Summary
Ribosomal RNA (rRNA) is transcribed from rDNA by RNA polymerase I (Pol I) to produce the 45S precursor of the 28S, 5.8S, and 18S rRNA components of the ribosome. Two transcription factors have been defined for Pol I in mammals, the selectivity factor SL1, and the upstream binding transcription factor (UBF), which interacts with the upstream control element to facilitate the assembly of the transcription initiation complex including SL1 and Pol I. In seven unrelated affected individuals, all suffering from developmental regression starting at 2.5–7 years, we identified a heterozygous variant, c.628G>A in UBTF, encoding p.Glu210Lys in UBF, which occurred de novo in all cases. While the levels of UBF, Ser388 phosphorylated UBF, and other Pol I-related components (POLR1E, TAF1A, and TAF1C) remained unchanged in cells of an affected individual, the variant conferred gain of function to UBF, manifesting by markedly increased UBF binding to the rDNA promoter and to the 5′- external transcribed spacer. This was associated with significantly increased 18S expression, and enlarged nucleoli which were reduced in number per cell. The data link neurodegeneration in childhood with altered rDNA chromatin status and rRNA metabolism.
Main Text
Neurodegenerative disorders that present in childhood are a group of heterogeneous conditions with a genetic basis that lead to a progressive decline in psychomotor function. The primary target in these disorders might involve any of the structural components of the nervous system (neurons and their projections, and supportive elements; astrocytes and glial cells). Earlier concepts considered neurodegenerative disorders as affecting the “grey matter” (e.g., neuronal ceroid lipofuscinosis [MIM 256730], Rett syndrome [MIM 312750]) or “white matter” (e.g., Krabbe Leukodystrophy [MIM 245200]) disorders. Advances in genomic medicine have begun to unravel the underlying causes for several of these conditions, it is clear that a number of cellular organelles and pathways are involved as targets for pathogenic mutations eventually leading to a degenerating neural network.
In childhood, these disorders present with developmental regression (loss of developmental milestones) affecting multiple domains (motor, social, cognitive) with or without the presence of seizures.1 Diagnostic delineation rests on the age of onset, deviations in head circumference (microcephaly, macrocephaly) findings on brain imaging (regional distribution of loss of gray or white matter (atrophy), and associated signal abnormalities that might involve the cortex, subcortical gray matter, and the white matter. In late stages of these disorders, the distinction between gray and white matter involvement is often blurred.2
The affected individuals, 6 females and 1 male, 8–23 years of age, originated from seven unrelated families; their clinical and radiologic data are presented in Table 1. In 6/7, the pregnancy was uneventful, whereas in one, gestational diabetes and polyhydramnios were reported. The delivery was uncomplicated in 6/7; one affected individual had C-section for failed induction and fetal distress. Six/7 were born at term, and one was born at 33 weeks of gestation. The perinatal course was uneventful and the initial psychomotor development was reported normal in 4/7 affected individuals; three affected individuals had mild developmental delay till 2 years of age. Developmental regression invariably occurred at 2.5–7 years and in 5 affected individuals was initially confined to motor domains with a gradual loss of ambulation, fine motor abilities, self-care skills, and ultimately feeding difficulties leading to failure to thrive. Within 1–3 years of regression onset, this was followed by obvious language regression and the appearance of an extrapyramidal movement disorder, which consisted of dystonia, chorea, parkinsonism, or rigidity. Pyramidal signs and ataxia were present in 4 and 3 affected individuals, respectively, but were not prominent. Two affected individuals had initial cognitive/language regression, which was followed by motor regression though eventually leading to the same endpoint of severe to profound intellectual disability and inability to ambulate. Focal and generalized seizures were reported for 3 affected individuals and one further affected individual had an abnormal EEG. Head circumference percentiles, normal at birth, declined over time. No other systems were involved in particular there was no evidence of dysmorphism (Figure 1), organomegaly, and hearing or visual impairment. No hematological or skeletal abnormalities were present. At the time of writing all the affected individuals were alive.
Table 1.
Clinical and Radiologic Findings in the Affected Individuals
| Affected individual -country | 1-USA | 2-Canada | 3-USA | 4-France | 5-Israel | 6-Russia | 7-USA |
|---|---|---|---|---|---|---|---|
| Sex/DOB/ no. healthy sibs | F/1994/1 | F/2000/ 2 | F/2001/3 | F/1998/ 1 | F/1998/ 1 | M/2006/1 | F/2009/1 |
| Current age in years | 23 | 17 | 16 | 19 | 19 | 11 | 8 |
| Presentation | Motor and language regression starting at 2.5 years. | Developmental delay till 2 years, motor regression starting at 5 years. | Developmental delay till 2 years; motor and language regression starting at 7 years. | Motor regression starting at 3 years. | Motor regression starting at 3 years. | Motor and language regression starting at 4 years | Developmental delay till 2 years, motor regression starting at 3.5 years. |
| Clinical features | Borderline microcephaly, profound ID, nonverbal, non-ambulatory, ataxia, dystonia. | Acquired microcephaly profound ID, nonverbal, non-ambulatory, spasticity, dystonia. | Acquired microcephaly, profound ID, nonverbal, non-ambulatory, spasticity, rigidity, ataxia. | Acquired microcephaly, profound ID, nonverbal, non-ambulatory, spasticity, dystonia, chorea. | Acquired microcephaly, profound ID, nonverbal, non-ambulatory, spasticity, parkinsonism. | Acquired microcephaly, profound ID, nonverbal, non-ambulatory, spasticity, dystonia. | Normal head circumference, profound ID, nonverbal, assisted walk of short distances, ataxia, spasticity. |
| Findings at last brain MRI | Severe supra-tentorial & cerebellar atrophy, diffuse WM T2 hyper-intensity. | Severe supra-tentorial & cerebellar atrophy, diffuse WM T2 hyper-intensity. | Severe supra-tentorial & cerebellar atrophy, diffuse WM T2 hyper-intensity. | Severe supra-tentorial & cerebellar atrophy, diffuse WM T2 hyper-intensity. | Severe supra-tentorial atrophy, diffuse WM T2 hyper-intensity. | Severe supra-tentorial atrophy, diffuse WM T2 hyper-intensity. | Severe supra-tentorial & cerebellar atrophy, diffuse WM T2 hyper-intensity. |
| Epilepsy | No. | Focal seizures at 15 years | Onset at 5 years. | Onset at 14 years | No. Normal EEG. | No, but abnormal EEG | No. Normal EEG. |
Figure 1.
Affected Individual 2 at Age 1 Year (A), 6 Years (B), and 10 Years (C)
Numerous routine diagnostic laboratory tests were performed without any specific abnormalities noted. Brain MRI revealed a non-specific pattern of diffuse cortical atrophy and T2 hyperintensity that was either localized to the periventricular white matter or more extensively affected the entire subcortical white matter in affected individuals. This was accompanied by cerebellar atrophy in 5 affected individuals. In affected individuals where sequential imaging was available, cortical atrophy was noted initially and appeared to precede the development of cerebellar atrophy (Figure 2). These findings are frequently encountered in inherited pediatric neurodegenerative disorders that primarily affect the gray matter (mitochondrial disorders, Menkes disease [MIM 309400], Niemann Pick disease type C [MIM 257220], etc.), and therefore would not be considered as specific to this condition.
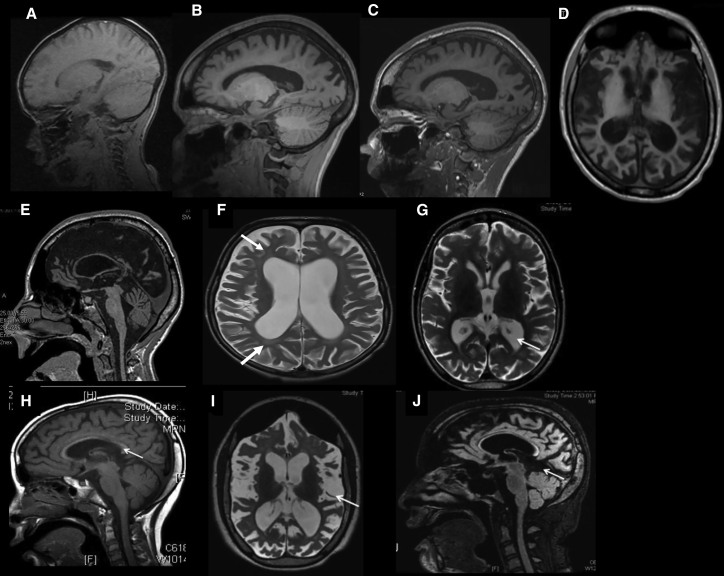
Figure 2.
Progressive Cortical Atrophy Observed in Serial Brain MRI
(A–C) Parasagittal T1-weighted brain MRI of affected individual 3 at age 4, 9, and 11 years, respectively. Note progressive cortical atrophy with slight cerebellar atrophy.
(D and E) Axial and Midsagittal T1 images of affected individual 4 at age 15 years. Note cortical and cerebellar atrophy.
(F) Axial T2 of affected individual 6 at age 8 years. Note cortical atrophy and periventricular white matter hyperintensity (arrows).
(G and H) T2 weighted axial and sagittal sequences. Early imaging on affected individual 2 showing modest ventricular dilatation ex-vacuo associated with a thin corpus callous (arrows).
(I and J) T2 Weighted axial and sagittal FLAIR sequences in a follow up imaging study repeated after 6 years showing dramatic cortical atrophy and ex-vacuo ventricular dilatation. Note is made of the subdural hygroma below the tentorium in (J).
For affected individuals 1 and 2 and their parents, WES was performed on exon targets captured using the Clinical Research Exome kit (Agilent Technologies, Santa Clara, CA). The full sequencing methodology and variant interpretation protocol has been previously described.3 The general assertion criteria for variant classification are publicly available on the GeneDx ClinVar submission page. For affected individual 3 and her parents WES was performed at the Broad Institute of Harvard and MIT (Cambridge, MA, USA) as described previously.4 WES of affected individual 4 and her parents was performed on exon targets captured using Roche SeqCap EZ MedExome. Sequencing was performed on NextSeq 500. Variant interpretation protocol has been previously described.5 WES of affected individual 5 and her parents and affected individual 6 was performed on exon targets captured using SureSelect Human All Exon 50 Mb Kit V.4 (Agilent Technologies, Santa Clara, CA, USA). Sequences were determined by HiSeq2000 (Illumina, San Diego, CA, USA). The full sequencing methodology and variant interpretation protocol were previously described.5 Clinical exome sequencing of affected individual 7 and her parents was performed at Ambry Genetics as previously described.6
All the procedures were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national); proper informed consent was obtained from all guardians.
In the WES of affected individuals 1–5 and 7, following reads alignment, variant calling, and filtration, all remaining variants were heterozygous, and all but one were inherited from the parents. This was Hg19.Chr17: 42290219G>A, NM_014233.3:c.628G>A, p.Glu210Lys in UBTF [MIM 600673]. We verified the finding by Sanger sequencing in the affected individuals (Figure S1), and confirmed its absence from the DNA of all the parents and the healthy sibs. The variant was not carried by any of the ∼140,000 individuals whose exome analyses were deposited at gnomAD but was carried by one of the affected individuals at the Hadassah WES database (2,100 exome analyses). This was affected individual 6 and the variant was confirmed de novo by Sanger sequencing.
UBTF encodes the 764 amino acids of UBF, which is a Pol I transcription factor and is instrumental in the generation of rRNA transcripts. The biogenesis of ribosome components is a delicate and balanced process, which is tightly coupled to cellular growth and proliferation rates and which occupies the cell transcription machinery to a large extent. Accordingly, in rapidly growing mammalian cells, rRNA synthesis accounts for 35%–60% of all RNA transcription.7, 8 Transcription of rDNA is mediated by Pol I to produce the 45S rRNA precursor of the 28S, 5.8S, and 18S rRNA components of the ribosome.9 Two transcription factors have been defined for Pol I in mammals: one is the selectivity factor SL1, which consists of the TATA-binding protein (TBP) and three TBP-associated factors (TAFs), and plays a critical role in recognition of the core promoter element, and the other is the UBF, which interacts with the upstream control element to facilitate the assembly of the transcription initiation complex including SL1 and Pol I.10 UBF belongs to the sequence nonspecific class of HMG (high mobility group) proteins. It has six HMG-box homology domains and has been proposed to interact with rDNA as a dimer and to induce six in-phase bends to generate a single 360°-loop structure that resembles the nucleosome. Glu210 is part of the second HMG-box homology domain and is conserved in all species which have UBTF. Because of the similarity of the clinical phenotype among the affected individuals, the rarity of this variant and its de novo recurrence, we assumed this is the disease-causing variant. Glu210 is followed by two lysine residues (211 and 212), therefore p.Glu210Lys variant would result in a string of three lysine residues. Such a highly positive charge region may lead to a stronger interaction with negatively charged DNA, conferring gain of function to the protein.
We first studied the levels of UBF, Ser388 phosphorylated UBF, as well as other Pol I-related components, including POLR1E, TAF1A, and TAF1C, in fibroblasts of affected individual 6 and a healthy control. Fibroblasts from passage 3–4 were grown in RPMI media with 10% FBS and 2 mM Glutamine and the antibodies used for western blots were UBF (Santa Cruz Biotechnology sc-13125), phospho-UBF Ser388 (Santa Cruz Biotechnology sc-21637), PolR1E (Santa Cruz Biotechnology sc-398270), TAF1C (Santa Cruz Biotechnology sc-367333), TAF1A (Santa Cruz Biotechnology sc-6572), and actin (Genetex gt5512). This experiment revealed that all proteins were present in the affected individual cells in a comparable abundance to those of the control cells (Figure 3A). Next, we investigated the transcription factor activity of UBF on rDNA. To this end, we studied UBF binding to rDNA promoters using chromatin immunoprecipitation (ChIP)-qPCR as previously described.11 Briefly, cells were crosslinked with 1% formaldehyde, and lysed in buffer containing 1% SDS. The lysate was sonicated and incubated overnight with 5 μg of UBF antibody (Santa Cruz Biotechnology sc-13125) and Protein A/G Sepharose (Santa Cruz Biotechnology). Following elution, DNA was purified using a PCR purification kit (Omega Bio-Tek) and subjected to real-time qPCR with PerfeCTa SYBR Green SuperMix (Quanta), using a CFX Connect Real-Time Cycler (BioRad). Previously described primers10 for rDNA Promoter (for: CCCGGGGGAGGTATATCTTT; rev: ACAGGTCGCCAGAGGACAG, rDNA ETS region (for: GAACGGTGGTGTGTCGTTC; rev: CGTCTCGTCTCGTCTCACTC), and rDNA NTS region (for: GATGGGTTTCGGGGTTCTAT; rev: GGCAGGCAAATGTAGAAGG) were used.
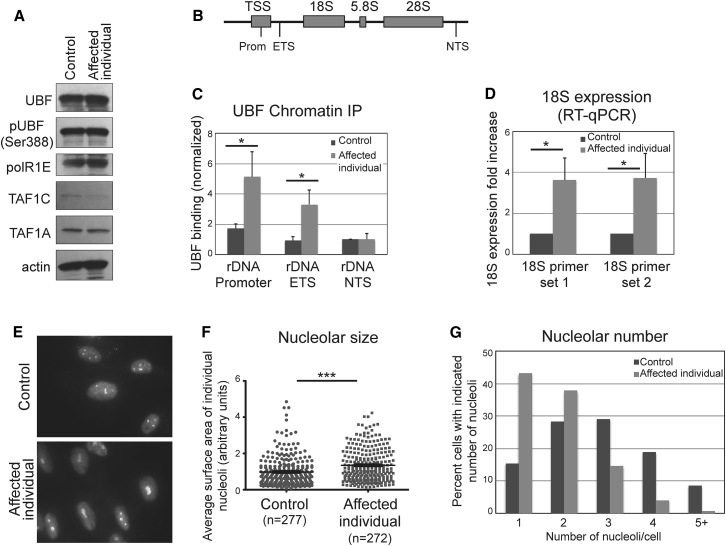
Figure 3.
Mutant UBF Is Hyperactive and Drives Increased Expression of rDNA Genes, Associated with Nucleolar Alterations
(A) Western blot analysis of cells of control and affected individual showing that the UBTF mutation does not affect UBF steady state levels, its phosphorylation status, or the levels of the Pol I subunits PolR1E, TAF1A, and TAFC.
(B) Schematic representation of the rDNA gene locus, indicating the location of each primer set used: Promoter, external transcribed spacer (ETS), and non-transcribed spacer (NTS).
(C) UBF ChIP-qPCR showing that UBF binding to the Promoter and ETS region is significantly increased in cells of the affected individual compared to control cells. Binding was quantified relative to input material, and shown as normalized to the NTS locus in control cells. Bars represent the average of three independent experiments, and error bars represent SD. The asterisk indicates statistical significance (p < 0.05) evaluated using the t test (two-tailed, equal variance).
(D) Quantification of 18S expression in cells of affected individual and control, using real-time qPCR, quantified relative to GAPDH expression. Two different 18S primer sets were used. The results are shown as normalized to control cells. Bars represent the average of three independent experiments, and error bars represent SD. The asterisk indicates statistical significance (p < 0.05) evaluated using the t test (two-tailed, equal variance).
(E) Representative micrographs of anti-nucleolin immunofluorescence showing altered nucloelar pattern in affected individual cells.
(F) Quantification of nucleolar size. Nucleolar area was quantified using ImageJ and statistical analysis was done with Prism (t test, two-tailed, equal variance). Asterisks indicate statistical significance (p < 0.0001). The mean ± SEM is also shown.
(G) Quantification of the number of nucleoli in each cell (117 control and 151 affected individual cells were analyzed).
It was already shown that UBF binds strongly to rDNA promoter, weakly to the 5′-ETS (external transcribed spacer), and does not bind at all to the NTS (non-transcribed spacer)10 (Figure 3B). In agreement, the ChIP-qPCR experiments revealed a significantly increased binding (about X3) of UBF to the rDNA promoter and ETS regions in the affected individual cells compared to control cells (Figure 3C). UBF binding to the NTS control region in the affected individual cells was comparable to that in the control cells.
Because of the increased binding of UBF to the rDNA promoter in the affected individual cells, we next verified that the expression of rDNA is increased in these cells by quantifying 18S expression by real-time qPCR. Total mRNA was purified from affected individual and control cells using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions, and subjected to reverse transcription using the RevertAid Reverse Transcriptase Kit (Thermo Fisher Scientific) with oligo dT primers. Real-time qPCR was performed as described above. The cDNA of GAPDH was obtained and analyzed in parallel for normalization. Primers used were: 18S set 1 (as in12) (for: GAAACTGCGAATGGCTCATTAAA; rev: CCACAGTTATCCAAGTAGGAGAGGA); 18S set 2 13 (for: TTCGAACGTCTGCCCTATCAA; rev: ATGGTAGGCACGGCGACTA); and GAPDH (for: TGCACCACCAACTGCTTAGC; rev: GGCATGGACTGTGGTCATGAG). This analysis showed that the 18S expression was about four-fold higher in the affected individual cells compared to control cells (Figure 3D). These results confirm that the UBF which carries the p.Glu210Lys variant functions as a hyperactive transcription factor, resulting in increased expression of rDNA.
The rDNA locus occupies a distinctive region of the genome and is located in the nucleolus, where its transcription is coupled to ribosomal biogenesis.14 Increased rDNA expression is associated with chromatin decondensation at the rDNA locus and enlarged nucleoli.15, 16 We therefore checked whether the mutant UBF affects nucleolar architecture. Cells of an affected individual and control were methanol-fixed and subjected to immunofluorescence staining using antibodies recognizing the nucleolar marker nucleolin (Abcam ab50279). In line with the increased rDNA expression, we observed that the affected individual cells showed increased nucleolar size (Figures 3E and 3F). Interestingly, this was coupled with a reduction in the overall number of nucleoli per cell (Figures 3E and 3G). This reorganization of the nucleolar material is suggestive of altered rDNA chromatin status.
Alteration of UBF level was already shown to propagate detrimental effect. Depletion of UBF was shown to silence rDNA and its total ablation is likely incompatible with life.17, 18 In agreement, UBTF is highly intolerant to both deleterious and missense variants; in the ExAC cohort, the number of UBTF variants markedly deviate from the expected by its length (Z = 5.61) with expected/observed number of missense variants 295/98 and of loss-of-function (LOF) variants 36/0.
Overexpression of UBTF in neonatal rat cardiomyocytes led to a proportional increase in rDNA transcription.19 In agreement, we demonstrate that the increased binding of UBF to the rDNA promoter is associated with a similar increase in 18 s rRNA level. Markedly elevated, normal rRNA transcripts, seems a unique phenomenon and several consequences could be envisioned, including scavenging of RNA binding proteins, scavenging of RNA disposal machinery, or altered ribosome biogenesis. It is of note that the expression of rRNA and ribosomal proteins is tightly co-regulated in response to growth and stress signals.20 Uncoupled overexpression of bacterial rDNA results in increased free ribosomal subunits with marked dominant negative effect.21 The effect of excessive rRNA on neural tissue could be deleterious and might even culminate in neurodegeneration; in adult rat hippocampal neurons, enhanced Pol I activation promoted neurite outgrowth, including abnormally increased total neurite length and branching,22 which in itself was shown to precede neurodegeneration.23
Cell mitosis and DNA damage repair could also be affected by excessive transcription of rDNA due to depletion of untranscribed rDNA. Untranscribed rDNA serve as binding sites for condensin,24 which is required for DNA damage repair and chromatid cohesion.25 Impaired binding of condensin to untranscribed rDNA is associated with premature sister chromatids separation, before damage repair is completed.26
Finally, UBF is an important regulator of chromatin structure at the rDNA locus; it was shown to outcompete linker Histone H1 resulting in decondensation of rDNA chromatin.17 Changes in chromatin status at the rDNA locus, coupled with nucleolar alterations, have been previously reported in Huntington disease [MIM 143100]16 and were regulated by UBF post-translational modification by acetylation at Lys-352 and methylation at Lys-232.16, 27 The abnormally increased nucleolar size and reduced number of nucleoli per cell which was observed in the cells of the affected individual, suggest that the mutant hyperactive UBF causes an alteration in rDNA chromatin structure. Such a dysregulation would have a profound impact, since rDNA heterochromatin status is tightly regulated during differentiation and has a broader effect on heterochromatin formation throughout the nucleus.17, 28 Obviously, the data and the consequent assumptions should be regarded as preliminary, because all derived from a single cell line of an affected individual and one healthy control.
In summary, in seven individuals with developmental regression and neurodegeneration in childhood, we identified the heterozygous c.628G>A variant in UBTF. In cells of an affected individual, the variant conferred gain of function to UBF with the subsequent production of markedly increased amount of rRNA and alteration of nucleoli size and number. This report links neuronal degeneration in childhood with aberrant rRNA metabolism.
Acknowledgments
We thank the affected individuals and their families for allowing us to share their data. C.P. and A.N.P. thank Samantha Colaiacovo for genetic counseling. L.S. thanks Erica Wahl for genetic counseling. P.B.A. was supported by NIH/NIAMS 1R01AR068429-01 and NICHD/NHGRI/NIH U19HD077671. Sequencing and analysis for family 3 was provided by the Broad Institute of MIT and Harvard Center for Mendelian Genomics (Broad CMG) and was funded by the National Human Genome Research Institute, the National Eye Institute, and the National Heart, Lung and Blood Institute grant UM1 HG008900 to Daniel MacArthur and Heidi Rehm. This work was supported in part by the Trudy Mandel Louis Charitable Trust to O.E. This work resulted in part from a successful GeneMatcher match.29 A.B. and B.B. are employees of GeneDx; Z.P. is an employee of Ambry Genetics.
Published: August 3, 2017
Footnotes
Supplemental Data include one figure and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.07.002.
Contributor Information
George-Lucian Moldovan, Email: gmoldovan@pennstatehealth.psu.edu.
Orly Elpeleg, Email: elpeleg@hadassah.org.il.
Web Resources
gnomAD Browser, http://gnomad.broadinstitute.org/
OMIM, http://www.omim.org/
Supplemental Data
References
- 1.Crumrine P.K. Degenerative disorders of the central nervous system. Pediatr. Rev. 2001;22:370–379. [PubMed] [Google Scholar]
- 2.Schiffmann R., van der Knaap M.S. Invited article: an MRI-based approach to the diagnosis of white matter disorders. Neurology. 2009;72:750–759. doi: 10.1212/01.wnl.0000343049.00540.c8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanaka A.J., Cho M.T., Millan F., Juusola J., Retterer K., Joshi C., Niyazov D., Garnica A., Gratz E., Deardorff M. Mutations in SPATA5 Are Associated with Microcephaly, Intellectual Disability, Seizures, and Hearing Loss. Am. J. Hum. Genet. 2015;97:457–464. doi: 10.1016/j.ajhg.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perić S., Glumac J.N., Töpf A., Savić-Pavićević D., Phillips L., Johnson K., Cassop-Thompson M., Xu L., Bertoli M., Lek M. A novel recessive TTN founder variant is a common cause of distal myopathy in the Serbian population. Eur. J. Hum. Genet. 2017;25:572–581. doi: 10.1038/ejhg.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ta-Shma A., Zhang K., Salimova E., Zernecke A., Sieiro-Mosti D., Stegner D., Furtado M., Shaag A., Perles Z., Nieswandt B. Congenital valvular defects associated with deleterious mutations in the PLD1 gene. J. Med. Genet. 2016 doi: 10.1136/jmedgenet-2016-104259. [DOI] [PubMed] [Google Scholar]
- 6.Farwell K.D., Shahmirzadi L., El-Khechen D., Powis Z., Chao E.C., Tippin Davis B., Baxter R.M., Zeng W., Mroske C., Parra M.C. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet. Med. 2015;17:578–586. doi: 10.1038/gim.2014.154. [DOI] [PubMed] [Google Scholar]
- 7.Moss T., Stefanovsky V.Y. At the center of eukaryotic life. Cell. 2002;109:545–548. doi: 10.1016/s0092-8674(02)00761-4. [DOI] [PubMed] [Google Scholar]
- 8.Cavanaugh A., Hirschler-Laszkiewicz I., Rothblum L. Ribosomal DNA transcription in mammals. In: Olson M.O.J., editor. The Nucleolus. Kluwer Academic; Plenum, New York: 2003. pp. 89–129. [Google Scholar]
- 9.McStay B., Grummt I. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu. Rev. Cell Dev. Biol. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- 10.Friedrich J.K., Panov K.I., Cabart P., Russell J., Zomerdijk J.C. TBP-TAF complex SL1 directs RNA polymerase I pre-initiation complex formation and stabilizes upstream binding factor at the rDNA promoter. J. Biol. Chem. 2005;280:29551–29558. doi: 10.1074/jbc.M501595200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woolnough J.L., Atwood B.L., Liu Z., Zhao R., Giles K.E. The Regulation of rRNA Gene Transcription during Directed Differentiation of Human Embryonic Stem Cells. PLoS ONE. 2016;11:e0157276. doi: 10.1371/journal.pone.0157276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hinds T.D., Jr., Stechschulte L.A., Cash H.A., Whisler D., Banerjee A., Yong W., Khuder S.S., Kaw M.K., Shou W., Najjar S.M., Sanchez E.R. Protein phosphatase 5 mediates lipid metabolism through reciprocal control of glucocorticoid receptor and peroxisome proliferator-activated receptor-γ (PPARγ) J. Biol. Chem. 2011;286:42911–42922. doi: 10.1074/jbc.M111.311662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iglesias-Bartolome R., Torres D., Marone R., Feng X., Martin D., Simaan M., Chen M., Weinstein L.S., Taylor S.S., Molinolo A.A., Gutkind J.S. Inactivation of a Gα(s)-PKA tumour suppressor pathway in skin stem cells initiates basal-cell carcinogenesis. Nat. Cell Biol. 2015;17:793–803. doi: 10.1038/ncb3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi T. A new role of the rDNA and nucleolus in the nucleus--rDNA instability maintains genome integrity. BioEssays. 2008;30:267–272. doi: 10.1002/bies.20723. [DOI] [PubMed] [Google Scholar]
- 15.Koh C.M., Gurel B., Sutcliffe S., Aryee M.J., Schultz D., Iwata T., Uemura M., Zeller K.I., Anele U., Zheng Q. Alterations in nucleolar structure and gene expression programs in prostatic neoplasia are driven by the MYC oncogene. Am. J. Pathol. 2011;178:1824–1834. doi: 10.1016/j.ajpath.2010.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J., Hwang Y.J., Ryu H., Kowall N.W., Ryu H. Nucleolar dysfunction in Huntington’s disease. Biochim. Biophys. Acta. 2014;1842:785–790. doi: 10.1016/j.bbadis.2013.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanij E., Poortinga G., Sharkey K., Hung S., Holloway T.P., Quin J., Robb E., Wong L.H., Thomas W.G., Stefanovsky V. UBF levels determine the number of active ribosomal RNA genes in mammals. J. Cell Biol. 2008;183:1259–1274. doi: 10.1083/jcb.200805146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamdane N., Stefanovsky V.Y., Tremblay M.G., Németh A., Paquet E., Lessard F., Sanij E., Hannan R., Moss T. Conditional inactivation of Upstream Binding Factor reveals its epigenetic functions and the existence of a somatic nucleolar precursor body. PLoS Genet. 2014;10:e1004505. doi: 10.1371/journal.pgen.1004505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hannan R.D., Stefanovsky V., Taylor L., Moss T., Rothblum L.I. Overexpression of the transcription factor UBF1 is sufficient to increase ribosomal DNA transcription in neonatal cardiomyocytes: implications for cardiac hypertrophy. Proc. Natl. Acad. Sci. USA. 1996;93:8750–8755. doi: 10.1073/pnas.93.16.8750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao L., Grove A. Coordination of Ribosomal Protein and Ribosomal RNA Gene Expression in Response to TOR Signaling. Curr. Genomics. 2009;10:198–205. doi: 10.2174/138920209788185261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gourse R.L., Takebe Y., Sharrock R.A., Nomura M. Feedback regulation of rRNA and tRNA synthesis and accumulation of free ribosomes after conditional expression of rRNA genes. Proc. Natl. Acad. Sci. USA. 1985;82:1069–1073. doi: 10.1073/pnas.82.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomes C., Smith S.C., Youssef M.N., Zheng J.J., Hagg T., Hetman M. RNA polymerase 1-driven transcription as a mediator of BDNF-induced neurite outgrowth. J. Biol. Chem. 2011;286:4357–4363. doi: 10.1074/jbc.M110.170134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saad Y., Segal D., Ayali A. Enhanced neurite outgrowth and branching precede increased amyloid-β-induced neuronal apoptosis in a novel Alzheimer’s disease model. J. Alzheimers Dis. 2015;43:993–1006. doi: 10.3233/JAD-140009. [DOI] [PubMed] [Google Scholar]
- 24.Johzuka K., Horiuchi T. RNA polymerase I transcription obstructs condensin association with 35S rRNA coding regions and can cause contraction of long repeat in Saccharomyces cerevisiae. Genes Cells. 2007;12:759–771. doi: 10.1111/j.1365-2443.2007.01085.x. [DOI] [PubMed] [Google Scholar]
- 25.Lam W.W., Peterson E.A., Yeung M., Lavoie B.D. Condensin is required for chromosome arm cohesion during mitosis. Genes Dev. 2006;20:2973–2984. doi: 10.1101/gad.1468806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang B.D., Butylin P., Strunnikov A. Condensin function in mitotic nucleolar segregation is regulated by rDNA transcription. Cell Cycle. 2006;5:2260–2267. doi: 10.4161/cc.5.19.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J., Hwang Y.J., Boo J.H., Han D., Kwon O.K., Todorova K., Kowall N.W., Kim Y., Ryu H. Dysregulation of upstream binding factor-1 acetylation at K352 is linked to impaired ribosomal DNA transcription in Huntington’s disease. Cell Death Differ. 2011;18:1726–1735. doi: 10.1038/cdd.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen le X.T., Raval A., Garcia J.S., Mitchell B.S. Regulation of ribosomal gene expression in cancer. J. Cell. Physiol. 2015;230:1181–1188. doi: 10.1002/jcp.24854. [DOI] [PubMed] [Google Scholar]
- 29.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.