A synergistic application of holography and deep learning enables rapid optical screening of anthrax spores and other pathogens.
Abstract
Establishing early warning systems for anthrax attacks is crucial in biodefense. Despite numerous studies for decades, the limited sensitivity of conventional biochemical methods essentially requires preprocessing steps and thus has limitations to be used in realistic settings of biological warfare. We present an optical method for rapid and label-free screening of Bacillus anthracis spores through the synergistic application of holographic microscopy and deep learning. A deep convolutional neural network is designed to classify holographic images of unlabeled living cells. After training, the network outperforms previous techniques in all accuracy measures, achieving single-spore sensitivity and subgenus specificity. The unique “representation learning” capability of deep learning enables direct training from raw images instead of manually extracted features. The method automatically recognizes key biological traits encoded in the images and exploits them as fingerprints. This remarkable learning ability makes the proposed method readily applicable to classifying various single cells in addition to B. anthracis, as demonstrated for the diagnosis of Listeria monocytogenes, without any modification. We believe that our strategy will make holographic microscopy more accessible to medical doctors and biomedical scientists for easy, rapid, and accurate point-of-care diagnosis of pathogens.
INTRODUCTION
Bacillus anthracis, a gram-positive spore-forming bacterium causing the disease anthrax, is one of the most destructive biological weapons, which is prone to be abused for bioterrorism (1). It is thus crucial to rapidly detect and identify anthrax spores for biodefense (2). Various biological, chemical, and optical fingerprinting methods have been studied to accelerate diagnosis of B. anthracis (3–5). Conventional culture-based methods take days and are often inaccurate. Polymerase chain reaction–based methods provide species-level specificity but still take hours and require heavy instrumentation with skilled personnel to operate the system (4). Photoluminescence and surface-enhanced Raman scattering methods take only minutes but require labeling with exogenous agents and cannot discriminate B. anthracis from other Bacillus species that are ubiquitous in nature (5). Most of these methods are limited by detection sensitivities that require minimum sample sizes of at least thousands of bacterial cells; thus, their applications in practical settings such as aerosolized spores require sample amplification processes that significantly limit the detection speed.
Recent developments of optical methods based on holographic microscopy combined with machine learning, which enables rapid and label-free identification of single cells, can be an important step to address the anthrax issue (6–18). These techniques were pioneered by Javidi’s group (6–14) and developed further by several groups (15–18). Holographic microscopy (19), or quantitative phase imaging (QPI) in a broader sense, measures optical field images (that is, nanometer-scale distortions of wavefronts passing through a sample) using laser-based interferometry. In addition to the amplitude images available from conventional intensity-based microscopy techniques, holographic microscopy quantitatively measures the optical phase delay maps dictated by the refractive index (RI) distribution of a sample (19). Because the endogenous RI distribution in a cell is strongly related to the structural and biochemical characteristics (20) of the target classes (for example, species or cell types), the measured field images of single cells and the corresponding class labels are passed to data-driven machine learning algorithms for systematic discovery of class-specific fingerprints encoded in the images. These approaches can be combined with flow cytometry and/or bioaerosol collection systems to achieve ultrafast identification of unlabeled cells and pathogens (16, 17). However, none of these methods achieved subgenus specificity required for discriminating B. anthracis from other Bacillus species ubiquitous in nature.
Here, we present a next-generation holographic screening method by adopting “deep learning,” a state-of-the-art machine learning technique based on deep multilayered neural networks (21, 22), to holographic microscopy. We designed a deep convolutional neural network (CNN), HoloConvNet, specialized in the classification of holographic images of living cells. After training with quantitative phase images of individual Bacillus spores, the network identified new anthrax spores with single-spore sensitivity and subgenus specificity. Its remarkable learning ability enables direct training from raw images by automatically recognizing key biological traits encoded in the images, including the dry mass of individual bacteria, and presents outstanding accuracy that outperforms previous approaches in all accuracy measures. As demonstrated below, this method is readily applicable to classification of various single cells, in addition to B. anthracis, without any modification.
RESULTS
The overall framework of our method is shown in Fig. 1. We used QPI unit (QPIU), a cost-effective palm-sized module that converts a conventional microscope into a holographic microscope (23), for phase imaging of individual Bacillus spores in an isolated biosafety level 3 (BSL-3) laboratory at the Agency for Defense Development, Korea. It is attached to the output port of an existing bright-field microscope to form a common-path interferometry for optical field imaging (Fig. 1, A to C). After imaging B. anthracis and four different Bacillus species with various levels of phylogenetic relatedness (see Materials and Methods), we trained our deep neural network named HoloConvNet as a species classifier using the phase images of individual spores and the corresponding species labels (training set). The learnable parameters of the deep neural network were iteratively adjusted by the error backpropagation algorithm (Fig. 1D) (21, 22). The performance of the trained HoloConvNet was tested by taking new images (test set), which were never seen before by the network, as the input to the network (Fig. 1E). The machine-predicted species labels were compared with the true classes to estimate identification accuracy.
Fig. 1. Holographic deep learning framework for screening of anthrax spores.
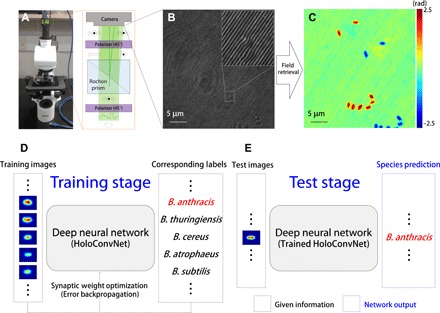
(A) Schematic diagram of QPIU for holographic imaging of individual Bacillus spores. (B) Interferogram formed by spatial modulation. It encodes quantitative phase images of individual spores, as shown in (C). (D) The measured phase images from multiple Bacillus species are used to train a deep neural network using the error backpropagation algorithm. (E) The trained network accurately predicts the corresponding species when independently measured phase images are shown.
The following five Bacillus species were used in this study. (i) B. anthracis Sterne is an attenuated strain of B. anthracis (24). The Sterne strain has the pXO1 plasmid encoding anthrax toxins. However, it lacks the pXO2 plasmid that encodes the polysaccharide capsule, whose role is to defend against phagocytosis by the immune cells in the vegetative state. Because we were interested in the sporulated state of B. anthracis, it was appropriate to choose the Sterne strain as a representative strain instead of fully virulent strains (for example, the Ames strain) for safety reasons. Although B. anthracis was the key target species in the present study, we selected other Bacillus species with various levels of relatedness to this species to assess the subgenus specificity of the proposed method (25). (ii) B. thuringiensis and (iii) B. cereus are the closest neighbors of B. anthracis (26). These three species form the so-called B. cereus group. Despite their genetic similarity, their pathogenic properties differ by species: the causative agent of anthrax, the source of powerful biological insecticides, and the cause of several food-borne illnesses, respectively. (iv) B. atrophaeus and (v) B. subtilis are the common simulants of B. anthracis (27). These two species are closely related to each other; however, they have some distance from the species mentioned above (28). These species have been the most prominent surrogate organisms for B. anthracis in biodefense programs. We also note that B. subtilis is one of the best-studied model microbes, which is considered the gram-positive counterpart of Escherichia coli. The detailed data on Bacillus spores are presented in table S1.
The quantitative nature of holographic microscopy captures subcellular phase delay distribution that could be exploited by machine learning algorithms to extract fingerprint information (15, 20). On the other hand, conventional techniques (for example, phase contrast microscopy) provide rough morphological information only (fig. S1). Simple morphological parameters such as spore size (fig. S2), which is consistent with previous reports (29, 30), are not enough for species discrimination due to high genetic similarities and large cell-to-cell variations (15, 23).
The endogenous RI distribution of Bacillus spores, which dictates the sample-induced phase delay imaged by QPIU, is strongly related to specific characteristics of each species (15, 20). However, because this relation is often indirect, it should be approximated using supervised learning. The precision of this function approximation obviously dominates the performance of the trained classifiers. Deep neural networks are universal approximators for virtually any arbitrarily nonlinear functions (22), whereas conventional machine learning techniques mostly rely on linear or only slightly nonlinear decision boundaries (15).
The network architecture of HoloConvNet is illustrated in Fig. 2 (and table S2). A phase image of a single spore is processed by multiple layers of convolution, nonlinearity, and pooling operations and then finally receives scored class labels through fully connected layers. The network makes its prediction by selecting the final-layer neuron with the strongest activation. The key functional block of this process is a convolutional layer followed by nonlinearity
| (1) |
where x and y are input and output vectors, respectively, and w and b are synaptic weights and biases, respectively. Equation 1 emulates integration of synaptic inputs by a biological neuron (21, 22) that fires only when the net input exceeds a certain threshold [more precisely, a population of neurons with an output firing rate modeled by a rectified linear unit (ReLU)]. Note that the entire processing by the network from images to class labels is a nonlinear mapping that corresponds to the approximating function explained above.
Fig. 2. Architecture of HoloConvNet.
When a phase image of an individual spore is taken as the input, the network first processes the images through three rounds of convolution, ReLU nonlinearity, and max pooling layers. Then, two fully connected (and ReLU) layers follow: (i) the last hidden layer under dropout regularization and (ii) the output layer with the class scores. These scores are used to calculate the loss function and to make species predictions in the training and test stages, respectively. Only 10 two-dimensional activation maps per layer are presented with layer-wise scaling for visualization (see table S2 for detailed architecture).
Training a deep neural network is essentially a large-scale nonlinear optimization of the synaptic weights (and biases) that govern the network behavior. The large number of the learnable parameters makes training process extremely difficult. However, CNNs such as HoloConvNet have markedly smaller number of parameters (21, 31) by using localized and shared receptive field structures inspired by physiological visual processing. Thus, the network can be trained using the error backpropagation algorithm that minimizes the mismatch between the machine-predicted and true labels (see Materials and Methods). HoloConvNet efficiently converges to a hierarchical representation of the images that gradually transforms the data space in which the classes are easily separable. This property is called the “representation learning” capability of deep learning (21) and enables direct training from raw images.
The performance of HoloConvNet is shown in Fig. 3 (and fig. S3 with different visualization). A well-trained neural network reflects the general relations between the input and output data so that it accurately predicts the class labels of new images (generalization property). First, the multiclass identification performance of the network for the five Bacillus species (B. anthracis, B. thuringiensis, B. cereus, B. atrophaeus, and B. subtilis), trained with five class labels representing individual species, is shown in Fig. 3A. HoloConvNet identifies B. anthracis spores from the other four species with high sensitivity and specificity (table S3), despite relatively less accurate classification between the other species that is irrelevant for the screening of anthrax spores.
Fig. 3. Performance of HoloConvNet.
(A to C) The test images are used to measure the performance of (A) multiclass classification of the five Bacillus species (B. anthracis, B. thuringiensis, B. cereus, B. atrophaeus, and B. subtilis), (B) binary classification of B. anthracis and the other four species (B. thuringiensis, B. cereus, B. atrophaeus, and B. subtilis), and (C) binary classification of B. anthracis and the two nonmember species of the B. cereus group (B. atrophaeus and B. subtilis). (D) The performance of the proposed method is compared to previous techniques (see the main text). Holographic microscopy and deep learning significantly improve the performance in all cases. (E to G) t-SNE visualization of the CNN codes at the last hidden layer, corresponding to the classification schemes of (A) to (C), which shows the representation learning capability of HoloConvNet. The error bars in (A) to (D) indicate the SD calculated from 10 classification models with different random initializations.
Because diagnosing anthrax spores from other species is our prime objective, the network was next trained with binary class labels (anthrax versus non-anthrax). With this method, the performance could be enhanced (Fig. 3B and table S4) by letting the optimization process focus on the characteristics distinguishing B. anthracis from others (B. thuringiensis, B. cereus, B. atrophaeus, and B. subtilis). When the problem was relaxed by excluding the two B. cereus group species (B. thuringiensis and B. subtilis), HoloConvNet achieved a remarkable accuracy of 96.3% (Fig. 3C and table S5). These results suggest the potential of deep learning–based holographic screening of anthrax spores in realistic settings.
In Fig. 3D, the performance of our method (“Holography + Deep”) was compared with the performance of several previous techniques: holographic microscopy with conventional machine learning (“Holography + Conventional”) (15), conventional microscopy with deep learning (“Conventional + Deep”) (training HoloConvNet with binary morphology images; see Materials and Methods), and conventional microscopy with conventional machine learning (“Conventional + Conventional”) (linear discriminant analysis with the morphological parameters in fig. S2). More extensive comparisons to various conventional classifiers, such as support vector machines, are presented in tables S6 and S7. HoloConvNet outperformed the previous methods in all performance measures with a substantial margin.
Representation learning by HoloConvNet, the fundamental improvement developed in this study, was further examined (Figs. 3, E to G, and 4). The network transforms the images into a representation in which the data points are linearly separable because a single layer of neurons is a linear classifier (22). We applied t-distributed stochastic neighbor embedding (t-SNE), a high-dimensional data visualization technique (32), to the activation of individual neurons in the last hidden layer (Fig. 3, E to G). The good separation observed indicates the great ability of HoloConvNet to learn the optimal representation of phase images without any predesigned features required by conventional machine learning techniques. The different degrees of separation in the three cases explain the different identification performance. Additionally, the relative distances between the species clusters shown in Fig. 3E are consistent with the phylogenetic relationship explained above; it should be noted here that the relationship was independently discovered by HoloConvNet through training.
Fig. 4. Representation learning by HoloConvNet: Dry mass as a key biological trait.
The interspecies difference in cellular dry mass is automatically recognized and used for screening of anthrax spores. (A to C) The activation of the “anthrax neuron” at the output layer shows a strong correlation with dry mass. a.u., arbitrary units. (D) Dry mass of individual Bacillus spores calculated from the quantitative phase images. (E) Computationally disabling the dry mass information significantly impairs the performance of HoloConvNet. Dry mass alone is not enough for full performance as well. Data in (D) are presented as box-and-whisker plots displaying median and interquartile ranges. The error bars in (E) indicate the SD calculated from 10 classification models with different random initializations.
The outstanding performance of the proposed method raises a question: What are the key biological traits that are measured and exploited for the identification of anthrax spores? We speculated that cellular dry mass (33), the mass of nonaqueous cellular components, is one of the most important traits. This hypothesis is based on the domain knowledge that there exists an additional outermost structure, called the exosporium, in the B. cereus group spores but not in the remaining two species (34). It was reasoned that structural distinction might result in an interspecies difference of dry mass that is inherently measured by holographic imaging with femtogram-level sensitivity (see Materials and Methods). A strong positive correlation was found between dry mass and activation of the “anthrax neuron” at the output layer (Fig. 4, A to C). This observation makes sense if the mean dry mass of B. anthracis is the heaviest among the five species, which turns out to be true (Fig. 4D). As expected, B. anthracis is slightly heavier than the other two B. cereus group species, and the remaining two species lacking exosporium have considerably lighter dry mass. The subtle difference within the B. cereus group might be due to species-dependent compositions and nanostructures of exosporium (35), although their contribution to dry mass should be confirmed by additional investigations. It was noted that the same order relation of dry mass was observed in all independent measurements (fig. S4), and the overall range of measured dry mass is consistent with previous studies (23, 36).
To confirm the causality between dry mass and species prediction by HoloConvNet, we used a computational disabling strategy. Detrimental effects on the performance were observed by computationally normalizing the phase images to remove the dry mass information. As shown in Fig. 4E, the network trained and tested with normalized phase images shows a significantly impaired performance, supporting the key role of dry mass. However, it does not mean that dry mass is the sole information extracted by the network; the performance of a single-feature linear discriminant classifier based solely on dry mass was also significantly worse. This suggests that other traits such as spatial distribution of subcellular components in the spores play roles in screening. From these observations, it can be concluded that the interspecies difference of dry mass is recognized and exploited through representation learning by HoloConvNet. Here, it should be emphasized that we never taught the network on how to calculate dry mass from phase images. On the other hand, a conventional machine learning algorithm cannot make use of dry mass unless it is manually selected by a researcher.
Finally, the generality of our method expected from the outstanding learning abilities was investigated. As a proof-of-concept example, HoloConvNet was trained for diagnosing the pathogen Listeria monocytogenes, the causative agent of listeriosis that is often fatal to neonates and the elderly (37), from five different Listeria species (see table S8 for data specification). The diagnostic accuracy was surprisingly high, showing higher than 85% (fig. S5). The architecture and learning rules were identical to those used for the diagnosis of Bacillus species. It is also noted that L. monocytogenes is not the species with the heaviest dry mass in this case (fig. S6). Accordingly, the other classifiers based only or mostly on dry mass show markedly lower performance compared to the equivalent comparison in the Bacillus problem (fig. S5, B and C). The superior performance of HoloConvNet suggests that, for the Listeria problem, the deep neural network discovers and exploits key biological traits other than dry mass. These results suggest that the holographic deep learning framework reported here has immediate and wide applicability, in contrast to problem-specific conventional machine learning approaches.
DISCUSSION
We proposed and experimentally demonstrated a novel method for screening of anthrax spores by combining holographic microscopy and deep learning for the first time. The new strategy enables rapid label-free identification of individual anthrax spores with subgenus specificity, extending our previous intergenus bacterial fingerprinting method based on conventional machine learning (15). In addition to the superior performance due to the extreme flexibility of deep neural networks, the transition from classical machine learning to deep learning fundamentally transforms holographic single-cell identification techniques by acquiring the representation learning capability. HoloConvNet automatically recognizes and then uses key biological characteristics that are species-dependent (for example, dry mass in the anthrax problem) from raw images. Additionally, the present method can be readily extended to other single-cell classification problems, such as the diagnosis of L. monocytogenes demonstrated in this study, without any modification. Thus, our method eliminates the need to manually design and optimize features based on trial and error for individual problems.
The next steps beyond this proof-of-concept study to achieve practical ultrafast screening of anthrax spores are straightforward. Above all, the proposed method should be combined with flow cytometry (16, 17) and bioaerosol collection (38) systems to fully exploit the single-spore and label-free nature of the method. Then, a large amount of holographic imaging data from the resultant high-throughput device would be used to train HoloConvNet for more species and strains under various environmental conditions to assure stable field performance. The performance could be further improved by adopting multimodal QPI [for example, spectral (39), polarimetric (40), or tomographic (41) images as the stacked input to the network] to increase the amount of raw information investigated by the network.
Despite the fast and label-free nature of holographic microscopy, the limited chemical specificity has left this tool overshadowed by fluorescence microscopy. Specific domain knowledge (for example, homogeneity of hemoglobin concentration in red blood cells and high RI of lipid droplets in eukaryotic cells) has been required for effective use of the technique. The method proposed in this paper solves this difficulty by using the powerful learning abilities of deep neural networks. As we demonstrated in this study, intelligent holographic microscopy can now actively recognize and exploit the class-specific fingerprints, encoded in the raw images of various biological samples, without any previous knowledge. We believe that our strategy will make holographic microscopy more accessible to medical doctors and biomedical scientists for easy, rapid, and accurate diagnosis of pathogens and facilitate exciting new applications.
MATERIALS AND METHODS
Experimental design
Preparation of Bacillus spores
B. anthracis Sterne (pXO1+ and pXO2−) was obtained from the Centers for Disease Control and Prevention, Korea (KCDC). B. thuringiensis BGSC 4AJ1 was obtained from the Bacillus Genetic Stock Center (BGSC). B. cereus ATCC 4342 was obtained from the American Type Culture Collection (ATCC). B. atrophaeus KCCM 11314 was obtained from the Korean Culture Center for Microorganisms (KCCM). B. subtilis 168 was obtained from the Korean Collection for Type Cultures (KCTC).
All experiments involving B. anthracis were approved by the institutional biosafety committee and conducted in a BSL-3 laboratory following the regulations in the Republic of Korea. Bacterial cells from frozen glycerol stocks were streaked onto Luria-Bertani (LB) agar plates and incubated at 30°C overnight. The next day, a single colony was inoculated into 5 ml of LB broth in a 50-ml CELLSTAR CELLreactor tube (Greiner Bio-One) and incubated at 30°C with shaking (200 rpm) for 8 hours. Then, 250 μl of the culture broth was transferred to 25 ml of GYS (glucose yeast salt) sporulation medium (42) in a 125-ml polycarbonate Erlenmeyer flask with a vent cap (Corning) and incubated at 30°C with shaking (200 rpm) for 48 hours. After sporulation was completed, spores were harvested by centrifugation (5420g, 4°C) and washed four times with phosphate-buffered saline (PBS) (Life Technologies). Finally, the spores were suspended in 5 ml of PBS and stored at 4°C until use. Note that we prepared all the species with the same procedure.
A small volume (approximately 10 μl) of the bacterial solution was placed in an imaging chamber composed of standard cover glasses (C024501, Matsunami Glass) and (optional) spacers with a thickness of 20 to 30 μm. Imaging was performed at room temperature after the spores settled down to the bottom and spread into a single layer.
Although the optimal conditions for cultivation and sporulation somewhat vary, all the species were prepared using the same protocol to guarantee that the screening system recognized only the species-dependent characteristics. Additionally, all the procedures for spore preparation and imaging were performed multiple times to ensure the independence of the training and test sets.
Holographic imaging
Because all anthrax experiments had to be conducted in a separate BSL-3 facility at the Agency for Defense Development, we used a compact and portable QPIU recently developed in our group (23), as the holographic imaging modality. It consists of two polarizers (LPVISE100-A, Thorlabs Inc.) and a Rochon prism (#68-824, Edmund Optics Inc.) inside an aluminum tube mounted in front of a charge-coupled device (CCD) camera (FL3-U3-88S2C-C, Point Grey). Inserting the unit into the output port of a conventional bright-field microscope (B-382PLi-ALC, Optika) converts it into a holographic microscope. The light source for illumination was a diode laser (CPS532, λ = 532 nm, 4.5 mW; Thorlabs Inc.), and the total magnification was ×100 determined by an objective lens (M-148; numerical aperture, 1.25; oil immersion; Optika). Acquisition time per interferogram was less than 20 ms, which could be reduced by many orders with high-intensity light sources and more sensitive cameras.
QPIU, shown in Fig. 1A, is a spatially modulated self-reference interferometry. When the light passing through the sample encounters the unit, it becomes linearly polarized by the front polarizer. Then, the following Rochon polarizing prism divides the beam into two duplicated beams with slightly different propagation directions. Finally, the orthogonal polarization states of the divided beams become parallel by the rear polarizer. Thus, the two beams of parallel polarization generate an interference pattern at the overlapped region on the CCD plane. The linear polarizers before and after the prism are adjusted so that the interferogram has a high visibility (Fig. 1B). The quantitative phase information is retrieved (Fig. 1C) from the measured interferogram using a standard field retrieval algorithm (43). The details on the principle of QPIU can be found elsewhere (23).
Convolutional neural networks
The network architecture of HoloConvNet shown in Fig. 2 is categorized as a CNN, which is the most popular deep learning framework for image classification and still has high interest in research (21, 31). Because the input for a deep learning algorithm is a raw image, instead of extracted features, the large number of learnable parameters (see Eq. 1 in the main text and filter dimensions shown in table S2) makes the training process extremely difficult. Using a CNN can resolve this issue by reducing the dimensionality through several constraints on the synaptic weights. Inspired by the classical work on early visual cortex (44), the synaptic weights of a CNN are localized and shared to mimic the receptive fields of biological vision. The resulting network exploits local spatial correlations to be robust under natural distortions, just as in physiological visual processing, with a significantly smaller number of parameters to be optimized. Although dimensionality reduction in conventional machine learning is done by manually designed feature extraction using problem-dependent domain expertise (10, 11, 15–18), biologically inspired intuition about the general properties of images mentioned above can have the same role in deep learning. This point is important because it enables direct training from raw images.
The Listeria experiments
The six major bacterial species of the genus Listeria, namely, L. monocytogenes (10403S), L. grayi (ATCC 19120), L. innocua (ATCC 33090), L. ivanovii (ATCC 19119), L. seeligeri (ATCC 35967), and L. welshimeri (ATCC 35897), were cultured in brain-heart infusion medium without antibiotics. After culturing overnight in a 37°C shaking incubator, the vegetative bacterial cells were washed and diluted with PBS based on the cultured concentration estimated by optical density measurements at 600 nm. The bacterial solution was placed and imaged in imaging chambers described above for the Bacillus experiments.
The holographic imaging of the prepared samples was done with a Mach-Zehnder interferometer (19) with varying illumination angles to exploit the high-resolution synthetic aperture imaging technique (45). Optical field reconstruction and image processing protocols were identical to those of the Bacillus experiments.
The same network architecture and learning rule for training the original HoloConvNet (for Bacillus spores) were used to train the network for Listeria. The only preprocessing was to adjust the size of the input images to match the input dimension of HoloConvNet.
Statistical analysis
Image analysis
All image analysis procedures were done with MatLab (R2014b, MathWorks Inc.). The reconstructed phase images containing multiple spores were segmented by phase thresholding to be separated into images of single spores. The isolated spores were computationally aligned at the centers of square backgrounds for further analysis. The segmented regions were considered as the morphologies of individual spores that could be measured with conventional microscopy techniques such as phase contrast microscopy. The representative morphological parameters plotted in fig. S2 were quantified with the regionprops function of MatLab. For implementing the conventional classifiers, the built-in functions in the Statistics and Machine Learning Toolbox of MatLab were used.
Calculation of the single-spore dry mass from phase images exploited the well-known proportionality between the optical phase delay and cellular dry mass (46). The total dry mass (m) can be calculated from the phase delay map () as follows
where λ is the illumination wavelength, S is the projection area of the cell surface, and α is the RI increment for nonaqueous molecules. Because the RI increment is known to be 0.18 to 0.21 ml/g for typical biological cells (46), we used α = 0.2 ml/g, and the results were consistent with those measured by other techniques (23, 36, 47). Note that we never explicitly taught the network about this relation.
Deep learning
HoloConvNet is a CNN designed for the classification of holographic images of single cells. We implemented HoloConvNet using the MatConvNet (48) framework (version 1.0, beta 20) because of its simplicity and compatibility with our experimental data primarily processed with MatLab. The final network architecture shown in Fig. 2 and table S2 was carefully chosen after comparing several variations. Motivated by the recent trend for “small receptive fields and deep layers,” the sizes of the receptive fields of the convolutional layers were chosen to be small (3 × 3), and thus, the total number of learnable parameters was relatively manageable (approximately 0.1 million). Because the architecture is substantially deep, we used the ReLU nonlinearity as the neuron model to avoid the vanishing gradient problem (49). Note that we only used phase images and not noisy amplitude images (due to the transparency and small sizes of the single spores) as the inputs to the network.
In addition to the traditional “weight decay” regularization (22), several recent techniques were used to reduce overfitting. We used the “dropout” technique, a regularization method based on efficient ensemble learning (50), for the last hidden layer with a dropout rate of 0.5. For further regularization and accelerated training speed, “batch normalization” was done at every interface between a convolutional layer and the following ReLU layer (51). Because of the large number of learnable parameters, we used “data augmentation” that enlarged the training data set (31) by a factor of 128. This is done by generating the new labeled images by rotating the original images by random angles sampled from a zero-mean Gaussian distribution with an SD of 10° and by flipping the images with a probability of 0.5.
During the training stage, the learnable parameters were updated toward the direction minimizing the loss function using the concept of error backpropagation (21, 22). We used cross-entropy loss based on softmax function, which quantifies the mismatch between the machine-predicted and true labels, as the objective function to be minimized. By calculating the partial derivatives of the loss function with respect to the elements of the synaptic weight tensors using the chain rule, we could update the parameters in a stochastic gradient descent (SGD) scheme (21, 22). The learning rule was conventional SGD assisted with a momentum of 0.5 (note that using recent learning rules instead could further improve the performance), and the training batch size was 1024. The weights were initialized from a zero-mean Gaussian distribution with layer-wise scaling based on the input sizes (52). The biases were initialized with the constant 0. We used an equal learning rate for all layers, which was attenuated by a factor of 5 per five epochs (22). The hyperparameters were selected by cross-validation; the grid searching process with the initial learning rate and weight decay regularization strength resulted in values of 0.05 and 0.0005, respectively. We used one graphics processing unit (GPU) (GeForce GTX 680, NVIDIA) and CUDA Toolkit 7.5 (NVIDIA), which increased the training speed typically by 5- to 10-fold. We note that it is possible with more computing resources to train multiple network models with different random initializations to compose a committee machine to further enhance the performance (31). Finally, the identification performance was estimated using separate test images that were never shown during the training stage. The error bars in Figs. 3 and 4 represent the SD calculated from 10 classification models with different random initializations.
The typical training time for 30 epochs (which were used for species prediction) was 25 min (multiclass identification with five Bacillus species, GTX 680). The typical time required for species prediction was less than 1 ms per cell (batch size of 445 cells). We found an approximately twofold increase of computing speed for GTX 980 Ti (NVIDIA) and expect more than a threefold increase when using the state-of-the-art GPUs such as GTX 1080 Ti (NVIDIA).
The visualization of HoloConvNet codes was performed by the unsupervised dimensionality reduction technique t-SNE, which embeds high-dimensional data in a low-dimensional space while preserving the pairwise distances of the data points, implemented in MatLab (32). The activation strengths of individual neurons at the last hidden layer by the test images were used as the raw variables. The parameters for the stochastic optimization for t-SNE were as follows: The perplexity was 30, and the dimension for initial principal components analysis was 30.
Acknowledgments
We thank M. Choi [Korea Advanced Institute of Science and Technology (KAIST)] and S. Kim (Samsung Electronics) for assisting the Listeria experiments; K. R. Choi (KAIST) and S. Yum (University of Texas Southwestern Medical Center) for biological insights; K. Lee, K. Kim, and S. Lee (KAIST) for discussions; and S.-Y. Lee (KAIST) for inspiring lectures on neural networks. Funding: This work was supported by KAIST, Agency for Defense Development (14-70-06-10), Asia Pacific Center for Theoretical Physics (APCTP), Tomocube Inc., and National Research Foundation of Korea (2015R1A3A2066550, 2014K1A3A1A09063027, 2014K1A3A1A09063027, and 2014M3C1A3052567). Y.J. acknowledges support from KAIST Presidential Fellowship. Author contributions: Y.J. and Y.P. conceived the original idea for holographic deep learning. S.Y.L. and Y.P. initiated and coordinated the project. Y.J. and H.J. developed and analyzed the HoloConvNet framework. J.J. and J.Y. implemented QPIU and analyzed the holographic data. S.P., under the supervision of M.C.C. and S.Y.L., conducted the Bacillus experiments. Y.J. and M.-h.K., under the supervision of S.-J.K. and Y.P., performed the Listeria experiments. Y.J., S.P., J.J., J.Y., and Y.P. wrote the initial version of the manuscript. All authors revised the manuscript. Competing interests: Y.P. has financial interests in Tomocube Inc., a company that commercializes optical diffraction tomography and phase imaging instruments and is one of the sponsors of this work. All other authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/8/e1700606/DC1
fig. S1. Representative images of individual Bacillus spores.
fig. S2. Morphological features of individual Bacillus spores.
fig. S3. Confusion matrices illustrating the performance of HoloConvNet.
fig. S4. Dry mass of individual Bacillus spores measured on different days.
fig. S5. Comparison of Listeria identification techniques.
fig. S6. Dry mass of individual bacteria from the Listeria species.
table S1. Detailed data on Bacillus spores.
table S2. Detailed architecture of HoloConvNet.
table S3. Performance of HoloConvNet on multiclass classification of the five Bacillus species.
table S4. Performance of HoloConvNet on binary classification of the five Bacillus species.
table S5. Performance of HoloConvNet on binary classification of the three Bacillus species.
table S6. Performance of conventional machine learning techniques in morphology-based identification of Bacillus spores.
table S7. Performance of conventional machine learning techniques in holographic identification of Bacillus spores.
table S8. Detailed description of the Listeria data.
REFERENCES AND NOTES
- 1.Brachman P. S., Bioterrorism: An update with a focus on anthrax. Am. J. Epidemiol. 155, 981–987 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Walt D. R., Franz D. R., Peer reviewed: Biological warfare detection. Anal. Chem. 72, 738A–746A (2000). [DOI] [PubMed] [Google Scholar]
- 3.King D., Luna V., Cannons A., Cattani J., Amuso P., Performance assessment of three commercial assays for direct detection of Bacillus anthracis spores. J. Clin. Microbiol. 41, 3454–3455 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hurtle W., Bode E., Kulesh D. A., Kaplan R. S., Garrison J., Bridge D., House M., Frye M. S., Loveless B., Norwood D., Detection of the Bacillus anthracis gyrA gene by using a minor groove binder probe. J. Clin. Microbiol. 42, 179–185 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X., Young M. A., Lyandres O., Van Duyne R. P., Rapid detection of an anthrax biomarker by surface-enhanced Raman spectroscopy. J. Am. Chem. Soc. 127, 4484–4489 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Javidi B., Moon I., Yeom S., Carapezza E., Three-dimensional imaging and recognition of microorganism using single-exposure on-line (SEOL) digital holography. Opt. Express 13, 4492–4506 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Moon I., Javidi B., Three-dimensional identification of stem cells by computational holographic imaging. J. R. Soc. Interface 4, 305–313 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moon I., Javidi B., 3-D visualization and identification of biological microorganisms using partially temporal incoherent light in-line computational holographic imaging. IEEE Trans. Med. Imaging 27, 1782–1790 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Moon I., Daneshpanah M., Javidi B., Stern A., Automated three-dimensional identification and tracking of micro/nanobiological organisms by computational holographic microscopy. Proc. IEEE 97, 990–1010 (2009). [Google Scholar]
- 10.Javidi B., Daneshpanah M., Moon I., Three-dimensional holographic imaging for identification of biological micro/nanoorganisms. IEEE Photonics J. 2, 256–259 (2010). [Google Scholar]
- 11.Moon I., Daneshpanah M., Anand A., Javidi B., Cell identification computational 3-D holographic microscopy. Opt. Photonics News 22, 18–23 (2011). [Google Scholar]
- 12.Anand A., Chhaniwal V., Patel N., Javidi B., Automatic identification of malaria-infected RBC with digital holographic microscopy using correlation algorithms. IEEE Photonics J. 4, 1456–1464 (2012). [Google Scholar]
- 13.Moon I., Anand A., Cruz M., Javidi B., Identification of Malaria-infected red blood cells via digital shearing interferometry and statistical inference. IEEE Photonics J. 5, 6900207 (2013). [Google Scholar]
- 14.Anand A., Moon I., Javidi B., Automated disease identification with 3-D optical imaging: A medical diagnostic tool. Proc. IEEE 105, 924–946 (2017). [Google Scholar]
- 15.Jo Y., Jung J. H., Kim M.-h., Park H. J., Kang S.-J., Park Y. K., Label-free identification of individual bacteria using Fourier transform light scattering. Opt. Express 23, 15792–15805 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Vercruysse D., Dusa A., Stahl R., Vanmeerbeeck G., de Wijs K., Liu C., Prodanov D., Peumansa P., Lagae L., Three-part differential of unlabeled leukocytes with a compact lens-free imaging flow cytometer. Lab Chip 15, 1123–1132 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Chen C. L., Mahjoubfar A., Tai L.-C., Blaby I. K., Huang A., Niazi K. R., Jalali B., Deep learning in label-free cell classification. Sci. Rep. 6, 21471 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park H. S., Rinehart M. T., Walzer K. A., Chi J.-T. A., Wax A., Automated detection of P. falciparum using machine learning algorithms with quantitative phase images of unstained cells. PLOS ONE 11, e0163045 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee K., Kim K., Jung J., Heo J. H., Cho S., Lee S., Chang G., Jo Y. J., Park H., Park Y. K., Quantitative phase imaging techniques for the study of cell pathophysiology: From principles to applications. Sensors 13, 4170–4191 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu P. Y., Chin L. K., Ser W., Chen H. F., Hsieh C.-M., Lee C.-H., Sung K.-B., Ayi T. C., Yap P. H., Liedberg B., Wang K., Bourouinaj T., Leprince-Wang Y., Cell refractive index for cell biology and disease diagnosis: Past, present and future. Lab Chip 16, 634–644 (2016). [DOI] [PubMed] [Google Scholar]
- 21.LeCun Y., Bengio Y., Hinton G., Deep learning. Nature 521, 436–444 (2015). [DOI] [PubMed] [Google Scholar]
- 22.S. Haykin, Neural Networks and Learning Machines (Pearson, 2009), vol. 3. [Google Scholar]
- 23.Jo Y., Jung J. H., Lee J. W., Shin D., Park H. J., Nam K. T., Park J.-H., Park Y. K., Angle-resolved light scattering of individual rod-shaped bacteria based on Fourier transform light scattering. Sci. Rep. 4, 5090 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green B. D., Battisti L., Koehler T. M., Thorne C. B., Ivins B. E., Demonstration of a capsule plasmid in Bacillus anthracis. Infect. Immun. 49, 291–297 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O. A. Økstad, A.-B. Kolstø, Genomics of Bacillus species, in Genomics of Foodborne Bacterial Pathogens, M. Wiedmann, W. Zhang, Eds. (Springer, 2011), pp. 29–53. [Google Scholar]
- 26.Helgason E., Økstad O. A., Caugant D. A., Johansen H. A., Fouet A., Mock M., Hegna I., Kolstø A.-B., Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis—One species on the basis of genetic evidence. Appl. Environ. Microbiol. 66, 2627–2630 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenberg D. L., Busch J. D., Keim P., Wagner D. M., Identifying experimental surrogates for Bacillus anthracis spores: A review. Investigative Genet. 1, 4 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibbons H. S., Broomall S. M., McNew L. A., Daligault H., Chapman C., Bruce D., Karavis M., Krepps M., McGregor P. A., Hong C., Park K. H., Akmal A., Feldman A., Lin J. S., Chang W. E., Higgs B. W., Demirev P., Lindquist J., Liem A., Fochler E., Read T. D., Tapia R., Johnson S., Bishop-Lilly K. A., Detter C., Han C., Sozhamannan S., Rosenzweig C. N., Skowronski E. W., Genomic signatures of strain selection and enhancement in Bacillus atrophaeus var. globigii, a historical biowarfare simulant. PLOS ONE 6, e17836 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carrera M., Zandomeni R., Fitzgibbon J., Sagripanti J.-L., Difference between the spore sizes of Bacillus anthracis and other Bacillus species. J. Appl. Microbiol. 102, 303–312 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Plomp M., Leighton T. J., Wheeler K. E., Malkin A. J., Architecture and high-resolution structure of Bacillus thuringiensis and Bacillus cereus spore coat surfaces. Langmuir 21, 7892–7898 (2005). [DOI] [PubMed] [Google Scholar]
- 31.A. Krizhevsky, I. Sutskever, G. E. Hinton, ImageNet classification with deep convolutional neural networks, in Advances in Neural Information Processing Systems 25 (NIPS 2012) (NIPS, 2012), pp.1097–1105. [Google Scholar]
- 32.van der Maaten L., Hinton G., Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008). [Google Scholar]
- 33.Popescu G., Park Y. K., Lue N., Best-Popescu C., Deflores L., Dasari R. R., Feld M. S., Badizadegan K., Optical imaging of cell mass and growth dynamics. Am. J. Physiol. Cell Physiol. 295, C538–C544 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henriques A. O., Moran C. P. Jr., Structure, assembly, and function of the spore surface layers. Annu. Rev. Microbiol. 61, 555–588 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Ball D. A., Taylor R., Todd S. J., Redmond C., Couture-Tosi E., Sylvestre P., Moir A., Bullough P. A., Structure of the exosporium and sublayers of spores of the Bacillus cereus family revealed by electron crystallography. Mol. Microbiol. 68, 947–958 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Carrera M., Zandomeni R. O., Sagripanti J.-L., Wet and dry density of Bacillus anthracis and other Bacillus species. J. Appl. Microbiol. 105, 68–77 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Low J., Donachie W., A review of Listeria monocytogenes and listeriosis. Vet. J. 153, 9–29 (1997). [DOI] [PubMed] [Google Scholar]
- 38.Després V. R., J. Huffman A., Burrows S. M., Hoose C., Safatov A. S., Buryak G., Fröhlich-Nowoisky J., Elbert W., Andreae M. O., Pöschl U., Jaenicke R., Primary biological aerosol particles in the atmosphere: A review. Tellus B 64, 15598 (2012). [Google Scholar]
- 39.Jung J.-H., Jang J., Park Y., Spectro-refractometry of individual microscopic objects using swept-source quantitative phase imaging. Anal. Chem. 85, 10519–10525 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Kim Y., Jeong J., Jang J., Kim M. W., Park Y., Polarization holographic microscopy for extracting spatio-temporally resolved Jones matrix. Opt. Express 20, 9948–9955 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Kim K., Yoon H. O., Diez-Silva M., Dao M., Dasari R. R., Park Y. K., High-resolution three-dimensional imaging of red blood cells parasitized by Plasmodium falciparum and in situ hemozoin crystals using optical diffraction tomography. J. Biomed. Opt. 19, 011005 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yousten A., Rogoff M., Metabolism of Bacillus thuringiensis in relation to spore and crystal formation. J. Bacteriol. 100, 1229–1236 (1969). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Debnath S. K., Park Y., Real-time quantitative phase imaging with a spatial phase-shifting algorithm. Opt. Lett. 36, 4677–4679 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Hubel D. H., Wiesel T. N., Receptive fields of single neurones in the cat’s striate cortex. J. Physiol. 148, 574–591 (1959). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee K., Kim H.-D., Kim K., Kim Y., Hillman T. R., Min B., Park Y. K., Synthetic Fourier transform light scattering. Opt. Express 21, 22453–22463 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Barer R., Interference microscopy and mass determination. Nature 169, 366–367 (1952). [DOI] [PubMed] [Google Scholar]
- 47.Godin M., Delgado F. F., Son S., Grover W. H., Bryan A. K., Tzur A., Jorgensen P., Payer K., Grossman A. D., Kirschner M. W., Manalis S. R., Using buoyant mass to measure the growth of single cells. Nat. Methods 7, 387–390 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.A. Vedaldi, K. Lenc, MatConvNet: Convolutional neural networks for MATLAB, in Proceedings of the 23rd ACM International Conference on Multimedia (ACM, 2015), pp. 689–692. [Google Scholar]
- 49.V. Nair, G. E. Hinton, Rectified linear units improve restricted Boltzmann machines, in Proceedings of the 27th International Conference on Machine Learning (ICML 2010) (ICML, 2010), pp. 807–814. [Google Scholar]
- 50.Srivastava N., Hinton G., Krizhevsky A., Sutskever I., Salakhutdinov R., Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 15, 1929–1958 (2014). [Google Scholar]
- 51.S. Ioffe, C. Szegedy, Batch normalization: Accelerating deep network training by reducing internal covariate shift, in Proceedings of the 32nd International Conference on Machine Learning (ICML 2015) (ICML, 2015), pp. 448–456. [Google Scholar]
- 52.K. He, X. Zhang, S. Ren, J. Sun, Delving deep into rectifiers: Surpassing human-level performance on ImageNet classification, in Proceedings of the IEEE International Conference on Computer Vision (ICCV) (IEEE, 2015), pp. 1026–1034. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/8/e1700606/DC1
fig. S1. Representative images of individual Bacillus spores.
fig. S2. Morphological features of individual Bacillus spores.
fig. S3. Confusion matrices illustrating the performance of HoloConvNet.
fig. S4. Dry mass of individual Bacillus spores measured on different days.
fig. S5. Comparison of Listeria identification techniques.
fig. S6. Dry mass of individual bacteria from the Listeria species.
table S1. Detailed data on Bacillus spores.
table S2. Detailed architecture of HoloConvNet.
table S3. Performance of HoloConvNet on multiclass classification of the five Bacillus species.
table S4. Performance of HoloConvNet on binary classification of the five Bacillus species.
table S5. Performance of HoloConvNet on binary classification of the three Bacillus species.
table S6. Performance of conventional machine learning techniques in morphology-based identification of Bacillus spores.
table S7. Performance of conventional machine learning techniques in holographic identification of Bacillus spores.
table S8. Detailed description of the Listeria data.


