Abstract
Vitamin D insufficiency is common, correctable, and influenced by genetic factors, and it has been associated with risk of several diseases. We sought to identify low-frequency genetic variants that strongly increase the risk of vitamin D insufficiency and tested their effect on risk of multiple sclerosis, a disease influenced by low vitamin D concentrations. We used whole-genome sequencing data from 2,619 individuals through the UK10K program and deep-imputation data from 39,655 individuals genotyped genome-wide. Meta-analysis of the summary statistics from 19 cohorts identified in CYP2R1 the low-frequency (minor allele frequency = 2.5%) synonymous coding variant g.14900931G>A (p.Asp120Asp) (rs117913124[A]), which conferred a large effect on 25-hydroxyvitamin D (25OHD) levels (−0.43 SD of standardized natural log-transformed 25OHD per A allele; p value = 1.5 × 10−88). The effect on 25OHD was four times larger and independent of the effect of a previously described common variant near CYP2R1. By analyzing 8,711 individuals, we showed that heterozygote carriers of this low-frequency variant have an increased risk of vitamin D insufficiency (odds ratio [OR] = 2.2, 95% confidence interval [CI] = 1.78–2.78, p = 1.26 × 10−12). Individuals carrying one copy of this variant also had increased odds of multiple sclerosis (OR = 1.4, 95% CI = 1.19–1.64, p = 2.63 × 10−5) in a sample of 5,927 case and 5,599 control subjects. In conclusion, we describe a low-frequency CYP2R1 coding variant that exerts the largest effect upon 25OHD levels identified to date in the general European population and implicates vitamin D in the etiology of multiple sclerosis.
Keywords: vitamin D, multiple sclerosis, GWAS, low-frequency genetic variants
Introduction
Vitamin D insufficiency affects approximately 40% of the general population in developed countries.1 This could have important public health consequences, given that vitamin D insufficiency has been associated with musculoskeletal consequences and several common diseases, such as multiple sclerosis (MIM: 126200), type 1 diabetes (MIM: 222100), type 2 diabetes (MIM: 125853), and several cancers.2 Further, repletion of vitamin D status can be achieved safely and inexpensively. Thus, understanding the determinants of vitamin D insufficiency, and their effects, can provide a better understanding of the role of vitamin D in disease susceptibility with potentially important public health benefits.
Approximately half of the variability in the concentration of the widely accepted biomarker for vitamin D status, 25-hydroxyvitamin D (25OHD), has been attributed to genetic factors in twin and family studies.3, 4 Four common (minor allele frequency [MAF] > 5%) genetic variants in loci near four genes known to be involved in cholesterol synthesis (DHCR7 [MIM: 602858]), hydroxylation (CYP2R1 [MIM: 608713]), vitamin D transport (GC [MIM: 139200]), and catabolism (CYP24A1 [MIM: 126065]) are strongly associated with 25OHD levels yet explain little of its heritability.5 Low-frequency and rare genetic variants (defined as those with a MAF ≤ 5% and 1%, respectively) have recently been found to have large effects on clinically relevant traits,6, 7, 8 providing an opportunity to better understand the biologic mechanisms influencing disease susceptibility in the general population.
Therefore, the principal objective of the present study was to detect low-frequency and rare variants with large effects on 25OHD levels through a large-scale meta-analysis and describe their biological and clinical relevance. Similar to an earlier genome-wide association study (GWAS) examining common (MAF ≥ 5%) genetic variation by the SUNLIGHT consortium,5 we sought to increase understanding of the genetic etiology of vitamin D variation within the general population; however, our current study focused on genetic variation with a MAF < 5%. This has only recently been made possible through whole-genome sequencing (WGS) and the use of improved genotype imputation for low-frequency and rare variants with the recent availability of large WGS reference panels.9 The second objective of this study was to better understand whether low-frequency genetic variants with large effects on 25OHD could predict a higher risk of vitamin D insufficiency in their carriers and whether vitamin D intake through diet might interact with such genetic factors to prevent, or magnify, vitamin D insufficiency. Finally, we sought to understand whether these genetic determinants of 25OHD levels are implicated in multiple sclerosis, a disease influenced by low 25OHD levels.10
To do so, we first undertook an association study of WGS data and deeply imputed genome-wide genotypes to identify novel genetic determinants of vitamin D in 42,274 individuals. We next tested if these genetic variants conferred a higher risk of vitamin D insufficiency in 8,711 subjects and whether this insufficiency showed effect modification by dietary intake. Last we assessed their effect on multiple sclerosis in a separate sample of 5,927 case and 5,599 control subjects.
Material and Methods
Cohorts
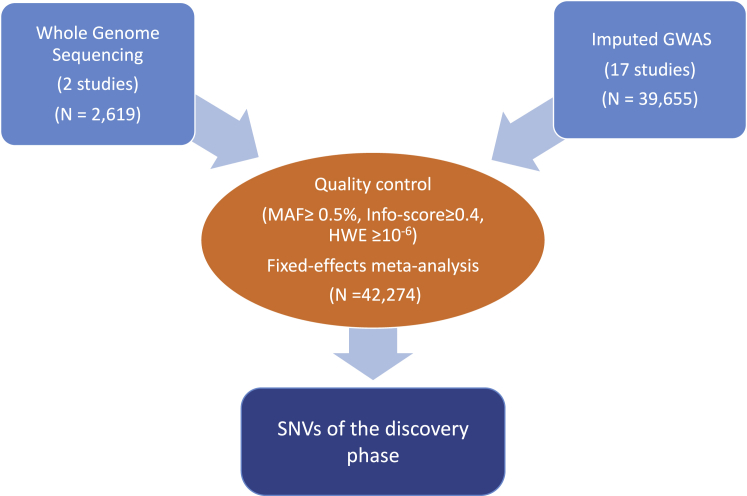
All human studies were approved by each respective institutional or national ethics review committee, and all participants provided written informed consent. To investigate the role of rare and low-frequency genetic variation on 25OHD levels in individuals of European descent, we used WGS data at a mean read depth of 6.7× in 2,619 subjects from two cohorts with available 25OHD phenotypes in the UK10K project11 (Table 1). We also used imputation reference panels to impute variants that were missing, or poorly captured, from previous GWASs of 39,655 subjects (Table 1 and Figure 1). The participating individuals were drawn from independent cohorts of individuals of European descent. A detailed description of each of the participating studies is provided in Table S1.
Table 1.
Participating Cohorts and Number of DNA Samples per Cohort
| Study | Imputed | Whole-Genome Sequenced |
|---|---|---|
| ALSPAC | 3,679 | 1,606 |
| TUK | 1,919 | 1,013 |
| Generation R | 1,442 | – |
| BPROOF | 2,514 | – |
| FHS | 5,402 | – |
| MrOS | 3,265 | – |
| RSI | 3,320 | – |
| RSII | 2,022 | – |
| RSIII | 2,913 | – |
| CHS | 1,792 | – |
| BMDCS | 863 | – |
| MrOS GBG | 945 | – |
| GOOD | 921 | – |
| MrOS Malmo | 893 | – |
| PIVUS | 943 | – |
| ULSAM | 1,095 | – |
| NEO | 5,727 | – |
| Total | 39,655 | 2,619 |
Figure 1.
Schematic of the Discovery Single-Variant Meta-analysis
25OHD Measurements
The methods applied for measuring 25OHD levels differed among the participating cohorts (Tables S1 and S6). The four methods used were tandem mass spectrometry (in the Bone Mineral Density in Childhood Study [BMDCS], Osteoporotic Fractures in Men USA [MrOS], and B-Vitamins for the Prevention of Osteoporotic Fractures [BPROOF]), combined high-performance liquid chromatography and mass spectrometry (in the Avon Longitudinal Study of Parents and Children [ALSPAC], BPROOF, Cardiovascular Health Study [CHS], Upssala Longitudinal Study of Adult Men [ULSAM], Netherlands Epidemiology of Obesity [NEO], and Generation R Study [Generation R]), chemiluminescence immunoassay (DiaSorin) (in TwinsUK [TUK], the Prospective Investigation of the Vasculature in Upssala Seniors [PIVUS], the Framingham Heart Study [FHS], Osteoporotic Fractures in Men Malmo [MrOS Malmo], Osteoporotic Fractures in Men Gothenburg [MrOS GBG], and Gothenburg Osteoporosis and Obesity Determinants [GOOD]), and electrochemiluminescence immunoassay (COBAS, Roche Diagnostics) (in Rotterdam Studies I [RSI], II [RSII], and III [RSIII]). Detection limits for the different methods are provided in Table S6.
WGS, Genotyping, and Imputation
ALSPAC WGS and TUK WGS cohorts had been sequenced at an average read depth of 6.7× through the UK10K consortium on the Illumina HiSeq platform and aligned to the GRCh37 human reference sequence with Burrows-Wheeler Aligner 31.12 Single-nucleotide variant (SNV) calls were completed with SAMtools/BCFtools,13 and VQSR14 and GATK were used to recall these variants. WGS for the ALSPAC and TUK cohorts has been described in detail in a previous publication from our group.7 Table S8 summarizes the data-generation method for sequencing-based cohorts.
Participating studies separately genotyped samples and imputed them to WGS-based reference panels. The most recent imputation panels, such as the UK10K and 1000 Genomes Project (v.3) combined panel (7,562 haplotypes from the UK10K project and 2,184 haplotypes from the 1000 Genomes Project9) and the Haplotype Reference Consortium (HRC) panel (64,976 haplotypes15), enabled more accurate imputation of low-frequency variants than the UK10K or 1000 Genomes reference panel alone.9 Specifically, 11 of the 17 participating cohorts were imputed to the combined UK10K and 1000 Genomes reference panel (total number of imputed individuals included in the meta-analysis = 25,589). Three of the participating cohorts were imputed with the HRC panel (n = 5,717). Finally, two cohorts were imputed to the 1000 Genomes panel (n = 7,536), and one cohort was imputed to the UK10K panel (n = 863) (Table S1). Details on genotyping methods and imputation for the 17 participating cohorts are presented in Table S6. Info scores for the imputed SNVs per participating cohort are presented in Table S7. To assess the quality of imputation, we tested the non-reference discordance rate for the low-frequency genome-wide-significant SNVs and found this to be 0% (Table S9).
Association Testing for 25OHD Levels and Meta-analysis
We conducted a GWAS separately for each cohort by using an additive genetic model for 25OHD levels. Because 25OHD concentrations were measured by different methods, log-transformed 25OHD levels were standardized to Z scores after adjustment for age, sex, BMI, and season of measurement. Specifically, the phenotype for each GWAS was prepared according to the following steps: (1) We log transformed 25OHD levels to ensure normality. (2) We used linear regression models to generate cohort-specific residuals of log-transformed 25OHD levels adjusted for covariates (age, sex, BMI, and season). Season was treated as a non-ordinal categorical variable (summer: July to September; fall: October to December; winter: January to March; and spring: April to June). (3) We added the mean of log-transformed 25OHD levels to the residuals to create the adjusted 25OHD phenotype. (4) We then normalized the above phenotype within each cohort (mean of 0 with 1 SD) to make the phenotype consistent across cohorts, given that our consortium has measured 25OHD levels in different cohorts by different methods. (5) Finally, we removed outliers beyond 5 SD from step 4.
For comparison purposes, we computed the average 25OHD levels, adjusted for age, sex, BMI, and season of measurement, in one cohort of our meta-analysis (TUK WGS) in carriers and non-carriers of the lead SNV(s).
The software used for each cohort’s GWAS is listed in Table S1. We performed single-variant tests for variants with MAF > 0.1% by using an additive effect of the minor allele at each variant in each cohort. The type of software employed for single-variant testing for each cohort is shown in Table S1. Studies with related individuals used software that accounted for relatedness. Cohort-specific genomic inflation factors (lambda values) are also shown in Table S1 (the mean lambda value was 1.015).
We then meta-analyzed association results from all discovery cohorts (n = 42,274). This stage included validation of the results file format, filtering files by the above quality-control (QC) criteria, comparison of trait distributions among different studies, and identification of potential biases (large beta values and/or standard errors, inconsistent effect allele frequencies, and/or extreme lambda values). Meta-analysis QC of the GWAS data included the following SNV-level exclusion criteria: (1) information score < 0.4, (2) Hardy-Weinberg equilibrium (HWE) p value < 10−6, (3) missingness > 0.05, and (4) MAF < 0.5%. SNV alignment across studies was done with the chromosome and position information for each variant according to genome build hg19 (UCSC Genome Browser). SNVs in the X chromosome were not included in the meta-analysis. Fixed-effects meta-analysis was performed with the software package GWAMA16 with adjustment for genomic control. We tested bi-allelic SNVs with MAF ≥ 0.5% for association and declared genome-wide statistical significance at p ≤ 1.2 × 10−8 for variants present in more than one study. This stringent p value threshold was set to adjust for all independent SNVs above the MAF threshold of 0.5%.17
Conditional analysis was undertaken for the four previously described lead vitamin D SNVs from the SUNLIGHT consortium with the Genome-wide Complex Trait Analysis (GCTA) package.18 This method uses an approximate conditional-analysis approach from summary-level statistics from the meta-analysis and inter-SNV linkage-disequilibrium corrections estimated from a reference sample. We used UK10K individuals as the reference sample to calculate the linkage disequilibrium of SNVs. The associated regions flanking within 400 kb of the top SNVs from SUNLIGHT were extracted, and the conditional analyses were conducted within these regions. Conditional analyses of individual variants presented in Tables 2 and S5 were conducted with GCTA v.0.93.9 and default parameters.
Table 2.
Association Results for Genome-wide-Significant Low-Frequency Variants from Discovery 25OHD Meta-analysis before and after Conditioning on the Lead Common CYP2R1 SNP, rs10741657, and the Lead Low-Frequency CYP2R1 Variant, rs117913124
| SNV | Chr | Position | EAa | EAFb | Candidate Gene | Function | Betac | p Value |
Conditional on rs10741657 |
Conditional on rs117913124 |
n | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Betac | p Value | Betac | p Value | ||||||||||
| rs117913124 | 11 | 14900931 | A | 0.025 | CYP2R1 | exon 4 (synonymous codon) | −0.43 | 1.5 × 10−88 | −0.39 | 2.4 × 10−78 | NA | NA | 41,336 |
| rs116970203 | 11 | 14876718 | A | 0.025 | CYP2R1d | intron 11 variant | −0.43 | 2.2 × 10−90 | −0.40 | 3.3 × 10−80 | NA | NA | 41,138 |
| rs117361591 | 11 | 14861957 | T | 0.014 | CYP2R1d | intron 11 variant | −0.44 | 9.1 × 10−51 | −0.40 | 2.2 × 10−44 | −0.05 | 0.017 | 38,286 |
| rs117621176 | 11 | 14861320 | G | 0.014 | CYP2R1d | intron 11 variant | −0.44 | 8.7 × 10−51 | −0.40 | 2.1 × 10−44 | −0.05 | 0.016 | 38,273 |
| rs142830933 | 11 | 14838760 | C | 0.014 | CYP2R1d | intron 5 variant | −0.44 | 1.4 × 10−48 | −0.40 | 1.7 × 10−42 | −0.05 | 0.03 | 37,541 |
| rs117672174 | 11 | 14746404 | T | 0.014 | CYP2R1d | intron 1 variant | −0.43 | 2.8 × 10−45 | −0.39 | 2.9 × 10−39 | −0.04 | 0.062 | 37,209 |
Abbreviations are as follows: Chr, chromosome; EA, effect allele; EAF, effect allele frequency; NA, not applicable; SNV, single-nucleotide variant.
Effect allele is the 25OHD decreasing allele.
Effect allele frequency.
Beta values represent changes in standard deviations of the standardized log-transformed 25OHD levels.
Nearest gene: PDE3B.
We used analyses of haplotype blocks for the candidate variants of interest by deriving phased haplotypes from 1,013 individuals from the TUK WGS cohort with a custom R package.
Effects on Vitamin D Insufficiency
To investigate the effect of genome-wide-significant SNVs on vitamin D insufficiency (defined as 25OHD levels below 50 nmol/L), we used data from four cohorts: TUK imputed, TUK WGS, BPROOF, and MrOS (n = 8,711). We performed logistic regression of this binary phenotype against the SNVs by adjusting for the following covariates: age, sex, BMI, and season of measurement. Meta-analysis of cohort-level summary statistics was performed in R19 with the epitools20 and metafor21 packages.
Interaction Analysis with Vitamin D Intake
We analyzed interactions between our candidate SNV(s) and vitamin D dietary intake (continuous and tertiles) in 9,224 individuals from five of the cohorts (FHS, PIVUS, ULSAM, BPROOF, and RSIII) participating in our discovery phase. A detailed description of the method for capturing vitamin D intake in each of the participating cohorts appears in Table S6. Linear regression was conducted in each of these studies under an additive genetic model. The following variables and co-variables were included in the model: log-transformed serum 25OHD as the dependent variable; SNV genotype (coded as 0, 1, or 2) as an independent variable; SNV (genotype) × dietary vitamin D intake (continuous or tertiles) as an interaction term; and age, sex, BMI, season of 25OHD measurement, dietary vitamin D intake (continuous or tertiles), supplemented vitamin D (yes or no), and total energy intake as covariates. The results from the five studies were meta-analyzed by a fixed-effects model with the metafor tool of the R statistical package.
Effects on Multiple Sclerosis
We tested the effect of the genome-wide-significant SNVs on the risk of multiple sclerosis in 5,927 case and 5,599 control samples by assuming an additive genetic model. Control samples were obtained from the UK Biobank22 by random selection of participants without multiple sclerosis. Case samples were obtained from the UK Biobank,22 previously published multiple sclerosis GWASs,23, 24 and newly genotyped UK subjects. Before genotype imputation of the genotyped case samples, we applied numerous QC criteria to ensure unbiased genotype calls between cohorts. These included retaining only SNVs with a MAF > 1% and excluding SNVs or samples with high missingness.25 Further, samples were assessed for population stratification with EIGENSTRAT,26, 27 and outliers were removed. Genotype data were then imputed by the Sanger Imputation Service15 with the combined UK10K and 1000 Genomes Phase 3 reference panels,9, 28 the same reference panel used for the UK Biobank control samples. Genotype data were phased with EAGLE229 and imputed with PBWT.30 SNPTEST31 was used for association testing on the combined case-control dataset, which included testing the additive effect of each allele on multiple sclerosis status and using the top ten principal components from EIGENSTRAT26, 27 to adjust for population stratification and batch effects.
Results
GWAS
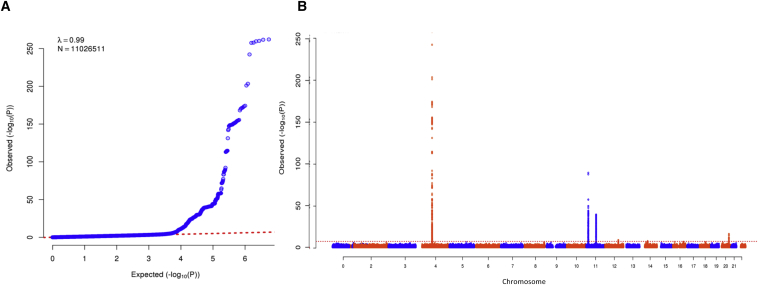
After strict QC, the genomic inflation factor for the meta-analysis of 19 GWASs was 0.99, suggesting a lack of bias due to population stratification (Figure 2). Through meta-analysis of 11,026,511 sequenced and imputed variants from our discovery cohorts (Table 1), we identified a signal at the chromosomal locus 11p.15.2, which harbors variants associated with 25OHD levels (lead low-frequency SNV g.14900931G>A [p.Asp120Asp] [rs117913124(A)] [GenBank: NC_000011.9]; MAF = 2.5%, allelic effect size = −0.43 SD of the standardized log-transformed 25OHD levels [SD], p = 1.5 × 10−88; Figure 3 and Table 2). The direction of effect was consistent across all discovery cohorts (Table 3 and Figure 3A), and the mean imputation information score for the imputed studies was 0.97. This low-frequency synonymous coding variant is in exon 4 of CYP2R1 and is ∼14 kb from the previously identified common CYP2R1 variant rs10741657 (r2 between these two SNVs = 0.03) (Figure 4). To our knowledge, rs117913124 has not previously been associated with any vitamin-D-related traits in humans.
Figure 2.
Discovery Single-Variant Meta-analysis
(A) Quantile-quantile plot for the single SNV meta-analysis.
(B) Manhattan plot of the meta-analysis depicts variants with MAF > 0.5% across the 22 autosomes against the −log10 p value from the meta-analysis of 19 cohorts, which included 42,274 individuals.
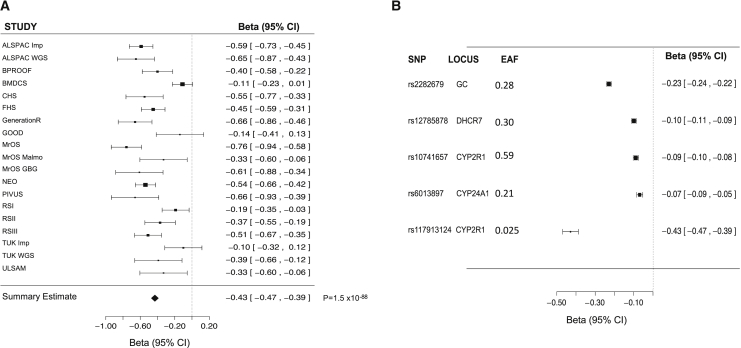
Figure 3.
Forest Plot by Cohort for rs117913124 and Forest Plot for rs117913124 and the Previously Described Common 25OHD-Related Variants from Discovery Meta-analysis
(A) Forest plot of estimates from all 19 studies for the low-frequency CYP2R1 variant rs117913124.
(B) Forest plot of the effect of the four common SUNLIGHT variants and the CYP2R1 low-frequency variant rs117913124 on log-transformed 25OHD levels.
Squares represent beta values in the 19 studies, and bars around the squares represent 95% confidence intervals (CIs).
Table 3.
Summary Statistics for the CYP2R1 Low-Frequency Variant, rs117913124, from 19 Studies
| Study | 25OHD Measurement Method | n | EAF (A Allelea) | Betab | Standard Error | p Value | Information Score |
|---|---|---|---|---|---|---|---|
| ALSPAC imputed | MS | 3,675 | 0.028 | −0.59 | 0.07 | 3.43 × 10−18 | 0.99 |
| ALSPAC WGS | MS | 1,606 | 0.028 | −0.65 | 0.11 | 8.23 × 10−10 | NA |
| BPROOF | MS | 2,512 | 0.027 | −0.4 | 0.09 | 4.99 × 10−6 | 0.97 |
| BMDCS | MS | 863 | 0.019 | −0.11 | 0.06 | 0.058 | 0.98 |
| CHS | MS | 1,581 | 0.022 | −0.55 | 0.11 | 5.15 × 10−7 | 0.88 |
| FHS | CLIA | 5,402 | 0.021 | −0.45 | 0.07 | 2.32 × 10−10 | 0.97 |
| GenerationR | MS | 1,442 | 0.033 | −0.66 | 0.1 | 1.78 × 10−6 | 1 |
| GOOD | CLIA | 921 | 0.028 | −0.14 | 0.14 | 0.31 | 0.96 |
| MrOS | MS | 3,265 | 0.018 | −0.76 | 0.09 | 5.63 × 10−16 | 0.96 |
| MrOS Malmo | CLIA | 893 | 0.033 | −0.33 | 0.14 | 0.016 | 0.94 |
| MrOS GBG | CLIA | 945 | 0.026 | −0.61 | 0.14 | 7.87 × 10−6 | 1 |
| NEO | MS | 5,727 | 0.025 | −0.54 | 0.06 | 2.73 × 10−19 | 1 |
| PIVUS | CLIA | 943 | 0.028 | −0.66 | 0.14 | 2.56 × 10−6 | 0.99 |
| RSI | ECLIA | 3,320 | 0.025 | −0.19 | 0.08 | 0.019 | 0.98 |
| RSII | ECLIA | 2,022 | 0.033 | −0.37 | 0.09 | 2.38 × 10−5 | 0.99 |
| RSIII | ECLIA | 2,913 | 0.027 | −0.51 | 0.08 | 4.61 × 10−10 | 0.98 |
| TUK imputed | CLIA | 1,919 | 0.021 | −0.1 | 0.11 | 0.35 | 0.98 |
| TUK WGS | CLIA | 1,013 | 0.025 | −0.39 | 0.14 | 0.006 | NA |
| ULSAM | MS | 1,095 | 0.025 | −0.33 | 0.14 | 0.02 | 1 |
Abbreviations are as follows: CLIA, chemiluminescence immunoassay; EAF, effect allele frequency; ECLIA, electrochemiluminescence immunoassay; MS, mass spectrometry; NA, not applicable; 25OHD, 25-hydroxyvitamin D.
Effect allele is the 25OHD decreasing allele.
Beta values represent changes in standard deviations of the standardized log-transformed 25OHD levels.
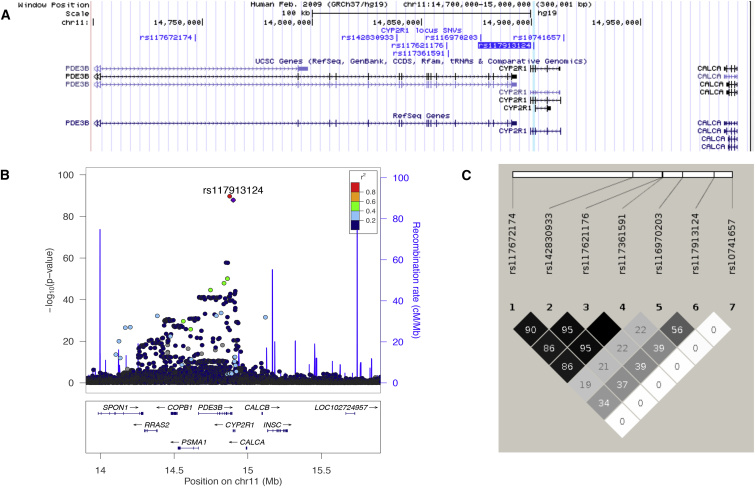
Figure 4.
Association Signals from 11p.15.2
(A) UCSC Genome Browser snapshot including the top low-frequency SNVs (see Table 2) and the lead common variant rs10741657 in CYP2R1. The position of rs117913124 is highlighted in light blue.
(B) Regional disequilibrium plot showing rs117913124 (purple dot), its perfect proxy rs11670203 (red dot), and the other genome-wide-significant SNVs in the same locus (blue and green dots). The plot depicts SNVs within 1 Mb of a locus’s lead SNV (x axis) and their associated meta-analysis p value (−log10) (see Table S10 for more details). SNVs are color coded according to r2 with the lead SNV (labeled; r2 was calculated from the UK10K WGS dataset). The recombination rate (blue line), position of genes and their exons, and direction of transcription are also displayed (below plot).
(C) Linkage-disequilibrium plot indicating the r2 values between the SNVs of Table 2 (top low-frequency variants) and between these low-frequency SNVs and the lead common variant (rs107416570) at the same CYP2R1 locus (r2 calculated from the 1000 Genomes dataset).
Figure S1 shows a comparison of the average 25OHD levels, adjusted for age, sex, BMI, and season of measurement, in non-carriers and heterozygous carriers of the A allele of rs117913124 in the TUK WGS cohort. The average 25OHD levels, adjusted for age, sex, BMI, and season of measurement were computed in 542 individuals from the TUK WGS cohort, among which 510 were not carriers and 32 were heterozygous carriers of the A allele of rs117913124 (no homozygous carriers were present in this cohort). After removing outliers (adjusted 25OHD levels ±3 SD from the mean), we included in our analysis 449 non-carriers and 30 heterozygous carriers (for a total of 479 individuals). A linear-regression model with the adjusted 25OHD levels as the dependent variable and the dose of the A allele of rs117913124 (numeric factor 1 or 0) as the independent variable demonstrated an 8.3 nmol/L decrease in the adjusted 25OHD levels per A allele. The mean adjusted 25OHD levels were 64.3 nmol/L in non-carriers and 56.0 nmol/L in heterozygous carriers.
Two-way conditional analysis between the CYP2R1 common (rs10741657) and low-frequency (rs117913124) variants revealed that the two association signals are largely independent. Specifically, after conditioning on rs10741657, rs117913124 remained strongly associated with 25OHD levels (pcond = 2.4 × 10−78); after conditioning on rs11791324, the effect of rs10741657 on 25OHD levels remained significant (pcond = 4.0 × 10−33 versus ppre-cond = 8.8 × 10−45; Tables 2 and S5). Further, no other low-frequency variant in the region remained significant after conditioning on rs117913124 (Table 2). To further disentangle the role of rs117913124 from that of rs10741657 on 25OHD levels, we undertook a haplotype analysis based on WGS data from 3,781 individuals from the TUK WGS and ALSPAC WGS cohorts. We found that the 25OHD decreasing A allele of rs117913124 was always transmitted in the same haplotype block with the 25OHD decreasing G allele of the common CYP2R1 variant rs10741657. By using 25OHD data from the TUK WGS cohort, we compared the 25OHD levels among carriers of the various haplotype blocks. We observed lower levels of 25OHD in carriers of the A allele of rs117913124 than in non-carriers, independently of the presence of the effect allele G of the common CYP2R1 variant (Table 4).
Table 4.
Effect of Different Haplotype Combinations of the Low-Frequency (rs117913124) and Common (rs10741657) CYP2R1 Variants on 25OHD Levels
| Haplotypea | Betab | p Value | n |
|---|---|---|---|
| GA GA | −0.02 | 0.79 | 156 |
| AG GA | −0.49 | 0.02 | 23 |
| AG GG | −0.3 | 0.13 | 27 |
| GA GG | 0.01 | 0.87 | 477 |
| GG GG | 0.05 | 0.58 | 330 |
Results are based on individuals from the TUK WGS cohort.
The first allele in each chromatid corresponds to the low-frequency variant rs117913124; the second allele corresponds to the common variant rs10741657. The two AG blocks contain the 25OHD decreasing allele (A) of the low-frequency variant, which is always inherited with the 25OHD decreasing allele (G) of the common variant.
Beta values represent changes in standard deviations of the standardized log-transformed 25OHD levels.
No other low-frequency or rare variants were identified in the three previously described vitamin-D-related loci at DHCR7, GC, and CYP24A1. The mean effect size of the four previously reported common (MAF ≥ 5%) genome-wide-significant SNVs from the SUNLIGHT consortium was −0.13 SD, and the largest effect size was −0.25 SD (for the GC variant) in our meta-analysis (Table S3 and Figure 3B). The effect size of rs10741657(G), the known common CYP2R1 variant, was −0.09 SD. Hence, the observed effect size of rs117913124 is 3-fold larger than the above mean, 4-fold larger than that of the common CYP2R1 variant, and almost twice that of the largest previously reported effect of the GC variant. Last, the percentage of the 25OHD phenotype variance explained by the low-frequency CYP2R1 variant (0.9%) was more than double the percentage of the variance explained by the CYP2R1 common variant (0.4%).
We also identified 18 genome-wide-significant low-frequency and rare SNVs on the same chromosome 11 region as rs117914124 in the neighboring PDE3B (MIM: 602047) (Tables 2 and S4 and Figure 4B). Signals from these SNVs in PDE3B were independent of the common variant at CYP2R1 (Table 2). We then created haplotype blocks with rs117913124 and SNVs at PDE3B on the basis of haplotype information from the 3,781 individuals from the TUK WGS and ALSPAC WGS cohorts (Table S2). We found that the 25OHD decreasing allele (A) of rs117913124 was always inherited with the 25OHD decreasing allele (A) of its perfect proxy rs116970203 (r2 = 1). Therefore, rs116970203 is not likely to have a distinct effect from that of rs117913124 on 25OHD levels. On the other hand, the 25OHD decreasing alleles of the remaining four low-frequency variants (all with a MAF of approximately 1.4%) were not always inherited in the same haplotype block as rs117913124 and rs116970203 and were in moderate linkage disequilibrium with rs117913124 (all r2 < 0.6; Figures 4B and 4C). Each of the four alleles was in almost perfect linkage disequilibrium with the remaining three (all r2 > 0.96). This implies that these four SNVs might influence 25OHD levels independently of rs117913124. Nevertheless, as mentioned above, after conditioning on the lead low-frequency CYP2R1 SNV rs117913124, the p values of the four PDE3B SNVs became non-significant and their beta values decreased substantially (Table 2), demonstrating that they probably do not represent an independent signal at the chromosome 11 locus.
rs117913124 and Risk of Vitamin D Insufficiency
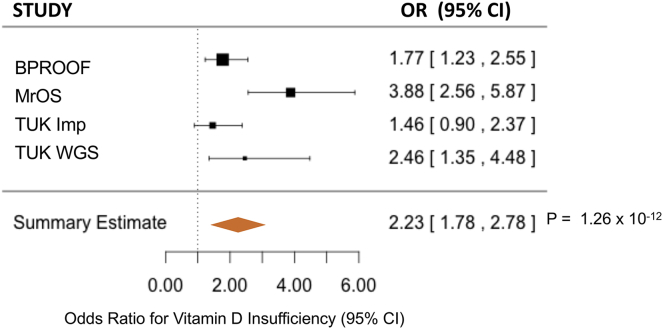
To further investigate the clinical significance of the low-frequency CYP2R1 variant rs117913124, we tested its effect on a binary outcome for vitamin D insufficiency (defined as 25OHD levels < 50 nmol/L) in 8,711 individuals from four studies (TUK WGS, TUK IMP, BPROOF, and MrOS). rs117913124 was strongly associated with an increased risk of vitamin D insufficiency (odds ratio [OR] = 2.20, 95% confidence interval [CI] = 1.8–2.8, p = 1.2 × 10−12) (Figure 5) after control for relevant covariates as described in the Material and Methods.
Figure 5.
Effect of rs117913124 on Vitamin D Insufficiency
Forest plot of the effect of the low-frequency CYP2R1 variant rs117913124 on vitamin D insufficiency in four studies. Squares represent odds ratios for vitamin D insufficiency in the four studies, and bars represent 95% CIs.
Common 25OHD-Associated SNVs
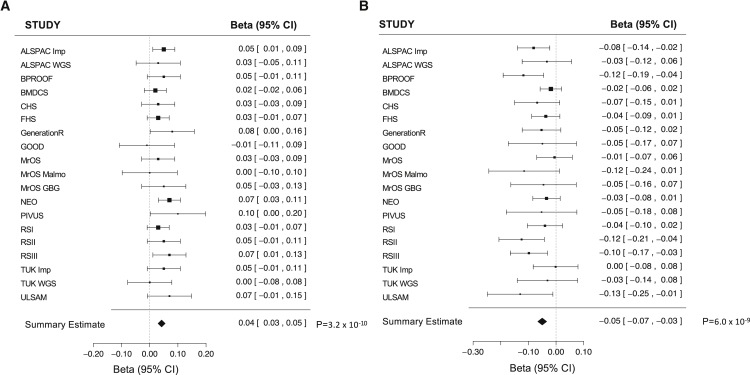
We report two additional loci associated with 25OHD levels (Table 5). Variants leading these associations were common and exerted a rather small effect on 25OHD: (1) a variant in chromosome 12 (rs3819817[C], intronic to HAL [MIM: 609457]) with a MAF of 45%, a beta value of 0.04, and a p value of 3.2 × 10−10; and (2) a variant in chromosome 14 (rs2277458[G], intronic to GEMIN2 [MIM: 602595]) with a MAF of 21%, a beta value of −0.05, and a p value of 6.0 × 10−9. Both variants were present in all 19 studies, and the direction of the effect was the same among the 19 studies (Figure 6). Neither the HAL nor the GEMIN2 locus is previously known to be associated with 25OHD levels. Of note, neither variant was present in the HapMap imputation reference used in the SUNLIGHT study.
Table 5.
Main Findings of the GWAS Meta-analysis
| SNP | Chr | Candidate Gene | EA | EAF | Betaa | p Value | n |
|---|---|---|---|---|---|---|---|
| rs117913124 | 11 | CYP2R1 | A | 0.025 | −0.43 | 1.5 × 10−88 | 41,336 |
| rs3819817 | 12 | HAL | C | 0.45 | 0.04 | 3.2 × 10−10 | 41,071 |
| rs2277458 | 14 | GEMIN2 | G | 0.21 | −0.05 | 6.0 × 10−9 | 39,746 |
Abbreviations are as follows: Chr, chromosome; EA, effect allele; EAF, effect allele frequency; SNP, single-nucleotide polymorphism.
Beta values represent changes in standard deviations of the standardized log-transformed 25OHD levels while controlling for age, sex, BMI, and season of measurement.
Figure 6.
Association Signals from Chromosomes 12 and 14
Forest plots with (A) estimates for the chromosome 12 common variant rs3819817 and (B) estimates for the chromosome 14 common variant rs2277458 from all 19 studies of the meta-analysis where both variants were present. Squares represent beta values in the 19 studies, and bars around the squares represent 95% CIs.
Interaction Analysis
CYP2R1 encodes the enzyme responsible for 25-hydroxylation of vitamin D in the liver,32 a necessary step in the conversion of dietary vitamin D and vitamin D oral supplements to the active metabolite, 1,25 dihydroxy-vitamin D. Therefore, we hypothesized that, in contrast with non-carriers, individuals heterozygous or homozygous for rs117913124 in CYP2R1 would not show a response in their 25OHD levels to vitamin D intake. In other words, we expected carriers of the effect allele of rs117913124 to have steadily lower 25OHD levels, independently of their vitamin D intake. To investigate this hypothesis, we tested the presence of interaction between rs117913124 and vitamin D dietary intake (continuous values and tertiles) on 25OHD levels in 9,224 individuals from five studies (Figure S2). We found no interaction between rs117913124 and dietary vitamin D intake (beta value = −0.0002 and interaction p value = 0.41 for continuous vitamin D intake; beta value = 0.012 and p value = 0.60 for tertiles of vitamin D intake). Given that the two common 25OHD-associated SNVs are located in genes (HAL and GEMIN2) with no known role in the processing of dietary vitamin D, we found no biological rationale for undertaking a gene-diet interaction analysis for these variants.
25OHD-Assosiated Variants and Risk of Multiple Sclerosis
We tested whether the CYP2R1 low-frequency variant rs117913124 and the common variants rsrs3819817 and rs2277458 in HAL and GEMIN2, respectively, influence the risk of multiple sclerosis. In 5,927 multiple sclerosis samples and 5,599 control samples, we found that the 25OHD decreasing allele at rs117913124[A] was associated with increased odds of multiple sclerosis (OR = 1.40; 95% CI = 1.19–1.64; p value = 2.6 × 10−5). By way of comparison, the OR of multiple sclerosis for the common CYP2R1 variant was 1.03 (95% CI = 0.97–1.08; p value = 0.03) in the same study and has previously been reported to be 1.05 (95% CI = 1.02–1.09; p value 0.004) in a separate study.33 Thus, the effect per allele of rs117913124 on multiple sclerosis was 12.4-fold larger than that attributed to the already known common variant at CYP2R1. With regard to the two common SNVs, the 25OHD decreasing allele (T) at the HAL variant rs3819817 was not clearly associated with risk of multiple sclerosis; however, there was a trend in the expected direction: OR = 1.05 (95% CI = 1.00–1.11; p value = 0.07). We found no association between the 25OHD decreasing allele (G) at the GEMIN2 variant rs2277458 and risk of multiple sclerosis: OR = 1.03 (95% CI = 0.96–1.11; p value = 0.34).
Discussion
In the largest GWAS meta-analysis of 25OHD levels in European populations to date, we have identified a low-frequency, synonymous coding genetic variant that has a large effect and strongly associates with 25OHD levels. This variant has an effect size 4-fold larger than that described for the common variant in the same gene (CYP2R1) and is associated with a 2-fold increase in risk of vitamin D insufficiency and a 40% increase in the odds of developing multiple sclerosis. The biological plausibility of these findings is supported by the fact that the low-frequency variant is located in CYP2R1, encoding the major hepatic 25-hydroxylase for vitamin D.32 These findings are of clinical relevance given that 5% of the general European population carries this variant in either the homozygous or heterozygous state, and it is associated with a clinically relevant increase in the risk of multiple sclerosis.
Our study was enabled by large imputation reference panels (UK10K-1000 Genomes and HRC) that offer at least 10-fold more European samples than the 1000 Genomes reference panel alone. We did not identify genome-wide-significant variants with a large effect on 25OHD in novel genes in Europeans, although we did find variants with smaller effects in two loci not previously known to be associated with 25OHD. We also identified in a known vitamin-D-related gene low-frequency variants with much larger effects than those of the previously described common variants.
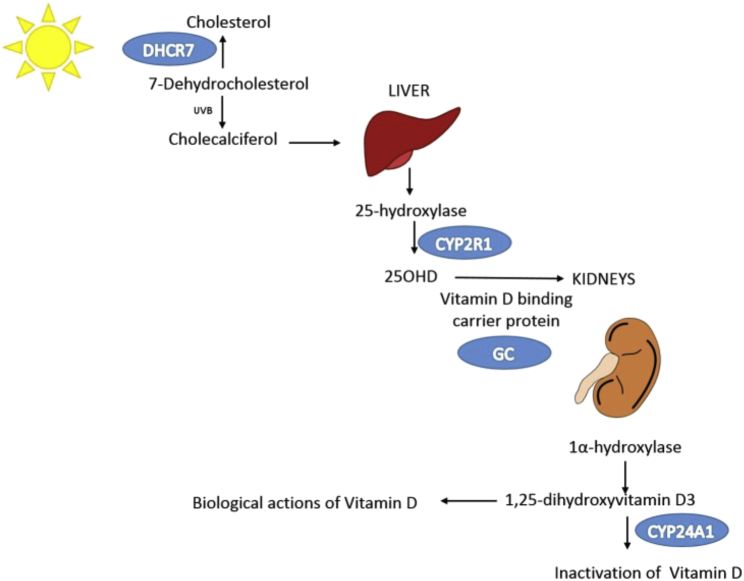
CYP2R1 encodes the enzyme that is responsible for 25-hydroxylation of vitamin D and is one of the two main enzymes responsible for vitamin D hepatic metabolism32 (Figure 7). Rare mutations in CYP2R1 have already been described to cause rickets (MIM: 27744).32, 34 Given the important role of CYP2R1 in the conversion of dietary vitamin D and vitamin D oral supplements to the active form of vitamin D, we hypothesized that carriers of the low-frequency CYP2R1 variant might respond poorly to vitamin D replacement therapy. We tested this hypothesis by undertaking an interaction analysis between the CYP2R1 low-frequency variant and dietary vitamin D intake, which showed no clear interaction. However, we note that studies of gene-environment interactions are generally underpowered, measurement error in dietary data is common, and this interaction was further limited by time differences between assessment of dietary intake and measurement of 25OHD levels. Therefore, whether this genetic variant influences 25OHD response to vitamin D administration requires further study.
Figure 7.
Schematic of the Vitamin D Metabolic Pathway
UVB, ultraviolet B rays.
Although the aim of the present study was to describe variants of low MAF and large effect on 25OHD, we report two common chromosome 12 (HAL) and 14 (GEMIN2) variants that have a small effect size and reached genome-wide significance in our meta-analysis. Although no existing evidence implicates GEMIN2 in vitamin-D-related physiological pathways, HAL is expressed in the skin and is involved in the formation of urocanic acid, a “natural sunscreen.”35, 36 Thus, this could constitute a plausible pathophysiologic mechanism implicating HAL in vitamin D synthesis in the skin. Additional functional follow-up of the signals in chromosomes 12 and 14 is needed to characterize the genes and/or mechanisms underlying these associations.
Our findings could have clinical relevance for several reasons. First, individuals carrying at least one copy of the low-frequency CYP2R1 variant have lower levels of 25OHD by a clinically relevant degree. Specifically, the risk of vitamin D insufficiency is doubled in these individuals. Second, their risk of multiple sclerosis is also increased in accordance with previous evidence supporting a causal role for vitamin D in the risk of multiple sclerosis.10 Third, these findings affect ∼5% of individuals of European descent. Fourth and finally, rs117913124 could be used along with the previously identified common vitamin-D-related variants as an additional genetic predictor of low 25OHD levels in Mendelian randomization studies investigating the causal role of low vitamin D levels in human disease.
Our study also has its limitations. First, although the scope of our study was detection of low-frequency and rare variants, we opted to include in our meta-analysis two WGS studies with a relatively low read depth of 6.7×, as well as three studies imputed to older imputation panels (1000 Genomes and UK10K). These studies have a limited capacity to capture very rare variants, which might explain why we failed to identify such associations. In addition to the limitations arising from the time difference between assessment of dietary vitamin D intake and 25OHD measurements, the analysis of the gene-diet interaction, as mentioned above, might have lacked statistical power. Because our analysis was restricted to populations of European ancestry, we cannot make any assumptions concerning the effect of rs117913124 in non-European populations. Nonetheless, according to the 1000 Genomes reference, this variant is rare in Africans (MAF = 0.3%) and has not been described in East Asians (MAF = 0%). Therefore, describing with any certainty the effect of this variant on 25OHD levels in these populations will require large sample sizes of these populations. Finally, in the absence of functional experiments showing the exact function of rs117913124 in CYP2R1 and given that this synonymous polymorphism does not affect protein sequence, we cannot unequivocally confirm that this low-frequency variant is causal; however, given that this is a coding variant in a well-documented 25OHD-associated gene, it seems likely that it exerts its effect on CYP2R1.
In conclusion, our findings demonstrate the utility of WGS-based discovery and deep imputation for enabling the characterization of genetic associations, offering an improved understanding of the pathophysiology of vitamin D, providing an enriched set of genetic predictors of 25OHD levels for future study, and enabling the identification of groups at increased risk for vitamin D insufficiency and multiple sclerosis.
Published: July 27, 2017
Footnotes
Supplemental Data include 2 figures, 10 tables, and Supplemental Acknowledgments and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.06.014.
Accession Numbers
The GWAS summary statistics reported in this paper have been deposited in the Genome-wide Repository of Associations between SNPs and Phenotypes (GRASP).
Web Resources
GRASP: Genome-wide Repository of Associations between SNPs and Phenotypes, https://grasp.nhlbi.nih.gov/Overview.aspx
OMIM, http://www.omim.org
UCSC Genome Browser, https://genome.ucsc.edu/
UK10K, http://www.uk10k.org
VQSLOD, http://www.broadinstitute.org/gsa/wiki/index.php/Variant_quality_score_recalibration
Supplemental Data
Highlighted are the 19 low-frequency and rare variants of large effect on 25OHD levels in the same chromosome 11 locus.
MAF, minor allele frequency.
NRD, non-reference discordance rate.
Highlighted is the lead SNV rs117913124.
References
- 1.Forrest K.Y., Stuhldreher W.L. Prevalence and correlates of vitamin D deficiency in US adults. Nutr. Res. 2011;31:48–54. doi: 10.1016/j.nutres.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Rosen C.J., Adams J.S., Bikle D.D., Black D.M., Demay M.B., Manson J.E., Murad M.H., Kovacs C.S. The nonskeletal effects of vitamin D: an Endocrine Society scientific statement. Endocr. Rev. 2012;33:456–492. doi: 10.1210/er.2012-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shea M.K., Benjamin E.J., Dupuis J., Massaro J.M., Jacques P.F., D’Agostino R.B., Sr., Ordovas J.M., O’Donnell C.J., Dawson-Hughes B., Vasan R.S., Booth S.L. Genetic and non-genetic correlates of vitamins K and D. Eur. J. Clin. Nutr. 2009;63:458–464. doi: 10.1038/sj.ejcn.1602959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Livshits G., Karasik D., Seibel M.J. Statistical genetic analysis of plasma levels of vitamin D: familial study. Ann. Hum. Genet. 1999;63:429–439. doi: 10.1046/j.1469-1809.1999.6350429.x. [DOI] [PubMed] [Google Scholar]
- 5.Wang T.J., Zhang F., Richards J.B., Kestenbaum B., van Meurs J.B., Berry D., Kiel D.P., Streeten E.A., Ohlsson C., Koller D.L. Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet. 2010;376:180–188. doi: 10.1016/S0140-6736(10)60588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sidore C., Busonero F., Maschio A., Porcu E., Naitza S., Zoledziewska M., Mulas A., Pistis G., Steri M., Danjou F. Genome sequencing elucidates Sardinian genetic architecture and augments association analyses for lipid and blood inflammatory markers. Nat. Genet. 2015;47:1272–1281. doi: 10.1038/ng.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng H.F., Forgetta V., Hsu Y.H., Estrada K., Rosello-Diez A., Leo P.J., Dahia C.L., Park-Min K.H., Tobias J.H., Kooperberg C., AOGC Consortium. UK10K Consortium Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature. 2015;526:112–117. doi: 10.1038/nature14878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen J.C., Kiss R.S., Pertsemlidis A., Marcel Y.L., McPherson R., Hobbs H.H. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 9.Huang J., Howie B., McCarthy S., Memari Y., Walter K., Min J.L., Danecek P., Malerba G., Trabetti E., Zheng H.F., UK10K Consortium Improved imputation of low-frequency and rare variants using the UK10K haplotype reference panel. Nat. Commun. 2015;6:8111. doi: 10.1038/ncomms9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mokry L.E., Ross S., Ahmad O.S., Forgetta V., Smith G.D., Goltzman D., Leong A., Greenwood C.M., Thanassoulis G., Richards J.B. Vitamin D and Risk of Multiple Sclerosis: A Mendelian Randomization Study. PLoS Med. 2015;12:e1001866. doi: 10.1371/journal.pmed.1001866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walter K., Min J.L., Huang J., Crooks L., Memari Y., McCarthy S., Perry J.R., Xu C., Futema M., Lawson D., UK10K Consortium The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCarthy S., Das S., Kretzschmar W., Delaneau O., Wood A.R., Teumer A., Kang H.M., Fuchsberger C., Danecek P., Sharp K., Haplotype Reference Consortium A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mägi R., Morris A.P. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics. 2010;11:288. doi: 10.1186/1471-2105-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu C., Tachmazidou I., Walter K., Ciampi A., Zeggini E., Greenwood C.M., UK10K Consortium Estimating genome-wide significance for whole-genome sequencing studies. Genet. Epidemiol. 2014;38:281–290. doi: 10.1002/gepi.21797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Team R.C. R Foundation for Statistical Computing; 2013. R: A language and environment for statistical computing. [Google Scholar]
- 20.Aragon, T.J., Wollschlaeger, D., and Omidpanah, A. (2017). epitools: Epidemiology Tools. https://cran.r-project.org/package=epitools.
- 21.Viechtbauer W. Conducting meta-analyses in R with the metafor package. J. Stat. Softw. 2010;36:1–48. [Google Scholar]
- 22.Sudlow C., Gallacher J., Allen N., Beral V., Burton P., Danesh J., Downey P., Elliott P., Green J., Landray M. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hafler D.A., Compston A., Sawcer S., Lander E.S., Daly M.J., De Jager P.L., de Bakker P.I., Gabriel S.B., Mirel D.B., Ivinson A.J., International Multiple Sclerosis Genetics Consortium Risk alleles for multiple sclerosis identified by a genomewide study. N. Engl. J. Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 24.Australia and New Zealand Multiple Sclerosis Genetics Consortium (ANZgene) Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat. Genet. 2009;41:824–828. doi: 10.1038/ng.396. [DOI] [PubMed] [Google Scholar]
- 25.Anderson C.A., Pettersson F.H., Clarke G.M., Cardon L.R., Morris A.P., Zondervan K.T. Data quality control in genetic case-control association studies. Nat. Protoc. 2010;5:1564–1573. doi: 10.1038/nprot.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 27.Patterson N., Price A.L., Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loh P.R., Danecek P., Palamara P.F., Fuchsberger C., A Reshef Y., K Finucane H., Schoenherr S., Forer L., McCarthy S., Abecasis G.R. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet. 2016;48:1443–1448. doi: 10.1038/ng.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Durbin R. Efficient haplotype matching and storage using the positional Burrows-Wheeler transform (PBWT) Bioinformatics. 2014;30:1266–1272. doi: 10.1093/bioinformatics/btu014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchini J., Howie B., Myers S., McVean G., Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 32.Cheng J.B., Levine M.A., Bell N.H., Mangelsdorf D.J., Russell D.W. Genetic evidence that the human CYP2R1 enzyme is a key vitamin D 25-hydroxylase. Proc. Natl. Acad. Sci. USA. 2004;101:7711–7715. doi: 10.1073/pnas.0402490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beecham A.H., Patsopoulos N.A., Xifara D.K., Davis M.F., Kemppinen A., Cotsapas C., Shah T.S., Spencer C., Booth D., Goris A., International Multiple Sclerosis Genetics Consortium (IMSGC) Wellcome Trust Case Control Consortium 2 (WTCCC2) International IBD Genetics Consortium (IIBDGC) Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat. Genet. 2013;45:1353–1360. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Casella S.J., Reiner B.J., Chen T.C., Holick M.F., Harrison H.E. A possible genetic defect in 25-hydroxylation as a cause of rickets. J. Pediatr. 1994;124:929–932. doi: 10.1016/s0022-3476(05)83184-1. [DOI] [PubMed] [Google Scholar]
- 35.Barresi C., Stremnitzer C., Mlitz V., Kezic S., Kammeyer A., Ghannadan M., Posa-Markaryan K., Selden C., Tschachler E., Eckhart L. Increased sensitivity of histidinemic mice to UVB radiation suggests a crucial role of endogenous urocanic acid in photoprotection. J. Invest. Dermatol. 2011;131:188–194. doi: 10.1038/jid.2010.231. [DOI] [PubMed] [Google Scholar]
- 36.Suchi M., Sano H., Mizuno H., Wada Y. Molecular cloning and structural characterization of the human histidase gene (HAL) Genomics. 1995;29:98–104. doi: 10.1006/geno.1995.1219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Highlighted are the 19 low-frequency and rare variants of large effect on 25OHD levels in the same chromosome 11 locus.
MAF, minor allele frequency.
NRD, non-reference discordance rate.
Highlighted is the lead SNV rs117913124.