Abstract
Clathrin-mediated endocytosis (CME) involves nanoscale bending and inward budding of the plasma membrane, by which cells regulate both the distribution of membrane proteins and the entry of extracellular species1,2. Extensive studies have shown that CME proteins actively modulate the plasma membrane curvature1,3,4. However, the reciprocal regulation of how the plasma membrane curvature affects the activities of endocytic proteins is much less explored, despite studies suggesting that membrane curvature itself can trigger biochemical reactions5–8. This gap in our understanding is largely due to technical challenges in precisely controlling the membrane curvature in live cells. In this work, we use patterned nanostructures to generate well-defined membrane curvatures ranging from +50 nm to −500 nm radius of curvature. We find that the positively curved membranes are CME hotspots, and that key CME proteins, clathrin and dynamin, show a strong preference toward positive membrane curvatures with a radius < 200 nm. Of ten CME related proteins we examined, all show preferences to positively curved membrane. By contrast, other membrane-associated proteins and non-CME endocytic protein, caveolin1, show no such curvature preference. Therefore, nanostructured substrates constitute a novel tool for investigating curvature-dependent processes in live cells.
Keywords: clathrin, dynamin, endocytosis, membrane curvature, curvature sensing, nanopillar, nanowire, nanostructure, live cell, plasma membrane
Membrane curvature is no longer seen merely as a passive feature of membranes, but has emerged as a highly active player in regulating protein activities9,10. To date, how membrane curvature affects protein binding and activity has been primarily studied using in vitro systems, such as supported lipid bilayers and lipid vesicles8,11,12. However, these findings need to be validated in live cells, which in the context of endocytosis requires controlling plasma membrane curvature at the scale of tens to hundreds of nanometer radius. This is a challenging task as the plasma membrane is a dynamic and complex system containing hundreds of different lipid and protein components. A recent study has shown that the plasma membrane can be deformed on nanoscale conical structures (nanocones), which triggers the recruitment of curvature-sensing N-BAR proteins13. However, as nanocones were variables in sizes and densely packed14, the extent of nanocone-induced membrane curvature was not well controlled and not individually discernible under an optical microscope13. Conversely, we and others have shown that vertically-aligned nanopillars that are evenly spaced and uniform in size can induce conformal plasma membrane wrapping in mammalian cells15–17. Based on these studies, we hypothesize that the shape and geometry of vertically-aligned nanostructures can be used to control local membrane curvature in live cells (Fig. 1a). In this work, we demonstrate that vertically-aligned nanostructures generate well-defined membrane curvature, which we exploit to probe how membrane curvature affects CME activities.
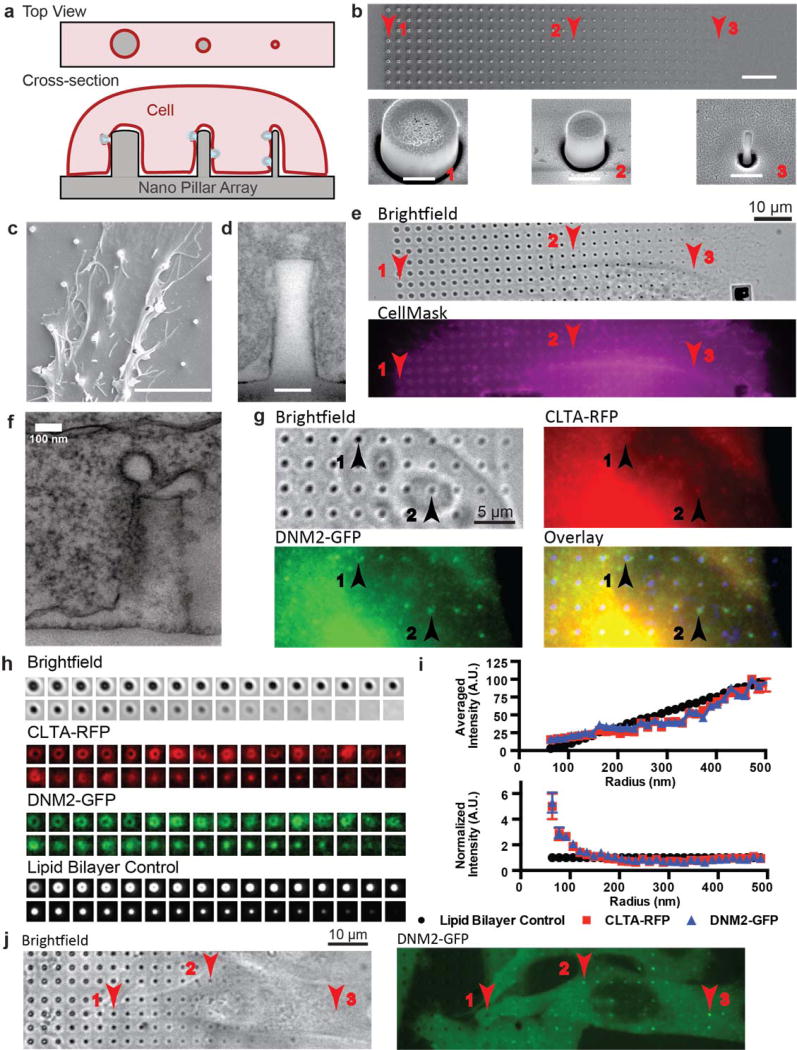
Figure 1. Vertical nanopillars generate well-defined membrane curvatures that induce local accumulation of endocytic proteins.
a. Schematic illustration of nanopillars with different radii (grey block) deforming the cell membrane to generate different membrane curvatures (red line). b. An SEM image of a gradient nanopillar array with 700 nm height, 3 µm center-to-center distance between nanopillars, and radii ranging from 500 nm (left) to 50 nm (right) with 14 nm increment. Scale bar: 10 µm. Bottom row: zoomed-in SEM images of individual nanopillars as indicated by corresponding locations in the top row (red with arrow). Scale bar: 400 nm. c. An SEM image showing a SK-MEL-2 cell deforming on nanopillars. Scale bar: 5 µm. d. A TEM image showing the cell membrane wrapping around a nanopillar. Scale bar: 100 nm. e. The plasma membrane stained with CellMask™ DeepRed shows increased membrane signal at nanopillar locations. Red arrows indicate locations of nanopillars with different radius. Scale bar: 10 µm. f. A TEM image of the plasma membrane wrapping around a nanopillar captures a clathrin-coated pit. Scale bar: 100 nm. g. Immunostaining of clathrin and dynamin2 in hCLTAEN/hDNM2EN cells shows accumulation of clathrin and dynamin2 at nanopillar locations. In the overlay image, the inverted brightfield image is converted to blue (bottom right). Scale bar: 10 µm. Numbered black arrows indicate locations of nanopillars with different radii. h. Quantification of the clathrin, dynamin2 and membrane signals at nanopillars of 32 different radii. Signals from many nanopillars of the same radius are averaged to obtain the averaged image. Averaged images were sorted from largest (top left) to smallest radius (bottom right). Four imaging channels are shown from top to bottom: bright field, clathrin, dynamin2 and lipid bilayer control for surface area increment. i. Intensities of clathrin-RFP, dynamin2-GFP and lipid membrane all increase with nanopillar radius (top graph). After normalizing against the membrane intensities, it is clear that the clathrin/membrane and dynamin2/membrane ratios increase significantly when the nanopillar radius is less than 200 nm (bottom graph). Each data point is an average over 90–191 nanopillars with error bar representing standard error of the mean. Detailed statistic analysis is in Supplementary Table S7. Similar trend is also observed with background calculated from the averaged small image (Supplementary Fig. S6) instead of the rolling ball algorithm used in i. j. The time-averaged image of a 4 min movie of dynamin2-GFP demonstrates that dynamin2-GFP exhibits strong preference to sharp nanopillars, but much less to large radius nanopillars in the same cell (Arrow 1 vs. Arrow 3). Scale bar: 10 µm.
To demonstrate that membrane curvature can be controlled using vertically-aligned nanopillars, we engineered a gradient array of SiO2 nanopillars with different radii from 50 nm to 500 nm with a 14 nm increment (scanning electron microscopy (SEM) see Fig. 1b). The size variations are within 10% of the nominal value (Supplementary Fig. S1, Table S1–4). The area along the side of each nanopillar is about 10 times the area of the top, so we primarily considered the membrane curvature along the side, a positive curvature denoted by the nanopillar radius. These curvatures cover the mid range of endocytic curvature progression (from flat to ~50 nm radius). SK-MEL-2 cells cultured on a poly-L-lysine coated substrate bend their membrane on nanopillars, as seen in the SEM image in Fig. 1c. This was substantiated by the cross-section visualization using TEM (Fig. 1d) and focused ion beam and scanning electron microscopy (FIB-SEM) (Supplementary Fig. S2–S3). The plasma membrane wraps around nanopillars with an average gap distance of 21.6±14.1 nm (mean ± s.d.), consistent with previous findings15. Membrane wrapping around nanopillars of all sizes was further confirmed by membrane staining (CellMask™ Deep Red, Fig. 1e), where rings appeared on large nanopillars and dots appeared on small nanopillars below the diffraction limit. The fluidity of the membrane around nanopillars was verified to be similar to that of flat areas using fluorescence recovery after photobleaching (Supplementary Fig. S4, Movie S1). These results confirm that vertically-aligned nanopillars can induce well-defined plasma membrane curvatures.
In TEM (Fig. 1f) and FIB-SEM (Supplementary Fig. 2e), we captured a few endocytic events as shown by the characteristic clathrin-coated pit profiles, indicating that CME does occur on nanopillar-curved membranes. To investigate CME further, we used gradient nanopillar arrays to examine how membrane curvature affects the distribution of two key CME proteins, clathrin and dynamin18,19. Immunostaining clearly showed increased clathrin and dynamin signals at nanopillars (arrows, Fig. 1g). To account for cell-to-cell variations, we averaged signals from over 2000 nanopillars in 41 cells to obtain averaged fluorescence images of clathrin and dynamin at each nanopillar radius (Fig. 1h). As a control for the surface area, we also measured the fluorescence signal of a supported lipid membrane (Fig. 1h, bottom panel), a membrane associated protein (GFP-CAAX), and CellMask staining on the same array (Supplementary Fig. S5, Table S5). The fluorescent signals for the membrane, clathrin and dynamin all increased with increasing nanopillar radius (Fig. 1i, top). By normalizing the protein intensities against the membrane ones (Fig. 1i, bottom), we found that the ratios of clathrin/membrane and dynamin/membrane are relatively constant for radii > 200 nm. However, for radii < 200 nm, both ratios increase significantly, indicating a clear preference of clathrin and dynamin for curvature radius < 200 nm. This strong preference for high curvature is also evident in single cells. In an averaged image from a 4-min movie (Fig. 1j), dynamin2-GFP exhibits stronger signals at small-radius nanopillars than at large-radius nanopillars in the same cell (Arrow 3 vs. Arrow 1 in Fig. 1j), despite their smaller membrane area. Similar conclusion was also obtained with co-transfected dynamin2-GFP and membrane marker mCherry-CAAX (Supplementary Fig. S7, Table S6).
The preferential accumulation of proteins on highly curved membranes is more compellingly demonstrated by nanostructures that generate a local combination of different curvatures. As illustrated in Fig. 2a, a single nanobar structure locally induces two different membrane curvatures – high curvature at the two ends and zero curvature in the middle. Fig. 2b shows an SEM image of a nanobar array. When cells were cultured on the nanobars, the plasma membrane was found to wrap evenly around the bars, outlining the nanobar shape as shown by CellMask staining (Fig. 2c). To probe protein dynamics, genome-edited SK-MEL-2 cells (hCLTAEN/hDNM2EN)19 that express clathrin-RFP and dynamin2-GFP were cultured on nanobars. Unlike the CellMask staining, both clathrin and dynamin2 showed strong preferences for the highly curved ends and very little accumulation with the middle region (Fig. 2d). The averaged images from over 167 nanobars (Fig. 2e) more clearly illustrated the strong accumulation of clathrin and dynamin at nanobar ends (Fig. 2f). Kymograph analysis of time-lapse movies demonstrated that both clathrin and dynamin2 were dynamic and had strong preference for nanobar ends (Fig. 2g, Supplementary Fig. S8, Movie S2). Endocytic events, as evidenced by the characteristic appearance of dynamin2 near the end of the clathrin lifetime19, are abundant at nanobar ends, while very few events are observed in the middle of the same nanobars. Similarly, the endocytic adaptor protein AP2 (AP2-RFP) also shows preferred accumulation on nanopillars with the appearance of dynamin2-GFP at the end of the AP2 lifetime (Supplementary Fig. S9, Movie S3).
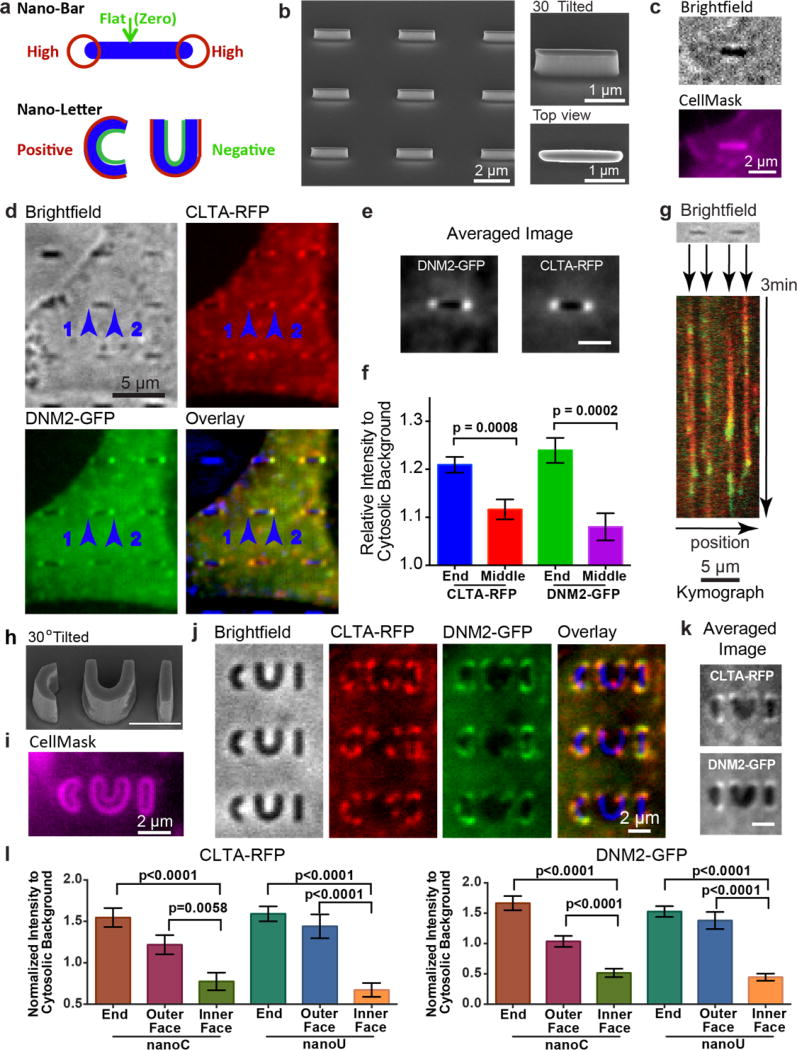
Figure 2. Engineered 3D nanostructures f or versatile control of membrane curvatures and endocytic protein accumulations.
a. Schematic illustration of nanobar and nano-letter design for inducing a range from flat membranes to high membrane curvatures (top, nanobar) and combined positive and negative membrane curvatures (bottom, nanoC and nanoU) by the same nanostructure. b. An SEM image of a nanobar array showing individual nanobars of 150 nm width, 2 µm length, 1 µm height and 5 µm pitch(left, scale bar: 2 µm.). c. When SK-MEL-2 cells were cultured on nanobar arrays, CellMask™ DeepRed staining demonstrated that the plasma membrane wrapped evenly around the nanobar. Scale bar: 2 µm. d. Averaged fluorescence images of a 4 min movie shows that clathrin and dynamin2 prefer the two highly curved ends of nanobars as compared with the sidewalls. This is in sharp contrast to the membrane staining shown in c. In the overlay image, bright field images of nanobars are shown in blue. Scale bar: 5 µm. e. Averaged image of clathrin and dynamin2 distribution on 167 nanobars. Scale bar: 2 µm. f. The quantitative values of fluorescence intensity at nanobar ends and nanobar center with the cytosolic background as reference. Error bar represents standard error of the mean. Statistic analysis shows significant difference between nanobar end and nanobar center (p value of unpaired t-test: 0.0008 for Clathrin-RFP and 0.0002 for Dynamin2-GFP). g. Kymograph plots of clathrin-RFP (red) and dynamin2-GFP (green) on two adjacent nanobars show the appearance of a dynamin2 peak near the end of the clathrin segment, which is characteristic of clathrin-mediated endocytosis. The dynamic events are observed at the ends of nanobars, with very little signal along the side walls or between nanobars. More kymographs on a larger area with more adjacent nanobars are in Supplementary Fig. S11. h. An SEM image of the nanoCUI structure in 30° tilted view. Scale bar: 2 µm. i. CellMask™ DeepRed staining of cells on nanoCUI array shows that the cell membrane wrapped around both the inside and the outside surfaces of the nanoC and nanoU structures. Scale bar: 2 µm. j. Averaged fluorescence images of a 4 min movie show that clathrin and dynamin2 prefer positive membrane curvatures at the ends and the outer surfaces of nanoC and nanoU, with much less protein signal on the inner surfaces. k. Averaged images of clathrin-RFP (top) and Dynamin2-GFP (bottom) on 51 pairs of nanoC and nanoU clearly show preferred accumulation on the ends and their outer faces of both nanoC and nanoU comparing to the inner faces. Scale bar: 2 µm. l. The quantified fluorescence intensity (in k) on the end, the outer face, and the inner face of both nanoC (left) and nanoU (right) with the cytosolic background as reference. Error bar represents standard error of the mean. Statistic analysis shows significant difference at the end/the outer face vs. the inner face of both nanoC and nanoU. The p value of unpaired t-test for each pair is indicated on the plot.
In addition to probing membrane curvatures of different magnitudes, we also probed curvatures of opposite signs. We engineered nanoC and nanoU to induce positive curvature at the outer face (Fig. 2a, red line) and negative curvature at the inner face (Fig. 2a, green line). In order to distinguish the outer and inner faces by optical microscopy, we made nanoC and nanoU structures of 500 nm width (Fig. 2h). Membrane staining by CellMask clearly illustrates the inner and the outer faces and confirms that nanoC and nanoU induced both positive and negative membrane curvatures (Fig. 2i). When hCLTAEN/hDNM2EN cells were cultured on nanoC and nanoU, both clathrin and dynamin2 exhibited accumulation on the outer surface of the nanostructures with positive membrane curvature. By contrast, very little protein signal appeared on the negative side (Fig. 2j). The averaged images from over 50 nanoCUI structures show that both clathrin and dynamin prefer high curvature locations at the ends of nanoC, nanoU and nanoI (Fig. 2k). For nanoC and nanoU, the fluorescence intensities at the outer surface are statistically higher than on the inner surface (Fig. 2l). These results demonstrate that endocytosis, a process of positive curvature progression, is favored by preformed positive, but not negative, curvature.
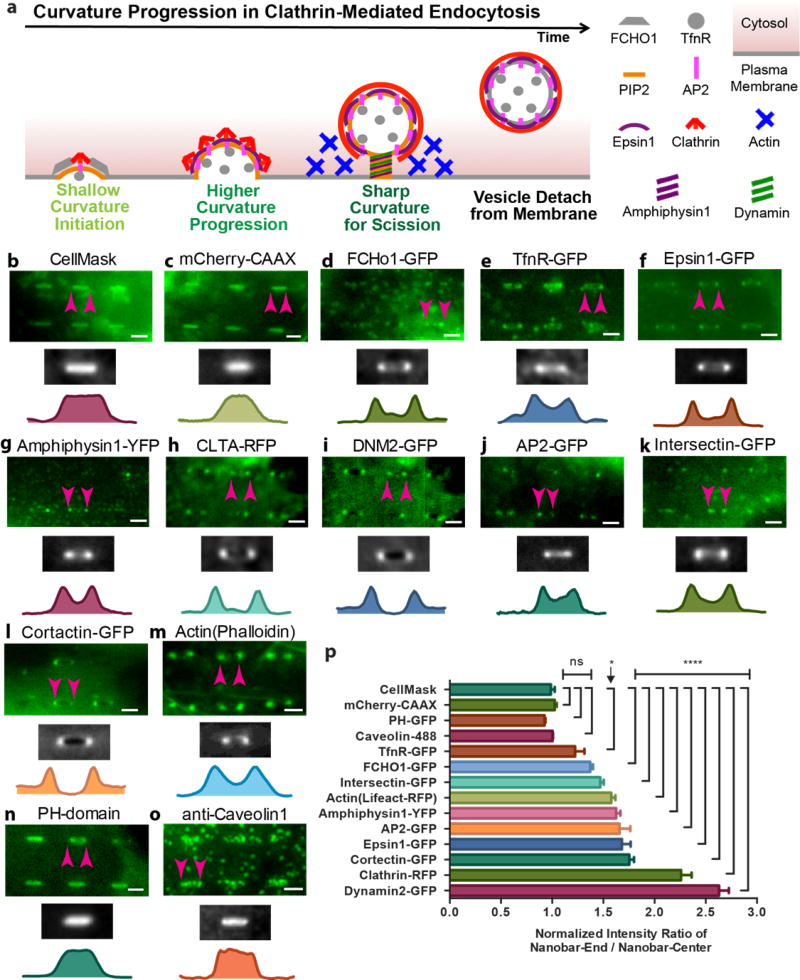
Besides clathrin and dynamin2, many other proteins and lipids participate in CME20 (schematic see Fig. 3a). We examined the curvature preference of twelve proteins on nanobars, including those with curvature-sensing domains (e.g., epsin with an ENTH domain21 and amphiphysin with an N-BAR domain22) and those not reported to have curvature-sensing domains (e.g., AP223). The high magnification fluorescence images in Fig. 3 b–o (green color images) show the spatial distributions of different proteins on six nanobars (the corresponding full images are shown in Supplementary Fig. S10–S11). For each protein, the averaged image over hundreds of nanobars is shown in grey-scale and the protein intensity is plotted along the length of the nanobar (Fig. 3 b–o). Their curvature preference was evaluated by the ratio of the intensity at the nanobar-end vs. nanobar-center (Fig. 3p, Supplementary Table S8).
Figure 3. Probing curvature sensitivity of various endocytic proteins using nanobar arrays.
a. Schematic illustration of proteins involved in different stages of clathrin-mediated endocytosis. The high magnification fluorescence images in b–o (green color images, scale bar: 2 µm) show the distributions of different proteins on six nanobars. The corresponding large images can be found in the supplementary information (Fig. S9 and S10). The averaged image of hundreds of nanobars and the intensity along the length of nanobars are shown beneath the fluorescent images. The spatial distributions of CellMask™ DeepRed staining the plasma membrane (b), and membrane-associated protein mCherry-CAAX (c) are distributed relatively evenly along the entire length of the nanobars. Eight other endocytic components involved in the clathrin-dependent endocytosis including epsin1-GFP (f), amphiphysin1-YFP (g), clathrin-RFP (h), dynamin2-GFP (i), AP2-GFP (j), intersectin (k), cortactin (l) and actin (m) show strong bias toward the ends of the nanobars with little accumulation along the sidewalls. In comparison, FCHo1-GFP (e), and transferrin receptor (TfnR-GFP) (f) show less preference to the nanobar ends than those above. On the other hand, PIP2 probed by PH-GFP (n) and caveolin1 (o), an essential component of the caveolin-dependent endocytosis, is evenly distributed along the entire length of nanobars. All the nanobars are 2 µm in length. p. Distribution of 14 different proteins/dyes on nanobars quantified by normalized intensity ratio of nanobar-end to nanobar-center. For each protein, the distribution is measured by averaging over 56–2072 nanobars. Error bar represents standard error of the mean. Statistical significance of each protein vs. CellMask was evaluated by unpaired t-test with Welch’s correction (details see Supplementary Table S8). p-value: **** < 0.0001, * < 0.05, ns >0.05.
As a reference for protein distribution, the plasma membrane was probed by CellMask (Fig. 3b) and a plasma membrane-associated protein mCherry-CAAX (Fig. 3c), both were homogeneous along the length of the nanobar. In sharp contrast, all 10 CME proteins showed spat ial preference for the highly curved ends (Fig. 3. d–m). Proteins involved in the early stages of CME such as FCHo-1-GFP24 (Fig. 3d) and transferrin receptor (TfnR-GFP)25 (Fig. 3e) showed moderate preference to nanobar ends. Proteins involved in the later stages of CME showed much stronger preference, including epsin1-GFP (Fig. 3f,), amphiphysin1-YFP (Fig. 3g,), CLTA-RFP (Fig. 3h,), DNM2-GFP (Fig. 3i,), AP2-GFP (Fig. 3j,), intersectin (Fig. 3k,), cortactin (Fig. 3l,), and actin (Fig. 3m). Their averaged images show a characteristic dumbbell shape and the intensity plots indicate two pronounced peaks at the two ends of the nanobars (Fig. 3f–m). Of all the CME proteins, AP2, intersectin, cortactin and actin are not reported to have curvature sensitive-domains. Their accumulation at nanobar ends strongly indicates that both curvature sensitive and non-sensitive CME proteins co-assemble at highly curved membrane for CME progression.
Although CME proteins show strong preference to highly curved membrane, a signaling lipid involved in the CME process, phospholipid phosphatidylinositol 4,5-biphosphate (PIP2)26,27, did not show such preference. PIP2 was probed using the PH domain of PLC-delta (PH-GFP) (Fig. 3n), which showed uniform distribution along the nanobar similar to CellMask. We also examined the curvature sensing of caveolin1, an essential protein in caveolae-dependent endocytosis28. As shown in Fig. 3o, caveolin1 puncta evenly distributed along the entire length of nanobars with no preference toward the ends. This indicates that caveolae-dependent and clathrin-mediated endocytosis are affected differently by membrane curvature.
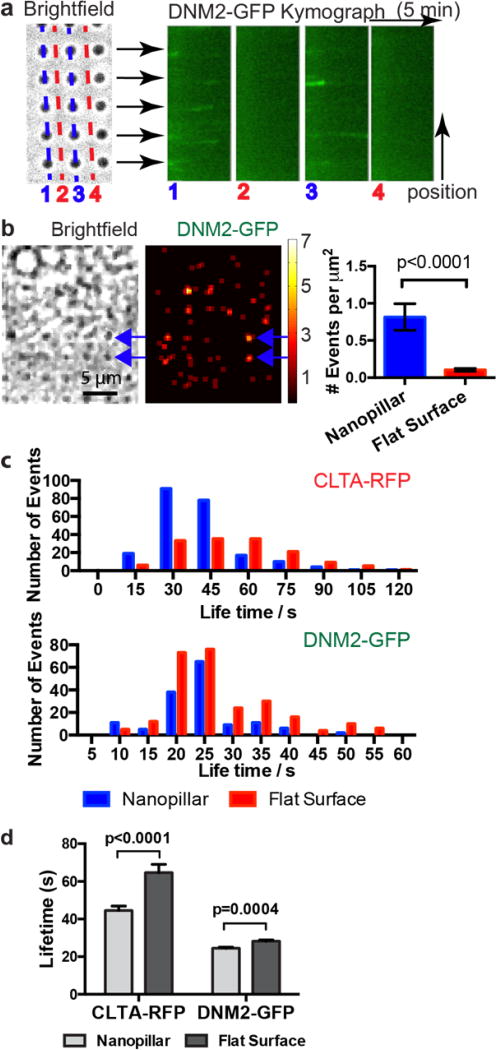
In addition to probing the spatial preference of CME proteins for membrane curvatures, we also investigated their dynamics. We compared the occurrence frequency of dynamin2 spots on nanopillars of 150 nm radius vs. on flat areas. Line kymographs crossing several nanopillars showed clear segments of dynamin2 traces aligned with nanopillars (Fig. 4a, blue lines), while very few dynamin2 events on adjacent flat areas (Fig. 4a, red lines). The rapid appearance and disappearance of a dynamin2 spot is a signature of an endocytic vesicle scission event19. A spatial map of dynamin2 events clearly shows hotspots for endocytosis at nanopillars (Fig. 4b). After normalizing for the membrane area, there is significantly more occurrence of dynamin2 clusters on nanopillars than on flat membranes (Fig. 4b), indicating that pre-curved membranes are preferred CME sites.
Figure 4. Pre-curved membranes are preferred sites for endocytosis.
a. Kymograph plots of dynamin2-GFP along lines of nanopillars show repeated appearance and disappearance of dynamin2 spots at nanopillar locations (line 1 and 3). Similar kymograph plots along lines between nanopillars show very little signal of dynamin2-GFP (line 2 and 4). b. Spatial mapping of the occurrence frequency of dynamin2-GFP puncta shows hot spots for endocytosis at nanopillar locations. We measured 65 dynamin2 blinking events on 75 nanopillars in 6 minutes. Nanopillar area is calculated to be 1 µm2 each. From the same set of movies, we measured 62 dynamin2 blinking events on 75 flat areas, 8 µm2 each. After normalizing against the calculated membrane areas, the endocytic event occurrence on curved membrane at nanopillars is significantly higher than that on flat. Error bars represent standard error of the mean and the P value of Kolmogorov–Smirnov test is < 0.0001. c. Lifetime distribution of clathrin-RFP (up) and dynamin2-GFP (bottom) puncta on nanopillars (blue) and on flat areas (red). For clathrin, 226 events were measured on nanopillars and 154 events were measured on flat areas. For dynamin2, 147 events were measured on nanopillars and 260 events were measured on flat areas. For comparison between nanopillar and flat, Kolmogorov–Smirnov test gives p-value of 0.0082 for dynamin2-GFP and <0.0001 for clathrin-RFP. The differences are significant. d. The average lifetimes of clathrin-RFP and dynamin2-GFP appear to be decreased on nanopillars (light grey) compared with on flat areas (dark grey). Unpaired t-test with equal SD gives p-value of 0.0004 for dynamin2-GFP and <0.0001 for clathrin-RFP. The differences are significant.
Besides frequency, we also measured the lifetime of dynamin2 and clathrin spots on nanopillars vs. on flat areas (Supplementary Movie S4). The dynamin2 lifetime distribution (Fig. 4c) shows two apparent populations on flat areas, one around 20 s and the other around 35 s. On nanopillars, the dynamin2 lifetime distribution also shows similar peaks, but the population with longer lifetimes is slightly decreased. The average lifetime of dynamin2 is slightly shorter on nanopillars (24±0.6 s vs. 28±0.7 s, mean±SEM) (Fig. 4d). On the other hand, the lifetime for clathrin spots appears to decrease significantly on nanopillars as compared to flat areas (45±2.3 s vs. 65±4.3 s, mean±SEM). It is plausible that pre-curved membrane reduces the energy barrier for the membrane bending and thus facilitates faster clathrin coat assembly. However, we note that the lifetime measurement may miss some long clathrin events as multiple endocytosis events sometimes overlap on nanopillars.
Our results demonstrate that CME proteins show spatial preference to highly positively curved over flat or negatively curved membranes in live cells. We also confirm that membrane curvature can directly modulate the biochemical activities of CME, as previously proposed5. Interestingly, actin also shows a strong preference for highly curved membranes. As actin and its regulators are involved in diverse cellular functions29,30, these results suggest that membrane curvature may affect many other cellular processes in addition to endocytosis. The ability to precisely manipulate membrane curvature in live cells opens up new avenues to study the influence of membrane curvature on a variety of cellular processes.
Methods (Online)
Nanostructure fabrication and characterization
Nanostructures used in this work were fabricated on square quartz coverslips (Technical Glass Products, Inc.) using electron-beam lithography similar to previously reported.31 In brief, the coverslips were spin-coated with 300nm of e-beam resist ZEP-520 (ZEON Chemicals), followed by a thin-layer (~20nm) of E-Spacer 300Z (Showa Denko) for charge dissipation. Desired patterns (dots for nanopillar, short line for nanobar and letter for nanoC and nanoU) were written by the Raith 150 electron-beam lithography system and subsequently developed in xylene. A metal mask was then created by sputter deposition of Cr of 100 nm thickness and lift-off in acetone. Nanostructures were subsequently generated by anisotropic reactive ion etching with a mixture of CHF3 and O2 (AMT 8100 etcher, Applied Materials) of the quartz substrate. The substrate was cleaned in O2 plasma and subsequently immersed in Chromium Etchant 1020 (Transene) to remove Cr masks. Before plating cells, the substrate was sterilized by UV irradiation and coated overnight in 0.2mg/ml poly-L-lysine (Sigma-Aldrich). The shape and dimensions of the nanostructures were measured by scanning electron microscopy (FEI Nova) after the substrate was sputter coated with 3nm Cr. Detailed dimensions for different structures are described in the text of each section.
Cell culture
SK-MEL-2 and genome-edited SK-MEL-2 line with hCLTAEN/hDNM2EN was described previously32. For culture, cells were plated on poly-L-Lysine coated nanostructure substrates (~ 2 × 104 / cm2) and maintained in the DMEM/F12/GlutaMAX™ medium (Gibco) supplemented with 10% fetal bovine serum (BioExpress) and 100 U/ml penicillin and 100 mg/ml streptomycin (Sigma-Aldrich). Plasmocin™ prophylactic (InvivoGen) was added at 2.5 mg/ml to the culture to prevent mycoplasma contamination. The cultures were maintained in a standard incubator at 37°C with 5% CO2.
SEM examination of the cell morphology on nanopillars arrays
To examine the morphology of the plasma membrane on nanostructures in high resolution, scanning electron microscopy (SEM) was used according to previously described protocols33. In preparation for SEM, cells were first cultured overnight on nanostructure substrates, followed by 30-min fixation in 2% glutaraldehyde and 4% paraformaldehyde in 0.1 M s odium cacodylate buffer (pH 7.3) at room temperature. For morphological visualization by SEM, SK-MEL-2 cells on gradient nanopillars arrays were washed twice in the same buffer and post-fixed in 1% reduced osmium tetroxide (1% osmium tetroxide with 0.8% potassium ferricyanide in 0.1 M sodium cacodylate). Then, the sample underwent dehydration in a graded ethanol series(Gold Shield) and followed by critical point drying. The sample was then sputter-coated with 3 nm Cr for final imaging using FEI Nova NanoSEM.
TEM examination of the cell membrane wrapping around nanopillars
For cross-section visualization using TEM, cells cultured on a nanopillar substrate underwent an extended overnight fixation in 4% paraformaldehyde and 2% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.3) at 4°C. The sample was then washed in 0.1M sodium cacodylate buffer with 0.2 M sucrose, and post-fixed in osmium (1% osmium tetroxide with 0.8% potassium ferricyanide in 0.1 M sodium cacodylate), all at 4°C. After washing three times with ice-cold distilled water, the sample was en-bloc stained with 2% uranyl acetate on ice, washed three times again with ice-cold distilled water, gradually dehydrated in an ethanol series (Gold Shield) to 100% ethanol, and then exchanged to acetonitrile. The cell sample was infiltrated with epoxy resin (Embed 812) and hardened after baking at 65°C for 24 hrs. Then, the bottom quartz coverslip was etched away by 49% hydrofluoric acid (Avantor). The sample was then re-embedded with fresh pure resin to refill the spaces left by the coverslip. The resulting resin block was then trimmed and sectioned on a Leica Ultracut ultramicrotome into 70 nm-thick sections. The sections were collected on 1 × 2 mm copper slot grids with carbon-formvar support film (Electron Microscopy Sciences) and post-stained for 30s in 1:1 uranyl acetate:acetone. The grids were imaged at 120 kV on a JEOL 1400 TEM.
FIB-SEM examination of the cell membrane wrapping around nanopillars
Protocol was adapted from another paper.34 Substrates with cells were rinsed with 0.1 M sodium cacodylate buffer (Electron Microscopy Sciences) and fixed with 3.2% glutaraldehyde (Sigma Aldrich) at 4°C overnight. Post-fixation was carried out with 4% osmium tetroxide and 2% potassium ferrocyanide (Electron Microscopy Sciences) for 1 hr, 1% thiocarbohyrazide (Electron Microscopy Sciences) for 20 mins, 2% aqueous osmium tetroxide for 30 mins. Cells were again rinsed twice with distilled water and then, finally, incubated overnight with syringe-filtered 4% aqueous uranyl acetate (Electron Microscopy Sciences, en bloc step). Gradual dehydration was carried out in an increasing ethanol series (10%–30%–50%–70%–90%–100%, 5–10 mins each). Specimens were infiltrated with an epoxy-based resin with increasing concentrations of resin in 100% ethanol, using these ratios: 1:3 (3 hrs), 1:2 (3 hrs), 1:1 (overnight), 2:1 (3 hrs), 3:1 (3 hrs). Samples were then infiltrated with 100% resin overnight at room temperature. The excess resin removal was removed and the polymerization was carried out at 60°C overnight.
Prepared samples were sputtered with a thin layer of metal and then loaded into the vacuum chamber of a dual-beam Helios Nanolab 600i FIB-SEM (FEI). For secondary SEM, a voltage in the range 3–5 kV and a current in the range 21 pA- 1.4nA were applied. For cross section imaging, a beam acceleration voltage between 2 kV–10 kV was selected, with the current ranging between 0.17–1.4 nA while using a backscattered electrons detector. Regions of interest were preserved by a double platinum layer with in situ sputtering. Tranches were created with an etching procedure fixing an acceleration voltage of 30 kV and currents in the range 9.1 nA – 0.74 nA. A fine polishing procedure of the resulting cross sections was carried out on the sections with a voltage of 30 kV and 80 pA.
Fluorescence imaging of the cell membrane on nanostructures
To visualize the plasma membrane by fluorescence imaging, two methods were used: 1) membrane staining with a lipophilic chemical (CellMask™ Deep Red, Life Technologies); and 2) transient expression of a fluorescence protein that was targeted to the plasma membrane by a K-Ras CAAX motif (GFP-CAAX or mCherry-CAAX)35. For CellMask™ staining, SK-MEL-2 cells cultured overnight on nanostructure substrates were incubated with 0.25 µg/ml CellMask™ Deep Red for 5 min prior to 4% paraformaldehyde fixation at room temperature for 20 min. After three washes with PBS, cells were imaged using an epi-fluorescence microscope (Leica DMI 6000B) within 1h. For GFP-CAAX or mCherry-CAAX transfection, U2OS cells were first starved in OPTI-MEM (Gibco) for 30 min. A transfection mixture was prepared separately by mixing 3µg plasmid with 1µl Lipofectamine® 2000 (Life Technologies) in OPTI-MEM and incubated at room temperature for 20 min. The cell medium was then exchanged to the transfection mixture and incubated at 37°C for 2 hrs. The transfection mixture was then replaced by culture medium and the cells were all owed to recover overnight before imaging by an epi-fluorescence microscope (Leica DMI 6000B). For Fluorescence Recovery After Photobleaching (FRAP), a subcellular area was selected using MetaMorph Software (Molecular Devices) and bleached with a 405 nm laser for 5 s. After photobleaching, the cell was imaged every second for 2 min using spinning disk confocal microscope (same set-up as described below in live cell imaging session). In both cases, the regularity of the nanopillar arrays makes it easy to identify nanopillar positions.
Immunostaining
The response of endocytic proteins was illustrated by clathrin and dynamin in SK-MEL-2 cells using immunostaining. To stain clathrin and dynamin, genome-edited cells (hCLTAEN/hDNM2EN)32 were fixed in 4% paraformaldehyde at room temperature for 20 min after overnight culture on gradient nanopillar array. After three washes with PBS, the cells were permeabilized in 1% Triton X-100/PBS, followed by blocking in 1% BSA / 0.1% Triton X-100/PBS for 30 min. After brief washing with PBS again, the sample was incubated with primary antibodies (anti-clathrin HC, Thermo Scientific MA1-065; anti-GFP, Abcam, ab6556) at 1:500 dilution for 1hr at room temperature. The sample was then washed with PBS 3 times for 15 min before incubation with secondary antibodies (goat anti-mouse IgG Alexa 488, Life Technologies A11001; goat anti-rabbit IgG Texas Red, Life Technologies T-2767) at 1:1000 dilution for 1 hr at room temperature. After three washes with PBS, the substrates were then imaged by epi-fluorescence microscope (Leica DMI 6000B).
Supported lipid bilayer on nanopillars
We used fluorescence-labeled supported lipid bilayer on gradient nanopillar arrays to reference the surface area at each radius. The supported lipid bilayer made of Egg-PC (Avanti) and doped with 0.5 mol% of Texas Red-1,2-Dihexadecanoyl-sn-Glycero-3-Phosphoethanolamine (Invitrogen) was prepared using a protocol as previously described36. Briefly, lipids were dissolved and mixed in chloroform. Then the lipid mixture was dried in clean glass tubes with nitrogen and desiccated overnight. To prepare lipid vesicles, 1mg of lipid mixture was suspended in 400uL PBS buffer and extruded through 100 nm Nucelpore® membrane filter using Avanti Polar Lipid Extruder and Hamilton syringes. Then the lipid vesicle solution was 5-time diluted in PBS buffer and applied to the nanostructure substrate freshly cleaned by 1hr air plasma. After 15 min of incubation with the vesicles, the substrate was washed with 0.5 mL PBS buffer to remove free vesicles before testing. Imaging was taken by epi-fluorescence microscope (Leica DMI 6000B).
Quantification of the clathrin and dynamin accumulation on nanopillars of different radius
Quantification of the clathrin and dynamin accumulation at nanopillars of different radii was performed by averaging many images obtained from the immunostaining of clathrin and dynamin. The background intensity of each image was subtracted by rolling ball algorithm in Fiji37 with 12 pixel radius. Custom-written Matlab (MathWorks) code was then used to create arrays of 3 µm by 3 µm square masks centered at nanopillars locations using the bright field image. Then, the same square masks were used to map out the square images in the corresponding images of clathrin, dynamin or lipid only, all centered on nanopillars. Those images from the same radius were averaged together for each channel. To quantify the fluorescence intensities on nanopillars, we integrated intensities of pixels inside a small circle around the center of each square images, where the circle size is adjusted according to each nanopillar radius. The signal intensities of different nanopillars radii were rescaled through dividing by the maximum intensity among different radii for easy comparison between different channels (data shown in Fig. 1i, top plot). Membrane area measured by supported lipid bilayer, GFP-CAAX, and CellMask all show a linear relationship with respect to the nanopillar diameter (Fig. S4). We normalized the clathrin and dynamin2 protein signal by the membrane area probed with lipid bilayer due to its low variation. Clathrin and dynamin intensities were normalized to lipid-only intensities of the same radius to reveal the curvature effect (data shown in Fig. 1i, bottom plot).
Live cell imaging
Live imaging of genome-edited SK-MEL-2 (hCLTAEN/hDNM2EN)32 cells on gradient nanopillar (same as above), nanobar (200 nm wide and 2 µm long) and nano-letter (nanoC, nanoU, 500 nm wide and 2 µm long) arrays was performed using spinning disk confocal microscopy on Zeiss Axio Observer equipped with a Yokagawa spinning disk unit as well as 488 nm and 561 nm laser lines. During imaging, cells were maintained at 37°C in an on-stage incubator (INUBSF-ZILCS, Tokai Hit) with DMEM w/o phenol red that was supplemented with 5% FBS and 10 mM HEPES. Movies were recorded with a dual emission filter (Chroma) alternating red and green excitation and detection. Samples were exposed for 200 ms per channel every 2 s for up to 4 min. The same microscope set-up was also used in FRAP analysis of GFP-CAAX in U2OS cells.
For data shown in Fig. 4 in the main text, live imaging of genome-edited SK-MEL-2 (hCLTAEN/hDNM2EN)32 cells on nanopillar array with 150nm radius for dynamics study was performed using Nikon TiE inverted microscope with Perfect Focus (PFS) mechanism and Yokogawa CSU-X1 spinning disk unit. Cells were imaged in growth medium at 37°C and 5% CO2 maintained in InVivo Scientific enclosure w/ environmental control. Movies of CLTA-RFP and DNM2-GFP were recorded alternatively every 3 s for up to 6 min.
Quantification of endocytic proteins on nanobar arrays
Different endocytic proteins participating in the clathrin-mediated endocytosis (schematic see Fig. 3a) was measured for their curvature preferences on nanobar arrays. Membrane markers, CellMask™ Deep Red and mCherry-CAAX were used as controls. Most endocytic proteins chosen in this study are probed by transfection similar to mCherry-CAAX, including transferrin receptor (TfnR-GFP), FCHo1 (FCHo1-GFP), PH-domain from PLC-delta (PH-GFP), AP2 (AP2-GFP), epsin1 (Epsin1-GFP) and amphiphysin 1(Amphiphysin1-YFP). The transfected cells were fixed and imaged by epi-fluorescence microscope (Leica DMI 6000B). Responses of clathrin and dynamin to nanobars were captured by averaging a 4-min movie of genome-edited SK-MEL-2 (hCLTAEN/hDNM2EN)32 cells on nanobar arrays. Actin was probed by Alexa Fluo®647 conjugated pholloidin staining (Life Technologies). Caveolin1 was probed by immunostaining using anti-caviolin-1 (Abcam, ab2910). The protein intensity at nanobar end and nanobar center was quantified using a custom-written Matlab code. We noticed that when protein expression levels were high, the fluorescence signal was mostly in the cytosol and its accumulation on nanobars was not observable (Supplementary Fig. S12). Those cells were therefore excluded from quantification.
Analysis of endocytic protein dynamics on nanopillar arrays
Time-lapse movie taken as describe in live cell imaging session above were first processed by a 3-frame median filter in FIJI37 to reduce random noise. Filtered movies were then analyzed using the Matlab package cmeAnalysis as previously described38, where the Gaussian PSF was estimated by data-based method, alpha was set to 0.001, tracking radius was 0.1 to 1, and tracking gap set to 1. Resulting tracks was manually validated to correct starting and end frames from buffer frames and to break unidentified merged tracks. Tracks shorter than 3 frames or last through the whole movie were excluded. Lifetimes of tracks on nanopillar position were identified by custom-written Matlab code. Histogram, scatter plot and statistical analysis of lifetimes on QNP and flat area were generated in GraphPad Prism.
Analysis of fluorescence intensity dynamics of AP2 and Dynamin2 on nanopillar arrays
We measured the fluorescence intensities of AP2-RFP and Dynamin2-GFP over time using custom-written software in ImageJ and Matlab. Movies of MDA-MB-231 cells endogenously expressing AP2-RFP and DNM2-GFP were corrected for photobleaching using an exponential fit. We selected regions of interest (ROIs) as lines crossing nanopillar arrays, and an ImageJ toolset made kymographs from maximum intensity projections across the 5-pixel width of each line region. From these kymographs we selected a second set of ROIs at the locations of each nanopillar. The ImageJ toolset saved the integrated intensity across each 7-pixel-wide line region as a line profile. A Matlab script normalized each intensity trace for the minimum and maximum intensity and plotted the traces. For the plot in Fig. S9c we selected line regions containing clear AP2-RFP and Dynamin2-GFP signal that were temporally isolated; the latter selection criterion rejected most of the tracks, since tracks tended to overlap in these movies. We aligned the selected tracks based on the time the Dynamin2-GFP signal dropped to 50% intensity at the end of the track.
Statistical analysis
For protein accumulation test, sample sizes were validated with statistical power of 0.8 and probability of 0.05 comparing to membrane control (e.g. CellMask or GFP-CAAX). Unpaired two-tailed t tests with Welch’s correction (not assuming equal SDs) were used to assess for significance unless otherwise stated in the figure legend. All tests were performed using Prism (GraphPad Software) and data are presented as mean ± s.e.m. or mean ± s.d. as indicated in the figures. All experiments were repeate d at least twice, unless otherwise explicitly stated in the figure legends.
Data availability
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
Code availability
Matlab script for image analysis on gradient nanopillar array, nanobar array and nanoC and nano U array are available from the corresponding author upon request. Matlab package cmeAnalysis for protein dynamic analysis was obtained from Danuser lab of Harvard University (http://lccb.hms.harvard.edu/software.html)38.
Supplementary Material
Acknowledgments
We thank Dr. Yansong Miao and Dr. Sun Hae Hong of the Drubin group in U.C. Berkeley for valuable discussion as well as helpful comments on genome-edited cell lines and endocytic lifetime analysis, Prof. Kang Shen in Stanford for generous support on spinning disk confocal microscopy, Dr. Milos Galic of the Tobias Meyer group in Stanford for suggestions and Amphiphysin1-YFP plasmid, Dr. Shunling Guo of the Bianxiao Cui group for constructing mCherry-CAAX plasmid, as well as Allister McGuire, Dr. Chong Xie and Dr. Ziliang Lin of the Bianxiao Cui group in Stanford for advice and help on the nanostructure fabrication. We also thank Qunxiang Ong and Luke Kaplan of Bianxiao Cui group for comments on the manuscript. Fabrication and characterization of nanostructures were conducted in Stanford Nanofabrication Center and Stanford Nano Shared Facilities (SNSF). Spinning disk confocal with perfect focus for lifetime measurement was conducted in Cell Science Imaging Facility (CSIF) at Stanford University. This work was supported by the NSF (CAREER award no. 1055112), the NIH (grant no. NS057906), a Searle Scholar award, a Packard Science and Engineering Fellowship (to B.C.), NIH fellowship 1F32 GM113379-01A1 (to J.R.M.), Studying Abroad Scholarship (to H.-Y. L.), Arnold O. Beckman Postdoctoral Fellowship (to M.A.), Heart Rhythm Research Fellowship (to F.S.) and the NIH grant R35GM118149 (to D.G.D.).
Footnotes
Author Contribution
W.Z., B.C., Y.C., and D.G.D conceived the study and designed the experiment. W.Z. fabricated the nanostructure substrates, and performed most of experiments. L.H. performed TEM measurements. F.S. conducted the FIB-SEM characterization. H.Y.L. performed most of the endocytic protein test on nanobar arrays and the quantification and statistic analysis. W.Z., P.C. and B.C. developed the Matlab code for the dynamic analysis. W.Z. analyzed the most of the data. M.A. analyzed the AP2/Dynamin2 movies. A.G., and J.M provided and characterized the genome-edited cell line. W.Z. and B.C. wrote the manuscript. All the authors discussed the results and commented on the manuscript.
Supplementary information is available in the online version of the paper.
Competing Financial Interest
The authors declare no competing financial interests.
References
- 1.McMahon HT, Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2011;12:517–533. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- 2.Di Fiore PP, Zastrow von M. Endocytosis, signaling, and beyond. Cold Spring Harb. Perspect. Biol. 2014;6 doi: 10.1101/cshperspect.a016865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johannes L, Wunder C, Bassereau P. Bending ‘on the rocks’--a cocktail of biophysical modules to build endocytic pathways. Cold Spring Harb. Perspect. Biol. 2014;6:a016741–a016741. doi: 10.1101/cshperspect.a016741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirchhausen T, Owen D, Harrison SC. Molecular Structure, Function, and Dynamics of Clathrin-Mediated Membrane Traffic. Cold Spring Harb. Perspect. Biol. 2014;6:a016725–a016725. doi: 10.1101/cshperspect.a016725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, Sun Y, Drubin DG, Oster GF. The mechanochemistry of endocytosis. PLoS Biol. 2009;7:e1000204. doi: 10.1371/journal.pbio.1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galic M, et al. Dynamic recruitment of the curvature-sensitive protein ArhGAP44 to nanoscale membrane deformations limits exploratory filopodia initiation in neurons. eLife. 2014;3:e03116. doi: 10.7554/eLife.03116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larsen JB, et al. Membrane curvature enables N-Ras lipid anchor sorting to liquid-ordered membrane phases. Nat Chem Biol. 2015;11:192–194. doi: 10.1038/nchembio.1733. [DOI] [PubMed] [Google Scholar]
- 9.Epand RM, D'Souza K, Berno B, Schlame M. Membrane curvature modulation of protein activity determined by NMR. Biochim. Biophys. Acta. 2015;1848:220–228. doi: 10.1016/j.bbamem.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Iversen L, Mathiasen S, Larsen JB, Stamou D. Membrane curvature bends the laws of physics and chemistry. Nat. Cell Bio. 2015;11:822–825. doi: 10.1038/nchembio.1941. [DOI] [PubMed] [Google Scholar]
- 11.Wu M, et al. Coupling between clathrin-dependent endocytic budding and F-BAR-dependent tubulation in a cell-free system. Nat. Cell Bio. 2010;12:902–908. doi: 10.1038/ncb2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee I-H, Kai H, Carlson L-A, Groves JT, Hurley JH. Negative membrane curvature catalyzes nucleation of endosomal sorting complex required for transport (ESCRT)-III assembly. Proc. Natl. Acad. Sci. U.S.A. 2015;112:15892–15897. doi: 10.1073/pnas.1518765113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galic M, et al. External push and internal pull forces recruit curvature-sensing N-BAR domain proteins to the plasma membrane. Nat. Cell Bio. 2012;14:874–881. doi: 10.1038/ncb2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeong S, McDowell MT, Cui Y. Low-Temperature Self-Catalytic Growth of Tin Oxide Nanocones over Large Areas. ACS Nano. 2011;5:5800–5807. doi: 10.1021/nn2015216. [DOI] [PubMed] [Google Scholar]
- 15.Hanson L, Lin ZC, Xie C, Cui Y, Cui B. Characterization of the cell-nanopillar interface by transmission electron microscopy. Nano Lett. 2012;12:5815–5820. doi: 10.1021/nl303163y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mumm F, Beckwith KM, Bonde S, Martinez KL, Sikorski P. A transparent nanowire-based cell impalement device suitable for detailed cell-nanowire interaction studies. Small. 2013;9:263–272. doi: 10.1002/smll.201201314. [DOI] [PubMed] [Google Scholar]
- 17.Santoro F, et al. Interfacing electrogenic cells with 3D nanoelectrodes: position, shape, and size matter. ACS Nano. 2014;8:6713–6723. doi: 10.1021/nn500393p. [DOI] [PubMed] [Google Scholar]
- 18.Avinoam O, Schorb M, Beese CJ, Briggs JAG, Kaksonen M. Endocytic sites mature by continuous bending and remodeling of the clathrin coat. Science. 2015;348:1369–1372. doi: 10.1126/science.aaa9555. [DOI] [PubMed] [Google Scholar]
- 19.Doyon JB, et al. Rapid and efficient clathrin-mediated endocytosis revealed in genome-edited mammalian cells. Nat. Cell Bio. 2011;13:331–337. doi: 10.1038/ncb2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor MJ, Perrais D, Merrifield CJ. A High Precision Survey of the Molecular Dynamics of Mammalian Clathrin-Mediated Endocytosis. PLoS Biol. 2011;9:e1000604. doi: 10.1371/journal.pbio.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ford MGJ, et al. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- 22.Peter BJ. BAR Domains as Sensors of Membrane Curvature: The Amphiphysin BAR Structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 23.Kelly BT, et al. AP2 controls clathrin polymerization with a membrane-activated switch. Science. 2014;345:459–463. doi: 10.1126/science.1254836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henne WM, et al. FCHo Proteins Are Nucleators of Clathrin-Mediated Endocytosis. Science. 2010;328:1281–1284. doi: 10.1126/science.1188462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dautry-Varsat A, Ciechanover A, Lodish HF. pH and the recycling of transferrin during receptor-mediated endocytosis. Proc. Natl. Acad. Sci. U.S.A. 1983;80:2258–2262. doi: 10.1073/pnas.80.8.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 27.Posor Y, et al. Spatiotemporal control of endocytosis by phosphatidylinositol-3,4-bisphosphate. Nature. 2013;499:233–237. doi: 10.1038/nature12360. [DOI] [PubMed] [Google Scholar]
- 28.Chaudhary N, et al. Endocytic Crosstalk: Cavins, Caveolins, and Caveolae Regulate Clathrin-Independent Endocytosis. PLoS Biol. 2014;12:e1001832. doi: 10.1371/journal.pbio.1001832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grassart A, et al. Actin and dynamin2 dynamics and interplay during clathrin-mediated endocytosis. J. Cell Biol. 2014;205:721–735. doi: 10.1083/jcb.201403041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dominguez R, Holmes KC. Actin Structure and Function. Annu. Rev. Biophys. 2011;40:169–186. doi: 10.1146/annurev-biophys-042910-155359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanson L, et al. Vertical nanopillars for in situ probing of nuclear mechanics in adherent cells. Nat. Nanotechnol. 2015;10:554–562. doi: 10.1038/nnano.2015.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doyon JB, et al. Rapid and efficient clathrin-mediated endocytosis revealed in genome-edited mammalian cells. Nat. Cell Bio. 2011;13:331–337. doi: 10.1038/ncb2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanson L, Lin ZC, Xie C, Cui Y, Cui B. Characterization of the cell-nanopillar interface by transmission electron microscopy. Nano Lett. 2012;12:5815–5820. doi: 10.1021/nl303163y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santoro F, et al. Revealing the cell-material interface by FIB-SEM. BioRxiv. 2017 doi: 10.1021/acsnano.7b03494. URL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gorfe AA, Hocker HJ. Membrane Targeting: Methods. John Wiley & Sons, Ltd; 2001. [Google Scholar]
- 36.Lin WC, Yu CH, Triffo S. Supported membrane formation, characterization, functionalization, and patterning for application in biological science and technology. Curr Protoc Chem Biol. 2010;2:235–269. doi: 10.1002/9780470559277.ch100131. [DOI] [PubMed] [Google Scholar]
- 37.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aguet F, Antonescu CN, Mettlen M, Schmid SL, Danuser G. Advances in analysis of low signal-to-noise images link dynamin and AP2 to the functions of an endocytic checkpoint. Dev. Cell. 2013;26:279–291. doi: 10.1016/j.devcel.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request.