Abstract
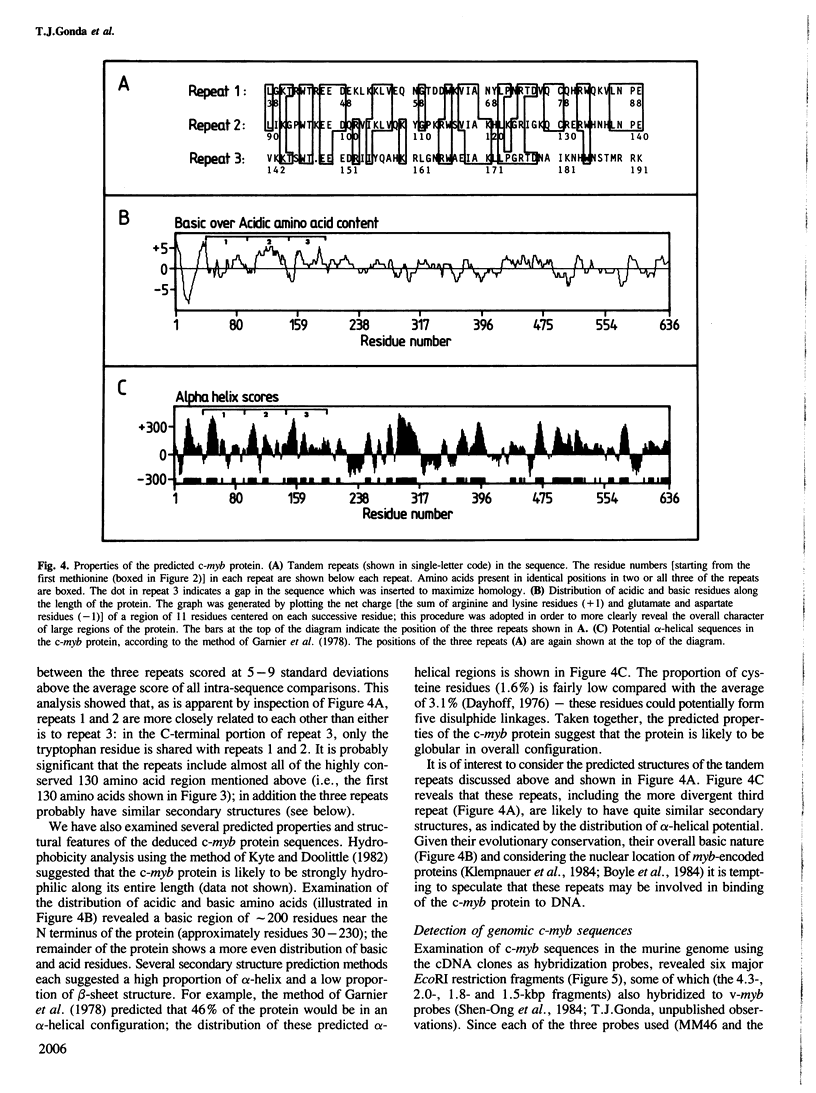
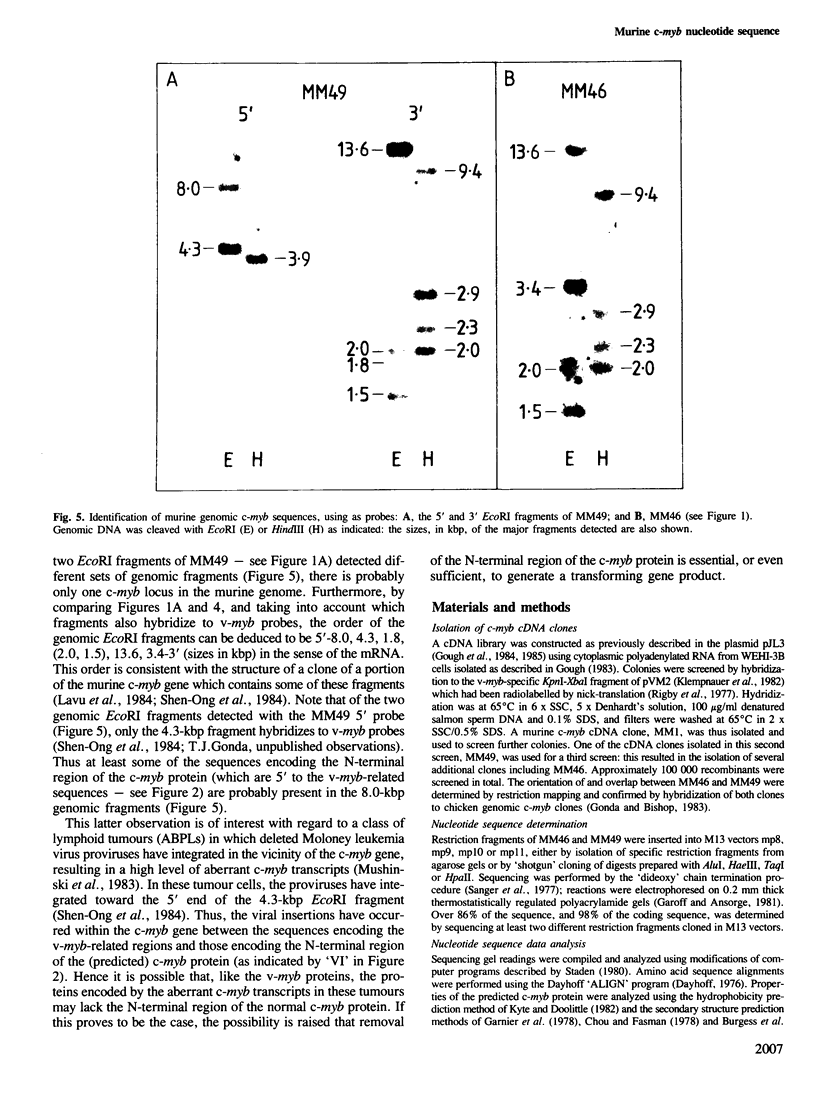
We have isolated cDNA clones of murine c-myb mRNA which contain approximately 2.8 kb of the 3.9-kb mRNA sequence. Nucleotide sequencing has shown that these clones extend both 5' and 3' to sequences homologous to the v-myb oncogenes of avian myeloblastosis virus and avian leukemia virus E26. The sequence contains an open reading frame of 1944 nucleotides, and could encode a protein which is both highly homologous, and of similar size (71 kd), to the chicken c-myb protein. Examination of the deduced amino acid sequence of the murine c-myb protein revealed the presence of a 3-fold tandem repeat of 52 residues near the N terminus of the protein, and has enabled prediction of some of the likely structural features of the protein. These include a high alpha-helix content, a basic region toward the N terminus of the protein and an overall globular configuration. The arrangement of genomic c-myb sequences, detected using the cDNA clones as probes, was compared with the reported structure of rearranged c-myb in certain tumour cells. This comparison suggested that the rearranged c-myb gene may encode a protein which, like the v-myb protein, lacks the N-terminal region of c-myb.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boyle W. J., Lampert M. A., Lipsick J. S., Baluda M. A. Avian myeloblastosis virus and E26 virus oncogene products are nuclear proteins. Proc Natl Acad Sci U S A. 1984 Jul;81(14):4265–4269. doi: 10.1073/pnas.81.14.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Craig R. W., Bloch A. Early decline in c-myb oncogene expression in the differentiation of human myeloblastic leukemia (ML-1) cells induced with 12-O-tetradecanoylphorbol-13-acetate. Cancer Res. 1984 Feb;44(2):442–446. [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Garoff H., Ansorge W. Improvements of DNA sequencing gels. Anal Biochem. 1981 Aug;115(2):450–457. doi: 10.1016/0003-2697(81)90031-2. [DOI] [PubMed] [Google Scholar]
- Gonda T. J., Bishop J. M. Structure and transcription of the cellular homolog (c-myb) of the avian myeloblastosis virus transforming gene (v-myb). J Virol. 1983 Apr;46(1):212–220. doi: 10.1128/jvi.46.1.212-220.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonda T. J., Metcalf D. Expression of myb, myc and fos proto-oncogenes during the differentiation of a murine myeloid leukaemia. Nature. 1984 Jul 19;310(5974):249–251. doi: 10.1038/310249a0. [DOI] [PubMed] [Google Scholar]
- Gonda T. J., Sheiness D. K., Bishop J. M. Transcripts from the cellular homologs of retroviral oncogenes: distribution among chicken tissues. Mol Cell Biol. 1982 Jun;2(6):617–624. doi: 10.1128/mcb.2.6.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonda T. J., Sheiness D. K., Fanshier L., Bishop J. M., Moscovici C., Moscovici M. G. The genome and the intracellular RNAs of avian myeloblastosis virus. Cell. 1981 Jan;23(1):279–290. doi: 10.1016/0092-8674(81)90292-0. [DOI] [PubMed] [Google Scholar]
- Gough N. M. Core and E antigen synthesis in rodent cells transformed with hepatitis B virus DNA is associated with greater than genome length viral messenger RNAs. J Mol Biol. 1983 Apr 25;165(4):683–699. doi: 10.1016/s0022-2836(83)80274-5. [DOI] [PubMed] [Google Scholar]
- Gough N. M., Gough J., Metcalf D., Kelso A., Grail D., Nicola N. A., Burgess A. W., Dunn A. R. Molecular cloning of cDNA encoding a murine haematopoietic growth regulator, granulocyte-macrophage colony stimulating factor. 1984 Jun 28-Jul 4Nature. 309(5971):763–767. doi: 10.1038/309763a0. [DOI] [PubMed] [Google Scholar]
- Gough N. M., Metcalf D., Gough J., Grail D., Dunn A. R. Structure and expression of the mRNA for murine granulocyte-macrophage colony stimulating factor. EMBO J. 1985 Mar;4(3):645–653. doi: 10.1002/j.1460-2075.1985.tb03678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S. H., Payvar F., Spector D., Schimke R. T., Robinson H. L., Payne G. S., Bishop J. M., Varmus H. E. Heterogeneity of genetic loci in chickens: analysis of endogenous viral and nonviral genes by cleavage of DNA with restriction endonucleases. Cell. 1979 Oct;18(2):347–359. doi: 10.1016/0092-8674(79)90054-0. [DOI] [PubMed] [Google Scholar]
- Klempnauer K. H., Gonda T. J., Bishop J. M. Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular progenitor c-myb: the architecture of a transduced oncogene. Cell. 1982 Dec;31(2 Pt 1):453–463. doi: 10.1016/0092-8674(82)90138-6. [DOI] [PubMed] [Google Scholar]
- Klempnauer K. H., Ramsay G., Bishop J. M., Moscovici M. G., Moscovici C., McGrath J. P., Levinson A. D. The product of the retroviral transforming gene v-myb is a truncated version of the protein encoded by the cellular oncogene c-myb. Cell. 1983 Jun;33(2):345–355. doi: 10.1016/0092-8674(83)90416-6. [DOI] [PubMed] [Google Scholar]
- Klempnauer K. H., Symonds G., Evan G. I., Bishop J. M. Subcellular localization of proteins encoded by oncogenes of avian myeloblastosis virus and avian leukemia virus E26 and by chicken c-myb gene. Cell. 1984 Jun;37(2):537–547. doi: 10.1016/0092-8674(84)90384-2. [DOI] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lavu S., Mushinski J. F., Shen-Ong G. L., Potter M., Reddy E. P. Structural organization of mouse c-myb locus and the mechanism of its rearrangement in ABPL-2 tumor line induced by pristane and Abelson murine leukemia virus. Curr Top Microbiol Immunol. 1984;113:37–40. doi: 10.1007/978-3-642-69860-6_8. [DOI] [PubMed] [Google Scholar]
- Mushinski J. F., Potter M., Bauer S. R., Reddy E. P. DNA rearrangement and altered RNA expression of the c-myb oncogene in mouse plasmacytoid lymphosarcomas. Science. 1983 May 20;220(4599):795–798. doi: 10.1126/science.6687762. [DOI] [PubMed] [Google Scholar]
- Nunn M., Weiher H., Bullock P., Duesberg P. Avian erythroblastosis virus E26: nucleotide sequence of the tripartite onc gene and of the LTR, and analysis of the cellular prototype of the viral ets sequence. Virology. 1984 Dec;139(2):330–339. doi: 10.1016/0042-6822(84)90378-7. [DOI] [PubMed] [Google Scholar]
- Radke K., Beug H., Kornfeld S., Graf T. Transformation of both erythroid and myeloid cells by E26, an avian leukemia virus that contains the myb gene. Cell. 1982 Dec;31(3 Pt 2):643–653. doi: 10.1016/0092-8674(82)90320-8. [DOI] [PubMed] [Google Scholar]
- Ralston R., Bishop J. M. The protein products of the myc and myb oncogenes and adenovirus E1a are structurally related. Nature. 1983 Dec 22;306(5945):803–806. doi: 10.1038/306803a0. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rushlow K. E., Lautenberger J. A., Papas T. S., Baluda M. A., Perbal B., Chirikjian J. G., Reddy E. P. Nucleotide sequence of the transforming gene of avian myeloblastosis virus. Science. 1982 Jun 25;216(4553):1421–1423. doi: 10.1126/science.6283631. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheiness D., Gardinier M. Expression of a proto-oncogene (proto-myb) in hemopoietic tissues of mice. Mol Cell Biol. 1984 Jul;4(7):1206–1212. doi: 10.1128/mcb.4.7.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Ong G. L., Potter M., Mushinski J. F., Lavu S., Reddy E. P. Activation of the c-myb locus by viral insertional mutagenesis in plasmacytoid lymphosarcomas. Science. 1984 Nov 30;226(4678):1077–1080. doi: 10.1126/science.6093260. [DOI] [PubMed] [Google Scholar]
- Shen-Ong G. L., Potter M., Mushinski J. F., Lavu S., Reddy E. P. Activation of the c-myb locus by viral insertional mutagenesis in plasmacytoid lymphosarcomas. Science. 1984 Nov 30;226(4678):1077–1080. doi: 10.1126/science.6093260. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Staden R. A new computer method for the storage and manipulation of DNA gel reading data. Nucleic Acids Res. 1980 Aug 25;8(16):3673–3694. doi: 10.1093/nar/8.16.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westin E. H., Gallo R. C., Arya S. K., Eva A., Souza L. M., Baluda M. A., Aaronson S. A., Wong-Staal F. Differential expression of the amv gene in human hematopoietic cells. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2194–2198. doi: 10.1073/pnas.79.7.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]




