Abstract
Specialized myocytes of the cardiac conduction system (CCS) are essential to coordinate sequential contraction of cardiac atria and ventricles. Anomalies of the CCS can result in lethal cardiac arrhythmias, including sick sinus syndrome, and atrial or ventricular fibrillation. To develop future therapies and regenerative medicine aimed at cardiac arrhythmias, it is important to understand formation and function of distinct components of the CCS. Essential to this understanding is the development of CCS specific markers. In this review, we briefly summarize available mouse models of CCS markers, and focus on those involving the hyperpolarization cation-selective nucleotide gated cation channel, HCN4, which selectively marks all components of the specialized CCS in adult heart. Recent studies have revealed, however, that HCN4 expression during development is highly dynamic in cardiac precursors. These studies have offered insights into the contributions of the first and second heart field to myocyte and conduction system lineages, and suggested the timing of allocation of specific conduction system precursors during development. Altogether, they have highlighted the utility of HCN4 as a cell surface marker for distinct components of the CCS at distinct stages of development, which can be utilized to facilitate purification and characterization of CCS precursors in mouse and human model systems and pave the way for regenerative therapies.
Introduction
Normal heart function is critically dependent on electrical impulses generated by the specialized cardiac conduction system (CCS), which ensures coordinated contraction of atria, then ventricles, to optimize blood delivery and return. The CCS is comprised of central nodal tissues, which include the sinoatrial node (SAN, pacemaker), and the atrioventricular node (AVN, conduction delayer), and rapid-conducting peripheral components, including the interatrial conduction tracts, the His Bundle (HB), Bundle Branches (BB), and Purkinje fibers (PF) (Fig.1).
Figure 1. The specialized Cardiac Conduction System (CCS).
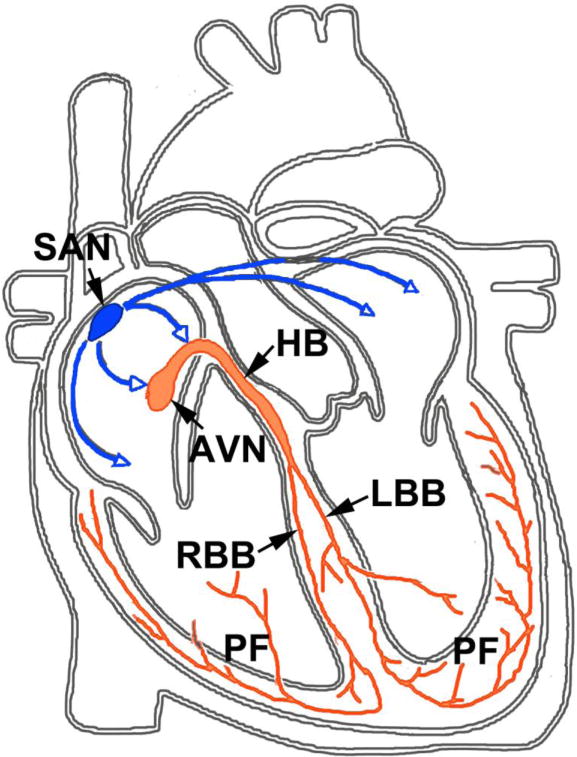
Abbreviations: Sinoatrial node (SAN), atrioventricular node (AVN) and the intermodal fibers, His-bundle (HB), bundle branches (RBB and LBB), and Purkinje fibers (PF).
Cardiac arrhythmias occur preferentially in areas derived from the CCS (Al-Khatib and Pritchett, 1999; Haissaguerre et al., 1998; Katritsis et al., 2002; Tsai et al., 2000), and abnormalities in CCS development may predispose individuals to arrhythmias and sudden death (Gourdie et al., 2003; Jongbloed et al., 2012). Thus, understanding genes and signaling pathways underlying CCS development is of critical importance to gain molecular insights into human cardiac arrhythmias and to develop more effective therapies.
Recent studies have given us great insight into molecular pathways required for CCS development and function, as recently reviewed in depth (Bakker et al., 2010a; Bakker et al., 2010b; Miquerol et al., 2011). The focus of this review will be to briefly summarize available mouse models for markers of the conduction system, with emphasis on recent studies highlighting the properties of the hyperpolarization cation-selective nucleotide gated cation channel, HCN4, as a marker for earliest differentiating myocytes during cardiogenesis, and the developing and adult CCS (Liang et al., 2013; Spater et al., 2013).
Mouse models and markers that facilitate studies of the CCS
During the past two decades, a number of transgenic or knockin mouse models have been generated with reporter genes or Cre recombinase being specifically or predominantly expressed in the CCS. These lines have been useful for visualization, isolation and characterization of CCS components, and have provided invaluable tools to delete genes of interest specifically in distinct components of the CCS (Table).
Table.
Mouse Models of the Cardiac Conduction System
| Mouse | Genetic Manipulation | Developmental Stages | Cardiac conduction system | Other cardiac expression | Refs | ||||
|---|---|---|---|---|---|---|---|---|---|
| SAN | AVN | HB | BB | PF | |||||
| minK-CreERT2 | transgenic | E9.5-adult | − | + | + | + | + | A, V, IVS | (Kondo et al., 2003; Kupershmidt et al., 1999) |
| Cx40-CreERT2 | knockin | E9.5-adult | − | +/− | + | + | + | A, EC, CA, | (Beyer et al., 2011) |
| HCN4-CreERT2 | knockin | E6.5-adult | + | + | + | + | + | LV, EC | (Hoesl et al., 2008; Liang et al., 2013; Spater et al., 2013) |
| Hcn4-CreERT2 | transgenic | E7.5-adult | + | + | + | + | + | LV, EC | (Wu et al., 2014) |
| Tbx2-Cre | knockin | E9.5-adult | − | + | − | − | − | AVC | (Aanhaanen et al., 2009; Aanhaanen et al., 2010) |
| cGATA6-Cre | transgenic | E8.5-neonates | − | + | + | − | − | A, LV | (Davis et al., 2001) |
| Tbx3-Cre | knockin | E11.5-adult | + | + | + | − | − | No | (Hoogaars et al., 2007) |
| Tbx18-Cre | knockin | E10.5-adult | + | − | − | − | − | EPI, EPDCs, SH | (Aanhaanen et al., 2010; Liang et al., 2013) |
| Isl1-Cre, | knockin | E8.5-adult | + | − | − | RBB | RPF | OFT, RV, A | (Liang et al., 2013; Sun et al., 2007) |
| Shox2-Cre | knockin | E9.0-adult | + | − | − | − | − | No | (Sun et al., 2013) |
| Shox2-LacZ | knockin | E9.5–12.5 | + | − | − | − | − | IVS | (Sun et al., 2013) |
| HCN4-nEGFP | knockin | E7.5-adult | + | + | + | + | + | LV (~ E12.5), EC | (Liang et al., 2013; Sun et al., 2007) |
| HCN4-nLacZ | knockin | E7.5-adult | + | + | + | + | + | LV (~ E12.5), EC | (Liang et al., 2013) |
| cGATA6-lacZ | transgenic | E7.5-neonate | − | + | + | − | − | A, LV | (Davis et al., 2001) |
| Troponin I-lacZ | transgenic | E9.5-adult | − | + | − | − | − | A, ring | (Di Lisi et al., 2000) |
| Des1-nLacZ | transgenic | E8–18 | − | − | − | + | + | No | (Li et al., 1993) |
| HF-1b-LacZ | knockin | E8.5-adult | − | + | + | + | + | RA | (Nguyen-Tran et al., 2000) |
| CCS-lacZ | transgenic | All stages | +/− | + | + | + | + | RA, VV | (Rentschler et al., 2001) |
| mink-lacZ | knockin | E8.5-Adult | − | + | + | + | + | OFT, IVS, AS, VV | (Kondo et al., 2003; Kupershmidt et al., 1999) |
| HOP-lacZ | knockin | E16.5-Adult | − | + | + | + | + | A, V | (Ismat et al., 2005) |
| Cntn2-lacZ | knockin | adult | + | + | + | + | + | No | (Pallante et al., 2010) |
| Cntn2-GFP | transgenic | adult | + | + | + | + | + | No | (Pallante et al., 2010) |
| Popdc1-LacZ | knockin | adult | + | ? | + | + | − | CMC | (Froese et al., 2012) |
| Popdc2-LacZ | knockin | adult | + | + | + | + | − | CMC | (Froese et al., 2012) |
| Cx40-EGFP | knockin | E9.5-adult | − | +/− | + | + | + | EC, A | (Miquerol et al., 2004) |
| Cx45-LacZ | knockin | E9.5-adult | + | + | + | + | − | A, V | (Kreuzberg et al., 2005; Kruger et al., 2000) |
| Cx30-LacZ | knockin | adult | + | − | − | − | − | No | (Gros et al., 2010) |
| Cx30.2-LacZ | knockin | E10.5-Adult | + | + | + | + | +? | No | (Kreuzberg et al., 2006; Kreuzberg et al., 2005) |
| Tbx3-GFP | transgenic | E9.5-adult | − | + | − | − | − | AVC | (Horsthuis et al., 2009) |
| Tbx18-GFP | knockin | E10.5-? | + | − | − | − | − | Epi, EPDCs, SH | (Wiese et al., 2009) |
| ISL1-nLacZ | knockin | E9.5-adult | + | − | − | − | − | OFT, RV, A | (Sun et al., 2007) |
| Irx3-tauLacZ | knockin | E10-adult | − | − | + | + | + | No | (Zhang et al., 2011) |
| mIrx3-EGFP | transgenic | E14.5-P3 | − | − | + | + | + | No | (Zhang et al., 2011) |
| Id2-LacZ | transgenic | transient E14.5 | − | − | + | + | − | No | (Moskowitz et al., 2007) |
Abbreviations: A, atria; AS, atrial septum; AVC, atrioventricular canal; AVN, atrioventricular node; BB, bundle branch; CMC, cardiomyocyte; EC, endothelial cell; CA, coronary artery; EPI, epicardium; EPDCs, epicardial-derived cells; His-bundle; IVS, interventricular septum; LV, left ventricle; OFT, outflow tract; PF, Purkinje fiber; RBB, right bundle branch; RPF, right Purkinje fiber; RA, right atria; RV, right ventricle; SAN, sinoatrial node; SH, sinus horn; V, ventricle; VV, venous valve.
In four widely used CCS-reporter mouse lines, Mink-LacZ, cGata6-LacZ, Troponin I-LacZ and CCS-LacZ (Davis et al., 2001; Di Lisi et al., 2000; Kondo et al., 2003; Kupershmidt et al., 1999; Rentschler et al., 2001), LacZ transgene expression has been documented throughout the conduction system, with the notable exception of the SAN. MinK is a potassium channel subunit which co-assembles with KvLQT1 to form the slowly activated, delayed rectifier K current that plays an important role in cardiac depolarization(Barhanin et al., 1996; Sanguinetti et al., 1996). Gata6 is a transcription factor whose function in the atrioventricular node has not yet been addressed, although the highly related transcription factor Gata4 plays a critical role in atrioventricular node function(Munshi et al., 2009). CCS-LacZ is a transgenic insertion of lacZ with complex genomic rearrangement resulting in expression within the CCS (Stroud et al., 2007).
Both CCS-LacZ and Cx40-GFP (Miquerol et al., 2013; Miquerol et al., 2004) facilitate visualization of Purkinje fibers. Connexin 40 (Cx40) is a relatively fast-conducting gap junction protein expressed in atrial myocytes and the ventricular conduction system, and is also required for normal conduction system function (Bevilacqua et al., 2000). Gene profiling of CCS-LacZ expressing Purkinje fibers identified Contactin2 as a new marker for Purkinje fibers (Pallante et al., 2010). Contactin2-LacZ or -EGFP labels Purkinje fibers and a subset of cells in central components of the CCS. Mink-LacZ has been used to perform gene profiling of isolated left bundle branch, resulting in identification of the transcription factor Id2 as a marker for bundle branches that is also required for normal bundle branch development (Moskowitz et al., 2007).
Transcription factors Shox2, Tbx18, Isl1, and Tbx3, and the gap junction channel Cx30 specifically mark the SAN, while Tbx3 additionally labels the atrioventricular conduction system (Gros et al., 2010; Horsthuis et al., 2009; Sun et al., 2013; Sun et al., 2007; Wiese et al., 2009). Shox2 is required for SAN differentiation and for preventing ectopic expression of atrial “working” myocardial genes within the SAN, including the transcription factor Nkx2.5, and the gap junction channel Cx40 (Blaschke et al., 2007; Espinoza-Lewis et al., 2009). Tbx18 is required for SAN growth, and overexpression of Tbx18 in non-conduction cardiomyocytes is sufficient for induction of SAN gene expression and pacemaker function (Christoffels et al., 2006; Kapoor et al., 2013; Wiese et al., 2009). A 160kb BAC containing the Tbx3 coding region and flanking sequences was used to generate a transgenic mouse line, and results in EGFP expression specifically in the AVN, but not in the SAN (Horsthuis et al., 2009). Gene profiling of AVN cells from Tbx3-GFP mice has revealed gene programs involved in formation and phenotype of the developing AVN (Horsthuis et al., 2009). Tbx3 is required for normal development and function of both SAN and AVN (Bakker et al., 2008; Frank et al., 2012; Wiese et al., 2009).
The hyperpolarization-activated cation-selective nucleotide-gated channel 4 (HCN4), encodes the “funny” current, required for pacemaking function in the heart (Bucchi et al., 2012; Stieber et al., 2003). Recently, a number of mutations in HCN4 have been associated with human arrhythmias, in particular sinus bradycardia (DiFrancesco, 2013). A number of HCN4 knockin and transgenic mouse lines have been generated, including HCN4-CreERT2, -LacZ and -GFP, and HCN4 reporters have been shown to delineate the entire CCS from E16.5 onward (Hoesl et al., 2008; Liang et al., 2013; Spater et al., 2013; Sun et al., 2007; Wu et al., 2014). In this review, we focus on HCN4 as a marker for progenitors of the first heart field and cardiac conduction system, and discuss the contribution of the first and second heart field progenitors to the formation of the pacemaker and cardiac conduction system.
Myocardial Lineages Derive from the First and Second Lineage
The formation of the heart is a complex morphogenetic process involving multiple cell types, including myocytes, endocardium, smooth muscle cells, fibroblasts, vascular endothelium, and vascular support cells. These cell types are derived from early mesodermal heart fields and two extrinsic populations, the cardiac neural crest and epicardium. Early retroviral labeling studies in avian species demonstrated that cells of the CCS derive from myocardial lineages (Mikawa and Fischman, 1996). Based on dye labeling and retrospective clonal analyses, myocardial lineages in the heart arise from two distinct lineages, the first and second myocardial lineage, based on their timing of differentiation and entry into the heart (Buckingham et al., 2005; Evans et al., 2010). Just prior to or at gastrulation, cardiogenic mesodermal cells are diversified into the first and second heart lineages that will give rise to the first and second heart field. The first heart field (FHF), derived from the first lineage, is comprised of the first myocardial cells to differentiate in the cardiac crescent, and will give rise to cells in the primitive heart tube and later cells of the left ventricle and portions of the atria (Buckingham et al., 2005; Kelly, 2012; Meilhac et al., 2004).
The second heart field (SHF), derived from the second lineage, is located in pharyngeal mesoderm medial and dorsal to the FHF at crescent stages, is proliferative, and exhibits a complex migratory pathway, where SHF progenitors that will contribute to the anterior pole migrate in an anterior direction to populate the second pharyngeal arch, and contribute to the outflow tract and right ventricle, and SHF progenitors that will contribute to the posterior pole migrate to contribute to both atria and the dorsal mesenchymal protrusion (Buckingham et al., 2005; Cai et al., 2003; Dyer and Kirby, 2009; Kelly, 2012). SHF progenitors are multipotent and can differentiate into cardiomyocytes, smooth muscle and endothelial cells(De Val et al., 2004; Moretti et al., 2006; Sun et al., 2007). Clonal lineage analysis has demonstrated that cells of the atrioventricular canal region are derived from both the first and second heart lineages (Harvey et al., 2009). At early forming heart tube stages, Tbx2 is expressed in a posterior domain, in cells that, by complementary dye labeling studies, will give rise to the atrioventricular canal region of the early looping heart (Aanhaanen et al., 2009; Dominguez et al., 2012; Meilhac et al., 2004). Tbx2 is actively expressed in the region of the atrioventricular canal, but is not observed in left ventricular myocardium. However, lineage studies with Tbx2Cre demonstrated a significant contribution of Tbx2 expressing atrioventricular myocardium to the left ventricular free wall by E10.5, indicating expansion of the atrioventricular myocardium into this region, demonstrating unexpected conversion of cells with atrioventricular canal identity to a working myocardial identity, and attesting to a plasticity of compartment boundaries during the course of cardiac morphogenesis.
Insights into lineage contributions of the SHF have been facilitated by a number of markers. The LIM homeodomain transcription factor Isl1 is actively expressed in all progenitors of the SHF, with its mRNA expression being downregulated upon differentiation (Cai et al., 2003; Sun et al., 2007). The secreted growth factor Fgf10 and an enhancer of the transcription factor Mef2C label the anterior SHF (Kelly et al., 2001; Verzi et al., 2005). Tbx1 is expressed in a subset of anterior SHF progenitors that contribute to the distal right ventricular outflow tract and pulmonary artery (Maeda et al., 2006). The discovery of specific SHF markers and development of related Cre mouse lines have provided invaluable tools to track, isolate and characterize these progenitors that have greatly enhanced our understanding of cardiac morphogenesis. Studies of the lineage potential and contributions of the FHF, however, have been hampered by lack of specific markers. This is particularly relevant, given that a majority of human heart diseases mainly affect left ventricle, and for more effective cardiac regenerative therapies, the progenitors of left ventricular cardiomyocytes are much needed.
Earliest Expression of HCN4 Marks the First Heart Field
HCN4 is a marker of the pacemaker of the heart, the SAN, both during development and adulthood (Ludwig et al., 1998; Stieber et al., 2003). Expression of HCN4 is first detected in the cardiac crescent and is maintained throughout the early heart tube (Garcia-Frigola et al., 2003). These observations suggested that HCN4, in addition to being a marker for pacemaker cells of the heart, might be a marker of the first heart field. To further investigate the contribution of HCN4 lineage cells to the heart and the dynamic expression of HCN4 during heart development, a number of HCN4 knockin mouse lines have been generated by our lab and others, including HCN4-nLacZ, -H2BGFP and -CreERT2 (Hoesl et al., 2008; Liang et al., 2013; Spater et al., 2013).
At cardiac crescent stages, in situ hybridization and coimmunostaining revealed that expression of HCN4 or HCN4 reporters (nLacZ or H2BGFP) partially overlaps with Nkx2-5 (marker of both the FHF and SHF), Tbx5 (preferential marker of the FHF at this stage), but not with Isl1 (SHF marker), suggesting that earliest HCN4 expression marks the FHF (Liang et al., 2013; Spater et al., 2013) (Fig. 2). In addition, expression of HCN4-nLacZ and -GFP at early developmental stages is also prominent in left ventricular myocardium and atria, consistent with HCN4 expression in the first heart field (Liang et al., 2013).
Figure 2. Early HCN4 expression marks the first heart field.
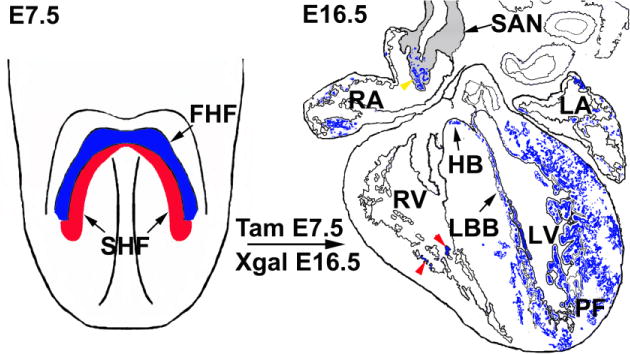
From an anterior view of the mouse embryo at E7.5, HCN4 is expressed in the first differentiating myocardial cells of the cardiac crescent, the first heart field (FHF), but at this stage is not expressed in the second heart field (SHF) as marked by the transcription factor Isl1. Tamoxifen induction of HCN4CreERT2; Rosa indicator mice from E6.0–E7.5 results in labeling of first heart field derived lineages, including myocytes within left ventricle (LV), His-bundle (HB), left bundle branch (LBB), Purkinje fibers (PF) within LV, and a subset within right ventricle (RV) (red arrowheads). A subset of pacemaker cells within the sinoatrial node (SAN) tail are also labeled (yellow arrowhead). It is important to note that although HCN4 is restricted to the FHF at E7.5, at later stages, HCN4 will be expressed in second heart field-derived cells within the SAN, and in other parts of the CCS which are derived from the second heart field (see Figs. 3 and 4).
Lineage studies with HCN4CreERT2 and lineage reporters confirmed the contribution of the earliest HCN4 expressing cells to the first heart field, as inductions from E6.0 to E7.5 result in preferential labeling of the left ventricle and parts of the atria (Hoesl et al., 2008; Liang et al., 2013; Spater et al., 2013) (Fig. 2). The first heart field, as labeled by HCN4CreERT2 at crescent stages, gives rise to myocardial lineages, but not endothelial or smooth muscle lineages, as evidenced by coimmunostaining of HCN4CreERT2 lineages induced at crescent stages with markers for endothelial (PECAM), myocardial (MF20, Tnnt2, Actn2) and smooth muscle (smMHC) cells (Liang et al., 2013; Spater et al., 2013). These data suggest that the FHF, in contrast to the SHF, is restricted to a myocardial fate in vivo. To investigate the in vitro potential of FHF cells, Spater et al (Spater et al., 2013) FACS purified HCN4CreERT2 lineage traced cells from E7.5–E8.0 embryos and performed clonal analyses. 18% of the cells plated showed limited expansion and almost exclusive differentiation into cardiomyogenic clones, with no positive staining for PECAM or smMHC observed. Human embryonic stem cell derived cardiomyocytes were FACS sorted utilizing antibody to HCN4 at day 6 or 7 of differentiation, and were found to comprise approximately 2% of the population at this stage, with quantitative PCR analyses demonstrating expression of HCN4, and enrichment for TBX5 and NKX2-5. Expression of SHF markers ISL1 and MEF2c were not significantly enriched, suggesting HCN4 may also be a marker of the FHF in human cardiomyocytes.
Later, Dynamic Expression of HCN4 in Cardiomyocytes and the CCS
Following its expression in the cardiac crescent, HCN4 is expressed throughout the early heart tube from E8.0 to E8.5 (Garcia-Frigola et al., 2003; Liang et al., 2013; Spater et al., 2013; Vedantham et al., 2013) (Fig. 3). At E9.5, HCN4 is expressed at low levels throughout the early looping heart tube, and is highly expressed in the sinus venosus region, where the early pacemaker is localized (Kamino et al., 1981; Van Mierop, 1967). From E10.5 to E12.5, HCN4, and HCN4 reporter transgenes are highly expressed in the coronary sinus, SAN, AVN, and left bundle branch. At E10.5 to E12.5, expression of HCN4 reporter transgenes, and to a lesser extent, endogenous HCN4 mRNA, is evident in left ventricular myocardium and atria (Fig. 3). By E16.5 until adult stages, HCN4 is highly expressed within all parts of the CCS, including the SAN, AVN, His-bundle, bundle branches, and Purkinje fibers (Fig. 3). Beginning at E12.5, HCN4 is also expressed in endothelial cells of the aorta and pulmonary artery, co-expressed with PECAM (Liang et al., 2013). Thus, HCN4 expression in the CCS is very dynamic. From E16.5 to adult stages, HCN4 is expressed in all components of the well delineated CCS (Liang et al., 2013).
Figure 3. Cardiac expression of HCN4 during mouse development.
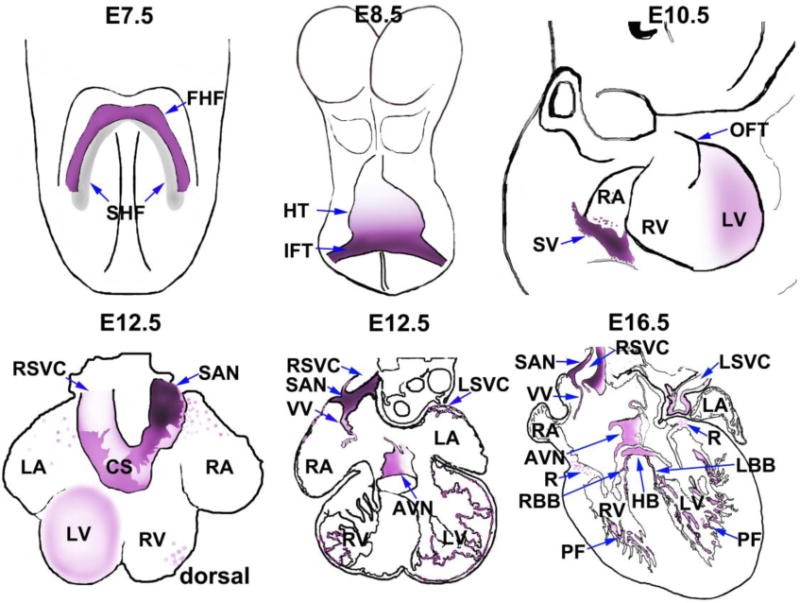
From an anterior view of the mouse embryo at E7.5, HCN4 expression is in the first differentiating myocardial cells of the cardiac crescent, the first heart field (FHF), but not the second heart field (SHF). From an anterior view at E8.5, HCN4 is strongly expressed in the inflow tract, and more weakly expressed throughout the more posterior heart tube (HT). At E10.5, from a right lateral viewpoint, showing the region of the embryo containing the heart, HCN4 is highly expressed in the sinus venosus (SV), including the junction with the caval vein, in the region of the prospective sinoatrial node. Faint expression is also still observed within the left ventricle (LV). Two views of the heart are diagrammed for E12.5, one a whole mount dorsal view, and the second a representative en face section from the heart. From the whole mount dorsal view, HCN4 expression is prominent in the coronary sinus (CS) and sinoatrial node (SAN), with fainter expression in the left ventricle, and scattered expression in other regions of the heart. From the section view, HCN4 is significantly expressed in the sinoatrial node (SAN), venous valves (VV), left superior vena cava (LSVC), atrioventricular node, and Purkinje fibers (PF) in right and left ventricle. HCN4 expression is also observed in the atrioventricular ring (R). From an enface section at E16.5, HCN4 is highly expressed within all parts of the CCS, including SAN, AVN, His-bundle (HB), right and left bundle branch (RBB, LBB), and Purkinje fibers (PF) of both ventricles. HCN4 expression is also found in the venous valves (VV), atrioventricular ring (R) and a subset of cells in the left superior vena cava (LSVC).
Tracking the Allocation of Temporally Distinct HCN4 Expressing Cells to the Cardiac Conduction System
Previous studies have shown that the SAN develops from sinus horn myocardium characterized by the expression of Tbx18, Tbx3, Shox2 and Isl1 (Munshi, 2012; Sizarov et al., 2011; Wiese et al., 2009), and that the AVN is derived from atrioventricular canal myocardium (Aanhaanen et al., 2009). Early retroviral labeling studies demonstrated that Purkinje fibers and working myocyte lineages share a common progenitor, findings recently reinforced by recent studies with Connexin40 mouse models (Gourdie et al., 2003; Miquerol et al., 2013; Miquerol et al., 2004).
Early expression of HCN4 in the first heart field, and dynamic expression of HCN4 in distinct subsets of conduction system precursors suggested that “pulse-chase” experiments with HCN4CreERT2 might provide insights into contributions of the first heart field to conduction system lineages, and also into the potential conduction system fate of temporally distinct HCN4 expressing populations. Tamoxifen inductions were performed at distinct time points and harvested at E16.5, when HCN4 is expressed throughout the conduction system, and gave insights into the progressive allocation of HCN4 expressing myocytes to distinct components of the CCS (Liang et al., 2013) (Fig. 4A).
Figure 4. Model for lineage origin of CCS precursors in developing heart.
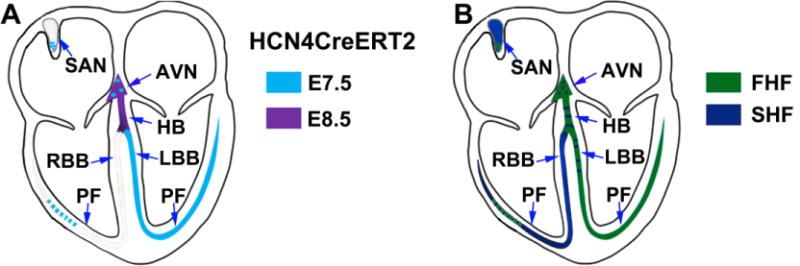
A) Timing of allocation of early HCN4 expressing cells to the CCS. Tamoxifen induction of HCN4CreERT2 embryos at E7.5, harvesting at E16, revealed contributions of HCN4 lineage labeled cells to left Purkinje fibers (PF), left bundle branch (LBB), and some cells in right bundle branch (RBB), atrioventricular node (AVN) and cells within the tail of the sinoatrial node (SAN) (blue). Tamoxifen inductions of HCN4CreERT2 embryos at E8.5, harvesting at E16, revealed the same populations as labeled by inductions at E7.5, with additional contributions of HCN4 expressing cells to His-bundle (HB) and AVN (purple). However, a majority of cells of the SAN and right ventricular conduction system are not derived from these early HCN4 lineages. Here, we are selectively examining contributions of HCN4 expressing cells to the CCS, but it should be noted that inductions at these early stages also label populations of working myocytes (see Fig. 2). B) Contributions of first and second heart field progenitors to the CCS. Lineage studies performed with HCN4CreERT2 at early stages to mark the first heart field were complemented by lineage studies utilizing Isl1Cre, and suggested first and second heart field contributions to the CCS. The SAN and right ventricular conduction system are derived from the second heart field (dark blue), whereas the AVN, HB and left ventricular conduction system are from the first heart field (green). Note that a small subset of cells in SAN and right PF are from the first heart field, and a small subset of cells in AVN, HB and LBB are also derived from the second heart field.
Tamoxifen inductions of HCN4CreERT2 at E7.5 resulted in labeling of a subset of cells in coronary sinus, SAN tail, venous valves, left bundle branch, Purkinje fibers, left ventricle, ventricular septum, and both atria. Infrequent labeling was observed in some Purkinje fibers of the right ventricle. Therefore the first heart field contains precursors of each of these lineages, consistent with previous data suggesting a common origin of working ventricular cardiomyocytes and Purkinje fibers. No labeling of right bundle branch or His-bundle was observed.
Inductions at E8.5 labeled the same populations as inductions at E7.5, and, in addition, the SAN head, AVN, His-bundle, and atrial septum. Inductions at E9.5 resulted in more restricted cell populations than at E8.5, with reduced labeling of Purkinje fibers, left ventricle, and atria, and strong labeling of the SAN, AVN, atrial septum, left superior vena cava, and coronary sinus. Inductions at E11.5 or E12.5 resulted in labeling of SAN, AVN, venous valves, left superior vena cava, and coronary sinus. A few cells in the His-bundle were labeled, but few or no labeled cells were observed in left bundle branch, Purkinje fibers, or left ventricle. As expected, inductions at E16.5, harvested at P1, resulted in labeling of all CCS components, including those not labeled by inductions at E12.5, indicating re-expression or de novo expression of HCN4 in distinct components of the CCS at later stages.
HCN4CreERT2 induction studies were also complemented by lineage studies using Isl1Cre (second heart field and SAN), Nkx2-5Cre (first and second heart field), and Tbx18Cre (posterior second heart field)(Bertrand et al., 2011; Dominguez et al., 2012; Lescroart et al., 2012) to examine contributions of these Cre lineages to the CCS. Results of these studies suggested a major contribution of the second heart field to the SAN, right bundle branches, and right Purkinje fibers, with minor contributions to other components of the CCS (Fig. 4B).
We found that the earliest HCN4-expressing cells of the first heart field contribute a few cells to the SAN tail, but later HCN4-expressing cells from the posterior second heart field contribute a majority of cells to the SAN (Fig. 4B). HCN4 is expressed very early in precursors of the left Purkinje fibers and left bundle branch, and slightly later also marks precursors of the His-bundle and AVN. A previous lineage study with Tbx2Cre revealed that a majority of atrial components of the conduction system including the AVN are derived from Tbx2+ atrioventricular canal myocardium, whereas the ventricular components of the conduction system including the His-bundle are derived from the ventricular myocardium (Aanhaanen et al., 2009). Together, these observations suggest that AVN and His-bundle may develop from distinct precursors that segregate early in development and that left bundle branch precursors differentiate before precursors of the AVN and His-bundle. Our lineage studies with HCN4CreERT2, as well as a previous study with Cx40CreERT2 (Beyer et al., 2011), reaffirm the lineage derivation of the Purkinje fibers from trabecular myocardium (Mikawa and Fischman, 1996). Furthermore, consistent with their derivation from the first heart lineage, a majority of cells within the AVN, His-bundle, left bundle branches and Purkinje fibers were selectively labeled by Nkx2-5Cre, but not Isl1Cre. In contrast, a majority of the SAN and right ventricular conduction system are derived from the second heart field, labeled by Isl1Cre. However, Tbx18Cre lineages that contribute a majority of cells to the SAN head, appeared not to contribute to AVN, His-bundle or ventricular conduction system.
HCN4 as a cell surface marker of the first heart field and CCS precursors
Recent studies utilized antibodies to the cell surface molecule CD166/Alcam to isolate a population of CD166+ pacemaker precursors from differentiating mouse ESCs that express high levels of genes involved in SAN development (Tbx18, Tbx3, Isl-1, Shox2) and function (Cx30.2, HCN4, HCN1, CaV1.3) (Scavone et al., 2013). In culture, CD166+ cells can differentiate into autorhythmic cells morphologically similar to, and with electrophysiological properties of murine SAN myocytes. Recent studies with hESCs indicate that HCN4 may serve as a cell surface marker to permit isolation of HCN4 expressing precursors from hESC or iPSCs (Spater et al., 2013).
Our data highlight the use of HCN4 as a marker for precursors of the CCS and emphasize that HCN4 expression marks distinct precursors or components of the CCS at distinct times during development. It is intriguing to note that HCN4 is expressed, then downregulated in several cardiac precursor populations, including those of the left ventricular working cardiomyocytes. Upregulation of HCN4 in ventricular myocytes in hypertrophic and failing human heart is associated with and has been postulated to contribute to cardiac arrhythmias(Herrmann et al., 2011), and may reflect reactivation of this developmentally silenced gene.
HCN4 in conjunction with other markers may serve to optimize protocols for generation of specific conduction system precursors. However, it must be noted that HCN4 expression occurs in several endothelial populations at later stages of heart development. HCN4 as a marker of the first heart field may allow for isolation of first heart field precursors, and identification of additional markers of these developmentally important cells.
Understanding developmental origins of CCS lineages, identifying markers that can be used for tracking these lineages, and for their isolation from human stem cells or induced pluripotent stem cells are essential steps toward elucidating the etiology of conduction system disease. Potential regenerative therapies aimed at the conduction system, including development of biological pacemakers, will also be facilitated by a clear understanding of CCS lineage development.
Acknowledgments
YFS was supported by grants from the Ministry of Science and Technology China (2013CB967400); SME by grants from NIH (HL-117649, HL119967). XQL by grants from the National Natural Science Foundation of China (31171393, 81370196).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
The authors confirm that there are no conflicts of interest.
References
- Aanhaanen WT, Brons JF, Dominguez JN, Rana MS, Norden J, Airik R, Wakker V, de Gier-de Vries C, Brown NA, Kispert A, Moorman AF, Christoffels VM. The Tbx2+ primary myocardium of the atrioventricular canal forms the atrioventricular node and the base of the left ventricle. Circulation research. 2009;104:1267–1274. doi: 10.1161/CIRCRESAHA.108.192450. [DOI] [PubMed] [Google Scholar]
- Al-Khatib SM, Pritchett EL. Clinical features of Wolff-Parkinson-White syndrome. Am Heart J. 1999;138:403–413. doi: 10.1016/s0002-8703(99)70140-7. [DOI] [PubMed] [Google Scholar]
- Bakker ML, Boukens BJ, Mommersteeg MT, Brons JF, Wakker V, Moorman AF, Christoffels VM. Transcription factor Tbx3 is required for the specification of the atrioventricular conduction system. Circulation research. 2008;102:1340–1349. doi: 10.1161/CIRCRESAHA.107.169565. [DOI] [PubMed] [Google Scholar]
- Bakker ML, Christoffels VM, Moorman AF. The cardiac pacemaker and conduction system develops from embryonic myocardium that retains its primitive phenotype. J Cardiovasc Pharmacol. 2010a;56:6–15. doi: 10.1097/FJC.0b013e3181e775d3. [DOI] [PubMed] [Google Scholar]
- Bakker ML, Moorman AF, Christoffels VM. The atrioventricular node: origin, development, and genetic program. Trends in cardiovascular medicine. 2010b;20:164–171. doi: 10.1016/j.tcm.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Barhanin J, Lesage F, Guillemare E, Fink M, Lazdunski M, Romey G. K(V)LQT1 and lsK (minK) proteins associate to form the I(Ks) cardiac potassium current. Nature. 1996;384:78–80. doi: 10.1038/384078a0. [DOI] [PubMed] [Google Scholar]
- Bertrand N, Roux M, Ryckebusch L, Niederreither K, Dolle P, Moon A, Capecchi M, Zaffran S. Hox genes define distinct progenitor sub-domains within the second heart field. Dev Biol. 2011;353:266–274. doi: 10.1016/j.ydbio.2011.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua LM, Simon AM, Maguire CT, Gehrmann J, Wakimoto H, Paul DL, Berul CI. A targeted disruption in connexin40 leads to distinct atrioventricular conduction defects. J Interv Card Electrophysiol. 2000;4:459–467. doi: 10.1023/a:1009800328836. [DOI] [PubMed] [Google Scholar]
- Beyer S, Kelly RG, Miquerol L. Inducible Cx40-Cre expression in the cardiac conduction system and arterial endothelial cells. Genesis. 2011;49:83–91. doi: 10.1002/dvg.20687. [DOI] [PubMed] [Google Scholar]
- Blaschke RJ, Hahurij ND, Kuijper S, Just S, Wisse LJ, Deissler K, Maxelon T, Anastassiadis K, Spitzer J, Hardt SE, Scholer H, Feitsma H, Rottbauer W, Blum M, Meijlink F, Rappold G, Gittenberger-de Groot AC. Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation. 2007;115:1830–1838. doi: 10.1161/CIRCULATIONAHA.106.637819. [DOI] [PubMed] [Google Scholar]
- Bucchi A, Barbuti A, Difrancesco D, Baruscotti M. Funny Current and Cardiac Rhythm: Insights from HCN Knockout and Transgenic Mouse Models. Frontiers in physiology. 2012;3:240. doi: 10.3389/fphys.2012.00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6:826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels VM, Mommersteeg MT, Trowe MO, Prall OW, de Gier-de Vries C, Soufan AT, Bussen M, Schuster-Gossler K, Harvey RP, Moorman AF, Kispert A. Formation of the venous pole of the heart from an Nkx2–5-negative precursor population requires Tbx18. Circ Res. 2006;98:1555–1563. doi: 10.1161/01.RES.0000227571.84189.65. [DOI] [PubMed] [Google Scholar]
- Davis DL, Edwards AV, Juraszek AL, Phelps A, Wessels A, Burch JB. A GATA-6 gene heart-region-specific enhancer provides a novel means to mark and probe a discrete component of the mouse cardiac conduction system. Mech Dev. 2001;108:105–119. doi: 10.1016/s0925-4773(01)00500-7. [DOI] [PubMed] [Google Scholar]
- De Val S, Anderson JP, Heidt AB, Khiem D, Xu SM, Black BL. Mef2c is activated directly by Ets transcription factors through an evolutionarily conserved endothelial cell-specific enhancer. Dev Biol. 2004;275:424–434. doi: 10.1016/j.ydbio.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Di Lisi R, Sandri C, Franco D, Ausoni S, Moorman AF, Schiaffino S. An atrioventricular canal domain defined by cardiac troponin I transgene expression in the embryonic myocardium. Anat Embryol (Berl) 2000;202:95–101. doi: 10.1007/s004290000102. [DOI] [PubMed] [Google Scholar]
- DiFrancesco D. Funny channel gene mutations associated with arrhythmias. J Physiol. 2013;591:4117–4124. doi: 10.1113/jphysiol.2013.253765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez JN, Meilhac SM, Bland YS, Buckingham ME, Brown NA. Asymmetric fate of the posterior part of the second heart field results in unexpected left/right contributions to both poles of the heart. Circ Res. 2012;111:1323–1335. doi: 10.1161/CIRCRESAHA.112.271247. [DOI] [PubMed] [Google Scholar]
- Dyer LA, Kirby ML. The role of secondary heart field in cardiac development. Developmental biology. 2009;336:137–144. doi: 10.1016/j.ydbio.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza-Lewis RA, Yu L, He F, Liu H, Tang R, Shi J, Sun X, Martin JF, Wang D, Yang J, Chen Y. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2–5. Developmental biology. 2009;327:376–385. doi: 10.1016/j.ydbio.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SM, Yelon D, Conlon FL, Kirby ML. Myocardial lineage development. Circulation research. 2010;107:1428–1444. doi: 10.1161/CIRCRESAHA.110.227405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DU, Carter KL, Thomas KR, Burr RM, Bakker ML, Coetzee WA, Tristani-Firouzi M, Bamshad MJ, Christoffels VM, Moon AM. Lethal arrhythmias in Tbx3-deficient mice reveal extreme dosage sensitivity of cardiac conduction system function and homeostasis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E154–163. doi: 10.1073/pnas.1115165109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Frigola C, Shi Y, Evans SM. Expression of the hyperpolarization-activated cyclic nucleotide-gated cation channel HCN4 during mouse heart development. Gene Expr Patterns. 2003;3:777–783. doi: 10.1016/s1567-133x(03)00125-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourdie RG, Harris BS, Bond J, Justus C, Hewett KW, O’Brien TX, Thompson RP, Sedmera D. Development of the cardiac pacemaking and conduction system. Birth Defects Res C Embryo Today. 2003;69:46–57. doi: 10.1002/bdrc.10008. [DOI] [PubMed] [Google Scholar]
- Gros D, Theveniau-Ruissy M, Bernard M, Calmels T, Kober F, Sohl G, Willecke K, Nargeot J, Jongsma HJ, Mangoni ME. Connexin 30 is expressed in the mouse sino-atrial node and modulates heart rate. Cardiovasc Res. 2010;85:45–55. doi: 10.1093/cvr/cvp280. [DOI] [PubMed] [Google Scholar]
- Haissaguerre M, Jais P, Shah DC, Takahashi A, Hocini M, Quiniou G, Garrigue S, Le Mouroux A, Le Metayer P, Clementy J. Spontaneous initiation of atrial fibrillation by ectopic beats originating in the pulmonary veins. N Engl J Med. 1998;339:659–666. doi: 10.1056/NEJM199809033391003. [DOI] [PubMed] [Google Scholar]
- Harvey RP, Meilhac SM, Buckingham ME. Landmarks and lineages in the developing heart. Circ Res. 2009;104:1235–1237. doi: 10.1161/CIRCRESAHA.109.199729. [DOI] [PubMed] [Google Scholar]
- Herrmann S, Fabritz L, Layh B, Kirchhof P, Ludwig A. Insights into sick sinus syndrome from an inducible mouse model. Cardiovascular research. 2011;90:38–48. doi: 10.1093/cvr/cvq390. [DOI] [PubMed] [Google Scholar]
- Hoesl E, Stieber J, Herrmann S, Feil S, Tybl E, Hofmann F, Feil R, Ludwig A. Tamoxifen-inducible gene deletion in the cardiac conduction system. Journal of molecular and cellular cardiology. 2008;45:62–69. doi: 10.1016/j.yjmcc.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Horsthuis T, Buermans HP, Brons JF, Verkerk AO, Bakker ML, Wakker V, Clout DE, Moorman AF, t Hoen PA, Christoffels VM. Gene expression profiling of the forming atrioventricular node using a novel tbx3-based node-specific transgenic reporter. Circ Res. 2009;105:61–69. doi: 10.1161/CIRCRESAHA.108.192443. [DOI] [PubMed] [Google Scholar]
- Jongbloed MR, Vicente Steijn R, Hahurij ND, Kelder TP, Schalij MJ, Gittenberger-de Groot AC, Blom NA. Normal and abnormal development of the cardiac conduction system; implications for conduction and rhythm disorders in the child and adult. Differentiation. 2012;84:131–148. doi: 10.1016/j.diff.2012.04.006. [DOI] [PubMed] [Google Scholar]
- Kamino K, Hirota A, Fujii S. Localization of pacemaking activity in early embryonic heart monitored using voltage-sensitive dye. Nature. 1981;290:595–597. doi: 10.1038/290595a0. [DOI] [PubMed] [Google Scholar]
- Kapoor N, Liang W, Marban E, Cho HC. Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat Biotechnol. 2013;31:54–62. doi: 10.1038/nbt.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katritsis D, Ioannidis JP, Giazitzoglou E, Korovesis S, Anagnostopoulos CE, Camm AJ. Conduction delay within the coronary sinus in humans: implications for atrial arrhythmias. J Cardiovasc Electrophysiol. 2002;13:859–862. doi: 10.1046/j.1540-8167.2002.00859.x. [DOI] [PubMed] [Google Scholar]
- Kelly RG. The second heart field. Curr Top Dev Biol. 2012;100:33–65. doi: 10.1016/B978-0-12-387786-4.00002-6. [DOI] [PubMed] [Google Scholar]
- Kelly RG, Brown NA, Buckingham ME. The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev Cell. 2001;1:435–440. doi: 10.1016/s1534-5807(01)00040-5. [DOI] [PubMed] [Google Scholar]
- Kondo RP, Anderson RH, Kupershmidt S, Roden DM, Evans SM. Development of the cardiac conduction system as delineated by minK-lacZ. J Cardiovasc Electrophysiol. 2003;14:383–391. doi: 10.1046/j.1540-8167.2003.02467.x. [DOI] [PubMed] [Google Scholar]
- Kupershmidt S, Yang T, Anderson ME, Wessels A, Niswender KD, Magnuson MA, Roden DM. Replacement by homologous recombination of the minK gene with lacZ reveals restriction of minK expression to the mouse cardiac conduction system. Circ Res. 1999;84:146–152. doi: 10.1161/01.res.84.2.146. [DOI] [PubMed] [Google Scholar]
- Lescroart F, Mohun T, Meilhac SM, Bennett M, Buckingham M. Lineage tree for the venous pole of the heart: clonal analysis clarifies controversial genealogy based on genetic tracing. Circ Res. 2012;111:1313–1322. doi: 10.1161/CIRCRESAHA.112.271064. [DOI] [PubMed] [Google Scholar]
- Liang X, Wang G, Lin L, Lowe J, Zhang Q, Bu L, Chen Y, Chen J, Sun Y, Evans SM. HCN4 dynamically marks the first heart field and conduction system precursors. Circ Res. 2013;113:399–407. doi: 10.1161/CIRCRESAHA.113.301588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig A, Zong X, Jeglitsch M, Hofmann F, Biel M. A family of hyperpolarization-activated mammalian cation channels. Nature. 1998;393:587–591. doi: 10.1038/31255. [DOI] [PubMed] [Google Scholar]
- Maeda J, Yamagishi H, McAnally J, Yamagishi C, Srivastava D. Tbx1 is regulated by forkhead proteins in the secondary heart field. Dev Dyn. 2006;235:701–710. doi: 10.1002/dvdy.20686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meilhac SM, Esner M, Kelly RG, Nicolas JF, Buckingham ME. The clonal origin of myocardial cells in different regions of the embryonic mouse heart. Developmental cell. 2004;6:685–698. doi: 10.1016/s1534-5807(04)00133-9. [DOI] [PubMed] [Google Scholar]
- Mikawa T, Fischman DA. The polyclonal origin of myocyte lineages. Annu Rev Physiol. 1996;58:509–521. doi: 10.1146/annurev.ph.58.030196.002453. [DOI] [PubMed] [Google Scholar]
- Miquerol L, Bellon A, Moreno N, Beyer S, Meilhac SM, Buckingham M, Franco D, Kelly RG. Resolving cell lineage contributions to the ventricular conduction system with a Cx40-GFP allele: a dual contribution of the first and second heart fields. Dev Dyn. 2013;242:665–677. doi: 10.1002/dvdy.23964. [DOI] [PubMed] [Google Scholar]
- Miquerol L, Beyer S, Kelly RG. Establishment of the mouse ventricular conduction system. Cardiovascular research. 2011;91:232–242. doi: 10.1093/cvr/cvr069. [DOI] [PubMed] [Google Scholar]
- Miquerol L, Meysen S, Mangoni M, Bois P, van Rijen HV, Abran P, Jongsma H, Nargeot J, Gros D. Architectural and functional asymmetry of the His-Purkinje system of the murine heart. Cardiovasc Res. 2004;63:77–86. doi: 10.1016/j.cardiores.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, Sun Y, Evans SM, Laugwitz KL, Chien KR. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- Moskowitz IP, Kim JB, Moore ML, Wolf CM, Peterson MA, Shendure J, Nobrega MA, Yokota Y, Berul C, Izumo S, Seidman JG, Seidman CE. A molecular pathway including Id2, Tbx5, and Nkx2–5 required for cardiac conduction system development. Cell. 2007;129:1365–1376. doi: 10.1016/j.cell.2007.04.036. [DOI] [PubMed] [Google Scholar]
- Munshi V. Gene regulatory networks in cardiac conduction system development. Circ Res. 2012;110:1525–1537. doi: 10.1161/CIRCRESAHA.111.260026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munshi V, McAnally J, Bezprozvannaya S, Berry JM, Richardson JA, Hill JA, Olson EN. Cx30.2 enhancer analysis identifies Gata4 as a novel regulator of atrioventricular delay. Development. 2009;136:2665–2674. doi: 10.1242/dev.038562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallante BA, Giovannone S, Fang-Yu L, Zhang J, Liu N, Kang G, Dun W, Boyden PA, Fishman GI. Contactin-2 expression in the cardiac Purkinje fiber network. Circ Arrhythm Electrophysiol. 2010;3:186–194. doi: 10.1161/CIRCEP.109.928820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentschler S, Vaidya DM, Tamaddon H, Degenhardt K, Sassoon D, Morley GE, Jalife J, Fishman GI. Visualization and functional characterization of the developing murine cardiac conduction system. Development. 2001;128:1785–1792. doi: 10.1242/dev.128.10.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanguinetti MC, Curran ME, Zou A, Shen J, Spector PS, Atkinson DL, Keating MT. Coassembly of K(V)LQT1 and minK (IsK) proteins to form cardiac I(Ks) potassium channel. Nature. 1996;384:80–83. doi: 10.1038/384080a0. [DOI] [PubMed] [Google Scholar]
- Scavone A, Capilupo D, Mazzocchi N, Crespi A, Zoia S, Campostrini G, Bucchi A, Milanesi R, Baruscotti M, Benedetti S, Antonini S, Messina G, DiFrancesco D, Barbuti A. Embryonic stem cell-derived CD166+ precursors develop into fully functional sinoatrial-like cells. Circulation research. 2013;113:389–398. doi: 10.1161/CIRCRESAHA.113.301283. [DOI] [PubMed] [Google Scholar]
- Sizarov A, Devalla HD, Anderson RH, Passier R, Christoffels VM, Moorman AF. Molecular analysis of patterning of conduction tissues in the developing human heart. Circ Arrhythm Electrophysiol. 2011;4:532–542. doi: 10.1161/CIRCEP.111.963421. [DOI] [PubMed] [Google Scholar]
- Spater D, Abramczuk MK, Buac K, Zangi L, Stachel MW, Clarke J, Sahara M, Ludwig A, Chien KR. A HCN4 + cardiomyogenic progenitor derived from the first heart field and human pluripotent stem cells. Nat Cell Biol. 2013;15:1098–1106. doi: 10.1038/ncb2824. [DOI] [PubMed] [Google Scholar]
- Stieber J, Herrmann S, Feil S, Loster J, Feil R, Biel M, Hofmann F, Ludwig A. The hyperpolarization-activated channel HCN4 is required for the generation of pacemaker action potentials in the embryonic heart. Proc Natl Acad Sci U S A. 2003;100:15235–15240. doi: 10.1073/pnas.2434235100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud DM, Darrow BJ, Kim SD, Zhang J, Jongbloed MR, Rentschler S, Moskowitz IP, Seidman J, Fishman GI. Complex genomic rearrangement in CCS-LacZ transgenic mice. Genesis. 2007;45:76–82. doi: 10.1002/dvg.20267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Zhang T, Liu C, Gu S, Chen Y. Generation of Shox2-Cre allele for tissue specific manipulation of genes in the developing heart, palate, and limb. Genesis. 2013;51:515–522. doi: 10.1002/dvg.22397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Liang X, Najafi N, Cass M, Lin L, Cai CL, Chen J, Evans SM. Islet 1 is expressed in distinct cardiovascular lineages, including pacemaker and coronary vascular cells. Dev Biol. 2007;304:286–296. doi: 10.1016/j.ydbio.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CF, Tai CT, Hsieh MH, Lin WS, Yu WC, Ueng KC, Ding YA, Chang MS, Chen SA. Initiation of atrial fibrillation by ectopic beats originating from the superior vena cava: electrophysiological characteristics and results of radiofrequency ablation. Circulation. 2000;102:67–74. doi: 10.1161/01.cir.102.1.67. [DOI] [PubMed] [Google Scholar]
- Van Mierop LH. Location of pacemaker in chick embryo heart at the time of initiation of heartbeat. Am J Physiol. 1967;212:407–415. doi: 10.1152/ajplegacy.1967.212.2.407. [DOI] [PubMed] [Google Scholar]
- Vedantham V, Evangelista M, Huang Y, Srivastava D. Spatiotemporal regulation of an Hcn4 enhancer defines a role for Mef2c and HDACs in cardiac electrical patterning. Dev Biol. 2013;373:149–162. doi: 10.1016/j.ydbio.2012.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzi MP, McCulley DJ, De Val S, Dodou E, Black BL. The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev Biol. 2005;287:134–145. doi: 10.1016/j.ydbio.2005.08.041. [DOI] [PubMed] [Google Scholar]
- Wiese C, Grieskamp T, Airik R, Mommersteeg MT, Gardiwal A, de Gier-de Vries C, Schuster-Gossler K, Moorman AF, Kispert A, Christoffels VM. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ Res. 2009;104:388–397. doi: 10.1161/CIRCRESAHA.108.187062. [DOI] [PubMed] [Google Scholar]
- Wu M, Peng S, Zhao Y. Inducible gene deletion in the entire cardiac conduction system using Hcn4-CreERT2 BAC transgenic mice. Genesis. 2014;52:134–140. doi: 10.1002/dvg.22730. [DOI] [PMC free article] [PubMed] [Google Scholar]


