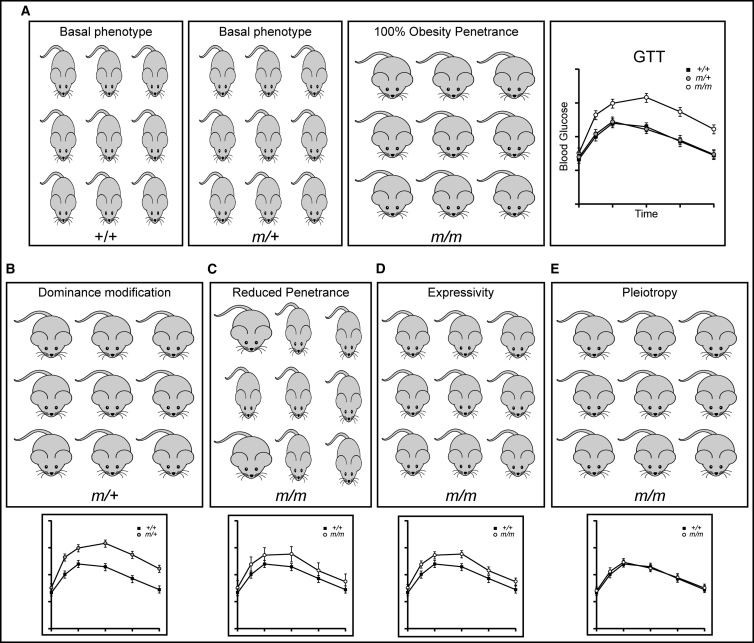
Figure 2.
Classes of Modifier Effects
(A) In this hypothetical example, the mutant allele m acts in a recessive manner to cause obesity and impaired glucose tolerance with complete penetrance in homozygotes.
(B) A dominance modifier allele causes heterozygous m/+ mice to develop obesity and glucose tolerance phenotypes indistinguishable from homozygotes.
(C) A genetic modifier of penetrance decreases the frequency of obesity and impaired glucose tolerance in m/m individuals. Some are unaffected, while others display phenotypes as extreme as the original m/m population. The resultant increased inter-individual variability within the population shifts the m/m glucose tolerance test (GTT) plot toward the basal phenotype but with an increase in standard deviation.
(D) An expressivity modifier allele decreases trait severity in all m/m individuals, producing a phenotype intermediate between the original +/+ and m/m populations. The m/m GTT plot shift has the same magnitude as with the penetrance modifier (C), but without the associated increase in standard deviation due to relative phenotypic homogeneity within the population.
(E) A genetic modifier of pleiotropy eliminates the impaired glucose tolerance phenotype without affecting obesity in m/m individuals.
Penetrance. Penetrance refers to the proportion of individuals carrying the causative target allele that are phenotypically affected. It is measured as a population characteristic across individuals that share the same target allele but are categorized as phenotypically affected or not. In mouse studies, penetrance modification is evident when the frequency of individuals affected by a mutant allele varies across distinct strain backgrounds or among segregating crosses. Similarly, individuals who are unaffected despite carrying a disease-causing allele are evidence of penetrance modification in humans.
Mutation of PRPF31 (pre-mRNA processing factor 31 [MIM: 606419]) can cause autosomal-dominant retinitis pigmentosa (adRP [MIM: 600138]) in humans, but not all individuals with mutations develop disease. Within affected families, some individuals develop complete blindness while others are unaffected despite inheriting identical PRPF31 mutant alleles.26 Genetic linkage analysis identified a region associated with variable protection that promotes higher expression from the wild-type PRPF31 allele in asymptomatic carriers.27 Through profiling of candidate genes in the region, it was discovered that CNOT3 (CCR4-NOT transcription factor subunit 3 [MIM: 604910]), which represses PRPF31 transcription, acts as a penetrance modifier whose expression level and sequence polymorphism affect clinical manifestation of adRP resulting from PRPF31 mutations.28
The curly tail (ct) mutation is a hypomorphic allele of the Grhl3 (grainyhead-like 3) gene that arose spontaneously on the GFF inbred mouse strain background.29 Homozygous mutants develop neural tube defects (NTDs), but variability in the proportion of affected individuals across strain backgrounds suggests that genetic variants modify penetrance. De Castro and colleagues used proteomic analysis to identify a variant of Lmnb1 (lamin B1) that reduces occurrence of spina bifida in this model.30 Penetrance decreased ∼3-fold, from 16% to 6%, in Grhl3Ct/Ct mice with an Lmnb1 allele encoding a more stable protein. Subsequent work identified destabilizing LMNB1 (MIM: 150340) variants in humans with NTDs,31 demonstrating the utility of mouse models for discovering modifier genes with translational relevance.
Expressivity. Expressivity describes the magnitude of a phenotype. The term applies to traits that vary quantitatively and is relevant only for individuals in which a trait is penetrant (i.e., affected individuals). Expressivity modifier genes may enhance or reduce target gene effects, and their influence can be detected as variation in phenotypic severity across genetic backgrounds. For example, a particular mutation may induce life-threatening conditions in some human populations or mouse strain backgrounds while producing only modest symptoms in others.
The clinical severity of congenital long-QT syndrome (MIM: 192500) resulting from a missense mutation in the KCNQ1 (potassium voltage-gated channel, subfamily Q, member 1 [MIM: 607542]) gene is modified by sequence variation at the NOS1AP (nitric acid synthase 1 adaptor protein [MIM: 605551]) gene.32 Family-based association analyses identified correlations between common allelic variants of NOS1AP and a longer QT interval in a population of South Africans harboring identical KCNQ1 mutations. These NOS1AP modifier alleles also increased risk of cardiac arrest and sudden death.
Nedd9 (neural precursor cell expressed, developmentally downregulated 9) was identified as a modifier of expressivity in a mouse model of polycystic kidney disease (PKD [MIM: 173900]) using a candidate approach.33 Pkd1 (polycystic kidney disease 1 homolog)-null mice develop PKD with 100% penetrance, and severity is increased with simultaneous deletion of Nedd9. While loss of Nedd9 function in itself produces no obvious kidney phenotype, co-deletion with Pkd1 promotes increased kidney size and cyst number in double-knockout Nedd9−/−Pkd1−/− mice compared to single-knockout Pkd1−/− mice.
Dominance. Dominance is a measure of the target gene dosage required for phenotypic manifestation of a trait and is assessed in heterozygous individuals. With complete dominance, phenotypes are indistinguishable between heterozygotes and homozygotes that carry two copies of the causal target allele. Intermediate or distinct phenotypes in heterozygotes indicate partial dominance, while absence of a heterozygous phenotype indicates recessive inheritance. If a single copy of a target allele is sufficient to induce a phenotype on certain genetic backgrounds but two copies are required on others, dominance modifiers are likely to be involved.
Missense mutations in HFE (hemochromatosis [MIM: 613609]) are the most common cause of hereditary hemochromatosis (MIM: 235200) in humans. Individuals homozygous for the HFEC282Y allele often develop severe iron overload, while heterozygotes are generally asymptomatic. Targeted sequencing analysis revealed, however, that when inherited together with a heterozygous loss-of-function mutation in HAMP (hepcidin antimicrobial peptide [MIM: 606464]), a single mutant HFE allele is sufficient to induce clinical iron overload.34
A spontaneous mutation in Atp2b2 (ATPase, Ca2+ transporting, plasma membrane 2), the deaf waddler (dfw) allele causes profound deafness in the homozygous state in several inbred mouse strains.35 In the heterozygous state, the mutation causes progressive hearing loss on some strain backgrounds but has no effect on others, indicating the presence of a dominance modifier gene. Noben-Trauth and colleagues used linkage analysis to map these modifier effects to a SNP within Cdh23 (cadherin 23), finding that hearing loss arises in Atp2b2+/dfw-2J heterozygotes only when a variant Cdh23753A allele is present in the homozygous state.36 This work led to the discovery of a similar functional interaction between CDH23 (MIM: 605516) and ATP2B2 (MIM: 108733) in human hearing loss.37
Pleiotropy. The combination of target gene-induced phenotypes that affected individuals display is referred to as pleiotropy, and it includes novel phenotypes induced only in the presence of modifier genes. The action of pleiotropy-modifier genes causes individuals that share the same target allele to show a range of phenotypes.
Bardet-Biedl syndrome (BBS [MIM: 209900, 600151, 605231, 615981–615996, 617406, 617119]) and Meckel-Gruber syndrome (MKS [MIM: 249000, 267010, 603194, 607361, 611561, 611134, 612284, 613885, 614175, 614209, 615397, 616258]) are human genetic disorders caused by dysfunctional cilia that can result from mutations in several genes. Due to the widespread importance of ciliary function, these syndromes are associated with a variety of clinical phenotypes, including obesity, vision loss, polydactyly, infertility, and renal failure. Using family-based association analysis, Leitch and colleagues found that mutation of MKS1 (Meckel syndrome type 1 [MIM: 609883]) was associated with seizures in BBS-affected subjects resulting from mutations in BBS1 (MIM: 209901), BBS9 (MIM: 607968), or BBS10 (MIM: 610148).38 As they are not typically associated with either BBS or MKS, the development of seizures in individuals harboring both MKS1 and BBS mutations represents a novel phenotype.
Deletion of the anti-apoptotic gene Bcl2 (B cell lymphoma/leukemia 2) in mice systemically activates cell death, resulting in several phenotypes that include hypopigmentation, lymphocytopenia, polycystic kidney disease, reduced body weight, and shortened lifespan.39 Deletion of the pro-apoptotic gene Bid (BH3 interacting domain death agonist), while having no detectable effects on its own, improves lymphocytopenia and survival in Bcl2−/− mice but does not mitigate low body weight, hypopigmentation, or kidney disease.40 This selective correction of some phenotypes but not others suggests tissue-specific effects and identifies Bid as a pleiotropy modifier.
Modifiers of Complex Traits
Most of the modifier genes discussed here, and indeed a majority of those described in the literature, influence simple traits where the phenotype of interest results primarily from a single genetic variant with large effects. In the strictest sense, all traits are complex. The impact of genetic variation is probabilistic rather than deterministic, and identical phenotypic manifestation within a population harboring a particular causal variant is exceedingly rare, if it occurs at all. Even for phenotypes with complete penetrance, characteristics such as severity or latency differ among affected individuals. By the most stringent definition, the existence of modifier effects (genetic or otherwise) precludes a “simple” Mendelian trait.
Most phenotypes, however, result from the complex actions of multiple genes and environmental factors.19 Presumably, modifier genes also affect these traits. But whether evidence of their action can be detected and whether they can be individually identified is less clear. Distinguishing conventional genetic variants such as QTLs from modifier genes is challenging in humans given the genetic and environmental variation among individuals, families, and populations.41 By contrast, the defined genetic backgrounds and controlled husbandry conditions available in model organisms such as laboratory mice simplify such studies. Usually modifiers are identified by studying multigenic background effects among a population of individuals that share a single target allele. For complex traits, this perspective is reversed and single genetic variants are tested for their capacity to modulate multigenic phenotypes (Figure 3). The examples presented below demonstrate how single variants, acting as modifiers, can have unusually large phenotypic effects on genetically complex models of human disease.
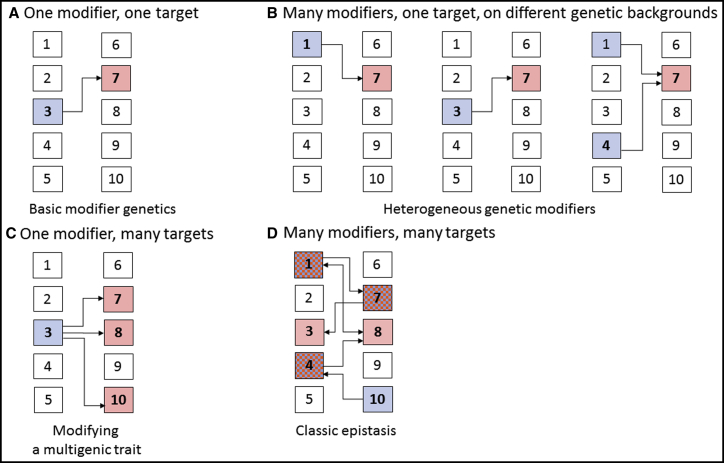
Figure 3.
Continuous Genetic Complexity of Modifiers and Targets in a Hypothetical 10-Gene Genome
(A) Classic one modifier (blue), one target (red) case.
(B) Three distinct genomes showing different modifiers (genes 1, 3, and 4) that modulate the phenotype associated with a gene 7 allele.
(C) A modifier (gene 3) acting on three targets (genes 7, 8, and 10).
(D) Example of complex epistasis with multiple genes (genes 1, 3, 4, 7, 8, and 10) that interact in various ways (some directional, others bidirectional). Red-blue checked boxes indicate genes that can act as both modifier and target.
The 129/Sv mouse strain develops spontaneous testicular germ cell tumors (TGCTs) at a rate of ∼7%.42 Genetic susceptibility in this strain is complex and involves multiple genes with low penetrance, as evidenced by the exceedingly low rate of tumorigenesis (one of ∼11,000 male offspring) in hybrids derived by intercrossing 129 mice with resistant strains, suggesting that many genes control susceptibility.43 Evidence from human studies similarly supports a multigenic basis for inherited TGCT risk.44 Several single-gene mutations modify TGCT penetrance when introgressed onto the 129/Sv genetic background. Kitl, Dnd1, Stra8, Eif2s2, Apobec1, A1cf, Ago2, Trp53, and other mutations each influence the frequency of affected males by a factor of ±2–3 when present in the heterozygous state.45, 46 In cases where homozygotes for these modifiers can be tested (i.e., where they do not result in embryonic lethality), exceptionally strong effects often emerge. For example, the Dnd1Ter mutant allele increases TGCT penetrance from the baseline rate of ∼7% in wild-type 129/Sv males to 17% in Dnd1Ter/+ heterozygotes and 94% in Dnd1Ter/Ter homozygotes.47, 48 On other strain backgrounds, however, Dnd1Ter/Ter homozygotes do not develop TGCTs,49 demonstrating that the mutant allele modifies the likelihood of tumor development only in genetically predisposed individuals and is not simply a general driver of tumorigenesis. Thus, these single gene mutations act in a dose-dependent manner to modify the penetrance of a highly complex multigenic trait.
The emergence of disease phenotypes often depends on both genetic predisposition and an environmental trigger. An example is the development of diet-induced metabolic syndrome (MetS) in laboratory mice. C57BL/6J but not A/J mice develop obesity, insulin resistance, hepatic steatosis, and liver cancer when fed a diet high in saturated fat.50, 51 Genome surveys using a panel of chromosome substitution strains (CSSs) in which each C57BL/6J chromosome has been individually replaced with the corresponding A/J chromosome52 showed that sequence variants on many chromosomes control susceptibility, with the effects of each acting in a highly non-additive manner.53, 54 For the majority of traits, the cumulative phenotypic effect (sum of the signed deviation from C57BL/6J) across all strains in the CSS panel was found to greatly exceed 100%, with an average cumulative effect of ∼800% across 90 blood, bone, and metabolic traits.53 Similar results for 54 traits in a panel of rat CSSs argue that pervasive and strong non-additive effects may be a general genetic phenomenon.53 Strong non-additivity clearly demonstrates a complex relationship between sequence variants contributing to diet-induced MetS susceptibility in these model systems.
A striking example of this complexity involves an obesity resistance QTL (Obrq2) that was mapped with a panel of congenic strains harboring different segments of A/J-derived chromosome 6 on a C57BL/6J genetic background. The A/J allele of Obrq2 significantly decreases weight gain induced with a high-fat diet, but the magnitude of this effect depends on the genetic source (A/J or C57BL/6J) of multiple additional QTLs on the same chromosome.53, 54, 55 In some cases, nearly complete protection from diet-induced obesity is found. Protection is lost, however, with various small changes in the strain origin of closely flanking chromosome segments. With the A/J-derived Obrq2 QTL, protection is provided despite at least 13 other obesity-promoting QTLs on the same chromosome and on at least 14 other chromosomes, and despite long-term exposure to high-risk (high-fat) diet.52, 55 In this model, naturally occurring protective A/J-derived alleles act in protective but context-dependent ways to modify the penetrance, expressivity, and pleiotropy of diet-induced MetS phenotypes on the genetically predisposed C57BL/6J background. Thus in this and other cases,56 QTLs act as genetic modifiers, with highly non-additive effects and strong dependence on genetic background. Context dependence probably explains the contrast in the prevalence of evidence for modifier effects in genetically heterogeneous populations such as the Resilience Project57 (see below) versus studies of human families and inbred strains of mice.20
From Modified Phenotypes to Modifier Genes
Strategies to Identify Genetic Modifiers in Humans
Several approaches can be used to study modifier genes directly in human populations, including comparative expression profiling, genome-wide association studies (GWASs), and family-based association analyses. To maximize the potential therapeutic benefit of any findings, studies are often designed to search for shared variants (candidate modifier genes) in individuals that are protected from a particular disease phenotype despite carrying a causal target allele. Two strategies are used to pursue such protective modifiers: (1) comprehensive sequence profiling of a population or family carrying the disease allele to identify variants that associate with phenotypic severity and (2) targeted sequencing of genes known to harbor disease-associated mutations in a healthy population to identify individuals who are phenotypically unaffected despite harboring causal alleles. Once these individuals are identified, mechanisms of resistance can be studied to identify candidate therapeutic targets for susceptible individuals.
The targeted sequencing strategy is exemplified by the Resilience Project, an effort to identify unaffected carriers of mutations known to cause highly penetrant and severe childhood diseases. Screening for 874 distinct Mendelian disease-causing mutations across 589,306 genomes, Chen and colleagues identified 13 asymptomatic adults carrying disease alleles.57 The low frequency of resilient individuals detected in this study results partly from the strict experimental parameters used. Mutations were selected for the screen based on their extreme rarity in the general population, induction of severe childhood conditions unlikely to be misdiagnosed, and prior clinical annotation of complete disease penetrance. Similar approaches using less stringent criteria are expected to identify a higher prevalence of candidate resilient individuals, albeit with associated increases in complexity. Nevertheless, this work provides proof-of-principle that individuals resistant to highly penetrant genetic diseases can be identified, paving the way for mechanistic studies to discover modifier genes that may be therapeutically manipulated to benefit susceptible individuals.
Studies of healthy elderly individuals illustrate the search for modifier alleles with whole-genome sequencing. Their long healthy life raises questions about whether some individuals are free of inherited disease-causing alleles because they won “Mendel’s lottery,” or whether they inherited both the usual variety of genetic variants that cause disease and dysfunction as well as appropriate modifier genes. Whole-genome sequences show that many healthy elderly individuals, including centenarians, carry deleterious genetic variants.58, 59 In fact, their mutational load is comparable to individuals who succumb to disease earlier in life.59, 60, 61 To paraphrase Jim Crow,62 “Given our mutational load, why aren’t we dead many times over?”—perhaps modifier genes are the answer. Interestingly, these surveys failed to find protective genetic variants that were shared across the well elderly. Although tests for general effects are reasonable, it seems more likely that modifiers act primarily in the context of specific target genes.16 We anticipate that larger, focused surveys of healthy elderly individuals that share specific disease alleles will reveal modifier effects resulting from inheritance of specific combinations of modifier and target alleles.
Another example of modifier identification through sequencing involves penetrance of nonsyndromic craniosynostosis (MIM: 617439), a condition in which the cranial sutures fuse prematurely. Exome sequencing identified recurrent loss-of-function SMAD6 (SMAD family member 6 [MIM: 602931]) mutations in patients, but sequencing of additional family members revealed a penetrance of only 24%.63 Strikingly, penetrance increased to 100% for individuals inheriting a common (42% allele frequency) SNP near the BMP2 (bone morphogenetic protein 2 [MIM: 112261]) locus along with a SMAD6 mutation. As expected given its high frequency in the general population, carriage of the BMP2-associated SNP alone was not associated with disease in any individuals from the study, demonstrating the context specificity of its influence on craniosynostosis development.
Identification of genetic modifiers through comparative expression profiling relies on large-scale transcriptomic, proteomic, and other -omic analyses of individuals that are differentially affected by a causal target allele. In the simplest case, populations carrying the causal allele can be separated into two groups—affected and unaffected individuals. The presence of distinct expression levels, protein or transcript isoforms, or other differences between groups may indicate functional modifiers. In more complex cases, the modified trait can be analyzed as a quantitative variable among individuals sharing the target allele.
PLS3 (plastin 3 [MIM: 300131]) was identified as a modifier gene for congenital spinal muscular atrophy (MIM: 253400) using transcriptome-wide differential gene expression profiling.64 Significantly higher PLS3 expression was detected in unaffected individuals as compared to affected family members sharing the same predisposing SMN1 (survival motor neuron 1 [MIM: 600354]) mutations. Functional studies in genetically modified mice and zebrafish validated the neuro-protective role of human PLS3 overexpression in the context of Smn1 loss of function.64
GWAS is a commonly used approach to identify sequence variants with significantly different prevalence among groups of individuals displaying distinct phenotypes. Normally, sequences are obtained for a representative set of marker loci distributed throughout the genome and imputation is used to predict genotypes at non-profiled sites based on assumed association within haplotypes. Increasingly, however, whole-exome and whole-genome sequencing approaches are being used for GWASs, yielding higher precision analyses. For modifier gene detection, a GWAS is typically performed on a large population of individuals sharing a causal target allele but with distinct phenotypic manifestations, testing for additional sequence variants that associate with particular phenotypes and their severities in the context of specific target alleles.
GWAS has been used effectively to identify modifier genes in patients with cystic fibrosis (CF [MIM: 219700]). An analysis of 6,365 patients with CFTR (cystic fibrosis transmembrane conductance regulator [MIM: 602421]) mutations identified five loci at which sequence variation was associated with severity of lung disease.65 Each associated region contained genes with biological relevance to lung function, increasing confidence that the identified loci are mechanistically involved in the manifestation of lung phenotypes induced by CFTR mutation. Another search for modifiers of lung infection in CF patients used an “extreme phenotype” study design to increase efficiency of modifier gene identification.66 Exome sequencing was performed on individuals with CFTR mutations specifically selected from the top and bottom of the distribution for age at onset of infection. From a set of just 91 affected subjects, missense mutations in DCTN4 (dynactin 4 [MIM: 614758]) were linked to early infection. A validation set of 696 affected subjects provided further support for a modifier effect of DCTN4 on susceptibility to airway infection conferred by CFTR mutation.
Family-based association studies utilize linkage analysis in consanguineous families for which individuals with the same inherited causal genetic variant display distinct phenotypes. As compared to wider population studies, this approach simplifies the search for modifier genes in three ways. Both the background genetic heterogeneity and the number of modifier genes are limited, which decreases the number of variable sequences within the study group and thereby increases the signal to noise ratio for detecting variants that actively modify the phenotype. Additionally, because the causal allele is usually identical across the study group, variability in the primary effects of the sequence variant on target gene function is reduced.
Mouton and colleagues studied modifiers of phenotypic severity in 27 South African families with inherited hypertrophic cardiomyopathy (MIM: 115195, 192600).67 Each patient carried one of three distinct causal mutant alleles in either MYH7 (myosin heavy chain 7 [MIM: 160760]) or TNNT2 (troponin T2, cardiac [MIM: 191045]). Sequence variation within MYBPH (myosin binding protein H [MIM: 160795]) was found to associate with severity of cardiac hypertrophy only in patients with a specific MYH7 mutation. No association was found for patients with a different causal mutation in MYH7 or TNNT2, suggesting that MYBPH modifies the phenotypic impact of MYH7 mutation in a specific manner.
Strategies to Identify Genetic Modifiers in Mice
With respect to the importance of genetic modifiers in human health, mouse models provide a powerful, controlled system in which to study modifier effects. They do not directly substitute for human studies, but instead provide complementary information that can be used to guide study design and interpretation of results. The defined nature of inbred laboratory mouse strains makes them an excellent resource for dissecting the genetic basis of modifier effects. By minimizing the complications of a heterogeneous genetic background, inbred strains facilitate experimental reproducibility, making it easier to detect modifier effects and attribute them to specific loci, and increasingly to individual allelic variants. Common strategies used to identify and map modifier genes in mice include linkage crosses, specialized inbred resources, and mutagenesis screens.
Linkage crosses are performed by breeding one phenotypically affected strain carrying a known causal target allele to another inbred strain, which could carry the same target allele depending on study design. The resulting F1 hybrids are either intercrossed with each other or backcrossed to one of the parental strains. Offspring that inherit the causal variant are then phenotyped and genotyped. Those that exhibit a distinct phenotype relative to the affected inbred parental strain may harbor modifiers. By analyzing sufficient mice, depending in part on the number and effect of segregating modifier genes, candidate genomic intervals predicted to contain modifier genes can be defined based on shared inheritance patterns among individuals exhibiting a particular modified phenotype. Further studies are generally required to identify and validate specific candidate genes within these intervals.
Specialized inbred resources such as congenic strains, in which the causal target allele or candidate modifier locus has been transferred to an alternative strain background,56 can be readily generated to identify genetic modifier effects and map the responsible loci. Genetic transfer can be achieved through selective breeding to isolate the variant on a distinct strain background or by direct genetic engineering of embryonic stem cells or oocytes and subsequent generation of founder mice.68 This approach is often used to follow up linkage cross studies in order to more precisely map candidate modifier gene-containing intervals and validate functional effects of specific allelic variants.
An example of modifier gene mapping with linkage crosses and specialized strains is provided by a series of studies designed to identify genetic modifiers of atherosclerosis. Genetic deficiency for Apoe (apolipoprotein E) promotes severe atherosclerosis on the C57BL/6 inbred background but produces a milder phenotype on the FVB/N strain background, indicating the presence of expressivity modifiers.69 To map genomic loci containing candidate modifiers, Dansky and colleagues bred susceptible C57BL/6J Apoe−/− mice to resistant FVB/NCr Apoe−/− mice, intercrossed resultant F1 offspring, and performed linkage analysis based on phenotypes and genotypes in F2 offspring.70 This screen identified several loci whose inheritance patterns associated with atherosclerosis susceptibility, including a highly significant region on chromosome 10 (chr10) for aortic lesion area. Subsequent generation of congenic Apoe−/− strains harboring FVB/NCr-derived regions of chr10 on an otherwise C57BL/6J background narrowed the modifier-containing interval to a 1 Mb region with five genes. Validation experiments implicated a SNP within the Raet1e (retinoic acid early transcript 1E) promoter that influences its expression.71 Through initial characterization of phenotypic differences in congenic C57BL/6 and FVB/N Apoe−/− strains, linkage analysis of a cross between these strains, generation of FVB/N-Chr10 congenic strains, and functional validation experiments, these studies successfully identified Raet1e as a modifier for atherosclerosis severity resulting from ApoE deficiency.
Mutagenesis screening is another strategy used to identify modifier genes in mice. Random induction of chemically or transposon-induced mutations throughout the genome in an existing mouse model with a clearly defined phenotype permits identification of genes that alter the phenotype when disrupted. This approach is typically used to identify phenotype-suppressing mutations that eliminate a particular target allele-induced trait.
The first modifier gene mutagenesis screen in mice was conducted to identify suppressors of platelet deficiency.72 Germline mutagenesis with N-ethyl-N-nitrosurea (ENU) was applied to the Mpl−/− (myeloproliferative leukemia virus oncogene) mouse model, which lacks a receptor required for normal platelet formation. Screening >1,500 mutant offspring identified two individuals with heritable suppression of the mutant Mpl phenotype, both of which had mutations that produced hypomorphic alleles of Myb (myeloblastosis oncogene). Validation experiments confirmed that partial loss of Myb function decreases the severity of Mpl−/−-induced platelet deficiency. A similar ENU-based mutagenesis screen identified modifier genes in the Mecp2−/− (methyl CpG binding protein 2) mouse model of Rett syndrome (MIM: 312750).73 As in humans, Mecp2 mutations cause a range of neurological defects in mice.74 The screen identified five independent loci at which mutation ameliorated distinct combinations of disease traits, implicating them as pleiotropy modifiers. One of the mitigating mutations introduced a premature stop codon in Sqle (squalene epoxidase), which encodes a key cholesterol biosynthesis enzyme. The authors showed that treatment of Mecp2−/− mice with cholesterol-lowering statin drugs significantly improved motor function and lifespan,73 suggesting that this widely used class of drugs may benefit individuals with Rett syndrome. A recently completed phase 2 clinical trial (NCT02563860) is evaluating this hypothesis, with results forthcoming (see Clinical Trials in Web Resources).
Features of Modifier Genes
Nature of Sequence Variation
The nature of modifier alleles could be instructive about the sensitivity of gene interactions and networks to particular kinds of DNA sequence variants. While a thorough review is premature because insufficient numbers of molecular variants have been identified and functionally characterized, current evidence suggests considerable heterogeneity in the types of sequence variation that produce modifier effects. The studies discussed in this review provide examples of modifier variants that cause partial or complete loss of gene function (e.g., missense mutation32, 66 or gene deletion33, 40), enhanced function (e.g., increased protein stability30), and gain of alternative function (e.g., novel protein-protein interaction75). They also demonstrate that modifier variants often occur within protein-coding exons, but may alternatively affect regulatory regions such as UTRs or promoters.71 There does not appear to be a general mechanism by which modifier alleles are generated. Rather, any sequence variation affecting a gene’s function in a way that impacts its interaction with other genes and networks may manifest as a modifier allele, depending on co-inheritance with an appropriate target allele and environmental context. As we discuss next, the diverse nature of modifier alleles is consistent with their specific and context-dependent functionality.
Nature of Modifier Functions
An important consideration is whether modifier genes tend to involve general regulatory processes such as SUMOylation, phosphorylation, ubiquitination, and RNA processing or whether they tend to act primarily on activities related to particular target genes. The functions of modifier genes that have been characterized suggest specific rather than general actions affecting a diversity of biological processes. For example, modifier alleles discussed in this review modulate metabolic pathway activity, intracellular transport processes, organelle stability, cell-cell interactions, and transcriptional activity. In most cases, these effects appear to be limited to functions of the target gene(s) responsible for the primary phenotype. Such functional specificity enhances the attractiveness of modifier genes as therapeutic targets, as it decreases the likelihood of undesired side effects.
Mutational Buffering
An established but more general mechanism for suppressing the adverse phenotypic effects of deleterious mutations involves chaperones (e.g., heat-shock proteins) that facilitate proper protein folding, particularly in the context of environmental or genetic stress.76 Under certain stress conditions, suppression of altered protein folding is relaxed presumably to release cryptic variation and increase the capacity to respond to ongoing challenges.77, 78 While we know of no examples where mapping studies have implicated chaperones as modifier genes (i.e., where genetic background effects on phenotypes are attributable to genetic variation within chaperone genes), it is clear they are capable of modifying the impact of genetic variation. The term “global modifier” has been proposed to describe genes that regulate the functional manifestation of genetic variation at several loci via general mechanisms, including chaperones, chromatin modifiers, and transcriptional regulators.79 Perhaps chaperones and other global modifiers provide a system to respond to generalized stress whereas modifier genes target specific molecular dysfunctions.
Modifiers as Mediators of Network Resilience
To understand modifier effects in a general way, we must consider how they might have evolved. Genetic modifiers might simply be an important consequence of inheriting combinations of gene variants that happen to compensate or exacerbate the phenotypic effect associated with target alleles. Alternatively, they may represent interaction networks that evolved to coordinate gene action during development and throughout adulthood. Coordination could involve direct molecular interactions or molecularly independent but functionally related pathways.80, 81 In some cases, modifier effects might result from ongoing activity that is functionally relevant for specific target alleles that are embedded in genetically defined networks. In other cases, developmental and physiological conditions in carriers of particular target alleles might induce activity of genes that provide modifier functions. In both scenarios, the phenotypic outcome is similar despite differences in the underlying network activity. Effective coordination would vary depending on genetic background and environmental exposures, suggesting that the structure and function of interaction networks is a dynamic rather than a fixed feature of organismal biology. According to this hypothesis, modifier genes are tipping points in these networks where phenotypes can be switched from one outcome to another depending on the structural and functional dynamics of gene interactions. It is difficult to imagine development and maintenance of organismal form and function without such networks. Their pervasive nature across phenotypes supports the concept that genetic modifiers are a common emergent property of gene interaction networks.
The mutation burden inherent in complex organisms may be an additional selective advantage for genetic modifiers. Each individual harbors both inherited genetic variants as well as new, often deleterious mutations that arise at every generation.82 Steady state is determined by the rate (per generation) at which new mutations arise and their rate of loss, which through drift and selection can take several generations.82 Based on large-scale genome-sequencing studies, steady state for the typical human genome has been estimated to be in the hundreds, including several variants with known disease associations.83, 84, 85 Comparable numbers are found in mice and other model organisms.86, 87, 88 How then are we able to survive as individuals and as a species given the cumulative burden of these deleterious genetic variants; why aren’t we “dead” many times over?62 Perhaps some genes are latent (cryptic) modifiers that reveal their potentiality depending on genetic background and environmental context.77, 78 These naturally occurring genetic variants that show modifier activity under appropriate circumstances could buffer individuals from persistent mutation burden. We currently know too little about the impact of genetic variation on network biology to predict which genes have the potential to be modifiers or the conditions under which their activity would be induced.80, 81
Whether genetic variants act as targets or their modifiers or as conventional QTLs with largely independent effects depends on the nature of the allelic variant, genetic background, and environmental context. Although comprehensive catalogs of genetic variation have been assembled, we generally lack similar catalogs of constant and variable network features across genetic backgrounds and environmental exposures.
Therapeutic Potential
Concept
Targeted manipulation of modifier gene activity and related interaction networks represents a new paradigm for preventing, reversing, or lessening disease phenotypes in genetically predisposed individuals.89, 90 This approach is complementary to the traditional strategy of identifying and targeting primary causal disease alleles and has great potential to improve treatment efficacy and safety. Protective modifier genes are discovered based on their ability to provide resistance to the adverse consequences of disease alleles, clearly demonstrating that manipulation of their activity can be effective. Lack of any apparent adverse phenotype in resistant individuals carrying a protective allele highlights the potential safety of this approach, although this aspect requires more thorough investigation. Each class of modifier effect can be highly relevant to disease genetics: reducing penetrance or dominance decreases incidence, weakening expressivity decreases severity, and restricting pleiotropy limits comorbidities. A deeper understanding of genetic modifier effects, along with the ability to manipulate their actions in specific circumstances, could provide access to safe and effective strategies to maintain health and to stabilize or reverse disease. Targeting modifiers could be especially beneficial in cases where the causal gene variant is unknown, complex, or not directly actionable. Several of the modifier effects discussed so far have the potential to inform clinical diagnostic or therapeutic strategies, such as the possibility of treating Rett syndrome-affected subjects with statins (see Clinical Trials in Web Resources).73 Below, we provide additional examples where clear opportunities for translational applications emerge from the characterization of modifier effects.
Examples
Mutations in NR2E3 (nuclear receptor subfamily 2, group E, member 3 [MIM: 604485]) cause enhanced S cone syndrome (ESCS [MIM: 268100]) in humans, which is characterized by retinal degeneration and a variety of vision problems. A similar phenotype is observed in mice lacking Nr2e3 expression,91 although disease penetrance is highly dependent on strain background. All mice exhibit retinal degeneration upon loss of Nr2e3 on a C57BL/6J strain background, but intercrossing with AKR/J, CAST/EiJ, or NOD.NON-H2nb1 strains was found to completely suppress disease in 28%–49% of homozygous mutants,92 indicating the presence of penetrance modifier genes. Fine-mapping of a candidate protective locus led to the identification of Nr1d1 (nuclear receptor subfamily 1, group D, member 1) as a modifier gene whose increased expression compensates for loss of Nr2e3 to eliminate the disease phenotype.93 Nr1d1 encodes the nuclear hormone receptor Rev-erbα, a target for which multiple synthetic agonists have been developed. Delivery of exogenous Nr1d1 by in vivo retinal electroporation rescued the phenotype of C57BL/6J Nr2e3 mutants,93 demonstrating the efficacy of increased Rev-erbα function and suggesting that such a strategy may benefit ESCS-affected individuals.
Charcot-Marie Tooth disease type 2D (CMT2D [MIM: 601472]) is a neurodegenerative disease caused by mutations in GARS (glycyl-tRNA synthetase [MIM: 600287]). Disease-associated mutations in humans and mouse models of CMT2D cause conformational changes in the GARS protein that confer the novel ability to bind NRP1 (neuropilin 1 receptor [MIM: 602069])75 and interfere competitively with VEGF (vascular endothelial growth factor) binding to NRP1, which is required for accurate motor neuron axon guidance. Genetic manipulation of Nrp1 or Vegf function in CMT2D mice was found to alter phenotypic severity,75 identifying them as modifier genes in the model and potential therapeutic targets for CMT2D-affected subjects. Notably, multiple modulators of VEGF activity are already used clinically to treat various other conditions.
Sickle cell disease (SCD [MIM: 603903]) results from mutations in HBB (hemoglobin subunit beta [MIM: 141900]) that produce an abnormal version of hemoglobin, causing deformation and destruction of red blood cells, obstruction of vessels, and deficient oxygen transport throughout the body. Because of its high affinity for oxygen, hydroxyurea-induced fetal hemoglobin (HbF) expression is used to treat adults with SCD,94 and interindividual variation of innate HbF expression is associated with clinical severity.95 Association studies in humans have identified sequence variants linked to HbF expression that may act as genetic modifiers of disease severity.96 Targeting genes that repress HbF expression in adults, and thereby modify disease phenotypes, has recently been proposed as a novel therapeutic approach for SCD-affected individuals.97
Modifying the Future
The strong, pervasive nature of modifier effects suggests new strategies to manage disease and maintain health.15, 20 Historically, logistical, analytical, and statistical issues have led to limited empirical evidence for interacting genes.15 However, new concepts, study designs, and genetic resources enable powerful global studies. In yeast, large-scale studies have begun to define gene interaction networks.98, 99 In mice and humans, a combination of empirical evidence and literature review has begun to define the circuitry of networks associated with disease.100, 101 In many species, modifier genes are readily identified for both simple and complex traits. With these emerging resources and technologies, specifying network components, their dependencies, and targets for interventions is increasingly feasible, especially for specific conditions and diseases. These networks coordinate biological activities throughout life and buffer form and function from genetic and environmental disruptions.80, 81 Buffering is essential because we are each born with inherited and new mutations that have deleterious effects. Perhaps generally good health despite our mutational burden shows that we are each good evidence for protective modifiers. By focusing on modifier genes and network solutions that nature has devised for deleterious genes and environmental exposures, perhaps we can develop safe and efficacious ways to stabilize, reverse, or prevent disease and dysfunction.
Contributor Information
Jesse D. Riordan, Email: jriordan@pnri.org.
Joseph H. Nadeau, Email: jnadeau@pnri.org.
Web Resources
ClinicalTrials.gov, https://clinicaltrials.gov/ct2/show/NCT02563860
OMIM, http://www.omim.org/
The Resilience Project, http://resilienceproject.com
References
- 1.Amberger J.S., Bocchini C.A., Schiettecatte F., Scott A.F., Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skarnes W.C., Rosen B., West A.P., Koutsourakis M., Bushell W., Iyer V., Mujica A.O., Thomas M., Harrow J., Cox T. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellen H.J., Levis R.W., He Y., Carlson J.W., Evans-Holm M., Bae E., Kim J., Metaxakis A., Savakis C., Schulze K.L. The Drosophila gene disruption project: progress using transposons with distinctive site specificities. Genetics. 2011;188:731–743. doi: 10.1534/genetics.111.126995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.C. elegans Deletion Mutant Consortium large-scale screening for targeted knockouts in the Caenorhabditis elegans genome. G3 (Bethesda) 2012;2:1415–1425. doi: 10.1534/g3.112.003830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giaever G., Nislow C. The yeast deletion collection: a decade of functional genomics. Genetics. 2014;197:451–465. doi: 10.1534/genetics.114.161620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varshney G.K., Lu J., Gildea D.E., Huang H., Pei W., Yang Z., Huang S.C., Schoenfeld D., Pho N.H., Casero D. A large-scale zebrafish gene knockout resource for the genome-wide study of gene function. Genome Res. 2013;23:727–735. doi: 10.1101/gr.151464.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hrabě de Angelis M., Nicholson G., Selloum M., White J.K., Morgan H., Ramirez-Solis R., Sorg T., Wells S., Fuchs H., Fray M., EUMODIC Consortium Analysis of mammalian gene function through broad-based phenotypic screens across a consortium of mouse clinics. Nat. Genet. 2015;47:969–978. doi: 10.1038/ng.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saleheen D., Natarajan P., Armean I.M., Zhao W., Rasheed A., Khetarpal S.A., Won H.H., Karczewski K.J., O’Donnell-Luria A.H., Samocha K.E. Human knockouts and phenotypic analysis in a cohort with a high rate of consanguinity. Nature. 2017;544:235–239. doi: 10.1038/nature22034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.East E.M. Inheritance in crosses between NICOTIANA LANGSDORFFII and NICOTIANA ALATA. Genetics. 1916;1:311–333. doi: 10.1093/genetics/1.4.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wright S. Evolution in Mendelian populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wright S. Surfaces of selective value revisited. Am. Nat. 1988;131:115–123. [Google Scholar]
- 14.Fisher R.A. Clarendon Press; 1930. The Genetical Theory of Natural Selection. [Google Scholar]
- 15.Mackay T.F. Epistasis and quantitative traits: using model organisms to study gene-gene interactions. Nat. Rev. Genet. 2014;15:22–33. doi: 10.1038/nrg3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sackton T.B., Hartl D.L. Genotypic context and epistasis in individuals and populations. Cell. 2016;166:279–287. doi: 10.1016/j.cell.2016.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams E.G., Auwerx J. The convergence of systems and reductionist approaches in complex trait analysis. Cell. 2015;162:23–32. doi: 10.1016/j.cell.2015.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grüneberg H. Cambridge University Press; 1943. The Genetics of the Mouse. [Google Scholar]
- 19.Haldane J.B.S. The relative importance of prinicpal and modifying genes in determining some human diseases. J. Genet. 1941;41:149–157. [Google Scholar]
- 20.Nadeau J.H. Modifier genes in mice and humans. Nat. Rev. Genet. 2001;2:165–174. doi: 10.1038/35056009. [DOI] [PubMed] [Google Scholar]
- 21.Boone C., Bussey H., Andrews B.J. Exploring genetic interactions and networks with yeast. Nat. Rev. Genet. 2007;8:437–449. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- 22.Pérez-Pérez J.M., Candela H., Micol J.L. Understanding synergy in genetic interactions. Trends Genet. 2009;25:368–376. doi: 10.1016/j.tig.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z., Sherwood D.R. Dissection of genetic pathways in C. elegans. Methods Cell Biol. 2011;106:113–157. doi: 10.1016/B978-0-12-544172-8.00005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mackay T.F. Epistasis for quantitative traits in Drosophila. Methods Mol. Biol. 2015;1253:47–70. doi: 10.1007/978-1-4939-2155-3_4. [DOI] [PubMed] [Google Scholar]
- 25.Levine A.J., Hu W., Feng Z., Gil G. Reconstructing signal transduction pathways: challenges and opportunities. Ann. N Y Acad. Sci. 2007;1115:32–50. doi: 10.1196/annals.1407.018. [DOI] [PubMed] [Google Scholar]
- 26.Evans K., al-Maghtheh M., Fitzke F.W., Moore A.T., Jay M., Inglehearn C.F., Arden G.B., Bird A.C. Bimodal expressivity in dominant retinitis pigmentosa genetically linked to chromosome 19q. Br. J. Ophthalmol. 1995;79:841–846. doi: 10.1136/bjo.79.9.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGee T.L., Devoto M., Ott J., Berson E.L., Dryja T.P. Evidence that the penetrance of mutations at the RP11 locus causing dominant retinitis pigmentosa is influenced by a gene linked to the homologous RP11 allele. Am. J. Hum. Genet. 1997;61:1059–1066. doi: 10.1086/301614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venturini G., Rose A.M., Shah A.Z., Bhattacharya S.S., Rivolta C. CNOT3 is a modifier of PRPF31 mutations in retinitis pigmentosa with incomplete penetrance. PLoS Genet. 2012;8:e1003040. doi: 10.1371/journal.pgen.1003040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Straaten H.W., Copp A.J. Curly tail: a 50-year history of the mouse spina bifida model. Anat. Embryol. (Berl.) 2001;203:225–237. doi: 10.1007/s004290100169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Castro S.C., Malhas A., Leung K.Y., Gustavsson P., Vaux D.J., Copp A.J., Greene N.D. Lamin b1 polymorphism influences morphology of the nuclear envelope, cell cycle progression, and risk of neural tube defects in mice. PLoS Genet. 2012;8:e1003059. doi: 10.1371/journal.pgen.1003059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson A., Partridge D., Malhas A., De Castro S.C., Gustavsson P., Thompson D.N., Vaux D.J., Copp A.J., Stanier P., Bassuk A.G., Greene N.D. Is LMNB1 a susceptibility gene for neural tube defects in humans? Birth Defects Res. A Clin. Mol. Teratol. 2013;97:398–402. doi: 10.1002/bdra.23141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crotti L., Monti M.C., Insolia R., Peljto A., Goosen A., Brink P.A., Greenberg D.A., Schwartz P.J., George A.L., Jr. NOS1AP is a genetic modifier of the long-QT syndrome. Circulation. 2009;120:1657–1663. doi: 10.1161/CIRCULATIONAHA.109.879643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nikonova A.S., Plotnikova O.V., Serzhanova V., Efimov A., Bogush I., Cai K.Q., Hensley H.H., Egleston B.L., Klein-Szanto A., Seeger-Nukpezah T., Golemis E.A. Nedd9 restrains renal cystogenesis in Pkd1-/- mice. Proc. Natl. Acad. Sci. USA. 2014;111:12859–12864. doi: 10.1073/pnas.1405362111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merryweather-Clarke A.T., Cadet E., Bomford A., Capron D., Viprakasit V., Miller A., McHugh P.J., Chapman R.W., Pointon J.J., Wimhurst V.L. Digenic inheritance of mutations in HAMP and HFE results in different types of haemochromatosis. Hum. Mol. Genet. 2003;12:2241–2247. doi: 10.1093/hmg/ddg225. [DOI] [PubMed] [Google Scholar]
- 35.Noben-Trauth K., Zheng Q.Y., Johnson K.R., Nishina P.M. mdfw: a deafness susceptibility locus that interacts with deaf waddler (dfw) Genomics. 1997;44:266–272. doi: 10.1006/geno.1997.4869. [DOI] [PubMed] [Google Scholar]
- 36.Noben-Trauth K., Zheng Q.Y., Johnson K.R. Association of cadherin 23 with polygenic inheritance and genetic modification of sensorineural hearing loss. Nat. Genet. 2003;35:21–23. doi: 10.1038/ng1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schultz J.M., Yang Y., Caride A.J., Filoteo A.G., Penheiter A.R., Lagziel A., Morell R.J., Mohiddin S.A., Fananapazir L., Madeo A.C. Modification of human hearing loss by plasma-membrane calcium pump PMCA2. N. Engl. J. Med. 2005;352:1557–1564. doi: 10.1056/NEJMoa043899. [DOI] [PubMed] [Google Scholar]
- 38.Leitch C.C., Zaghloul N.A., Davis E.E., Stoetzel C., Diaz-Font A., Rix S., Alfadhel M., Lewis R.A., Eyaid W., Banin E. Hypomorphic mutations in syndromic encephalocele genes are associated with Bardet-Biedl syndrome. Nat. Genet. 2008;40:443–448. doi: 10.1038/ng.97. [DOI] [PubMed] [Google Scholar]
- 39.Veis D.J., Sorenson C.M., Shutter J.R., Korsmeyer S.J. Bcl-2-deficient mice demonstrate fulminant lymphoid apoptosis, polycystic kidneys, and hypopigmented hair. Cell. 1993;75:229–240. doi: 10.1016/0092-8674(93)80065-m. [DOI] [PubMed] [Google Scholar]
- 40.Ni H.M., Chen X., Chen L., DiFrancesca D., Harada H., Yin X.M. The impact of genetic background and Bid on the phenotype of Bcl-2-deficiency in mice. Apoptosis. 2008;13:53–62. doi: 10.1007/s10495-007-0147-8. [DOI] [PubMed] [Google Scholar]
- 41.Lupski J.R., Belmont J.W., Boerwinkle E., Gibbs R.A. Clan genomics and the complex architecture of human disease. Cell. 2011;147:32–43. doi: 10.1016/j.cell.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang L.I., Nadeau J.H. 129/Sv mice--a model system for studying germ cell biology and testicular cancer. Mamm. Genome. 2001;12:89–94. doi: 10.1007/s003350010257. [DOI] [PubMed] [Google Scholar]
- 43.Stevens L.C. Genetic influences on teratocarcinogenesis and parthenogenesis. Prog. Clin. Biol. Res. 1981;45:93–104. [PubMed] [Google Scholar]
- 44.Litchfield K., Thomsen H., Mitchell J.S., Sundquist J., Houlston R.S., Hemminki K., Turnbull C. Quantifying the heritability of testicular germ cell tumour using both population-based and genomic approaches. Sci. Rep. 2015;5:13889. doi: 10.1038/srep13889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carouge D., Nadeau J.H. Mouse models of testicular germ cell tumors. In: Matin A., editor. Germ Cell Tumor. InTech; 2012. pp. 75–106. [Google Scholar]
- 46.Heaney J.D., Nadeau J.H. Testicular germ cell tumors in mice: new ways to study a genetically complex trait. Methods Mol. Biol. 2008;450:211–231. doi: 10.1007/978-1-60327-214-8_15. [DOI] [PubMed] [Google Scholar]
- 47.Noguchi T., Noguchi M. A recessive mutation (ter) causing germ cell deficiency and a high incidence of congenital testicular teratomas in 129/Sv-ter mice. J. Natl. Cancer Inst. 1985;75:385–392. [PubMed] [Google Scholar]
- 48.Youngren K.K., Coveney D., Peng X., Bhattacharya C., Schmidt L.S., Nickerson M.L., Lamb B.T., Deng J.M., Behringer R.R., Capel B. The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature. 2005;435:360–364. doi: 10.1038/nature03595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noguchi M., Watanabe C., Kobayashi T., Kuwashima M., Sakurai T., Katoh H., Moriwaki K. The ter mutation responsible for germ cell deficiency but not testicular nor ovarian teratocarcinogenesis in ter/ter congenic mice. Dev. Growth Differ. 1996;38:59–69. doi: 10.1046/j.1440-169X.1996.00008.x. [DOI] [PubMed] [Google Scholar]
- 50.Hill-Baskin A.E., Markiewski M.M., Buchner D.A., Shao H., DeSantis D., Hsiao G., Subramaniam S., Berger N.A., Croniger C., Lambris J.D., Nadeau J.H. Diet-induced hepatocellular carcinoma in genetically predisposed mice. Hum. Mol. Genet. 2009;18:2975–2988. doi: 10.1093/hmg/ddp236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Surwit R.S., Feinglos M.N., Rodin J., Sutherland A., Petro A.E., Opara E.C., Kuhn C.M., Rebuffé-Scrive M. Differential effects of fat and sucrose on the development of obesity and diabetes in C57BL/6J and A/J mice. Metabolism. 1995;44:645–651. doi: 10.1016/0026-0495(95)90123-x. [DOI] [PubMed] [Google Scholar]
- 52.Singer J.B., Hill A.E., Burrage L.C., Olszens K.R., Song J., Justice M., O’Brien W.E., Conti D.V., Witte J.S., Lander E.S., Nadeau J.H. Genetic dissection of complex traits with chromosome substitution strains of mice. Science. 2004;304:445–448. doi: 10.1126/science.1093139. [DOI] [PubMed] [Google Scholar]
- 53.Shao H., Burrage L.C., Sinasac D.S., Hill A.E., Ernest S.R., O’Brien W., Courtland H.W., Jepsen K.J., Kirby A., Kulbokas E.J. Genetic architecture of complex traits: large phenotypic effects and pervasive epistasis. Proc. Natl. Acad. Sci. USA. 2008;105:19910–19914. doi: 10.1073/pnas.0810388105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spiezio S.H., Amon L.M., McMillen T.S., Vick C.M., Houston B.A., Caldwell M., Ogimoto K., Morton G.J., Kirk E.A., Schwartz M.W. Genetic determinants of atherosclerosis, obesity, and energy balance in consomic mice. Mamm. Genome. 2014;25:549–563. doi: 10.1007/s00335-014-9530-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yazbek S.N., Buchner D.A., Geisinger J.M., Burrage L.C., Spiezio S.H., Zentner G.E., Hsieh C.W., Scacheri P.C., Croniger C.M., Nadeau J.H. Deep congenic analysis identifies many strong, context-dependent QTLs, one of which, Slc35b4, regulates obesity and glucose homeostasis. Genome Res. 2011;21:1065–1073. doi: 10.1101/gr.120741.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shao H., Sinasac D.S., Burrage L.C., Hodges C.A., Supelak P.J., Palmert M.R., Moreno C., Cowley A.W., Jr., Jacob H.J., Nadeau J.H. Analyzing complex traits with congenic strains. Mamm. Genome. 2010;21:276–286. doi: 10.1007/s00335-010-9267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen R., Shi L., Hakenberg J., Naughton B., Sklar P., Zhang J., Zhou H., Tian L., Prakash O., Lemire M. Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nat. Biotechnol. 2016;34:531–538. doi: 10.1038/nbt.3514. [DOI] [PubMed] [Google Scholar]
- 58.Erikson G.A., Bodian D.L., Rueda M., Molparia B., Scott E.R., Scott-Van Zeeland A.A., Topol S.E., Wineinger N.E., Niederhuber J.E., Topol E.J., Torkamani A. Whole-genome sequencing of a healthy aging cohort. Cell. 2016;165:1002–1011. doi: 10.1016/j.cell.2016.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gierman H.J., Fortney K., Roach J.C., Coles N.S., Li H., Glusman G., Markov G.J., Smith J.D., Hood L., Coles L.S., Kim S.K. Whole-genome sequencing of the world’s oldest people. PLoS ONE. 2014;9:e112430. doi: 10.1371/journal.pone.0112430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beekman M., Nederstigt C., Suchiman H.E., Kremer D., van der Breggen R., Lakenberg N., Alemayehu W.G., de Craen A.J., Westendorp R.G., Boomsma D.I. Genome-wide association study (GWAS)-identified disease risk alleles do not compromise human longevity. Proc. Natl. Acad. Sci. USA. 2010;107:18046–18049. doi: 10.1073/pnas.1003540107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sebastiani P., Solovieff N., Dewan A.T., Walsh K.M., Puca A., Hartley S.W., Melista E., Andersen S., Dworkis D.A., Wilk J.B. Genetic signatures of exceptional longevity in humans. PLoS ONE. 2012;7:e29848. doi: 10.1371/journal.pone.0029848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Crow J.F. The origins, patterns and implications of human spontaneous mutation. Nat. Rev. Genet. 2000;1:40–47. doi: 10.1038/35049558. [DOI] [PubMed] [Google Scholar]
- 63.Timberlake A.T., Choi J., Zaidi S., Lu Q., Nelson-Williams C., Brooks E.D., Bilguvar K., Tikhonova I., Mane S., Yang J.F. Two locus inheritance of non-syndromic midline craniosynostosis via rare SMAD6 and common BMP2 alleles. eLife. 2016;5:5. doi: 10.7554/eLife.20125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oprea G.E., Kröber S., McWhorter M.L., Rossoll W., Müller S., Krawczak M., Bassell G.J., Beattie C.E., Wirth B. Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy. Science. 2008;320:524–527. doi: 10.1126/science.1155085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Corvol H., Blackman S.M., Boëlle P.Y., Gallins P.J., Pace R.G., Stonebraker J.R., Accurso F.J., Clement A., Collaco J.M., Dang H. Genome-wide association meta-analysis identifies five modifier loci of lung disease severity in cystic fibrosis. Nat. Commun. 2015;6:8382. doi: 10.1038/ncomms9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Emond M.J., Louie T., Emerson J., Zhao W., Mathias R.A., Knowles M.R., Wright F.A., Rieder M.J., Tabor H.K., Nickerson D.A., National Heart, Lung, and Blood Institute (NHLBI) GO Exome Sequencing Project. Lung GO Exome sequencing of extreme phenotypes identifies DCTN4 as a modifier of chronic Pseudomonas aeruginosa infection in cystic fibrosis. Nat. Genet. 2012;44:886–889. doi: 10.1038/ng.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mouton J.M., van der Merwe L., Goosen A., Revera M., Brink P.A., Moolman-Smook J.C., Kinnear C. MYBPH acts as modifier of cardiac hypertrophy in hypertrophic cardiomyopathy (HCM) patients. Hum. Genet. 2016;135:477–483. doi: 10.1007/s00439-016-1649-7. [DOI] [PubMed] [Google Scholar]
- 68.Harms D.W., Quadros R.M., Seruggia D., Ohtsuka M., Takahashi G., Montoliu L., Gurumurthy C.B. Mouse genome editing using the CRISPR/Cas cystem. Curr. Protoc. Hum. Genet. 2014;83:1–27. doi: 10.1002/0471142905.hg1507s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dansky H.M., Charlton S.A., Sikes J.L., Heath S.C., Simantov R., Levin L.F., Shu P., Moore K.J., Breslow J.L., Smith J.D. Genetic background determines the extent of atherosclerosis in ApoE-deficient mice. Arterioscler. Thromb. Vasc. Biol. 1999;19:1960–1968. doi: 10.1161/01.atv.19.8.1960. [DOI] [PubMed] [Google Scholar]
- 70.Dansky H.M., Shu P., Donavan M., Montagno J., Nagle D.L., Smutko J.S., Roy N., Whiteing S., Barrios J., McBride T.J. A phenotype-sensitizing Apoe-deficient genetic background reveals novel atherosclerosis predisposition loci in the mouse. Genetics. 2002;160:1599–1608. doi: 10.1093/genetics/160.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rodríguez J.M., Wolfrum S., Robblee M., Chen K.Y., Gilbert Z.N., Choi J.H., Teupser D., Breslow J.L. Altered expression of Raet1e, a major histocompatibility complex class 1-like molecule, underlies the atherosclerosis modifier locus Ath11 10b. Circ. Res. 2013;113:1054–1064. doi: 10.1161/CIRCRESAHA.113.302052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carpinelli M.R., Hilton D.J., Metcalf D., Antonchuk J.L., Hyland C.D., Mifsud S.L., Di Rago L., Hilton A.A., Willson T.A., Roberts A.W. Suppressor screen in Mpl-/- mice: c-Myb mutation causes supraphysiological production of platelets in the absence of thrombopoietin signaling. Proc. Natl. Acad. Sci. USA. 2004;101:6553–6558. doi: 10.1073/pnas.0401496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Buchovecky C.M., Turley S.D., Brown H.M., Kyle S.M., McDonald J.G., Liu B., Pieper A.A., Huang W., Katz D.M., Russell D.W. A suppressor screen in Mecp2 mutant mice implicates cholesterol metabolism in Rett syndrome. Nat. Genet. 2013;45:1013–1020. doi: 10.1038/ng.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guy J., Hendrich B., Holmes M., Martin J.E., Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat. Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- 75.He W., Bai G., Zhou H., Wei N., White N.M., Lauer J., Liu H., Shi Y., Dumitru C.D., Lettieri K. CMT2D neuropathy is linked to the neomorphic binding activity of glycyl-tRNA synthetase. Nature. 2015;526:710–714. doi: 10.1038/nature15510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Taipale M., Jarosz D.F., Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 77.Paaby A.B., Rockman M.V. Cryptic genetic variation: evolution’s hidden substrate. Nat. Rev. Genet. 2014;15:247–258. doi: 10.1038/nrg3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tokuriki N., Tawfik D.S. Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature. 2009;459:668–673. doi: 10.1038/nature08009. [DOI] [PubMed] [Google Scholar]
- 79.Schell R., Mullis M., Ehrenreich I.M. Modifiers of the genotype-phenotype map: Hsp90 and beyond. PLoS Biol. 2016;14:e2001015. doi: 10.1371/journal.pbio.2001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wilkins A.S. Sinauer Associates; 2002. The Evolution of Developmental Pathways. [Google Scholar]
- 81.Wagner A. Princeton University Press; 2007. Robustness and Evolvability in Living Systems. [Google Scholar]
- 82.Henn B.M., Botigué L.R., Bustamante C.D., Clark A.G., Gravel S. Estimating the mutation load in human genomes. Nat. Rev. Genet. 2015;16:333–343. doi: 10.1038/nrg3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A., 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.MacArthur D.G., Balasubramanian S., Frankish A., Huang N., Morris J., Walter K., Jostins L., Habegger L., Pickrell J.K., Montgomery S.B., 1000 Genomes Project Consortium A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Campbell C.D., Eichler E.E. Properties and rates of germline mutations in humans. Trends Genet. 2013;29:575–584. doi: 10.1016/j.tig.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Keightley P.D., Ness R.W., Halligan D.L., Haddrill P.R. Estimation of the spontaneous mutation rate per nucleotide site in a Drosophila melanogaster full-sib family. Genetics. 2014;196:313–320. doi: 10.1534/genetics.113.158758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lynch M. Mutation and human exceptionalism: our future genetic load. Genetics. 2016;202:869–875. doi: 10.1534/genetics.115.180471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Uchimura A., Higuchi M., Minakuchi Y., Ohno M., Toyoda A., Fujiyama A., Miura I., Wakana S., Nishino J., Yagi T. Germline mutation rates and the long-term phenotypic effects of mutation accumulation in wild-type laboratory mice and mutator mice. Genome Res. 2015;25:1125–1134. doi: 10.1101/gr.186148.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nadeau J.H., Topol E.J. The genetics of health. Nat. Genet. 2006;38:1095–1098. doi: 10.1038/ng1006-1095. [DOI] [PubMed] [Google Scholar]
- 90.Harper A.R., Nayee S., Topol E.J. Protective alleles and modifier variants in human health and disease. Nat. Rev. Genet. 2015;16:689–701. doi: 10.1038/nrg4017. [DOI] [PubMed] [Google Scholar]
- 91.Akhmedov N.B., Piriev N.I., Chang B., Rapoport A.L., Hawes N.L., Nishina P.M., Nusinowitz S., Heckenlively J.R., Roderick T.H., Kozak C.A. A deletion in a photoreceptor-specific nuclear receptor mRNA causes retinal degeneration in the rd7 mouse. Proc. Natl. Acad. Sci. USA. 2000;97:5551–5556. doi: 10.1073/pnas.97.10.5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Haider N.B., Zhang W., Hurd R., Ikeda A., Nystuen A.M., Naggert J.K., Nishina P.M. Mapping of genetic modifiers of Nr2e3 rd7/rd7 that suppress retinal degeneration and restore blue cone cells to normal quantity. Mamm. Genome. 2008;19:145–154. doi: 10.1007/s00335-008-9092-2. [DOI] [PubMed] [Google Scholar]
- 93.Cruz N.M., Yuan Y., Leehy B.D., Baid R., Kompella U., DeAngelis M.M., Escher P., Haider N.B. Modifier genes as therapeutics: the nuclear hormone receptor Rev Erb alpha (Nr1d1) rescues Nr2e3 associated retinal disease. PLoS ONE. 2014;9:e87942. doi: 10.1371/journal.pone.0087942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lanzkron S., Strouse J.J., Wilson R., Beach M.C., Haywood C., Park H., Witkop C., Bass E.B., Segal J.B. Systematic review: Hydroxyurea for the treatment of adults with sickle cell disease. Ann. Intern. Med. 2008;148:939–955. doi: 10.7326/0003-4819-148-12-200806170-00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Platt O.S., Thorington B.D., Brambilla D.J., Milner P.F., Rosse W.F., Vichinsky E., Kinney T.R. Pain in sickle cell disease. Rates and risk factors. N. Engl. J. Med. 1991;325:11–16. doi: 10.1056/NEJM199107043250103. [DOI] [PubMed] [Google Scholar]
- 96.Lettre G., Sankaran V.G., Bezerra M.A., Araújo A.S., Uda M., Sanna S., Cao A., Schlessinger D., Costa F.F., Hirschhorn J.N., Orkin S.H. DNA polymorphisms at the BCL11A, HBS1L-MYB, and beta-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc. Natl. Acad. Sci. USA. 2008;105:11869–11874. doi: 10.1073/pnas.0804799105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Suzuki M., Yamamoto M., Engel J.D. Fetal globin gene repressors as drug targets for molecular therapies to treat the β-globinopathies. Mol. Cell. Biol. 2014;34:3560–3569. doi: 10.1128/MCB.00714-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Costanzo M., VanderSluis B., Koch E.N., Baryshnikova A., Pons C., Tan G., Wang W., Usaj M., Hanchard J., Lee S.D. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016;353:353. doi: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.van Leeuwen J., Pons C., Mellor J.C., Yamaguchi T.N., Friesen H., Koschwanez J., Ušaj M.M., Pechlaner M., Takar M., Ušaj M. Exploring genetic suppression interactions on a global scale. Science. 2016;354:354. doi: 10.1126/science.aag0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lim J., Hao T., Shaw C., Patel A.J., Szabó G., Rual J.F., Fisk C.J., Li N., Smolyar A., Hill D.E. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 101.Noebels J. Pathway-driven discovery of epilepsy genes. Nat. Neurosci. 2015;18:344–350. doi: 10.1038/nn.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]