Summary
In most sexually reproducing plants, a single somatic, sub-epidermal cell in an ovule is selected to differentiate into a megaspore mother cell, which is committed to giving rise to the female germline. However, it remains unclear how intercellular signaling among somatic cells results in only one cell in the sub-epidermal layer differentiating into the megaspore mother cell. Here we uncovered a role of the THO complex in restricting the megaspore mother cell fate to a single cell. Mutations in TEX1, HPR1 and THO6, components of the THO/TREX complex, led to the formation of multiple megaspore mother cells, which were able to initiate gametogenesis. We demonstrated that TEX1 repressed the megaspore mother cell fate by promoting the biogenesis of TAS3-derived ta-siRNA, which represses ARF3 expression. The TEX1 protein was present in epidermal cells but not in the germline, and through TAS3-derived ta-siRNA, restricted ARF3 expression to the medio domain of ovule primordia. Expansion of ARF3 expression into lateral epidermal cells in a TAS3 ta-siRNA-insensitive mutant led to the formation of supernumerary megaspore mother cells, suggesting that TEX1- and TAS3-mediated restriction of ARF3 expression limits excessive megaspore mother cell formation non-cell autonomously. Our findings reveal the role of a small RNA pathway in the regulation of female germline specification in Arabidopsis.
Introduction
Germline specification is a crucial step in sexual reproduction. In plants, the reproductive lineage does not arise early in development, as it does in animals; rather, the germline is specified during flower development [1]. Within the female reproductive organ that houses one to many ovules depending on the plant species, a single somatic, sub-epidermal cell in each ovule is selected to differentiate into the megaspore mother cell (MMC), which is committed to giving rise to the haploid, multi cellular female gametophyte. The specification of the MMC is of critical importance as it is the first committed cell of the female germline lineage. In light of this, how signaling among somatic cells, or between somatic cells and the potential MMC, occurs to ensure the specification of only one MMC is of much interest.
Recent studies have begun to identify components of the molecular network involved in MMC differentiation. Emerging data suggest that small RNAs play an important role in MMC formation. In maize, ago104 encodes an ARGONAUTE (AGO) protein critical for small RNA-mediated gene regulation. An ago104 mutation results in the failure of MMC meiosis, leading to the formation of functional, unreduced gametes. The AGO104 protein is present in the sub-epidermal cells surrounding the developing MMC but not in the MMC, suggesting that cells surrounding the MMC produce an ago104-dependent mobile signal influencing MMC differentiation and/or behavior [2]. Maize dmt103 encodes a methyltransferase homologous to DOMAINS REARRANGED METHYL TRANSFERASE (DRMs) in Arabidopsis, which along with small interfering RNAs (siRNAs) and other proteins are involved in both DNA and histone methylation via the process known as RNA-directed DNA methylation (RdDM) [3]. It is expressed in ovules in a restricted zone in and around the specified MMC. Down-regulation of dmt103 leads to additional cells near the MMC to enlarge, perhaps reflecting that they take on partial MMC character [4]. In rice, ARRESTED AT LEPTOTENE1 (MEL1) encodes a member of the AGO protein family, and is probably an ortholog of Arabidopsis AGO5 (AtAGO5). MEL1 is located mostly in the cytoplasm of germ cells, and associates preferentially with 21-nucleotide, phased siRNAs (phasiRNAs) [5]. In rice mel1 mutants, some MMCs fail to undergo meiosis leading to the absence of the female gametophyte in some ovules [6].
Arabidopsis AGO9, which binds 24-nt small RNAs derived from transposable elements (TEs) [7], and RNA-DEPENDENT RNA POLYMERASE 6 (RDR6) encoding an RNA-dependent RNA polymerase involved in the biogenesis of trans-acting siRNAs and other siRNAs [8–10] restrict MMC formation to a single hypodermal cell in the ovule. Loss-of-function mutations in these genes lead to supernumerary MMCs in ovule primordia [7]. The AGO9 protein is present in the cytoplasm in the epidermal layer (L1) of ovules [7], but accumulates in the nucleus of the MMC [11]. In both ago9 and rdr6 loss-of-function mutants, the supernumerary MMC phenotype is not completely penetrant, i.e., only a certain percentage of ovules show such phenotype, suggesting that the MMC specification program has built-in genetic redundancy.
Despite the afore mentioned genetic data implicating the involvement of small RNAs in MMC specification, the small RNA species involved in this process remains unknown. The incomplete penetrance of the mutants also indicates the existence of additional regulatory mechanisms. To obtain more insights into the genetic and molecular bases underlying MMC specification in Arabidopsis, and to further elucidate the regulatory function of small RNAs in this process, we carried out an ethyl methanesulfonate (EMS) screen in an rdr6 background to identify additional factors with roles in regulating MMC specification.
In this study, we uncovered a critical role of the Arabidopsis THO complex in preventing ectopic MMC formation. We showed that the THO complex and RDR6 control MMC differentiation through the TAS3-derived ta-siRNA, tasiR-ARF, which spatially restricts ARF3 expression in ovule primordia. TasiR-ARF-resistant ARF3 was expressed in a pattern similar to that of wild-type ARF3 in the tex1 mutant, and led to supernumerary MMCs. Furthermore, the expression patterns of the THO complex component TEX1 as well as those of ARF3 implicate signaling from L1 to L2 in MMC specification. Our findings reveal the identity of a small RNA species that serves as a critical player in MMC specification in Arabidopsis.
Results
Isolation of an rdr6 enhancer mutant
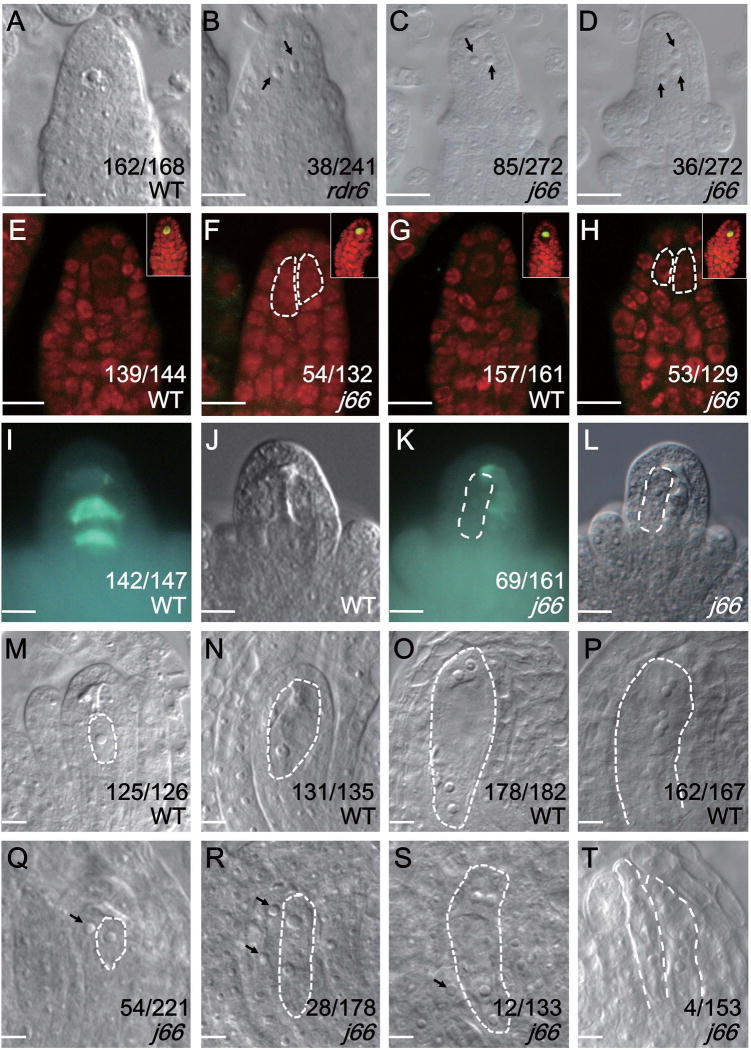
To identify additional genes functioning in MMC differentiation, we conducted an EMS mutagenesis screen for enhancers and suppressors of the rdr6–11 mutant, which harbors a premature stop codon that would truncate three-fourths of the protein [8]. This mutant exhibited two MMCs in pre-meiotic ovules at a higher frequency than that in wild type [7] (15.8% in rdr6–11 vs. 3.6% in wild type under our growth conditions, Figure 1A, B). We isolated one mutant, j66, which produced more than one (two or even three) MMC-like cells at a percentage of 44.5 %, higher than that in rdr6 (Figure 1C, D).
Figure 1. Supernumerary MMCs are formed in j66.
The numbers in the figures indicate the number of ovules with the phenotypes represented by the images out of the total number of ovules examined. The differences between WT and j66 were significant as determined by student’s t-test (p<0.05). Bars, 10 µm.
(A–D) MMC or MMC-like cells in pre-meiotic ovules in various genotypes. (A) A wide type (WT) ovule showing one MMC. (B) An rdr6 ovule showing two MMC-like cells. (C) A j66 ovule showing two MMC-like cells. (D) A j66 ovule showing three MMC-like cells. The MMC-like cells in B–D are indicated by black arrows.
(E–H) DMC1 (E, F) and ASY1 (G, H) immunolocalization in wide type (WT) and j66 pre-meiotic ovules. The images in the upper right insets of E–H represent immuno-positive meiotic ovules from wide type (WT) and j66 in the same immunolocalization experiment. The MMC-like cells are outlined with white dashed lines in F and H.
(I–L) Callose deposition in wide type (WT) and j66. (I, K) Callose staining by aniline blue. (J, L) Morphology of the ovules in I and K, respectively.
(M–P) Female gametophyte development in wide type (WT) at FG1, FG3, FG4 and FG6 stages, respectively. The gametophytes are outlined with white dashed lines.
(Q–T) Female gametophyte development in j66 at FG1, FG3, FG4 and FG6 stages, respectively. The abnormally enlarged cells in Q–S are indicated by black arrows. The gametophytes in K–T are outlined with white dashed lines.
To exclude the possibility that the two cells are products of meiosis I rather than two MMCs, we examined whether the enlarged MMC-like cells appear prior to meiosis I. We performed immunostaining for the meiosis I markers DMC1 [12, 13] and ASY1[14], which are specifically expressed in the megasporocyte undergoing meiosis. The positive controls (ovules at the meiotic stage) showed specific signals in the MMC, allowing us to confidently identify pre-meiotic ovules by the absence of signals (Figure 1E – H). Further examination of the pre-meiotic ovules found the presence of MMC-like cells in j66, suggesting that they appeared in the ovule primodia at the pre-meiotic stage (Figure 1E – H). Thus, the enlarged cells are not products of meiosis I.
To determinate if one or all MMC-like cells in j66 were capable of undergoing meiosis, we analyzed callose deposition, a reliable marker for MMCs undergoing meiosis [15], in wild type and j66 ovules. Consistent with previous observations, callose was deposited in the transverse walls between the functional megaspore and its degenerated sister cells in wild type ovules during meiosis (Figure 1I, J). In post-meiotic j66 ovules, callose was only detected in the adjacent walls between the single functional megaspore (FM) and the degenerated neighboring cells, but not at the additional, abnormally enlarged cell (Figure 1K, L). Therefore, despite the fact that several cells differentiated into enlarged cells like the MMC before meiosis in j66 ovules, only a single one underwent meiosis.
Female gametophyte development consists of two main phases: megasporogenesis followed by megagametogenesis. During Arabidopsis megasporogenesis, the diploid MMC undergoes meiosis and gives rise to haploid megaspores. During megagametogenesis, one of the megaspores, i.e. the functional megaspore (FM), through three rounds of mitosis, develops into the mature female gametophyte (FG) that contains one egg cell, one central cell, two synergids and three antipodal cells (Figure 1M – P) [16]. At the onset of megagametogenesis, i.e. at the stage of FM formation, only one FM and the remnants of the three degenerated megaspores could be observed in wild type ovules. But in j66, the FM was flanked by abnormally enlarged cells at a frequency of 24.5% (Figure 1Q). Such enlarged cells were also observed in the ovule at subsequent stages in j66 (Figure 1R, S). The number of ovules with abnormally enlarged cells decreased with the progression of ovule development. Despite this decline, mature j66 FGs exhibited twin embryo sacs at a low frequency (2.5%) (Figure1T). This phenotype was never observed in wild type (0 out of 323).
In addition to the defects in female gametophyte development, we also observed a variety of abnormalities in other aspects of development in j66. Compared with WT, j66 showed wider and serrated leaves, protruding pistils, shrunken siliques and aborted seeds (Figure 2A–F). To elucidate the cause for the reduced fertility, we carried out reciprocal crosses between wide type and j66. Pollination of j66 plants with wild type pollen produced nearly full-length siliques, and vice versa (Table S1), suggesting that both male and female gametophytes in j66 were fully functional. To further confirm this, we examined embryo sac morphology and pollen activity in j66. In j66, 96.3% of ovules harbored a normal embryo sac with obvious synergids, an egg cell and a central cell as in WT (98.2%) (Figure 2G, H). Pollen viability in j66 was normal as shown by Alexander’s staining (Figure 2I, J). Thus, the reduced fertility in j66 was not attributable to abnormal gametophyte development. Rather, it might be due to the failure of pollen to reach the pistil stigma because stamens were shorter than pistils. In conclusion, the supernumerary MMC phenotype in j66 did not affect ovule fertility.
Figure 2. Developmental defects in j66.
See also Table S1.
(A, B) Seedlings of wide type (WT) and j66.
(C) An inflorescence from j66 showing the protruding pistils in unopened flowers.
(D) Siliques of variable lengths from j66. A wide type-like silique is shown at the top and two shorter siliques are shown below.
(E) Opened siliques of j66 showing variable seed sets. A silique with full seed set is shown at the top and one with reduced seed set is shown below.
(F) Percentage of siliques with full seed set in WT and j66. Significant difference between WT and j66 was found by student’s t-test (p<0.05).
(G, H) Mature female gametophytes from wide type (WT) and j66. EC: Egg cell; CC: Central cell. The numbers in (G) and (H) indicate the number of ovules with the phenotypes represented by the images out of the total number of ovules examined. Bars, 20 µm.
(I, J) Alexander’s staining of pollen from wide type (WT) and j66. The red color indicates viable pollen.
TEX1 represses MMC and gametophytic cell fate
To pinpoint the causal mutation, j66 was crossed to the parental genotype rdr6–11. Bulked segregants from F2 of the backcross were used for next-generation sequencing for candidate gene identification. According to the method reported by Zhu et al [17], we got 18 SNPs. Among these, 12 were in intergenic regions; 1 was in a TE; 2 were in intron regions (not splicing acceptor or donor sites); 2 were in exons of two genes and 1 disrupted an intron acceptor site of TEX1. The two exon SNPs altered single amino acids (glycine to arginine for one SNP and arginine to cysteine for the other one), while the SNP in TEX1 was expected to result in a frame shift followed by premature translation termination, which made it the best candidate SNP. The G-to-A mutation at the splice acceptor site in the sixth intron of the TEX1 (AT5G56130) gene, a component of the Arabidopsis THO/TREX complex [18, 19], was designated as tex1–5 (Figure S1A). The mutation resulted in a single base deletion in exon 7 due to the use of an alternative splice acceptor site, as validated by RT-PCR followed by sequencing (Figure S1C). The deletion caused a frame shift from amino acid 171 followed by translation termination soon after.
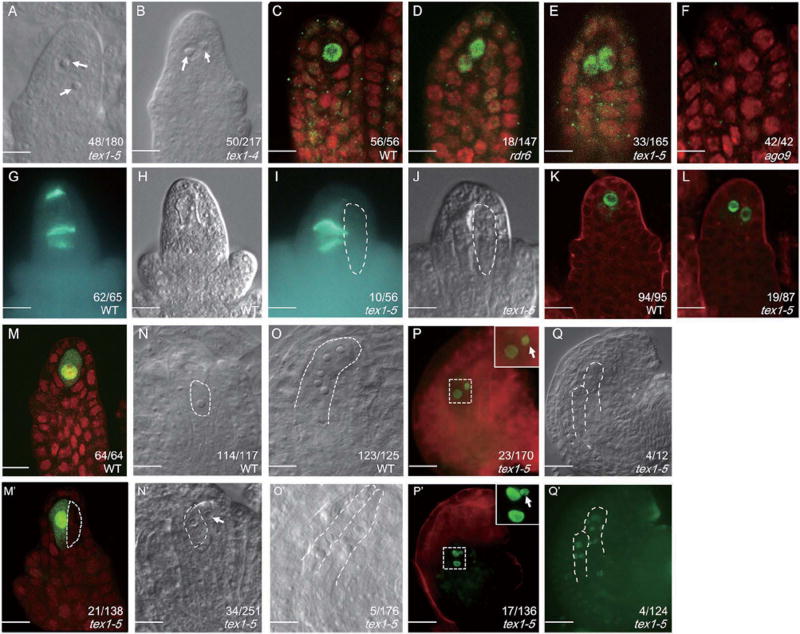
To verify if the mutation in TEX1 was indeed the causal mutation, we crossed j66 to wide type and separated tex1–5 from rdr6–11 (Figure S1D) and examined the MMC phenotype of tex1–5. We found that tex1–5 ovules displayed more than one MMC-like cells at a frequency of 27.8% (Figure 3A). In addition, a T-DNA insertion line in the Columbia (Col) background (SALK_100012; hereafter referred to as tex1–4) [19] was obtained from the Arabidopsis Biological Resource Center [20]. In tex1–4, the T-DNA insertion site was located between the second and third exons of TEX1 (Figure S1A). Pre-meiotic ovules in tex1–4 showed multiple MMC-like cells at a frequency of 23.7% (Figure 3B), significantly higher than that in wide type. The pTEX::TEX-GFP fusion construct complemented the MMC defects of tex1–5 (Figure S2), indicating that TEX1 plays a critical role in MMC differentiation.
Figure 3. Supernumerary MMCs are formed in tex1 ovules.
The numbers in the figure panels indicate the number of ovules with phenotypes represented by the images out of the total number of ovules examined. The differences between WT and the mutants were significant as determined by student’s t-test (p<0.05). Bars in (A-M’), 10 µm; Bars in (N-R’), 20 µm. See also Figure S1, S3, S4 and Table S2.
(A, B) Pre-meiotic tex1–5 and tex1–4 ovules showing two MMC-like cells (indicated by arrows).
(C–F) AGO9 immunolocalization in pre-meiotic WT, rdr6, tex1 and ago9 ovules.
(G–J) Callose deposition in wide type (WT) and tex1 ovules. (G, I) Callose staining with aniline blue. (H, J) Morphology of the ovules in (G, I). The abnormally enlarged cell without callose signals is outlined with white dashed lines.
(K, L) pKNU::KNU-Venus expression patterns in wide type (WT) and tex1–5 ovules at the MMC stage.
(M, M’) DMC1 immunolocalization in wide type (WT) and tex1 ovules. The abnormally enlarged cell without DMC1 signal in tex1 is outlined by white dashed lines.
(N, N’) Female gametophytes in wide type (WT) and tex1–5 ovules at the FG1 stage. The functional megaspores are outlined by the white dashed lines and the abnormally enlarged cell is indicated by a white arrow.
(O–O’) Female gametophytes in wide type (WT) and tex1–5 ovules at the FG5–6 stages. The female gametophytes are outlined by white dashed lines.
(P, P’) pAKV::H2B–YFP expression patterns in tex1–5 ovules at the FG1 (P) and FG2 (P’) stages. The nuclei of abnormally enlarged cells are indicated by white arrows. Images of the regions in the squares are shown in higher magnification in the insets in the upper right-hand corners.
(Q, Q’) pAKV::H2B–YFP expression patterns in tex1–5 ovules at the FG5–6 stages. (Q) Morphology of the ovule in (Q’). The female gametophytes are outlined by white dashed lines.
To determinate if the supernumerary MMC-like cells in tex1 acquired MMC identity, firstly, we examined the expression of pKUN::KNU-Venus, a marker of MMC identity [21], in tex1–5. KNU-Venus was detected in the single MMC of wide type ovule primordia (Figure 3K). In tex1–5, KNU-Venus was observed in multiple cells in 22% of ovules, suggesting that the additional, enlarged MMC-like cells in pre-meiotic tex1 ovules acquired MMC identity (Figure 3L)
Furthermore, we carried out whole-mount immunolocalization using anti-AGO9 antibody. AGO9 was shown to localize in the cytoplasm in cells surrounding the MMC but in the nucleus of the MMC (Figure 3C), thus the nuclear localization of AGO9 serves as a marker for MMC [22]. We observed that in 12.2% of rdr6–11 ovules, nuclear localization of AGO9 was detected in more than one cell (Figure 3D), consistent with previous observations [22]. Similarly, nuclear AGO9 localization was also observed in several cells in 20% of tex1–5 ovules (Figure 3E), implying that the enlarged MMC-like cells gained MMC identity in tex1–5 ovule primodia. The AGO9 localization analysis and the KNU-Venus expression analysis gave nearly identical results.
To examine whether the ectopic MMC-like cells present in tex1 were capable of undergoing meiosis, we determined callose deposition. Callose was only observed in the intermediate walls of daughter cells from one MMC in tex1, but not in the additional MMC-like cells (Figure 3G–J). In addition, we carried out immunolocalization using antibody against DMC1, which is specifically expressed in the megasporocyte undergoing meiosis in WT ovules (Figure 3M) [12, 13]. In ovules of tex1–5, the DMC1 signal was only detected in one MMC, not in the ectopic MMC-like cells (Figure 3M’). These results indicated that although several cells differentiated into MMC-like cells in pre-meiotic tex1 ovules, only a single one underwent meiosis.
Abnormally enlarged cells were also observed adjacent to the functional megaspore (FM) in post-meiotic tex1–5 ovules (Figure 3N, N'). Two narrowed embryo sacs were observed in a small subset of mature ovules, phenocopying j66 (Figure 3O, O'). To determine if the enlarged cells in tex1–5 ovules at megagametogenesis acquired gametophytic cell identity, we examined the expression of pAKV:H2B–YFP [23], a marker expressed in the nuclei of the functional megaspore and the developing female gametophyte, but not in the MMC or in the three meiotically-derived, degenerated megaspores (Figure S3). In tex1–5 ovules, the abnormally enlarged cells adjacent to FM displayed the expression of the marker gene (Figure 3P). Ectopic YFP signals were also detected in ovules at subsequent stages ((Figure 3P’). YFP signals were even detectable in numerous nuclei seemingly localized in different embryo sacs (Figure 3Q, Q’). These results indicated that the aberrant cells present in tex1 acquired gametophytic cell identity without undergoing meiosis.
In addition to female gametophytic defects, we also observed other abnormalities in tex1, including wider leaves, protruding pistils and aborted seeds, similar to those in j66 (Figure S4A–E). Seed set was normal in reciprocal crosses between wide type and tex1–5 (Table S2), suggesting that the male and female gametophytes were functional in tex1–5. This was consistent with normal embryo sac morphology and pollen viability in tex1–5 (Figure S4F, G). Besides, tex1–4 also exhibited phenotypes similar to tex1–5 (Figure S4H–K).
Dynamic expression patterns of TEX1 in ovules
We studied the localization of the TEX1 protein using a TEX1-green florescence protein (GFP) fusion driven by the TEX1 promoter. The pTEX1:: TEX1-GFP fusion protein complemented the leaf, inflorescence and MMC number defects of the tex1–5 mutant, indicating that the fusion protein was functional (Figure S2).
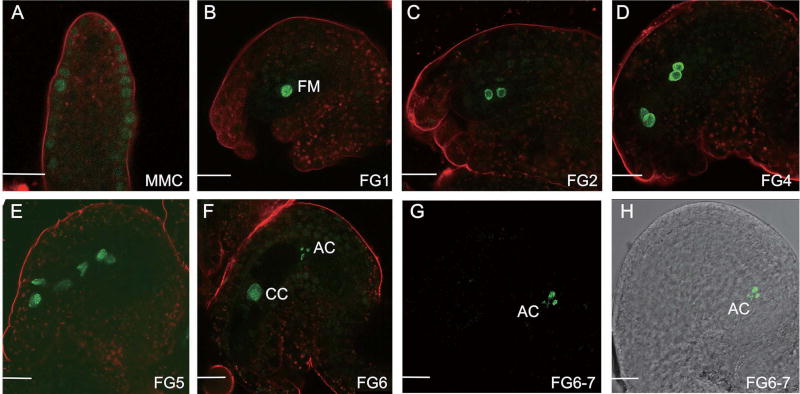
At the megasporogenesis stage, TEX1 accumulated in the nuclei of the epidermal cell layer of ovules (Figure 4A). When ovules entered megagametogenesis, the TEX1-GFP fusion protein was located in the nuclei of the FM at the FG1 stage, when the FM first became visible (Figure 4B). The nuclear GFP signal in the FM could be detected up to FG5, when the third mitotic division was completed (Figure 4C–E). At the FG6 stage, when two polar nuclei fused into a central nucleus, GFP was detected in the nuclei of the central cell and antipodal cells, but not in the egg cell and synergids (Figure 4F). At FG6-FG7 stages, the signal in the central cell disappeared and the GFP signal was only detected in antipodal cells (Figure 4G, H). With the maturation of ovules, GFP was not detectable in ovules perhaps as a consequence of the degeneration of antipodal cells. Therefore, the results suggested that after cellularization of the embryo sac, the expression of TEX1 ceased in a step-wise manner along the micropylar-chalazal axis.
Figure 4. Expression patterns of TEX1 in ovules shown by a native promoter driven TEX1-GFP transgene.
See also Figure S2.
The developmental stages of the ovules are indicated.
FM: Functional megaspore; CC: Central cell; AC: Antipodal cells.
Bar in (A), 10 µm; Bars in (B–H), 20 µm.
The THO/TREX complex is involved in MMC differentiation
TEX1 encodes a component of the THO complex identified first in yeast, in which the complex is composed of four proteins (HPR1, THO2, THP1 and MFT1) and is associated with TEX1 and the mRNA export factors SUB2 and YRA1 forming a larger complex termed TREX (Transcription-Export) [24, 25]. Although TEX1 is not generally considered a core component of the THO complex, it is tightly associated with the THO complex in yeast [26]. THO/TREX is conserved in all eukaryotes. The human TREX (hTREX) complex is composed of the multimeric THO (hTHO) complex, containing hHPR1/THOC1, THOC2, THOC5, THOC6, THOC7 and hTEX1/THOC3, the DEAD-box RNA helicase SUB2 and the mRNA export adaptor protein YRA1/THOC4 [27]. In Arabidopsis, through immunoprecipitation of HA-tagged TEX1 followed by mass spectrometry, peptides from all other THO complex core subunits (HPR1, THO2, THO5, THO6 and THO7) were identified, indicating that these proteins also form a protein complex in plants [19].
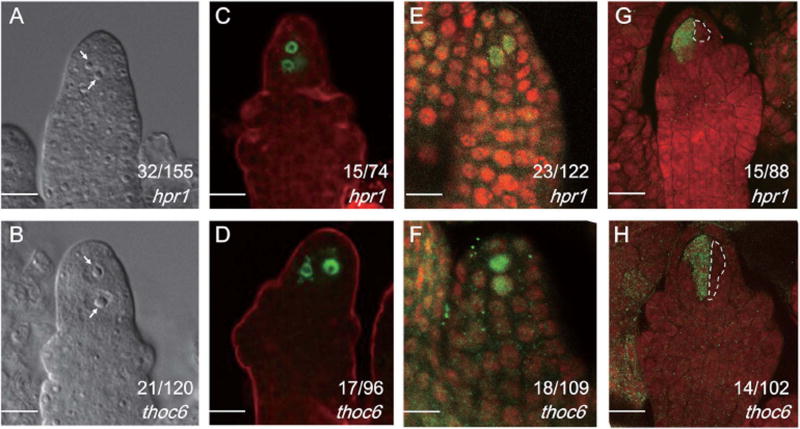
To test whether TEX1 exerts its function in MMC specification through the THO/TREX complex, we examined the MMC phenotype in mutants of Arabidopsis genes encoding homologs of HPR1and THO6. If the THO/TREX complex is involved in MMC specification, these mutants are expected to phenocopy the tex1 alleles described above. The rationale for selecting these two genes rather than other subunits for analysis was that they are present as single copies in the Arabidopsis genome such that our genetic analysis is not complicated by genetic redundancy. The hpr1–6 mutant carried a G-to-A mutation in the 16th exon of HPR1; this mutation introduced a premature stop codon (Figure S1B; unpublished results). The tho6 mutant, namely thoc6–1, harbored a T-DNA insertion in the first exon (SALK_051022). These mutants both showed enlarged cells adjacent to the MMC at a higher frequency than that in WT (Figure 5A, B). The findings that supernumerary MMC-like cells were present in tex1, hpr1 and tho6 indicated that a complex including the THO core is required for MMC differentiation in Arabidopsis.
Figure 5. Supernumarary MMCs are formed in hpr1 and thoc6 ovules.
The numbers in the figure panels indicate the number of ovules with the phenotypes represented by the images out of the total number of ovules examined. The differences between WT and the mutants were significant as determined by student’s t-test (p<0.05). Bars, 10 µm.
(A, B) Images of ovules with supernumerary MMC-like cells. The MMC-like cells are indicated by white arrows.
(C, D) pKNU::KNU-Venus expression patterns in hpr1 and thoc6 ovules at the MMC stage.
(E, F) AGO9 immunolicalization in pre-meiotic ovules.
(G, H) DMC1 immunolicalization in pre-meiotic ovules. The MMC-like cell without DMC1 signals were outlined by the white dashed lines.
To examine if one or all of the enlarged cells in hpr1 and thoc6 acquired MMC identity, and if they had the capability to undergo meiosis, we examined the expression of pKNU::KNU-Venus in the ovules from these mutants, and performed immunolocalization using antibodies against AGO9 and DMC1. The results showed that the enlarged cells adjacent to the MMC exhibited MMC identity as they expressed KNU-Venus and showed nuclear AGO9 (Figure 5C–F), whereas only one cell underwent meiosis, as evident by DMC1 staining (Figure 5G, H), consistent with the observations in tex1.
Loss of function in TAS3 results in supernumerary MMC formation
It is known that the THO complex is involved in ta-siRNA biogenesis [18, 19]. Ta-siRNAs are a class of endogenous siRNAs that regulate target genes other than their originating loci [28]. In Arabidopsis, there are four TAS family genes (TAS1–TAS4) encoding transcripts targeted for cleavage by different miRNAs. The TAS1 genes (TAS1A, TAS1B and TAS1C) and TAS2 are targets of miR173; TAS3A, TAS3B and TAS3C are targets of miR390; and TAS4 is a target of miR828 [8, 29–31]. The TAS transcripts undergo miRNA-guided cleavage and the cleavage fragments are stabilized by SUPPRESSOR OF GENE SILENCING3 and converted into dsRNAs by RDR6 [32], and the dsRNAs are further diced into ta-siRNAs. The THO/TREX complex is thought to transport ta-siRNA precursors from the nucleus to the subcellular location where their processing into siRNAs takes place [19].
TAS3 plays critical roles in the morphogenesis of organs with polarity. The TAS3-derived ta-siRNA, tasiR-ARF, targets the AUXIN RESPONSE FACTOR (ARF) family members ARF2, ARF3 and ARF4, which regulate leaf polarity, floral stem cell maintenance and lateral root growth [33–37].
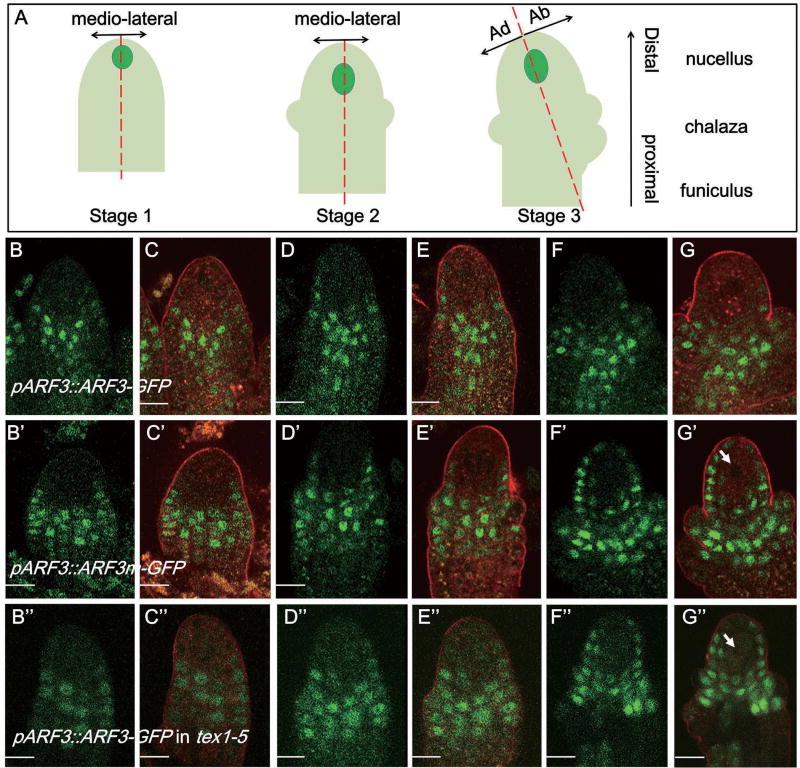
Ovules emerge from the placental tissue inside the gynoecium of flowers. At the early stage, ovules grow as radially symmetrical, cylindrical primordia, which further differentiate along proximal-distal, medio-lateral and adaxial-abaxial axes. Three structural elements, funiculus, chalaza, and nucellus can be distinguished along the proximal–distal axis. The asymmetric initiation of the outer integument marks the switch to adaxial–abaxial development, which leads to the formation of a bilaterally symmetrical (instead of radially symmetrical) ovule (Figure 7A) [38].
Figure 7. Expression patterns of ARF3::ARF3-GFP and ARF3::ARF3m–GFP in ovules.
Bars, 10 µm. See also Figure S6.
(A) Diagrams showing the polarities of ovules at three pre-meiotic stages. The axes are marked with arrows.
(B–G) Expression patterns of ARF3-GFP in ovules at stages 1 (B–C), 2 (D–E), and 3 (F–G).
(B’–G’) Expression patterns of ARF3::ARF3m–GFP in ovules at stages 1 (B’–C’), 2 (D’–E’), and 3 (F’–G’).
(B”–G”) Expression patterns of ARF3::ARF3-GFP in tex1–5 ovules at stages 1 (B”–C”), 2 (D”–E”), and 3 (F”–G”).
Panels B–B”, D–D” and F–F” are GFP fluorescence. Panels C–C”, E-E” and G–G” show the same ovules as in the panels to their left but with FM 4–64 staining. The MMC is marked with a white arrow in G’.
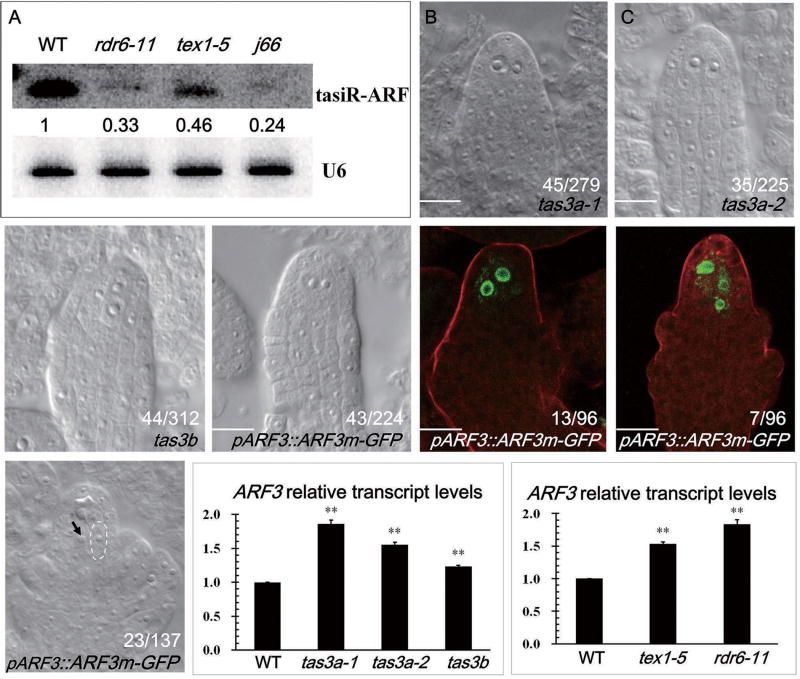
Considering the information above, we investigated whether TAS3 could account for the role of the THO complex and RDR6 in suppressing the number of MMCs. First of all, we performed northern blotting to determine the levels of tasiR-ARF in rdr6, tex1 and j66 inflorescences. As expected, tasiR-ARF was reduced in tex1 and rdr6 and further reduced in j66 (Figure 6A). We next examined whether loss-of-function mutants in TAS3 phenocopy tex1 and rdr6 in having supernumerary MMCs. There are three TAS3 loci in Arabidopsis, TAS3A (AT3G17185), TAS3B (AT5G49615) and TAS3C (AT5G57735). To determine which one might function in MMC specification, we examined the expression of these genes in pistils at the MMC stage by real-time RT-PCR. TAS3A and 3B were expressed in pistils, whereas TAS3C was not (Figure S5A). For TAS3A, we identified two mutant lines GK-621G08 (tas3a-1) and GK-723C07 (tas3a-2), in which the TAS3a transcript levels were reduced to 10% and 50% respectively, in comparison with wild type plants (Figure S5B). For TAS3B, one mutant GK-649H12 (tas3b) was identified, in which the TAS3B transcript was not detectable (Figure S5C). All these tas3 mutants showed supernumerary MMC-like cells in ovule primordia (Figure 6B–D). Therefore, TAS3 is involved in the regulation of MMC differentiation.
Figure 6. Supernumarary MMCs are formed in tas3a, tas3b and ARF3::ARF3m–GFP ovules.
The numbers in (B) to (H) indicate the number of ovules with the phenotypes represented by the images out of the total number of ovules examined. The differences between WT and the mutants were significant as determined by student’s t-test (p<0.05). Bars, 10 µm. See also Figure S5.
(A) Northern blotting to detect tasiR-ARF in tex1–1, rdr6 and j66. U6 was used as an internal control. The abundance of tasiR-ARF relative to wild type (WT) is indicated by the numbers. Two biological replicates were performed and gave similar results; only one is shown.
(B–E) Supernumerary MMC-like cells in tas3a-1, tas3a-2, tas3b and ARF3::ARF3m–GFP ovules.
(F,G) pKNU::KNU-Venus expression patterns in ARF3::ARF3m–GFP ovules at the MMC stage.
(H) The presence of abnormally enlarged cells in ARF3::ARF3m–GFP ovules at FG1. The functional megaspore is outlined by white dashed lines and the abnormally enlarged cell is indicated by the black arrow.
(I, J) Expression levels of ARF3 in tas3a, tas3b, tex1 and rdr6 as determined by real-time RT-PCR. The transcript levels of ARF3 in the mutants are significantly higher (p < 0.05, Student’s t-test) than those in WT, which are set as 1. Error bars represent standard deviation as determined by two technical replicates for each of three biological replicates. ACTIN2 was used to normalize ARF3 mRNA levels.
Ectopic expression of ARF3 phenocopies the MMC defects in tex1, rdr6 and tas3
As TAS3-derived tasiR-ARF targets ARF2, ARF3 and ARF4 [30], we hypothesized that the supernumerary MMC-like cells in tas3 mutants were due to deficiencies in the regulation of the ARFs. To test our hypothesis, we examined the MMC phenotype in an ARF3::ARF3m–GFP line that expresses a tasiR-ARF-insensitive mutant with silent mutations in the “A” and “B” target sites in ARF3 [39]; these mutations do not affect the amino acid sequence of the protein, but eliminate or diminish the ability of tasiR-ARF to repress ARF3 [37]. This line exhibited ectopic MMC-like cells in ovule primordia at a frequency of 19.3% (Figure 6E). Moreover, the expression of KNU-Venus in multiple cells suggests that the enlarged cells acquired MMC identity (Figure 6F, G). Besides, we also observed abnormally enlarged cells adjacent to the functional megaspore (FM) at later stages in ovule development (Figure 6H). These results suggested that the MMC defects of tas3 mutants could be due to the de-repression of the targets of TAS3-derived tasiR-ARF.
Thus, we tested whether defects in tasiR-ARF biogenesis in tex1 and rdr6 could account for the MMC defects in these mutants. We determined the expression level of ARF3 in tex1 and rdr6 mutant pistils at the MMC stage by real-time RT-PCR. We observed that ARF3 transcripts accumulated to higher levels in tex1 and rdr6; similar results were also observed in the tas3a-1, tas3a-2 and tas3b mutants (Figure 6I, J). Therefore, tex1, rdr6, and tas3 mutants all exhibited de-repressed ARF3 expression, which was consistent with reduced tasiR-ARF levels. We reasoned that if the de-repression of ARF3 was indeed the cause of the multiple MMC phenotype in tex1, rdr6 and tas3, loss of function in ARF3 should rescue the MMC defects in these mutants. To test this, we crossed an arf3 mutant (SALK_005658C) to tex1 and obtained the arf3 tex1 double mutant. The double mutant showed multiple MMCs in ovules at a frequency of 5.2% (n=216), which was similar to that in wide type and lower than the frequency in tex1. Taken together, these results indicate that the THO complex controls MMC differentiation through the repression of ARF3 by TAS3-derived tasiR-ARF in Arabidopsis.
ARF3 distribution is spatially patterned by TAS3 in ovules
To elucidate the function of TAS3 in MMC specification, we analyzed the expression patterns of ARF3::ARF3-GFP and ARF3::ARF3m–GFP, which were sensitive and resistant to tasiR-ARF, respectively. In order to elaborate the expression patterns clearly, we divided the MMC phase of ovule development into three stages: stage 1, characterized by the lack of integument initiation; stage 2, marked by inner integument initiation only; and stage 3, distinguished by having both inner and outer integuments (Figure 7A).
At stage 1, ovule emerges as a radially symmetrical structure with proximal-distal (P–D) and mediolateral polarities. Along the proximal-distal axis, ARF3-GFP and ARF3m–GFP were both detected in the central chalazal region. Along the mediolateral axis, ARF3-GFP was restricted to the medio side of ovules, whereas ARF3m–GFP was detected on both the medio and lateral sides (Figure 7B – C and B’–C’).
At stage 2, along the proximal-distal axis, ARF3-GFP and ARF3m–GFP were both expressed in the central chalazal domain. Along the mediolateral axis, similar to the expression pattern at stage 1, a more expanded domain of ARF3m–GFP signal from the medio side to the lateral side was observed, as compared to that of ARF3-GFP (Figure 7D – E and D’–E’).
At stage 3, along the proximal-distal axis, ARF3-GFP was expressed mainly in the central chalaza of ovules as in stages 1 and 2. However, ARF3m–GFP was detected both in the central chalaza and the distal nucellus [40], including the cells adjacent to the MMC, but not in the MMC. Along the mediolateral axis, the expression patterns of ARF3-GFP and ARF3m–GFP were similar to those of stages 1and 2. The ovule at this stage was characterized by adaxial-abaxial polarity with the asymmetrical initiation of the outer integument. ARF3-GFP and ARF3m–GFP showed no differences along the adaxial-abaxial axis (Figure 7F – G and F’–G ‘).
To sum up, ovules initiate as radially symmetrical, cylindrical primordia. At the early stage, tasiR-ARF regulates ARF3 expression in the chalazal region, the central part of ovule along the proximal–distal axis. They restrict ARF3 expression to the mediochalazal domain. At a later stage, with the initiation of integuments, tasiR-ARFs not only confer the medio distribution of ARF3 in the chalazal region, but also restrict ARF3 expression to the chalazal region along the proximal-distal axis by preventing its expression in the nucellus. Moreover, it should be noted that ARF3 shows no distribution bias along the adaxial-abaxial axis at the MMC phase of ovule development. Since ARF3::ARF3m–GFP plants displayed supernumerary MMCs at the early stage, it is likely that suppression of the expression of ARF3 in lateral epidermal cells is important for limiting excessive MMC formation.
Furthermore, we examined the expression pattern of ARF3-GFP in tex1. As shown in Figure 7(B”-G”), at the early stage of ovule development, ARF3-GFP was detected in the central chalaza of an ovule along the proximal-distal axis in tex1 as in wild type. However, along the medio-lateral axis, the expression of ARF3-GFP in tex1 extended laterally to the epidermal cells, which was similar to the pattern of ARF3m–GFP.
Discussion
Germline specification is a crucial step in the life cycle of all sexual organisms. Within the female reproductive organs in plants, the formation of the MMC marks the developmental switch from the sporophytic ("somatic") fate to the reproductive or "germline" fate. How somatic cells interact among themselves to ensure that only one cell in the sub-epidermal layer differentiates into the MMC remains unknown. In this study, we uncovered a role of the THO complex in restricting the MMC fate and revealed that THO acts through the TAS3-ARF3 module.
TEX1 acts redundantly with RDR6 to regulate MMC specification
It was reported that the loss-of-function mutant rdr6–11 produces ectopic MMCs and supernumerary female gametophytes at a certain frequency much less than 100% [7], suggesting that there must be redundancy in the genetic mechanisms that restrict MMC specification to only one cell in the sub-epidermal layer. Using EMS mutagenesis and genetic screening in the rdr6–11 background, we found that the ectopic MMC phenotype was much more severe in the rdr6–11 tex1 double mutant than in tex1 or rdr6–11 single mutants, suggesting that TEX1 acts redundantly with RDR6 in regulating MMC specification.
Both RDR6 and TEX1 have been demonstrated to function in the biogenesis of ta-siRNAs [8, 9, 18]). In this study, we found that TEX1 regulates MMC specification through the TAS3-ARF3 module. Since mutations in the two genes behaved in an additive manner in terms of the MMC specification defects, this implied that the two mutations also might act in an additive manner in terms of tasiR-ARF biogenesis. The rdr6–11 allele is predicted to encode an RDR6 protein only one fourth of the full-length protein, so rdr6–11 should be a null allele. It was reported that tasiR-ARF was absent in rdr6–11 [41], which was different from the results in this study, since a weak tasiR-ARF signal could be detected in our assay (Figure 6A). It is worth noting that tasiR-ARF was examined in inflorescences in our study but in seedlings or leaves in previous studies [39, 41]. It is possible that, to a certain extent, some other RDRs (such as RDR2) take the place of RDR6 in the biogenesis of tasiR-ARF in inflorescences when RDR6 function is absent. We found that the mutation in TEX1 induced a further decrease in tasiR-ARF levels in the rdr6–11 background; the additive effects of the tex1 and rdr6 mutations in tasiR-ARF levels is consistent with our model that TEX1 and RDR6 regulate MMC specification redundantly through the TAS3-ARF module.
TAS3 patterns cell type specific expression of ARF3 to prevent excessive MMC formation
TasiR-ARF, encoded by TAS3, targets ARF2, ARF3, and ARF4 to play critical roles in leaf polarity specification, lateral root formation, and floral stem cell termination [33–36]. In this study, we found that TAS3 patterns the distribution patterns of ARF3 in ovule primordia to regulate MMC specification.
We found that loss of function of TAS3A or TAS3B, or expression of TAS3-resistant ARF3, resulted in multiple MMC-like cells in ovule primordia. This provided strong evidence for a role of tasiR-ARF in preventing ectopic MMC specification. However, tasiR-ARF biogenesis requires DICER-LIKE4 (DCL4), and it was reported that supernumerary MMCs were not found in dcl4 mutants [7]. We speculate that the absence of MMC defects in dcl4 is likely owing to DCL2 and DCL3, which are known to contribute to RDR6-dependent ta-siRNA production in the absence of DCL4 [42].
TAS3 patterns the cell type specific expression of ARF3. At an early stage of ovule development, when MMC specification takes place, tasiR-ARF regulates ARF3 expression along the mediolateral axis in the chalazal area. Without the control by TAS3, ARF3 expression extends laterally to the epidermal cells, and this is accompanied by ectopic MMC formation in the sub-epidermal layer. It seems that the restriction of ARF3 expression to medio chalaza, i.e., prevention of ARF3 expression in the epidermal cells, is important for proper MMC specification. Here we propose a model of germline formation regulated by ARF3 in developing ovules of Arabidopsis (Figure S6). In wide type, epidermal (L1) cells in a young ovule primordium produce a signal "Y", which serves as a determinant to prevent the L2 cells adjacent to the MMC from differentiating into MMCs (Figure S6A). The expression of ARF3 in epidermal cells, due to insensitivity to tasiR-ARF or reduced tasiR-ARF levels, represses the production of "Y", therefore releasing the inhibition on the cells next to the MMC (Figure S6B). This model parallels the one proposed for AGO9, which also restricts ectopic MMC specification. AGO9 is thought to associate with 24-nt siRNAs produced in epidermal cells to prevent the cells adjacent to the MMC from acquiring MMC identity [7]. Thus, epidermal cells might have an important function in the specification of MMC identity.
Auxin is a key phytohormone that controls numerous developmental processes in plants [40, 43]. It was reported that auxin is concentrated in the apical epidermal cell layer of the nucellus in ovule primordia [44]. The supernumerary MMCs induced by ectopic expression of ARF3 and the polar distribution of auxin in ovule primordia at the MMC stage reported previously hint at a role of auxin in regulating MMC specification.
Star Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Xuemei Chen (xuemei.chen@ucr.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
A. thaliana (L.) ecotype Columbia (Col-0) was used as wild type. The mutants and transgenic plants used in the study were in the Col-0 background, except for ARF3::ARF3-GFP and ARF3::ARF3m–GFP, which were in the Landsberg erecta (Ler) background. Plants were grown in soil under controlled conditions (16 hr light/8 hr dark at 22°C).
METHOD DETAILS
RNA extraction and quantitative RT-PCR
Quantitative RT-PCR (RT-qPCR) was performed to determine to the expression levels of genes in pistils at the MMC stage. Pistils for each sample were harvested from about 50 plants. RNA was extracted using TRI reagent (MRC, Cat#TR118), a nd DNA was removed using DNase I (Roche, Cat#04716728001). Oligo-dT primer (Fermentas, Cat#S0131) and reverse transcriptase (Fermentas, Cat#EP0441) were used to synthesize cDNA. All procedures used were according to the manufacturers’ instructions. Quantitative PCR was performed on a Bio-Rad IQ cycler apparatus using iQ SYBR Green Supermix (Bio-Rad, Cat#170–8882). The primers used for RT-qPCR are listed in Table S3. The PCR cycle conditions used for quantitative PCR were as follows: 95°C for 5 min followed by 35 cycles of 95°C for 10 s, 60°C for 15 s, and 72°C for 15 s. ACTIN2 was used to normalize mRNA levels in RT-qPCR reactions reported in this study. For each qPCR analysis, six reactions were performed (two technical replicates for three biological replicates).
DIC observation of ovule structure
For phenotypic analysis of ovules, inflorescences from 6–8 plants of wild type, mutants or transgenic line were fixed in FAA overnight or longer, then the pistils were dissected from the flowers at stages 8 to 13 [45] in a drop of chloral hydrate solution (chloral hydrate: H2O: glycerol =8:2:1). Images of the cleared ovules were taken with an Olympus (BX63) microscope with DIC optics using a 40× objective.
Preparation of ovules for confocal laser scanning microscopy
Ovules from stages 1-III to 3-I [46] were dissected, stained in 40% glycerol with 5 µM FM4–64 for 5 min, and analyzed using a Leica TCS SP5 microscope. For the expression pattern analysis of pTEX1::TEX1-GFP, more than five independent transgenic lines were observed. All of them showed the same patterns.
Callose detection
Aniline blue staining for callose detection in the megaspores was carried out as previously described [13] with minor modifications. Inflorescences from 5–8 plants for each genotype were fixed in FAA for 16 h, and stained in 0.1% aniline blue in 100 mM Tris (pH 8.5) for 8 to 12 h. Stained pistils were dissected on a slide, mounted in 30% glycerol and observed with an Olympus (BX63) microscope under UV light (365-nm excitation and 420-nm emission). Two biological replicates were performed.
Whole-mount immunolocalization with ovules
Ovules were dissected from pistils of stages 9–11 flowers, fixed, and processed according to the recently published protocol[47]. The AGO9 primary antibody (Agrisera, AS10673), DMC1 primary antibody and ASY1 primary antibody (from Yingxiang Wang’s Lab) were used at a dilution of 1:100. The secondary antibody AlexaFluor® 488(Molecular Probes) was used at a dilution of 1:300. The samples were stained with propidiumiodide (500µg/mL) before mounting. Images were captured using a confocal microscope (Leica TCS SP5). For PI detection, excitation and emission were at 568 nm and 575–615 nm, respectively. For Alexa Fluor® 488 detection, excitation was at 488 nm and emission was at 500–550 nm. Laser intensity and gain were set at the same levels for all genotypes observed.
DMC1 primary antibody was provided by ABclonal Technology. For the preparation of DMC1 antibody, the whole AtDMC1 protein with 344aa was used as antigen and the recombinant protein was purified with GST Agarose (ABclonal Technology, Cat#AS044).
30–40 pistils from each genotype were subjected to immunostaining with each antibody (i.e. AGO9, DMC1 or ASY1). Two biological replicates were performed for the immunostaining with each antibody.
Plasmid Construction and plant transformation
For the construction of pTEX1::TEX1-GFP, 2 kb sequence upstream of TEX1 was amplified as the promoter from wild type genomic DNA using the primers TEX1_pro_F and TEX1_pro_ R. For the CDS region, a 948 bp cDNA sequence was amplified using primers TEX1_CDS_F and TEX1_CDS_R. The pTEX1::TEX1 fragment was generated by PCR using a mixture of promoter and CDS PCR products as the template. The pTEX1::TEX1 fragment was then cloned into the pDONR 207 vector (Invitrogen). The insert was then recombined into the destination vector pGWB504 [48] using LR Clonase II (Invitrogen).
Small RNA northern blot analysis
To determine the levels of tasiR-ARF in rdr6, tex1 and j66, inflorescences from approximately 15 plants for each genotype were collected and used to extract RNA. Two biological replicates were performed. For the northern blot analysis, ten microgram total RNA from the inflorescences above was resolved in a denaturing polyacrylamide gel and transferred to membrane. An oligonucleotide complementary to tasiR-ARF was end-labeled with γ- 32P to detect mature tasiR-ARF. U6 was used as the loading control. The signal intensities of the blots were quantified using the ImageJ processing and analysis software (http://rsbweb.nih.gov/ij/).
QUANTIFICATION AND STATISTICAL ANALYSIS
For phenotypic quantification, the numbers of ovules with defects (as shown in the figure panels of Figure 1, Figure 2G and 2H, Figure 3, Figure 5, Figure 6, Figure S2D and Figure S4F) as well as the total numbers of ovules examined were recorded.
Statistical analysis was performed using Student’s t test, with p values less than 0.05 being considered significant for any set of data.
Supplementary Material
Acknowledgments
We are grateful to Dr. Xigang Liu (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for the kind gift of ARF3::ARF3-GFP and ARF3::ARF3m–GFP, and to Dr. W. Yang (Institute of Genetics and Developmental Biology, CAS) for sharing the pAKV:H2B–YFP marker. This work was supported by grants from NIH (GM061146), NSFC (91440105), and Guangdong Innovation Research Team Fund (2014ZT05S078) to X.C.; NSFC (31522009; 31470284; U1605212) to Y.Q.; Fujian Innovative Center for Germplasm Resources and Cultivation of Woody plants (No. 125/KLA15001E) and FAFU International collaboration project (3161101458) to Y.Q and NSFC (31600249) to L. Z.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Z.S. and L.Z. conducted histological analysis. Z.S. performed bioinformatics analyses, small RNA northern blot analysis and wrote the paper. Y.Z. isolated the tex1 and hpr1 mutants. S-Y.W. performed the EMS mutagenesis. S.L. identified the hpr1 mutation. H.C. generated the transgenic lines. L.W., Z.L. and P.C. performed genotyping and genetic analyses. X.C. conceived the study. X.C. and Y. Q. designed the overall experimental scheme and revised the paper.
References
- 1.Yang WC, Shi DQ, Chen YH. Female gametophyte development in flowering plants. Annual review of plant biology. 2010;61:89–108. doi: 10.1146/annurev-arplant-042809-112203. [DOI] [PubMed] [Google Scholar]
- 2.Singh M, Goel S, Meeley RB, Dantec C, Parrinello H, Michaud C, Leblanc 0, Grimanelli D. Production of Viable Gametes without Meiosis in Maize Deficient for an ARGONAUTE Protein. The Plant cell. 2011;23:443–458. doi: 10.1105/tpc.110.079020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huettel B, Kanno T, Daxinger L, Bucher E, van der Winden J, Matzke AJ, Matzke M. RNA-directed DNA methylation mediated by DRD1 and Pol IVb: a versatile pathway for transcriptional gene silencing in plants. Biochimica et biophysica acta. 2007;1769:358–374. doi: 10.1016/j.bbaexp.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Aguilar M, Michaud C, Leblanc O, Grimanelli D. Inactivation of a DNA methylation pathway in maize reproductive organs results in apomixis-like phenotypes. The Plant cell. 2010;22:3249–3267. doi: 10.1105/tpc.109.072181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Komiya R, Ohyanagi H, Niihama M, Watanabe T, Nakano M, Kurata N, Nonomura K. Rice germline-specific Argonaute MEL1 protein binds to phasiRNAs generated from more than 700 lincRNAs. Plant J. 2014;78:385–397. doi: 10.1111/tpj.12483. [DOI] [PubMed] [Google Scholar]
- 6.Nonomura KI, Morohoshi A, Nakano M, Eiguchi M, Miyao A, Hirochika H, Kurata N. A germ cell-specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. The Plant cell. 2007;19:2583–2594. doi: 10.1105/tpc.107.053199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olmedo-Monfil V, Duran-Figueroa N, Arteaga-Vazquez M, Demesa-Arevalo E, Autran D, Grimanelli D, Slotkin RK, Martienssen RA, Vielle-Calzada JP. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature. 2010;464:628–632. doi: 10.1038/nature08828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes & development. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes & development. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Himber C, Dunoyer P, Moissiard G, Ritzenthaler C, Voinnet O. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. The EMBO journal. 2003;22:4523–4533. doi: 10.1093/emboj/cdg431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodríguez-Leal D, Leon-Martinez G, Abad-Vivero U, Vielle-Calzada J-P. Natural variation in epigenetic pathways affects the specification of female gamete precursors in Arabidopsis. The Plant cell. 2015;27:1034–1045. doi: 10.1105/tpc.114.133009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klimyuk VI, Jones JD. AtDMCl, the Arabidopsis homologue of the yeast DMC1 gene: characterization, transposon-induced allelic variation and meiosis-associated expression. The Plant journal : for cell and molecular biology. 1997;11:1–14. doi: 10.1046/j.1365-313x.1997.11010001.x. [DOI] [PubMed] [Google Scholar]
- 13.Siddiqi I, Ganesh G, Grossniklaus U, Subbiah V. The dyad gene is required for progression through female meiosis in Arabidopsis. Development. 2000;127:197–207. doi: 10.1242/dev.127.1.197. [DOI] [PubMed] [Google Scholar]
- 14.Sanchez-Moran E, Santos JL, Jones GH, Franklin FC. ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev. 2007;21:2220–2233. doi: 10.1101/gad.439007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Webb MC, Gunning BE. Embryo sac development in Arabidopsis thaliana. Sexual Plant Reproduction. 1990;3:244–256. [Google Scholar]
- 16.Yadegari R, Drews GN. Female gametophyte development. The Plant cell. 2004;16:S133–S141. doi: 10.1105/tpc.018192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Y, Mang HG, Sun Q, Qian J, Hipps A, Hua J. Gene Discovery Using Mutagen-Induced Polymorphisms and Deep Sequencing: Application to Plant Disease Resistance. Genetics. 2012;192:139–U171. doi: 10.1534/genetics.112.141986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jauvion V, Elmayan T, Vaucheret H. The conserved RNA trafficking proteins HPR1 and TEX1 are involved in the production of endogenous and exogenous small interfering RNA in Arabidopsis. The Plant cell. 2010;22:2697–2709. doi: 10.1105/tpc.110.076638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yelina NE, Smith LM, Jones AM, Patel K, Kelly KA, Baulcombe DC. Putative Arabidopsis THO/TREX mRNA export complex is involved in transgene and endogenous siRNA biosynthesis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13948–13953. doi: 10.1073/pnas.0911341107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 21.Payne T, Johnson SD, Koltunow AM. KNUCKLES (KNU) encodes a C2H2 zinc-finger protein that regulates development of basal pattern elements of the Arabidopsis gynoecium. Development. 2004;131:3737–3749. doi: 10.1242/dev.01216. [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez-Leal D, Leon-Martinez G, Abad-Vivero U, Vielle-Calzada JP. Natural variation in epigenetic pathways affects the specification of female gamete precursors in Arabidopsis. The Plant cell. 2015;27:1034–1045. doi: 10.1105/tpc.114.133009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt A, Wuest SE, Vijverberg K, Baroux C, Kleen D, Grossniklaus U. Transcriptome analysis of the Arabidopsis megaspore mother cell uncovers the importance of RNA helicases for plant germline development. PLoS biology. 2011;9:e1001155. doi: 10.1371/journal.pbio.1001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- 25.Chavez S, Beilharz T, Rondon AG, Erdjument-Bromage H, Tempst P, Svejstrup JQ, Lithgow T, Aguilera A. A protein complex containing Tho2, Hprl, Mftl and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. Embo J. 2000;19:5824–5834. doi: 10.1093/emboj/19.21.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pena A, Gewartowski K, Mroczek S, Cuellar J, Szykowska A, Prokop A, Czarnocki-Cieciura M, Piwowarski J, Tous C, Aguilera A, et al. Architecture and nucleic acids recognition mechanism of the THO complex, an mRNP assembly factor. The EMBO journal. 2012;31:1605–1616. doi: 10.1038/emboj.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R. Recruitment of the human TREX complex to mRNA during splicing. Genes & development. 2005;19:1512–1517. doi: 10.1101/gad.1302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaucheret H. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes & development. 2006;20:759–771. doi: 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- 29.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionary fluid set of microRNAs in Arabidopsis thaliana. Genes & development. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 31.Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crete P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Molecular cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 32.Kumakura N, Takeda A, Fujioka Y, Motose H, Takano R, Watanabe Y. SGS3 and RDR6 interact and colocalize in cytoplasmic SGS3/RDR6-bodies. FEBS letters. 2009;583:1261–1266. doi: 10.1016/j.febslet.2009.03.055. [DOI] [PubMed] [Google Scholar]
- 33.Chitwood DH, Nogueira FT, Howell MD, Montgomery TA, Carrington JC, Timmermans MC. Pattern formation via small RNA mobility. Genes & development. 2009;23:549–554. doi: 10.1101/gad.1770009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pekker I, Alvarez JP, Eshed Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. The Plant cell. 2005;17:2899–2910. doi: 10.1105/tpc.105.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia D, Collier SA, Byrne ME, Martienssen RA. Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Current Biology. 2006;16:933–938. doi: 10.1016/j.cub.2006.03.064. [DOI] [PubMed] [Google Scholar]
- 36.Liu X, Dinh TT, Li D, Shi B, Li Y, Cao X, Guo L, Pan Y, Jiao Y, Chen X. AUXIN RESPONSE FACTOR 3 integrates the functions of AGAMOUS and APETALA2 in floral meristem determinacy. Plant J. 2014;80:629–641. doi: 10.1111/tpj.12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marin E, Jouannet V, Herz A, Lokerse AS, Weijers D, Vaucheret H, Nussaume L, Crespi MD, Maizel A. miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. The Plant cell. 2010;22:1104–1117. doi: 10.1105/tpc.109.072553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sieber P, Gheyselinck J, Gross-Hardt R, Laux T, Grossniklaus U, Schneitz K. Pattern formation during early ovule development in Arabidopsis thaliana. Dev Biol. 2004;273:321–334. doi: 10.1016/j.ydbio.2004.05.037. [DOI] [PubMed] [Google Scholar]
- 39.Hunter C, Willmann MR, Wu G, Yoshikawa M, de la Luz Gutierrez-Nava M, Poethig SR. Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development. 2006;133:2973–2981. doi: 10.1242/dev.02491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature. 2005;433:39–44. doi: 10.1038/nature03184. [DOI] [PubMed] [Google Scholar]
- 41.Wu L, Mao L, Qi Y. Roles of dicer-like and argonaute proteins in TAS-derived small interfering RNA-triggered DNA methylation. Plant physiology. 2012;160:990–999. doi: 10.1104/pp.112.200279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 43.Cheng Y, Dai X, Zhao Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. The Plant cell. 2007;19:2430–2439. doi: 10.1105/tpc.107.053009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ceccato L, Masiero S, Roy DS, Bencivenga S, Roig-Villanova I, Ditengou FA, Palme K, Simon R, Colombo L. Maternal Control of PIN1 Is Required for Female Gametophyte Development in Arabidopsis. Plos One. 2013;8 doi: 10.1371/journal.pone.0066148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. The Plant cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneitz K, Hulskamp M, Pruitt RE. Wild-Type Ovule Development In Arabidopsis-Thaliana-a Light-Microscope Study Of Cleared Whole-Mount Tissue. Plant J. 1995;7:731–749. [Google Scholar]
- 47.Escobar-Guzman R, Rodriguez-Leal D, Vielle-Calzada JP, Ronceret A. Whole-mount immunolocalization to study female meiosis in Arabidopsis. Nature protocols. 2015;10:1535–1542. doi: 10.1038/nprot.2015.098. [DOI] [PubMed] [Google Scholar]
- 48.Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, et al. Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Bioscience, biotechnology, and biochemistry. 2007;71:2095–2100. doi: 10.1271/bbb.70216. [DOI] [PubMed] [Google Scholar]
- 49.Abramoff MD, Magalhães PJ, Ram SJ. Image processing with ImageJ. Biophotonics international. 2004;11:36–42. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.