Abstract
Massively parallel sequencing has revealed many de novo mutations in the etiology of developmental and epileptic encephalopathies (EEs), highlighting their genetic heterogeneity. Additional candidate genes have been prioritized in silico by their co-expression in the brain. Here, we evaluate rare coding variability in 20 candidates nominated with the use of a reference gene set of 51 established EE-associated genes. Variants within the 20 candidate genes were extracted from exome-sequencing data of 42 subjects with EE and no previous genetic diagnosis. We identified 7 rare non-synonymous variants in 7 of 20 genes and performed Sanger sequence validation in affected probands and parental samples. De novo variants were found only in SLC1A2 (aka EAAT2 or GLT1) (c.244G>A [p.Gly82Arg]) and YWHAG (aka 14-3-3γ) (c.394C>T [p.Arg132Cys]), highlighting the potential cause of EE in 5% (2/42) of subjects. Seven additional subjects with de novo variants in SLC1A2 (n = 1) and YWHAG (n = 6) were subsequently identified through online tools. We identified a highly significant enrichment of de novo variants in YWHAG, establishing their role in early-onset epilepsy, and we provide additional support for the prior assignment of SLC1A2. Hence, in silico modeling of brain co-expression is an efficient method for nominating EE-associated genes to further elucidate the disorder’s etiology and genotype-phenotype correlations.
Keywords: epileptic encephalopathy, whole-exome sequencing, de novo variants, YWHAG, SLC1A2
Main Text
Developmental and epileptic encephalopathies (EEs) are a genetically heterogeneous group of severe epilepsy syndromes characterized by early-onset, treatment-resistant seizures and developmental slowing or regression.1 There are currently 51 established EE-associated genes, in which many causal mutations arise de novo.2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13 However, with the advent of high-throughput sequencing, an increasing number of candidate genes are emerging with only a single reported de novo variant.4, 5 The interpretation of these findings remains challenging given that on average ∼0.5 de novo exonic mutations are expected per individual.14, 15, 16 Co-segregation analyses in large pedigrees, which usually provide strong genetic evidence for pathogenicity, are not applicable in the case of de novo mutations. Rather, identification of recurrent mutations or different mutations in the same gene helps provide evidence for causality. Such candidate genes can also be prioritized in silico if they belong to the same biological networks, including ion channels and synaptic and regulatory pathways.
Of 179 candidate genes with a single de novo variant reported,4 Oliver et al.17 prioritized 19 on the basis of brain-specific co-expression with a reference gene set of 29 established EE-associated genes. Five of the 19 candidate genes have since been confirmed as disease causing, demonstrating the validity of this approach. Using an updated reference gene set of 51 established EE-associated genes, Oliver et al.13 recently highlighted six more genes that are co-expressed. Here, we evaluate rare coding variability in all co-expressed candidate genes (n = 20) nominated by Oliver et al.13, 17 in 42 EE-affected individuals (14 males and 28 females) selected from 149 epileptic individuals hospitalized and/or attending BC Children’s Hospital neurology, medical genetic, and biochemical clinics and enrolled in the Epilepsy Genomics (EPGEN) Study. Eligible subjects had a clinical diagnosis of epilepsy1 with onset by the age of 5 years and an unknown cause after appropriate investigations, including electroencephalography (EEG), head MRI, neurometabolic testing, and chromosome microarray. Seizure types and electroclinical syndromes were classified according to the International League Against Epilepsy (ILAE).1 Individuals with self-limiting benign electroclinical syndromes, such as childhood absence epilepsy (onset > 4 years), were excluded on the basis of its more likely multifactorial inheritance. Individuals were identified to have an epileptic encephalopathy if the epileptic activity itself was determined to or was at risk of (e.g., new onset of infantile spasms) contributing to severe cognitive and behavioral impairments above and beyond what might be expected from the underlying pathology alone.1 The average age of seizure onset for the 42 selected EE subjects was 20.6 ± 18.9 months (1 week to 5 years). Focal seizures with impaired awareness were the most common seizure type (20 subjects [47.6%]), followed by generalized myoclonic seizures (16 subjects [40%]). Informed consent was obtained for all subjects in accordance with site-specific institutional review boards.
Our cohort represents a highly selected group of individuals without mutations in genes known to be associated with epilepsy (n = 545), including the 51 established EE-associated genes. Variants within co-expressed candidate genes (n = 20; Table S1) were extracted from exome sequencing data. On average, 96% (83%–100%) of the coding region was sequenced >10-fold (Table S1). Variant annotation was performed with ANNOVAR,18 which integrated data from a variety of public databases. Highly penetrant de novo mutations are not expected in healthy individuals, and we assessed only extremely rare variants (≤1 carrier in the Exome Aggregation Consortium [ExAC] Browser), including small indels and missense, nonsense, and splicing changes (±5 bp). Sanger sequencing in probands and both parents (when available) validated a total of seven non-synonymous variants in seven different genes (Table 1). Variants in four genes, GABBR2 (MIM: 607340), ANK3 (MIM: 600465), NBEA (MIM: 604889), and ACOT4 (MIM: 614314), were inherited from unaffected parents, so pathogenicity is doubtful. A variant of unknown significance was identified in PLXNA1 (MIM: 601055); whereas the mother is not a carrier, paternal DNA was not available. The remaining two mutations were de novo: (1) c.244G>A (p.Gly82Arg) in SLC1A2 (solute carrier family 1 member 2 [MIM: 600300; GenBank: NM_004171.3]) was identified in a 6-year-old boy (subject A) with seizure onset at 6 weeks and (2) c.394C>T (p.Arg132Cys) in YWHAG (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein gamma, also known as 14-3-3γ [MIM: 605356; GenBank: NM_012479.3]) was identified in an 18-year-old woman (subject B) with seizure onset at 12 months (Table 2).
Table 1.
Rare Coding Variants
| ID | Genea | hg19 Position | rsID | MAF | Nucleotide Change | Amino Acid Change | CADD | REVEL | Inheritance | Study |
|---|---|---|---|---|---|---|---|---|---|---|
| 109 | ACOT4 | chr14: 74,059,006 | rs762048021 | 4.09 × 10−6 | c.343G>A | p.Glu115Lys | 13.3 | 0.100 | paternal | this study (n = 42) |
| 044 | ANK3 | chr10: 61,828,433 | NA | NA | c.12206A>G | p.Glu4069Gly | 13.3 | 0.111 | paternal | this study (n = 42) |
| 015 | GABBR2 | chr9: 101,073,426 | NA | NA | c.1955 A>G | p.His652Arg | 23.2 | 0.830 | maternal | this study (n = 42) |
| 062 | NBEA | chr13: 35,770,172 | NA | NA | c.5099T>C | p.Met1700Thr | 13.2 | 0.079 | paternal | this study (n = 42) |
| 133 | PLXNA1 | chr3: 126,752,801 | rs149124469 | 8.51 × 10−6 | c.5632C>T | p.Arg1878Trp | 33.0 | 0.149 | unknown (not maternal) | this study (n = 42) |
| Subject A | SLC1A2 | chr11: 35,336,636 | NA | NA | c.244 G>A | p.Gly82Arg | 32.0 | 0.864 | de novo | this study (n = 42) |
| Subjects B, E, and F | YWHAG | chr7: 75,959,244 | NA | NA | c.394C>T | p.Arg132Cys | 33.0 | 0.791 | de novo | this study (n = 42), DDD19 (n = 4,293), GeneMatcher (NA) |
| Subject C | SLC1A2 | chr11: 35,314,059 | NA | NA | c.866C>G | p.Pro289Arg | 26.9 | 0.911 | de novo | DDD19 (n = 4,293) |
| Subject D | YWHAG | chr7: 75,988,082 | NA | NA | c.44A>C | p.Glu15Ala | 27.2 | 0.417 | de novo | GeneMatcher (NA) |
| AU027A | YWHAG | chr7: 75,959,490 | NA | NA | c.148A>C | p.Lys50Gln | 25.4 | 0.597 | de novo | De Rubeis et al.20 (n = 2,270) |
| ND27637 | YWHAG | chr7: 75,959,251 | NA | NA | c.387C>G | p.Asp129Glu | 26.9 | 0.524 | de novo | Epi4K4 (n = 264) |
| DDD4K.00159 | YWHAG | chr7: 75,959,240 | NA | NA | c.398A>C | p.Tyr133Ser | 27.1 | 0.911 | de novo | DDD19 (n = 4,293) |
Abbreviations are as follows: MAF, minor allele frequency from gnomAD; CADD, Combined Annotation Dependent Depletion (v.1.3); REVEL, Rare Exome Variant Ensemble Learner; NA, not available.
GenBank accession numbers: ACOT4, NM_152331.3; ANK3, NM_001204403.1; GABBR2, NM_005458; NBEA, NM_015678.4; PLXNA1, NM_032242.3; SLC1A2, NM_004171.3; and YWHAG, NM_012479.3.
Table 2.
Clinical Features of Subjects with SLC1A2 and YWHAG Mutations
|
SLC1A2 |
YWHAG |
|||||
|---|---|---|---|---|---|---|
| Subject A | Subject C | Subject B | Subject D | Subject E | Subject F | |
| Genetic findings | c.244G>A (p.Gly82Arg) | c.866C>G (p.Pro289Arg) | c.394C>T (p.Arg132Cys) | c.44A>C (p.Glu15Ala) | c.394C>T (p.Arg132Cys) | c.394C>T (p.Arg132Cys) |
| Sex | male | male | female | female | female | female |
| Seizure onset | 6 weeks | 5 days | 12 months | 6 months | <6 months | <6 years (unknown) |
| Age | 6 years | 10 years | 18 years | 10 years | 22 years | 15 years |
| Diagnosis | developmental and EE (treatment resistant) | developmental and EE (treatment resistant) | developmental and EE | unclassified epilepsy | unclassified or genetic generalized epilepsy | unclassified or genetic generalized epilepsy |
| Seizure type at onset | focal motor (left leg, right arm) initially without impaired awareness | focal motor (face, eyes, and right foot) | generalized myoclonic | prolonged focal motor seizure with fever | myoclonic | absence, eyelid myoclonia, myoclonic |
| Course of seizures | focal motor ± impaired awareness evolving to bilateral tonic-clonic | multiple seizure types | generalized myoclonic, atypical absence, GTC, no febrile seizure |
second prolonged seizure with fever and then two episodes of status epilepticus associated with regression and hemiparesis | prolonged GTC with fever, generalized myoclonic, absence, GTC | persistence of absence seizures |
| Ictal EEG | not available, but capture of right arm jerks and chin twitching showed no EEG correlate | spike-and-wave discharges (age 7 weeks) | myoclonic jerks, generalized spike-and-wave discharge | not available | spike-and-slow wave, polyspike, and slow-wave discharges | not available |
| Interictal EEG | 2 months: paroxysmal delta and frequent bilateral mid-central parietal spikes and sharp waves, occasional right temporal spikes; 4 years: slow dysrhythmic background, multifocal epileptiform discharges | very high amplitude (>500 μV), 0.7–3.3 Hz delta and very slow waves with intermixed generalized sharp waves (age 8 months) | 2 years: dysrhythmic background, generalized atypical spike wave, frequent bifrontal spikes; 14 years: dysrhythmic background, rare sharp waves in bianterior quadrants | not available | 21 months: generalized 3 Hz spike wave with absence seizures; 10 years: generalized polyspike wave with myoclonic seizures; 14 years: occasional spike wave | 8 years: bilateral frontotemporal spikes, generalized spike waves |
| Timing of seizure occurrence | diurnal or nocturnal | not provided | diurnal | not provided | frequently early morning | not provided |
| Head circumference | 45.1 cm at 8 months (50%), 52 cm at 5 years (50th percentile) | 44 cm at 4 months (91st percentile) | normal | ∼+1 SD (mother has macrocephaly) |
53.7 cm at 15 years (20th percentile) | normal (50th percentile) |
| Neurologic findings | visually inattentive with sluggish pupils, axial hypotonia, mild limb spasticity, dyskinetic movements of arms | hypotonia with up-going plantars and severe thoraco-lumbar kyphoscoliosis | action tremor in upper extremities and mild coordination difficulties | hypotonia, joint hyperlaxity | ataxia, thoracolumbar scoliosis | thoracolumbar kyphoscoliosis |
| Response to treatment | partial response to phenobarbital, levetiracetam, and clobazam; longest seizure-free period was 16 months |
partial response to ketogenic diet and phenytoin, no response to other antiseizure medications | responded to clonazepam, which was stopped at 8 years; lamotrigine at 10 years was ineffective for atypical absence and GTC seizure; seizure free since 11 years on divalproex sodium and ethosuximide; ethosuximide was weaned off by 17 years | epilepsy under control with divalproex sodium | partial response to divalproex sodium and stiripentol | epilepsy under control with divalproex sodium and lamotrigine |
| ID | yes (profound) | yes (severe) | mild to moderate ID | yes (IQ ca. 55) | yes (moderate to severe) | WPPSI III (6 years), VIQ 73, PIQ 58 |
| Development | delayed (severe), nonverbal, unable to sit unsupported | yes (severe) | early language delay, fine motor difficulties with tremor, ADHD | delayed | delayed | mildly delayed |
| Brain MRI | 6 weeks: normal; 2 years: incomplete myelination, generalized atrophy, thin corpus callosum, abnormal signal in thalami, putamen, and caudate, and abnormal peritrigonal white-matter signal; 4 years: progressive generalized atrophy, persistent abnormal signal in basal ganglia and thalami, and periventricular white-matter signal abnormalities | 2 months: normal; 2.5 years: severely delayed myelination, cerebral atrophy, and bilateral T2 prolongation in caudate heads and putamina | 3 years: asymmetric brainstem not thought to be significant | generalized atrophy with diffuse loss of white matter | 10 years: normal | normal |
Abbreviations are as follows: ADHG, attention-deficit hyperactivity disorder; EE, epileptic encephalopathy; EEG, electroencephalography; GTC, generalized tonic-clonic; ID, intellectual disability; PIQ, performance IQ; VIQ, verbal IQ; WPPSI, Wechsler Preschool and Primary Scale of Intelligence.
In an attempt to identify additional carriers of de novo variants in SLC1A2 and YWHAG, we queried biomedical literature on PubMed and three online resources: (1) GeneMatcher,21 a freely accessible tool that connects clinicians and researchers with interests in the same gene; (2) DECIPHER, which contains exome sequencing data from nearly 4,300 families affected by developmental disorders;22, 23 and (3) AnnEX, a collaborative platform that interrogates exomes specializing in neurologic and neurodegenerative disorders. We identified seven additional probands: subject C (SLC1A2 c.866C>G [p.Pro289Arg]), subject D (YWHAG c.44A>C [p.Glu15Ala]), AU027A with autism (YWHAG c.148A>C [p.Lys50Gln]),20 ND27637 with Lennox-Gastaut syndrome (YWHAG c.387C>G [p.Asp129Glu]),4 subjects E and F (YWHAG c.394C>T [p.Arg132Cys]), and DDD4K.00159 with a neurodevelopmental disorder (YWHAG c.398A>C [p.Tyr133Ser])19 (Table 1). All mutations were de novo and resulted in substitutions that involve highly conserved amino acids and are predicted to be damaging by multiple in silico algorithms (Combined Annotation Dependent Depletion [CADD] and Rare Exome Variant Ensemble Learner [REVEL]). None have been reported in public databases (gnomAD Browser), and one mutation (YWHAG c.394C>T) is recurrent in three subjects (B, E, and F). We used denovolyzeR24 to evaluate the excess of de novo mutations in YWHAG. Assuming that 6,869 individuals were included, we observed a highly significant enrichment of heterozygotes (0.2 expected versus 7 observed; p = 5.1 × 10−9; 30.8×; Table 1). Although the number of subjects in GeneMatcher was not defined, these results remained significant (p = 0.009) even when 100,000 individuals were included.
Table 2 summarizes the main clinical features of the six affected individuals who have not previously been described in the literature. All probands underwent WES and had no other pathogenic or likely pathogenic mutations in genes known to be associated with seizure disorders. Detailed case reports are provided in the Supplemental Note.
SLC1A2 encodes the excitatory amino acid transporter EAAT2 (also known as GLT-1), a glutamate transporter that is primarily expressed in astrocytes and is responsible for ∼95% of glutamate uptake activity in the mammalian brain. Lethal spontaneous seizures and progressive neuronal death due to excitotoxicity were reported in a Slc1a2 knockout mouse model.25 Elevated extracellular glutamate levels have also been reported in animal models of epilepsy and epilepsy patients alike, particularly in individuals with temporal lobe epilepsy.26, 27, 28, 29
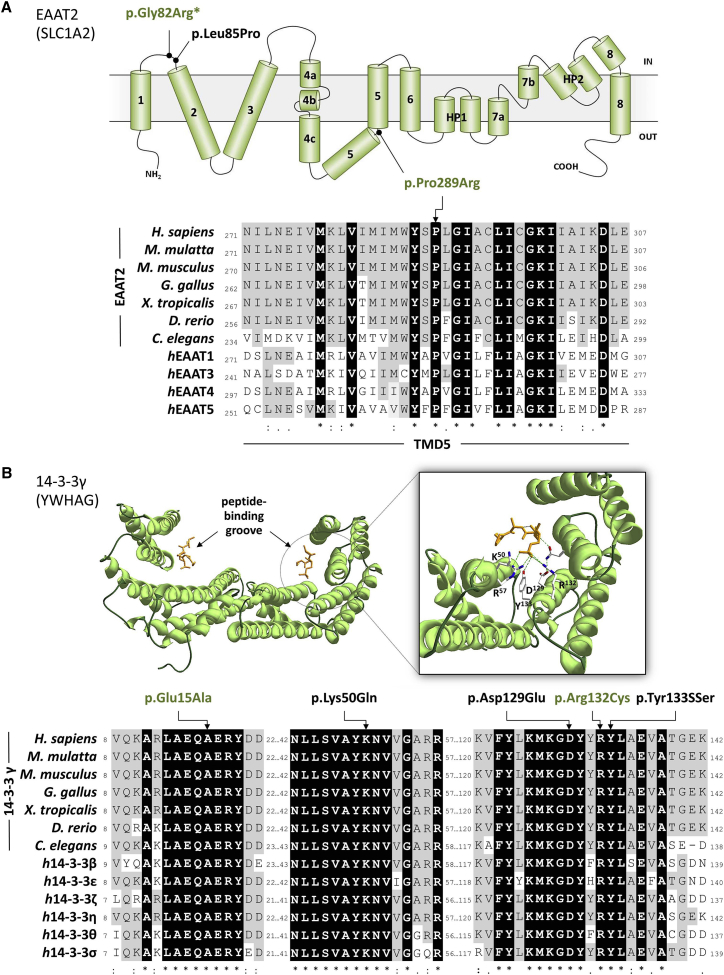
EAAT2 p.Gly82Arg (subject A) substitutes a small hydrophobic and neutrally charged glycine with hydrophilic, positively charged arginine within the cytoplasmic loop joining the first pair of transmembrane domains (Figure 1). The same substitution is reported in the Epi4K-EPGP dataset in a subject with infantile spasms4 and two additional individuals with a severe EE, and de novo variants (EAAT2 p.Gly82Arg and p.Leu85Pro) were reported33 during the preparation of this manuscript.
Figure 1.
De Novo SLC1A2 and YWHAG Mutations in Individuals with Early-Onset Epilepsy
(A) Top: transmembrane topology of EAAT2 (adapted from Yernool et al.30) and localization of the variants identified in this study (green) or previously reported (black) (the asterisk indicates a recurrent variant). Bottom: partial sequence alignment of EAAT2 orthologs and different human EAAT proteins surrounding p.Pro289Arg (indicated by an arrow). Identical residues across all proteins are shown in black, and residues identical to the human EAAT2 are in gray. GenBank accession numbers are as follows: Homo sapiens, NP_004162.2; Macaca mulatta, NP_001248598.1; Mus musculus, NP_001070982.1; Gallus gallus, NP_001012917.1; Xenopus tropicalis, XP_002937340.1; Dario rerio, NP_956273.1; Caenorhabditis elegans, NP_001024393.1; human EAAT1, NP_004163.3; human EAAT3, NP_004161.4; human EAAT4, NP_005062.1; and human EAAT5, NP_006662.3.
(B) Top: crystal structure of 14-3-3γ (PDB: 5D3E). Left: dimeric 14-3-3γ is shown as green ribbons, and the phosphopeptide ligand is shown as an orange stick. Right: close-up view of the binding groove, side chains of the residues crucial for the phosphopeptide binding, and hydrogen bonds (green dashed lines). Images were generated with Swiss-Pdb Viewer v.4.10.31 Bottom: partial sequence alignment of 14-3-3γ orthologs and different human 14-3-3 proteins surrounding the mutated residues identified in this study (green) or previously reported (black). Identical residues across all proteins are shown in black, and residues identical to the human 14-3-3γ are in gray. GenBank accession numbers are as follows: Homo sapiens, NP_036611.2; Macaca mulatta, NP_001181365.1; Mus musculus, NP_061359.2; Gallus gallus, NP_001026648.1; Xenopus tropicalis, NP_001072309.1; Dario rerio, NP_998187.1; Caenorhabditis elegans, AAA61872.1; human 14-3-3β, NP_003395.1; human 14-3-3ε, NP_006752.1; human 14-3-3 ζ, NP_001129174.1; human 14-3-3 η, NP_003396.1; human 14-3-3 θ, NP_006817.1; and human 14-3-3 NP_006133.1.
Sequences were aligned with CLUSTAL Omega.32 Asterisks indicate positions with a single fully conserved residue, colons indicate conservation between groups with strongly similar properties, and periods indicate conservation between groups with weakly similar properties.
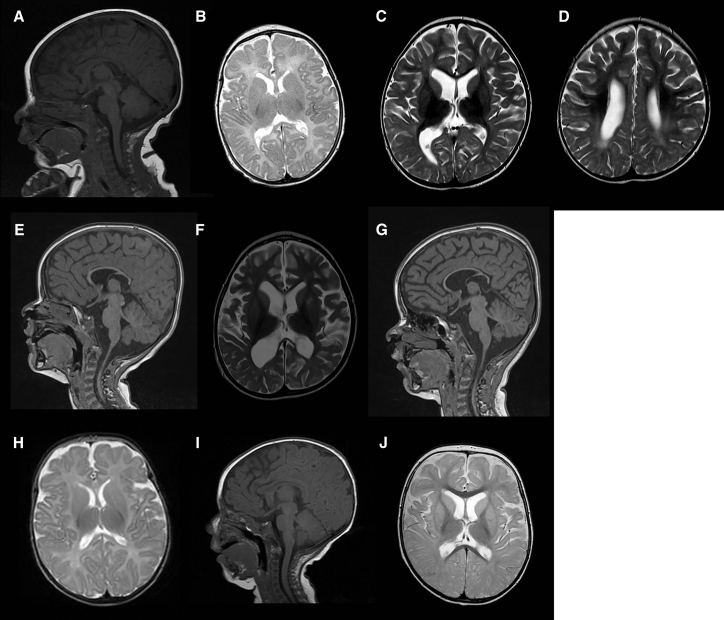
EAAT2 p.Pro289Arg (subject C) affects a highly conserved residue in all human EAATs (EAAT1–EAAT5) and their ancestral homolog. Pro289 leads to a hinge in the middle of the fifth transmembrane domain (Figure 1). Its substitution to positively charged arginine is predicted to cause a major structural change of the protein. The homologous mutation (c.1047C>G [p.Pro290Arg]) in SLC1A3 (MIM: 600111), encoding EAAT1, has been described in a subject with episodic ataxia, seizures, migraine, and alternating hemiplegia (MIM: 612656).34 In addition to mediating secondary-active glutamate uptake, EAATs also function as anion channels.35, 36, 37, 38 The EAAT1 p.Pro290Arg variant decelerates a conformational change associated with Na+ binding, causing a decrease in glutamate transport rates and a significant increase in EAAT1-associated anion current amplitudes.39 EAAT1 and EAAT2 have similar expression profiles (mainly expressed in astrocytes and localized to plasmalemma), share basic mechanisms of glutamate transport, and exhibit similar anion conduction properties. Therefore, it is likely that the EAAT2 p.Pro289Arg variant would have an effect similar to that of the homologous mutation.33 Similar to three previously reported individuals with SLC1A2 mutations, our subjects have a severe phenotype consisting of early-onset epilepsy and severe development delay. Four of five individuals have multiple seizure types, but subject A (who has the same mutation as the subject reported to have infantile spasms)4 primarily has focal motor seizures. This subject also experienced regression with an episode of lethargy and irritability, whereas no definite regression occurred in the other subjects. Neuroimaging revealed brain atrophy and abnormalities in white matter and basal ganglia in subjects A and C (Figure 2). The atrophy is possibly due in part to glutamate-induced excitotoxicity.25
Figure 2.
MRI Findings in Individuals with SLC1A2 Mutations
(A–G) Subject A. Sagittal T1 (A) and axial T2 (B) at 6 weeks revealed a thin corpus callosum but within normal limits for age. Axial T2 (C and D) and sagittal T1 (E) at 2 years revealed volume loss and a thin corpus callosum, increased T2 white-matter signal intensity, delayed myelination, decreased T2 signal of thalami, and increased T2 signal of putamen and caudate. At 4 years, axial T2 (F) and sagittal T1 (G) showed progressive volume loss; persistent abnormal signal in thalami, putamen, and caudate; a focal area of increased T2 signal in the left lentiform nucleus; and a thin corpus callosum.
(H–J) Subject C. Axial T2 (H) and sagittal T1 (I) at 2 months were normal. At 2.5 years, axial T2 (J) revealed atrophy, delayed myelination, and bilateral T2 prolongation in caudate heads and putamina.
Moreover, we identified four subjects (B and D–F) with de novo variants in YWHAG. The 14-3-3 family has seven conserved isoforms that are highly expressed in the mammalian brain. They are involved in a wide variety of biological processes, including intracellular signaling, cell-cycle control, apoptosis, and protein trafficking.40 Knockdown of 14-3-3γ in zebrafish resulted in delayed brain development, reduced brain size, and increased diameter of the heart tube.41 Curiously, in mice, a decrease or increase in 14-3-3γ leads to delayed neuronal migration of pyramidal neurons in the cerebral cortex.42, 43 Hence, normal neuronal migration in the developing brain is exquisitely sensitive to 14-3-3γ levels. Atypical neuronal migration has previously been implicated in epilepsy. Haploinsufficiency of YWHAG has been proposed as a potential cause of infantile spasms in individuals with Williams-Beuren syndrome (WBS [MIM: 194050]),41 a multisystem developmental disorder caused by a recurrent de novo hemizygous deletion of ∼1.5 Mb in chromosomal region 7q11.23.44, 45 Atypical distal 7q11.23 deletions ranging in size from 180 kb to 19.6 Mb have been described in several individuals presenting with a more severe phenotype that includes developmental delay, autistic features, and epilepsy. Three genes—MAGI2 (MIM: 606382), HIP1 (MIM: 601767), and YWHAG—have been suggested as possible candidates for these atypical features.46, 47, 48, 49, 50, 51, 52, 53 Ramocki et al.51 suggested that haploinsufficiency of HIP1 is sufficient to alter neuronal homeostasis and cause focal and generalized epilepsies and cognitive dysfunction. However, their findings do not exclude the possibility that YWHAG loss of function is sufficient to cause neurological phenotypes alone, as proven by our results. Epilepsy and infantile spasms were also described in WBS individuals harboring typical 7q11.23 deletions,54 albeit in relatively few subjects.55 Moreover, a recent paper56 described a subject harboring a WBS-typical 1.41 Mb deletion and presenting with additional infantile-onset refractory EE and severe developmental delay caused by a second, independent de novo mutation in GABRA1 (MIM: 137160). Hence, atypical aspects seen in individuals with a typical 7q11.23 deletion might also be caused by a second co-occurring but unrelated genetic defect.
Subject D harbors a de novo 14-3-3γ p.Glu15Ala variant. Native 14-3-3 exists in monomeric and dimeric states as homo- and heterodimers, respectively, although 14-3-3γ is almost entirely dimeric.57 Glu15 is part of a triad of residues (Leu13, Ala14, and Glu15) necessary for 14-3-3γ dimerization,58, 59 and its ability to complex with other proteins is potentially inhibited by substitution at this site.
The same recurrent de novo 14-3-3γ p.Arg132Cys variant was identified in subjects B, E, and F. The arginine residue at position 132 is part of a highly conserved triad of two arginines and a tyrosine (Arg132-Arg57-Tyr133) (Figure 1) that normally form a positively charged pocket within a binding groove for interacting phosphopeptides.57, 60 An individual (ND27637) with Lennox-Gastaut syndrome and a de novo p.Asp129Glu variant is present in the Epi4K-EPGP dataset.4 The Asp129 residue is also located within the 14-3-3γ binding groove (Figure 1) and plays an important role in determining the orientation of the phosphorylated peptides.57 A de novo mutation affecting position p.Tyr133Ser has been reported in a Deciphering Developmental Disorders cohort individual (DDD4K.00159) severely affected by a neurodevelopmental disorder,19 but we were unable to contact the clinician who followed this subject. Finally, a p.Lys50Gln de novo variant was identified in a subject with autism (AU027A).20 The Lys50 residue is located on the charged face of the binding groove (together with Arg57, Arg61, Arg132, and Tyr133) (Figure 1) and is essential for ligand binding.61, 62, 63
All four subjects (B and D–F) from this study are females with seizure onset in the first year of life, although a precise age of onset for subject F is unknown. Subjects B, E, and F have the same mutation and similar epilepsy phenotype, which includes generalized epilepsy with myoclonic, (atypical) absence, and generalized tonic-clonic (GTC) seizures. Subject E also has prolonged GTC seizures and fever. Subject D’s epilepsy is characterized by episodes of status epilepticus with or without fever. However, no subject had infantile spasms, which have been reported in some WBS individuals with atypical deletions encompassing YWHAG. The majority (three of four) have experienced seizure control on medication. All individuals have developmental delay with intellectual disability, and subject E was also diagnosed with autism spectrum disorder. Subjects B and E have tremor, coordination difficulties, or ataxia, and subjects E and F have scoliosis. Neuroimaging was unremarkable in subjects B, E, and F. The different and less severe clinical presentation of subject D might be explained by a partially different molecular mechanism associated with her specific mutation. Impairment of phosphopeptide binding is only a secondary consequence of impaired protein dimerization.
Empirically, we have demonstrated the validity of brain-specific co-expression for prioritizing EE candidate genes.13, 17 We have identified likely pathogenic mutations in 2 of 20 genes, which enabled us to make a genetic diagnosis in 5% (2/42) of our subjects. However, we do not rule out that mutations in the remaining 18 candidate genes have a role in EE, because more individuals must still be assessed. With the identification of both known and previously unreported mutations, our study provides further evidence for SLC1A2 mutations in EE and suggests a gain-of-function mechanism for this rather severe presentation. Furthermore, our data indicate that YWHAG de novo mutations cause early-onset epilepsy, including EE and intellectual disability, and provide the rationale for therapeutic development to enhance 14-3-3γ dimerization and/or ligand binding. All identified de novo mutations are predicted to impair dimerization and phosphopeptide binding and were ultimately found in seven unrelated subjects, three of whom present with similar epilepsy phenotypes. Nevertheless, it is important to consider both developmental and epileptic components, especially in YWHAG encephalopathy, given protein function and the observed phenotypic heterogeneity.
Consortia
Investigators in the Epilepsy Genomics (EPGEN) Study include Shelin Adam, Cyrus Boelman, Corneliu Bolbocean, Tara Candido, Patrice Eydoux, Gabriella Horvath, Linda Huh, Tanya N. Nelson, Graham Sinclair, Clara van Karnebeek, and Suzanne Vercauteren.
Conflicts of Interest
M.B.C. received research grants and/or speakers honoraria from Union Chimique Belge, Novartis, Biocodex, Eisai, and Sage Therapeutics. All honoraria have been donated to the Epilepsy Research and Development Fund. She also received research grants from the Canadian Institute for Health Research (CIHR) and the Alva Foundation. She is co-chair of the Canadian Paediatric Epilepsy Network. M.J.F. received research support from the Canadian Federal Government (Canada Excellence Research Chair, Canada Foundation for Innovation, and CIHR) and the Cunhill and Alva Foundations. He is a founder and CEO of Neurocode Laboratories, which currently provides clinically accredited exome sequencing to the Province of British Columbia.
Acknowledgments
We thank the parents and children involved. We gratefully acknowledge Dr. Suzanne Vercauteren, MD, PhD (director), Dr. William Gibson, MD, PhD (chair and member of the Biospecimen Advisory Committee), Tamsin Tarling (BioBank administrative manager), and the technical expertise of Katelin N. Townsend of the BC Children’s Hospital BioBank, which is supported by Mining for Miracles through the BC Children’s Hospital Foundation. We also thank the BC Children’s Hospital Department of Pathology & Laboratory Medicine and the BC Children’s Hospital Electroencephalogram Department. Clinical research was performed as part of the Epilepsy Genomics (EPGEN) program on the genetics of early-onset epilepsy at BC Children’s Hospital and the University of British Columbia. We appreciate the support of the Canada Excellence Research Chair (M.J.F.), the Alva Foundation, and the Rare Disease Foundation.
Published: August 3, 2017
Footnotes
Supplemental Data include a Supplemental Note and one table and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.07.004.
Contributor Information
Michelle Demos, Email: mdemos@cw.bc.ca.
Matthew J. Farrer, Email: mfarrer@can.ubc.ca.
Epilepsy Genomics Study:
Shelin Adam, Cyrus Boelman, Corneliu Bolbocean, Tara Candido, Patrice Eydoux, Gabriella Horvath, Linda Huh, Tanya N. Nelson, Graham Sinclair, Clara van Karnebeek, and Suzanne Vercauteren
Accession Numbers
The ClinVar accession number for the data reported in this paper was not available from ClinVar as of the date this article was finalized for press; please contact the corresponding authors for the number.
Web Resources
AnnEX, https://annex.can.ubc.ca
Combined Annotation Dependent Depletion (CADD), http://cadd.gs.washington.edu/
DECIPHER, https://decipher.sanger.ac.uk
ExAC Browser, http://exac.broadinstitute.org/
GeneMatcher, http://genematcher.org
OMIM, http://omim.org
RCSB Protein Data Bank, https://www.rcsb.org/pdb/home/home.do
REVEL: Rare Exome Variant Ensemble Learner, https://sites.google.com/site/revelgenomics/
UCSC Genome Browser, http://genome.ucsc.edu/
Supplemental Data
References
- 1.Scheffer I.E., Berkovic S., Capovilla G., Connolly M.B., French J., Guilhoto L., Hirsch E., Jain S., Mathern G.W., Moshé S.L. ILAE classification of the epilepsies: Position paper of the ILAE Commission for Classification and Terminology. Epilepsia. 2017;58:512–521. doi: 10.1111/epi.13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carvill G.L., Heavin S.B., Yendle S.C., McMahon J.M., O’Roak B.J., Cook J., Khan A., Dorschner M.O., Weaver M., Calvert S. Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nat. Genet. 2013;45:825–830. doi: 10.1038/ng.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Claes L., Del-Favero J., Ceulemans B., Lagae L., Van Broeckhoven C., De Jonghe P. De novo mutations in the sodium-channel gene SCN1A cause severe myoclonic epilepsy of infancy. Am. J. Hum. Genet. 2001;68:1327–1332. doi: 10.1086/320609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen A.S., Berkovic S.F., Cossette P., Delanty N., Dlugos D., Eichler E.E., Epstein M.P., Glauser T., Goldstein D.B., Han Y., Epi4K Consortium. Epilepsy Phenome/Genome Project De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.EuroEPINOMICS-RES Consortium. Epilepsy Phenome/Genome Project. Epi4K Consortium De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies. Am. J. Hum. Genet. 2014;95:360–370. doi: 10.1016/j.ajhg.2014.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kodera H., Nakamura K., Osaka H., Maegaki Y., Haginoya K., Mizumoto S., Kato M., Okamoto N., Iai M., Kondo Y. De novo mutations in SLC35A2 encoding a UDP-galactose transporter cause early-onset epileptic encephalopathy. Hum. Mutat. 2013;34:1708–1714. doi: 10.1002/humu.22446. [DOI] [PubMed] [Google Scholar]
- 7.Lemke J.R., Hendrickx R., Geider K., Laube B., Schwake M., Harvey R.J., James V.M., Pepler A., Steiner I., Hörtnagel K. GRIN2B mutations in West syndrome and intellectual disability with focal epilepsy. Ann. Neurol. 2014;75:147–154. doi: 10.1002/ana.24073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakamura K., Kodera H., Akita T., Shiina M., Kato M., Hoshino H., Terashima H., Osaka H., Nakamura S., Tohyama J. De Novo mutations in GNAO1, encoding a Gαo subunit of heterotrimeric G proteins, cause epileptic encephalopathy. Am. J. Hum. Genet. 2013;93:496–505. doi: 10.1016/j.ajhg.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nava C., Dalle C., Rastetter A., Striano P., de Kovel C.G., Nabbout R., Cancès C., Ville D., Brilstra E.H., Gobbi G., EuroEPINOMICS RES Consortium De novo mutations in HCN1 cause early infantile epileptic encephalopathy. Nat. Genet. 2014;46:640–645. doi: 10.1038/ng.2952. [DOI] [PubMed] [Google Scholar]
- 10.Syrbe S., Hedrich U.B., Riesch E., Djémié T., Müller S., Møller R.S., Maher B., Hernandez-Hernandez L., Synofzik M., Caglayan H.S., EuroEPINOMICS RES De novo loss- or gain-of-function mutations in KCNA2 cause epileptic encephalopathy. Nat. Genet. 2015;47:393–399. doi: 10.1038/ng.3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Torkamani A., Bersell K., Jorge B.S., Bjork R.L., Jr., Friedman J.R., Bloss C.S., Cohen J., Gupta S., Naidu S., Vanoye C.G. De novo KCNB1 mutations in epileptic encephalopathy. Ann. Neurol. 2014;76:529–540. doi: 10.1002/ana.24263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Veeramah K.R., O’Brien J.E., Meisler M.H., Cheng X., Dib-Hajj S.D., Waxman S.G., Talwar D., Girirajan S., Eichler E.E., Restifo L.L. De novo pathogenic SCN8A mutation identified by whole-genome sequencing of a family quartet affected by infantile epileptic encephalopathy and SUDEP. Am. J. Hum. Genet. 2012;90:502–510. doi: 10.1016/j.ajhg.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oliver K.L., Lukic V., Freytag S., Scheffer I.E., Berkovic S.F., Bahlo M. In silico prioritization based on coexpression can aid epileptic encephalopathy gene discovery. Neurol Genet. 2016;2:e51. doi: 10.1212/NXG.0000000000000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francioli L.C., Polak P.P., Koren A., Menelaou A., Chun S., Renkens I., van Duijn C.M., Swertz M., Wijmenga C., van Ommen G., Genome of the Netherlands Consortium Genome-wide patterns and properties of de novo mutations in humans. Nat. Genet. 2015;47:822–826. doi: 10.1038/ng.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong A., Frigge M.L., Masson G., Besenbacher S., Sulem P., Magnusson G., Gudjonsson S.A., Sigurdsson A., Jonasdottir A., Jonasdottir A. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Veltman J.A., Brunner H.G. De novo mutations in human genetic disease. Nat. Rev. Genet. 2012;13:565–575. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- 17.Oliver K.L., Lukic V., Thorne N.P., Berkovic S.F., Scheffer I.E., Bahlo M. Harnessing gene expression networks to prioritize candidate epileptic encephalopathy genes. PLoS ONE. 2014;9:e102079. doi: 10.1371/journal.pone.0102079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deciphering Developmental Disorders Study Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Rubeis S., He X., Goldberg A.P., Poultney C.S., Samocha K., Cicek A.E., Kou Y., Liu L., Fromer M., Walker S., DDD Study. Homozygosity Mapping Collaborative for Autism. UK10K Consortium Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deciphering Developmental Disorders Study Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samocha K.E., Robinson E.B., Sanders S.J., Stevens C., Sabo A., McGrath L.M., Kosmicki J.A., Rehnström K., Mallick S., Kirby A. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka K., Watase K., Manabe T., Yamada K., Watanabe M., Takahashi K., Iwama H., Nishikawa T., Ichihara N., Kikuchi T. Epilepsy and exacerbation of brain injury in mice lacking the glutamate transporter GLT-1. Science. 1997;276:1699–1702. doi: 10.1126/science.276.5319.1699. [DOI] [PubMed] [Google Scholar]
- 26.Cavus I., Kasoff W.S., Cassaday M.P., Jacob R., Gueorguieva R., Sherwin R.S., Krystal J.H., Spencer D.D., Abi-Saab W.M. Extracellular metabolites in the cortex and hippocampus of epileptic patients. Ann. Neurol. 2005;57:226–235. doi: 10.1002/ana.20380. [DOI] [PubMed] [Google Scholar]
- 27.Coulter D.A., Eid T. Astrocytic regulation of glutamate homeostasis in epilepsy. Glia. 2012;60:1215–1226. doi: 10.1002/glia.22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.During M.J., Spencer D.D. Extracellular hippocampal glutamate and spontaneous seizure in the conscious human brain. Lancet. 1993;341:1607–1610. doi: 10.1016/0140-6736(93)90754-5. [DOI] [PubMed] [Google Scholar]
- 29.Haglid K.G., Wang S., Qiner Y., Hamberger A. Excitotoxicity. Experimental correlates to human epilepsy. Mol. Neurobiol. 1994;9:259–263. doi: 10.1007/BF02816125. [DOI] [PubMed] [Google Scholar]
- 30.Yernool D., Boudker O., Jin Y., Gouaux E. Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature. 2004;431:811–818. doi: 10.1038/nature03018. [DOI] [PubMed] [Google Scholar]
- 31.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 32.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Epi4K Consortium De Novo Mutations in SLC1A2 and CACNA1A Are Important Causes of Epileptic Encephalopathies. Am. J. Hum. Genet. 2016;99:287–298. doi: 10.1016/j.ajhg.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jen J.C., Wan J., Palos T.P., Howard B.D., Baloh R.W. Mutation in the glutamate transporter EAAT1 causes episodic ataxia, hemiplegia, and seizures. Neurology. 2005;65:529–534. doi: 10.1212/01.wnl.0000172638.58172.5a. [DOI] [PubMed] [Google Scholar]
- 35.Fairman W.A., Vandenberg R.J., Arriza J.L., Kavanaugh M.P., Amara S.G. An excitatory amino-acid transporter with properties of a ligand-gated chloride channel. Nature. 1995;375:599–603. doi: 10.1038/375599a0. [DOI] [PubMed] [Google Scholar]
- 36.Wadiche J.I., Amara S.G., Kavanaugh M.P. Ion fluxes associated with excitatory amino acid transport. Neuron. 1995;15:721–728. doi: 10.1016/0896-6273(95)90159-0. [DOI] [PubMed] [Google Scholar]
- 37.Melzer N., Biela A., Fahlke C. Glutamate modifies ion conduction and voltage-dependent gating of excitatory amino acid transporter-associated anion channels. J. Biol. Chem. 2003;278:50112–50119. doi: 10.1074/jbc.M307990200. [DOI] [PubMed] [Google Scholar]
- 38.Kovermann P., Machtens J.P., Ewers D., Fahlke C. A conserved aspartate determines pore properties of anion channels associated with excitatory amino acid transporter 4 (EAAT4) J. Biol. Chem. 2010;285:23676–23686. doi: 10.1074/jbc.M110.126557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hotzy J., Schneider N., Kovermann P., Fahlke C. Mutating a conserved proline residue within the trimerization domain modifies Na+ binding to excitatory amino acid transporters and associated conformational changes. J. Biol. Chem. 2013;288:36492–36501. doi: 10.1074/jbc.M113.489385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aghazadeh Y., Papadopoulos V. The role of the 14-3-3 protein family in health, disease, and drug development. Drug Discov. Today. 2016;21:278–287. doi: 10.1016/j.drudis.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 41.Komoike Y., Fujii K., Nishimura A., Hiraki Y., Hayashidani M., Shimojima K., Nishizawa T., Higashi K., Yasukawa K., Saitsu H. Zebrafish gene knockdowns imply roles for human YWHAG in infantile spasms and cardiomegaly. Genesis. 2010;48:233–243. doi: 10.1002/dvg.20607. [DOI] [PubMed] [Google Scholar]
- 42.Wachi T., Cornell B., Marshall C., Zhukarev V., Baas P.W., Toyo-oka K. Ablation of the 14-3-3gamma Protein Results in Neuronal Migration Delay and Morphological Defects in the Developing Cerebral Cortex. Dev. Neurobiol. 2016;76:600–614. doi: 10.1002/dneu.22335. [DOI] [PubMed] [Google Scholar]
- 43.Cornell B., Wachi T., Zhukarev V., Toyo-Oka K. Overexpression of the 14-3-3gamma protein in embryonic mice results in neuronal migration delay in the developing cerebral cortex. Neurosci. Lett. 2016;628:40–46. doi: 10.1016/j.neulet.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 44.Pober B.R. Williams-Beuren syndrome. N. Engl. J. Med. 2010;362:239–252. doi: 10.1056/NEJMra0903074. [DOI] [PubMed] [Google Scholar]
- 45.Galaburda A.M., Wang P.P., Bellugi U., Rossen M. Cytoarchitectonic anomalies in a genetically based disorder: Williams syndrome. Neuroreport. 1994;5:753–757. doi: 10.1097/00001756-199403000-00004. [DOI] [PubMed] [Google Scholar]
- 46.Edelmann L., Prosnitz A., Pardo S., Bhatt J., Cohen N., Lauriat T., Ouchanov L., González P.J., Manghi E.R., Bondy P. An atypical deletion of the Williams-Beuren syndrome interval implicates genes associated with defective visuospatial processing and autism. J. Med. Genet. 2007;44:136–143. doi: 10.1136/jmg.2006.044537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marshall C.R., Young E.J., Pani A.M., Freckmann M.L., Lacassie Y., Howald C., Fitzgerald K.K., Peippo M., Morris C.A., Shane K. Infantile spasms is associated with deletion of the MAGI2 gene on chromosome 7q11.23-q21.11. Am. J. Hum. Genet. 2008;83:106–111. doi: 10.1016/j.ajhg.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martens M.A., Wilson S.J., Reutens D.C. Research Review: Williams syndrome: a critical review of the cognitive, behavioral, and neuroanatomical phenotype. J. Child Psychol. Psychiatry. 2008;49:576–608. doi: 10.1111/j.1469-7610.2008.01887.x. [DOI] [PubMed] [Google Scholar]
- 49.Mizugishi K., Yamanaka K., Kuwajima K., Kondo I. Interstitial deletion of chromosome 7q in a patient with Williams syndrome and infantile spasms. J. Hum. Genet. 1998;43:178–181. doi: 10.1007/s100380050064. [DOI] [PubMed] [Google Scholar]
- 50.Morimoto M., An B., Ogami A., Shin N., Sugino Y., Sawai Y., Usuku T., Tanaka M., Hirai K., Nishimura A. Infantile spasms in a patient with williams syndrome and craniosynostosis. Epilepsia. 2003;44:1459–1462. doi: 10.1046/j.1528-1157.2003.34703.x. [DOI] [PubMed] [Google Scholar]
- 51.Ramocki M.B., Bartnik M., Szafranski P., Kołodziejska K.E., Xia Z., Bravo J., Miller G.S., Rodriguez D.L., Williams C.A., Bader P.I. Recurrent distal 7q11.23 deletion including HIP1 and YWHAG identified in patients with intellectual disabilities, epilepsy, and neurobehavioral problems. Am. J. Hum. Genet. 2010;87:857–865. doi: 10.1016/j.ajhg.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Earhart B.A., Williams M.E., Zamora I., Randolph L.M., Votava-Smith J.K., Marcy S.N. Phenotype of 7q11.23 duplication: A family clinical series. Am. J. Med. Genet. A. 2017;173:114–119. doi: 10.1002/ajmg.a.37966. [DOI] [PubMed] [Google Scholar]
- 53.Fusco C., Micale L., Augello B., Teresa Pellico M., Menghini D., Alfieri P., Cristina Digilio M., Mandriani B., Carella M., Palumbo O. Smaller and larger deletions of the Williams Beuren syndrome region implicate genes involved in mild facial phenotype, epilepsy and autistic traits. Eur. J. Hum. Genet. 2014;22:64–70. doi: 10.1038/ejhg.2013.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nicita F., Garone G., Spalice A., Savasta S., Striano P., Pantaleoni C., Spartà M.V., Kluger G., Capovilla G., Pruna D. Epilepsy is a possible feature in Williams-Beuren syndrome patients harboring typical deletions of the 7q11.23 critical region. Am. J. Med. Genet. A. 2016;170A:148–155. doi: 10.1002/ajmg.a.37410. [DOI] [PubMed] [Google Scholar]
- 55.Samanta D. Infantile spasms in Williams-Beuren syndrome with typical deletions of the 7q11.23 critical region and a review of the literature. Acta Neurol. Belg. 2017;117:359–362. doi: 10.1007/s13760-016-0635-0. [DOI] [PubMed] [Google Scholar]
- 56.Popp B., Trollmann R., Büttner C., Caliebe A., Thiel C.T., Hüffmeier U., Reis A., Zweier C. Do the exome: A case of Williams-Beuren syndrome with severe epilepsy due to a truncating de novo variant in GABRA1. Eur. J. Med. Genet. 2016;59:549–553. doi: 10.1016/j.ejmg.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 57.Yang X., Lee W.H., Sobott F., Papagrigoriou E., Robinson C.V., Grossmann J.G., Sundström M., Doyle D.A., Elkins J.M. Structural basis for protein-protein interactions in the 14-3-3 protein family. Proc. Natl. Acad. Sci. USA. 2006;103:17237–17242. doi: 10.1073/pnas.0605779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Valente C., Turacchio G., Mariggiò S., Pagliuso A., Gaibisso R., Di Tullio G., Santoro M., Formiggini F., Spanò S., Piccini D. A 14-3-3γ dimer-based scaffold bridges CtBP1-S/BARS to PI(4)KIIIβ to regulate post-Golgi carrier formation. Nat. Cell Biol. 2012;14:343–354. doi: 10.1038/ncb2445. [DOI] [PubMed] [Google Scholar]
- 59.Zhou Y., Reddy S., Murrey H., Fei H., Levitan I.B. Monomeric 14-3-3 protein is sufficient to modulate the activity of the Drosophila slowpoke calcium-dependent potassium channel. J. Biol. Chem. 2003;278:10073–10080. doi: 10.1074/jbc.M211907200. [DOI] [PubMed] [Google Scholar]
- 60.Skjevik A.A., Mileni M., Baumann A., Halskau O., Teigen K., Stevens R.C., Martinez A. The N-terminal sequence of tyrosine hydroxylase is a conformationally versatile motif that binds 14-3-3 proteins and membranes. J. Mol. Biol. 2014;426:150–168. doi: 10.1016/j.jmb.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang L., Wang H., Liu D., Liddington R., Fu H. Raf-1 kinase and exoenzyme S interact with 14-3-3zeta through a common site involving lysine 49. J. Biol. Chem. 1997;272:13717–13724. doi: 10.1074/jbc.272.21.13717. [DOI] [PubMed] [Google Scholar]
- 62.Jagemann L.R., Pérez-Rivas L.G., Ruiz E.J., Ranea J.A., Sánchez-Jiménez F., Nebreda A.R., Alba E., Lozano J. The functional interaction of 14-3-3 proteins with the ERK1/2 scaffold KSR1 occurs in an isoform-specific manner. J. Biol. Chem. 2008;283:17450–17462. doi: 10.1074/jbc.M709185200. [DOI] [PubMed] [Google Scholar]
- 63.Nefla M., Sudre L., Denat G., Priam S., Andre-Leroux G., Berenbaum F., Jacques C. The pro-inflammatory cytokine 14-3-3ε is a ligand of CD13 in cartilage. J. Cell Sci. 2015;128:3250–3262. doi: 10.1242/jcs.169573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.