Abstract
Selective downregulation of the human antigen R (HuR) protein by siRNA may provide a powerful approach for treating lung cancer. To this end, we investigated the efficacy of transferrin receptor-targeted liposomal nanoparticle-based HuR siRNA (HuR-TfNP) therapy and compared to control siRNA (C)-TfNP therapy both, in vitro and in vivo using lung cancer models.
In vitro studies showed HuR-TfNP but not C-TfNP efficiently downregulated HuR and HuR-regulated proteins in A549, and HCC827 lung cancer cells resulting in reduced cell viability, inhibition of cell migration and invasion, and induction of G1 cell-cycle arrest culminating in apoptosis. However, HuR-TfNP activity in normal MRC-9 lung fibroblasts was negligible. In vivo biodistribution study demonstrated that fluorescently labeled HuR-siRNA or ICG dye loaded TfNP localized in tumor tissues. Efficacy studies showed intratumoral or intravenous administration of HuR-TfNP significantly inhibited A549 (>55% inhibition) and HCC827 (>45% inhibition) subcutaneous tumor growth compared to C-TfNP. Furthermore, HuR-TfNP treatment reduced HuR, Ki67, and CD31 expression and increased caspase-9 and PARP cleavage and TUNEL positive staining indicative of apoptotic cell death in tumor tissues compared to C-TfNP treatment. The antitumor activity of HuR-TfNP was also observed in an A549-luc lung metastatic model, as significantly fewer tumor nodules (9.5±3.1; p<0.001; 88% inhibition) were observed in HuR-TfNP-treated group compared with the C-TfNP-treated group (77.7±20.1). Significant reduction in HuR, Ki67, and CD31 expression was also observed in the tumor tissues of HuR-TfNP-treatment compared to C-TfNP treatment.
Our findings highlight HuR-TfNP as a promising nano-therapeutic system for lung cancer treatment.
Keywords: HuR, Lung Cancer, Nanoparticle, siRNA, Transferrin
Introduction
The effectiveness of conventional therapies for lung cancer has been challenged by ever increasing tumor heterogeneity and undesired side effects [1]. This scenario emphasizes the need for newer therapeutic strategies that overcome these limitations. The therapeutic potential of anti-cancer agents can be tremendously improved by means of targeted delivery towards cancer cells that overexpress surface molecules [2, 3]. Such molecules often act as receptors for ligands or antibodies with high specificity.
The iron-binding serum protein transferrin (Tf) has specific affinity towards transferrin receptors (TfR) expressed in the cell membrane, which is a basic requirement for iron transport in cells [4]. A significant number of lung cancers overexpress transferrin receptors; moreover, their expression in patients with non-small cell lung cancer (NSCLC) has shown significant prognostic value [5]. Therefore, TfR-targeted drug delivery may be a promising strategy to improve the efficiency of lung cancer therapy.
Recent studies have shown that high levels of the human antigen R (HuR), an RNA binding protein, is expressed in many cancer types [6–8], and regulates the expression of several pathologically important mRNAs, particularly in lung cancer [9,10]. Further, HuR regulates cancer cell proliferation, migration, angiogenesis, and metastasis [7–9, 11, 12]. HuR has also been associated to play a role in chemotherapeutic resistance [13–14]. Recent studies from our laboratory demonstrated that HuR downregulation inhibited cell growth in non-small cell lung cancer (NSCLC) in vitro and altered the cells’ ability to undergo migration and invasion [12,15]. Further, we recently showed HuR inhibition radiosensitized human breast cancer cells [16]. All of these studies indicate HuR is a molecular target for cancer therapy and advancing preclinical testing of HuR-targeted therapies could ultimately lead to clinical testing.
RNA interference (RNAi)-based gene silencing provides a promising strategy to knockdown target genes involved in cancer pathology. Downregulation of the HuR gene using complimentary small interfering RNA (siRNA) is an effective method that has demonstrated therapeutic efficiency in cancer cells [8, 12]. Because of its poor cell uptake and low serum stability, delivery of naked siRNA often utilizes nanocarriers, such as liposomes/lipid nanoparticles (NPs) [8, 15, 17] polymer nanoparticles [18, 19], or nanoparticles made from inorganic materials [20, 21]. In the present study we have used a lipid-based nanoformulation using DOTAP (1, 2-dioleoyl-3-trimethylammonium-propane chloride (DOTAP) and cholesterol as the carrier components for HuR siRNA delivery in vitro and in vivo [22–24].
For effective in vivo lung cancer therapy using HuR siRNA, we created a targeted delivery system by modifying the DOTAP:Chol nanoparticle platform with transferrin as a targeting ligand (DSPE-PEG-Tf). The targeting moiety, Tf, was chosen based on the expression levels of its receptors (TfR or CD71) in solid and metastatic lung tumor models. Initially, we tested the efficiency of HuR-TfNP versus HuR-NP (non-targeted) in vitro. Based on the results obtained, we hypothesized that TfNP-based targeted delivery of HuR siRNA would reduce the tumor burden in mouse models of lung cancer. We studied TfNP's biodistribution in mouse models and, relying on this data, we evaluated the target specificity and efficacy of HuR-TfNP versus C-TfNP (control siRNA) in solid tumor models. Finally, we used in vivo live imaging, tumor nodule counts, and immunohistochemistry of specific molecular markers to investigate tumor growth inhibition and the anti-metastatic activity of HuR-TfNP in a mouse model of metastatic A549-luc lung cancer.
Materials and Methods
Chemicals
1,2-dioleoyl-3-trimethylammonium-propane chloride (DOTAP), cholesterol, and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[(polyethylene glycol)-2000] (DSPE PEG2000) were purchased from Avanti Polar Lipids (Alabaster, AL). Transferrin and desferrioximine were purchased from Sigma-Aldrich Chemicals (St. Louis, MO). RPMI-1640 medium, EMEM medium, and fetal bovine serum (FBS) were purchased from GIBCO BRL Life Technologies (New York, NY). siGLO, Control siRNA (5’ UAA GGC UAU GAA GAGAUA C 3’), and HuR-siRNA (5’ UCA AAG ACG CCA ACU UGU A 3’) were purchased from Dharmacon (Lafayatte, CO).
Cell lines and cell culture
The human non-small cell lung cancer (NSCLC) A549 and HCC827 cell lines and normal MRC-9 lung fibroblasts were purchased from American Type Culture Collection (ATCC, Manassas, VA) and were authenticated via Single Tandem Repeat (STR; IDEXX Laboratories) profiling before the experiments. Mycoplasma testing by PCR was routinely performed using specific oligonucleotides (IDT, Chicago, IL). The passage number for tumor cells and normal lung fibroblasts used in the study was from 8–35 and from 4–12 respectively. Tumor cells and normal cells were cultured in RPMI-1640 and EMEM respectively, supplemented with 10% FBS and 1% penicillin/streptomycin.
Synthesis of Tf-NP
Preparation of DSPE-PEG-Tf
Briefly, transferrin (Tf; 150 nmol) was converted to its thiolated form by reacting Tf with Traut's reagent (2-iminothiolane) in sodium phosphate-EDTA buffer (pH 8.0). The crude product (Tf-SH) was then purified from unreacted reagents using a PD-10 (Sephadex-G25) desalting column with sodium phosphate-EDTA eluent buffer (pH 7.1). The purified Tf-SH (150 nmol) was allowed to react with DSPE-PEG-Maleimide (300 nmol) in sodium phosphate buffer (pH 7.1) for 24 h, reacting at 40°C to form a stable conjugate, DSPE-PEG-Tf. The product was then purified by dialysis against ultra-pure water overnight.
Preparation of TfNP
Liposomes (20 mM DOTAP:Chol) were synthesized and extruded stepwise through polycarbonate filters of different pore sizes (1.0, 0.45, 0.,2 and 0.1 nm), as previously described [12, 25]. For preparation of siRNA:liposome complexes, DOTAP:Chol (20 mM) stock solution and siRNA solution diluted in 5% dextrose in water (D5W) were mixed in equal volumes to give a final concentration of 4 mM DOTAP:Chol-siRNA in a 300-ul final volume. DSPE-PEG-Tf ligands were inserted into preformed liposomes using the post insertion technique [26]. Briefly, DOTAP:Chol-siRNA was mixed with an aqueous dispersion of Tf-PEG-DSPE (from 0.01%, 0.03%, 0.05%, and 0.1 mol% of total lipid) and was vigorously rinsed up and down in a pipette tip. This complex was incubated at RT for 60 min. In the case of unmodified DOTAP:Chol, D5W was added instead of Tf-PEG-DSPE solution. Both modified and unmodified liposomes were dialyzed against distilled water overnight at 4°C. Modified liposomes containing control siRNA and HuRsiRNA will henceforth be designated as C-TfNP and HuR-TfNP respectively.
Nanoparticle characterization
Confirmation of Tf binding to NP
Conjugation of Tf into DSPE-PEG and DSPE-PEG-Tf into DOTAP:Chol was confirmed by dot blot, as described previously [27].
Nanoparticle size, zeta potential, and morphology
The average hydrodynamic radius and zeta potential of NP and TfNP in solution was determined by dynamic light scattering (DLS) using a Zeta PALS Zeta potential and particle size analyzer (Brookhaven Instruments, Holtsville, NY). The shape and structure was analyzed using transmission electron microscopy (TEM) at the Oklahoma Medical Research Foundation (OMRF) core facility. The shape and size distribution of HuR-TfNP was observed under a Hitachi H600 transmission electron microscope (TEM, Hitachi, Tokyo, Japan).
Protection of siRNA
The stability of encapsulated siRNA in the presence of serum was monitored by incubating TfNP loaded with siRNA in 50% FBS at 37 °C. Aliquots of 20 µl were withdrawn at 0 min and 60 min, and subjected to gel electrophoresis using agarose (1.2%) gel at 100V for 20 min in TAE buffer (pH 8.0). The efficiency of siRNA condensation by nanoparticles was determined against the free siRNA control [18]. The electrophoresis gel stained with ethidium bromide (SIGMA St. Louis MO) was visualized and imaged using a Syngene gel documentation system.
SiRNA release profile
TfNP loaded with siGLO were suspended in tissue culture medium (pH 7.4), acetate buffer (ABS, pH 5.5) or tissue culture medium with 10% fetal bovine serum and were incubated at 37 °C with gentle shaking at 120 rpm. The release of siRNA was monitored at several time intervals, as described previously [21].
Optimization of Tf-PEG2000-DSPE on DOTAP:Chol
We evaluated the cellular uptake of TfNP in A549 cells with respect to modification ratio. A549 (2×105/well) cells seeded in a six-well plate were transfected with a siGLO-TfNP complex containing 0.01%, 0.03%, 0.05%, or 0.1 mol% of Tf-PEG2000-DSPE to the total lipid in serum-free medium for six hours. After transfection, cells were replenished with RPMI1640 containing 10% FBS, and incubation was continued. Cells were harvested at 24 h after transfection, and fluorescence expression was determined using an Envision® plate reader (Perkin Elmer, Santa Clara, CA). Cells that were not transfected or transfected without free ligand served as controls.
Receptor-mediated endocytosis
A549 (2×105 cells/well) were seeded in a six-well plate and were treated with siGLO-TfNP. Untreated cells served as controls. After NP treatment, the plates with cells were incubated in either 37°C or 4°C [24]. The cells were harvested after 1 h and 4 h of incubation and the fluorescence intensity determined using an Envision® plate reader.
Specificity studies
Receptor stimulation studies
A549 cells (2×105 /well) were seeded in six-well plates. After 24 h of incubation, the medium was replaced with 1 ml of serum-free medium, with or without 100 µM of desferrioxamine (DFO). After 24 h of incubation, the media was replaced with siGLO-TfNP containing media. After 6 h, this siGLO-TfNP containing media was replaced with fresh 2% serum containing media and the cells were incubated overnight. Next day the cells were harvested and the fluorescence intensity of siGLO was measured using Envision® plate reader.
Receptor blocking studies
A549 cells (2×105 cells/well) seeded in six-well plates were pretreated with or without human transferrin (HTf; 1 mg) and were incubated at 37 °C with 5% CO2 for 1 h. The cells were subsequently treated with siGLO-TfNP in serum-free medium. Incubation continued for an additional 1, 4, and 24 h. At the end of the incubation period, the cells were subjected to fluorescence intensity measurement.
In another set of experiments, A549 cells (2×105 cells/ well) seeded in six-well plates were pretreated with or without 10 µl of human transferrin antibody and were incubated at 37 °C with 5% CO2 for 1 h. The cells were subsequently treated with siGLO-TfNP in serum-free medium. Incubation continued for an additional 1 h and 4 h. At the end of the incubation period, the cells were subjected to fluorescence intensity measurement.
Cellular uptake studies
Cellular internalization of TfNP loaded with fluorescently labelled siRNA was measured quantitatively with an Envision® plate reader. A549, HCC827, and MRC-9 cells were separately seeded in six-well plates at 2×105 cells/ well. The next day, cells were transfected with either siGLO-TfNP or siGLO-NP in serum-free medium. Cells were harvested after 1 h, 4 h, and 24 h of incubation, and were analyzed for the mean fluorescence intensity.
Cell viability assay
A549, HCC827, and MRC-9 cells were seeded in a six-well plate at 2×105 in 2 ml of RPMI and EMEM media containing 10% FBS. After 24 h of incubation in a humidified atmosphere of 5% CO2 at 37 °C, the medium was replaced with 1 ml of serum-free medium and was treated with TfNP loaded with control siRNA (C-TfNP) or HuR-siRNA (HuR-TfNP). The final concentration of siRNA used in these experiments was 100 nM. After 6 h of incubation, the medium was replaced with RPMI and EMEM medium containing 2% serum and was incubated for an additional 24 h and 48 h at 37 °C. Cells were harvested and cell viability was determined with trypan blue exclusion assay [28].
Western blotting
Total cell lysates prepared from cultured cells or tumor tissues receiving various treatments were prepared using RIPA buffer and were subjected to western blot analysis as previously described [24, 29, 30]. Primary antibodies against human HuR, Bcl2, Cyclin D1, Cyclin E, and p27 (Santa Cruz Biotechnology, Dallas, TX); caspase 9 and PARP (Cell Signaling, Cambridge, MA), and beta actin (Sigma Chemicals) were all purchased and used as recommended by the manufacturers. Proteins were detected using the appropriate horseradish peroxidase- (HRP)-tagged secondary antibodies and differences in protein expression determined as previously described [24, 29, 30].
Quantitative real-time polymerase chain reaction (qRT-PCR)
qRT-PCR assay for HuR, Bcl2, and p27 was performed using specific oligonucleotide primers [Supplementary Table 1] as previously described [24, 25]. Briefly, first-strand complementary (c)DNA was synthesized from 2 µg of total RNA using Quant script cDNA synthesis kit (Bio-Rad, Richmond CA). The cDNA was subsequently used to perform qRT-PCR (Bio-Rad CFX96 Touch Real-Time PCR Detection System) with SYBR chemistry using iQTM SYBR Green super mix (Bio-Rad). The relative gene expression values were quantified by the 2−ΔΔCT method [30, 31].
Cell cycle analysis
C-TfNP and HuR-TfNP-treated cells were collected at 24 h and 48 h after treatment and stained with propidium iodide and were subjected to flow-cytometric analysis as previously described [24, 25, 32]. Untreated cells served as controls.
Migration and invasion assay
The cell migration and invasion assay was carried out as previously described [12, 24, 30]. Briefly, C-TfNP and HuR-TfNP-treated A549 cell were seeded in the upper chambers of the migration and invasion inserts. Cells receiving no treatment served as controls. The lower chamber was filled with 20% FBS-containing culture medium. After 24 h and 48 h of incubation, the number of migrated and invaded cells were counted and the results represented as previously described [24, 30].
Annexin V assay
A549 cells (2×105) seeded in a six well plate were treated with C-TfNP and HuR-TfNP and stained with annexinV conjugated to Alexa Fluor 488 dye and propidium iodide (PI) using Dead cell apoptosis kit (Life Technologies, Eugene, OR) according to manufacturer’s protocol. Briefly, the cells were harvested at 48 h after treatment and suspended in annexinV binding buffer at a concentration of 100,000 cells/ml. 100 µl of cell suspension was incubated with 5 µl of annexinV and 1µl of 100 µg/ml of PI for 15 min at room temperature. After the incubation period, the cells were analyzed in FACSCalibur flow cytometer using Cell Quest software (BD Biosciences) at excitation 488 nm and emission 530 nm. The number of of cells undergoing apoptosis was determined using the dual staining. Three subsets of cells were distinguished: Viable cells (annexin V and PI negative, early apoptotic cells (annexin V positive and PI negative), and dead cells (annexin V and PI positive). The results thus obtained were plotted as percentage of cells undergoing apoptosis after 48 h of treatment.
In vivo studies
Subcutaneous tumor xenograft and experimental lung metastasis models
Four-to-six-week-old female nude mice (Nu-Nu) were purchased from Charles River Laboratories (Wilmington, MA). The studies were conducted with the approval of and in accordance with guidelines of the Institutional Animal Care and Use Committee at the University of Oklahoma Health Sciences Center, Oklahoma City, OK. For establishing subcutaneous (s.c.) tumors, mice were anesthetized with 2% isoflurane and injected with A549 and HCC827 tumor cells (5×106 cells in 100 µl of PBS) subcutaneously in the lower right flank of mice. For studies involving bioluminescence imaging, A549 stably expressing luciferase (luc; A549-luc) was used. When the tumors reached a size of about 50–100 mm3 the mice were randomized and used for biodistribution and efficacy studies as described below.
For establishing experimental lung metastasis, A549-luc cells (5×106 cells in 100 µl of PBS) were injected intravenously (i.v.) via tail-vein. Tumor formation in the lungs was detected by intraperitoneally (i.p.) injecting D-luciferin (D-Luc; 150 mg/kg) and measuring bioluminescence using the IVIS® Spectrum Imaging System (Perkin Elmer). When the tumor bioluminescence signals reached approximately average radiance of 5000 p/s/cm²/sr, mice were randomly assigned to treatment groups and efficacy studies conducted.
Biodistribution study
For determining the TfNP biodistribution in vivo, indocyanine green (an amphiphilic carbocyanine green dye; Sigma-Aldrich, Missouri) that fluoresces at the near infra-red (NIR) region was encapsulated in TfNPs. These NPs were prepared following the same method for encapsulating the siRNA as described above for in vitro studies with minor modifications. Briefly, an equal volume of DOTAP:Chol (20 mM) stock solution and ICG solution (1 mg/ml in water) was mixed, vortexed, and incubated at room temperature for 20 min. DSPE-PEG-Tf ligands were inserted into preformed liposomes using the post insertion technique. The solutions were then dialyzed against water overnight using a 10 kDa MW cut-off membrane, and were collected and labelled as ICG-TfNP.
ICG-TfNP (30 µg of ICG) was administered i.v. via tail vein to mice bearing subcutaneous A549 and HCC827 tumors or to non-tumor-bearing naïve mice. Free ICG-injected Subcutaneous tumor-bearing mice were used as controls. Mice were imaged (Ex: 785 nm, Em: 820 nm) at 10 min, 30 min, 4 h, and 24 h intervals post-injection using IVIS® Spectrum Imaging System. Similarly, for determining siRNA biodistribution in vivo, fluorescently labeled DY657HuRsiRNA contained in TfNP (DY647HuR-TfNP, 100 µg siRNA) was injected i.v. via tail vein to HCC827 subcutaneous tumor-bearing mice and imaged (Ex: 785 nm, Em: 820 nm).
For determining the biodistribution in mice bearing experimental lung metastasis, ICG-TfNP (30 µg ICG) was injected i.v. via tail vein of mice bearing A549-luc expressing lung tumors. Prior to injecting ICG-TfNP, mice were injected with D-Luc (150 mg/Kg) i.p. and after 10 min were imaged to localize tumors in the lung. Subsequently ICG-TfNP was administered to mice and imaged and the localization of ICG-TfNP in the luc-expressing lung tumors determined by overlaying the images.
siRNA detection and quantitation in HuR-TfNP-treated tumors
For determining HuRsiRNA in HuR-TfNP-treated tumors, droplet digital PCR (ddPCR) QX100 (BioRad) assay using a custom-designed TaqMan® siRNA assay (Applied Biosystems) was employed. A549 and HCC827 tumor tissues from mice that were treated with a single dose of HuR-TfNP (n=2) or treated with C-TfNP (n=2) was harvested and total RNA was isolated. For use as positive control, a tumor lysate prepared from a mouse that did not receive any treatment was spiked with 100 µg of HuR siRNA and total RNA was isolated from it. Total RNA samples isolated from HuR-TfNP, C-TfNP and untreated control samples were diluted to 10 ng before the samples were used for the assay. Reverse transcription (RT) was performed with the diluted total RNA using specific stem-loop RT primers (Applied Biosystems) for human HuR siRNA. ddPCR was used to detect and quantify the siRNA in each tumor, following the manufacturer’s protocol. Briefly, the single-strand cDNA, TaqMan probes, ddPCR buffer, and water were mixed to obtain a 20:l ratio of the ddPCR reaction mix. Using a BioRad droplet generator, droplets were generated with the ddPCR reaction mix and droplet-making oil. The droplets were transferred into a 96-well plate. After sealing the plate with foil at 180 °C, 40 cycles of PCR amplification were carried out in a BioRad T100 iCycler. Each droplet was quantified for TaqMan probe fluorescence using the BioRad QX100 droplet counter. Absolute copies per µl were calculated using the Poisson distribution algorithm. The copy number of spiked siRNA detected by ddPCR was considered as 100% input siRNA, and relative percent intake was further calculated in the treated tumor tissues and results expressed as percent intake of siHuR/100% input siHuR.
Efficacy studies in subcutaneous tumor model
Nude mice bearing A549 and HCC827 subcutaneous tumors were randomized into two groups (n=5/group/tumor type) when the tumors reached a size of 50–100 mm3. Mice were treated intratumorally with C-TfNP and HuR-TfNP containing 100 µg of scrambled siRNA (C-TfNP) or HuRsiRNA (HuR-TfNP). The animals were treated three times a week for two weeks. Tumor volumes were measured at regular intervals as previously described [33, 34]. The experiment was terminated on day 25 after initiation of treatment and tumor tissues were harvested and used for molecular and immunohistochemical studies.
For determining the therapeutic efficacy of systemic HuR-TfNP treatment, mice bearing A549-luc and HCC827 subcutaneous tumors were divided into two groups (n=8/group/tumor model) when the tumors reached a size of 50–100 mm3. The mice were treated with C-TfNP and HuR-TfNP by intravenous tail-vein administration. All other experimental conditions including number of treatments and tumor measurement except the experiment duration remained the same as described above for intratumoral treatments. For determining the therapeutic effect of HuR-TfNP, two mice from each treatment group was euthanized after two treatments and the tumor tissues harvested for molecular analysis. At the end of the study, tumor tissues from all of the animals from the two treatment groups were harvested and used for molecular and immunohistochemical studies.
For determining growth of A549-luc tumors, we also conducted bioluminescence imaging and tumor growth calculated by measuring luminescence on days 1, 15 and 33. Luminescence was determined by injecting D-Luciferin (150 mg/Kg) i.p. and 10 min later subjecting the mice to imaging using IVIS® Spectrum Imaging System. The results were calculated and expressed as average radiance (photons/sec/cm2/sr) and subjected to statistical analysis. Imaging beyond day 33 could not be performed as the mice in the C-TfNP-treatment were euthanized due to large tumor burden and observance of signs of necrosis.
Efficacy studies in experimental lung metastasis model
A549-luc cells (5×106 suspended in 100 µl of PBS) were injected i.v. into the tail veins of nude mice. Once bioluminescence signals in the lungs reached approximately 3000 p/s/cm²/sr average photon radiance, they were randomly divided into two groups (n=10/group) and treated with C-TfNP and HuR-TfNP containing 100 µg of siRNA. TfNPs were injected i.v. via the lateral tail vein three days a week for two weeks for a total of six treatments. Tumor growth was measured by evaluating Luc expression by imaging. Mice were imaged on day zero (before treatment) using an IVIS® Spectrum Imaging System within 10 min after the injection of D-Luc (150 mg/kg). The imaging was performed every two weeks for a total period of 6 weeks. Living image 2.5 software was used to determine the regions of interest (ROI), and average radiance (p/s/cm2/sr) was measured for each mouse. The average ROI for each treatment group was measured, and the difference in the ROI between the groups was noted. All animals were sacrificed at week 6 and lungs from five animals in each group was collected and used for molecular and immunhistochemical studies.
The remaining 5 mice per treatment group were used for counting extrapulmonary lung nodules as previously described [25, 33, 35]. Briefly, 10 ml of Bouin’s fluid was intratracheally injected into the lungs of sacrificed mice. The inflated lungs were removed and washed with PBS. The extrapulmonary tumor nodules appearing as white nodules in each lobe of the lung were counted under the dissection microscope. The difference in the average number of nodules per lung from each treatment group was calculated and subjected to statistical analysis
Immunohistochemistry
Immunohistochemical (IHC) staining on tumor tissues harvested from C-TfNP and HuR-TfNP-treated mice was performed using an automated immunostainer (Leica, Bond-III, Leica, Buffalo Grove, IL) with the following primary polyclonal antibodies: HuR (Santa Cruz), Ki67 and mouse CD31 (Abcam, Cambridge, MA). Appropriate positive and negative controls were included. The stained tissue sections were analyzed using the Aperio ScanScope Image Analysis System (Aperio, Vista, CA) [36]. HuR and Ki-67 stained tumor tissues were analyzed using a positive nuclear count algorithm with the Aperio ImageScope viewer. The number of positive nuclei from regions-of-interest (ROIs) was determined for each animal from C-TfNP and HuR-TfNP treated groups. For the CD31 marker staining, we determined the number of microvessels per mm2 of the tissue using the Aperio microvessel analysis algorithm. The results obtained for each of the marker in the two treatment groups were subjected to statistical analysis.
TUNEL Assay
TUNEL Assay was performed using in situ cell death detection kit from Roche Diagnostics (Indianapolis, IN), according to the manufacturer’s protocol. Briefly, tumor tissue sections were incubated with proteinase K for 10 min, blocked and incubated with TUNEL reaction mixture for 60 min at 37 °C. Apoptotic nuclei were labeled by the addition of anti-fluorescein alkaline phosphatase conjugate. Stained tumor sections were analyzed using the Aperio Scanscope Image and the number of TUNEL positive nuclei counted using the positive nuclei count algorithm with the Aperio ImageScope viewer.
Statistical Analysis
The SAS 9.2 software was used for statistical analyses. All data were provided as mean ± standard deviation (SD). Univariate statistical significance was determined by one-way analysis of variance (ANOVA) with Tukey’s adjustment for pairwise comparisons. Differences between groups were obtained using a linear mixed effects model with Tukey’s adjustment. A P value of less than 0.05 was considered statistically significant.
Results
Physicochemical characterization of TfNP
Cationic lipid-based nanoparticles are the most commonly used carriers for gene therapeutics, due to their effective gene transfection ability in vitro and in vivo. Hence, we chose a cationic liposome platform, DOTAP:Chol, which demonstrated gene delivery potential in our previous studies [12, 22–24], in formulating TfR-targeted nanoparticles (TfNP). HuR-siRNA was allowed to interact electrostatically with cationic DOTAP:Chol liposomes to form HuR-NP. Table 1 shows the average particle sizes and zeta potentials of all formulations used in the present study. The unmodified nanoparticles carrying control siRNA (C-NP) were near spherical in structure, as observed with transmission electron microscopy (TEM) [Supplementary Fig. S1(a)].
Table 1.
Particle size and zeta potential of siRNA containing NPs
| Components | Diameter (Mean ± SD, nm) | Zeta potential (Mean ± SD, mV) |
|---|---|---|
| NP | 180.54 ± 9.6 | 33.98 ± 3.9 |
| HuR-NP | 202.59 ± 3.3 | −0.57 ± 1.9 |
| C-TfNP | 295.74± 16.4 | 9.53 ± 2.5 |
| HuR-TfNP | 313.43 ± 6.5 | 10.28 ± 1.4 |
The scheme of HuR-TfNP synthesis is shown in the Fig. 1A. The optimal Tf ligand density in NPs that favored an increased cell uptake of fluorescently labelled siRNA (siGLO) was determined to be 0.05% compared to 0.01%, 0.03% and 0.1% Tf ligand and no ligand [Supplementary Fig. S2; p <0.05]. The observation that 0.01% and 0.03% Tf ligand density on NP showed relatively lower fluorescence and cell uptake compared to no ligand on NP was somewhat unexpected and likely due to insufficient number of ligand attached to NP. Additionally, the presence of neutral polymer PEG which is post-inserted in DOTAP:Chol lipid bilayer as DSPE-PEG-Tf might have influenced the strength of electrostatic binding of the NP to the cell membrane causing a reduced cell uptake [37]. The insufficient Tf ligand concentration and contributory effect of PEG are likely overcome when 0.05% Tf ligand concentration is used resulting in enhanced cell uptake. Our results clearly demonstrate that ligand concentration on NP surface is an important determining factor for the optimal affinity of NP to the cell surface receptors and uptake and concurs with previous report [38.].
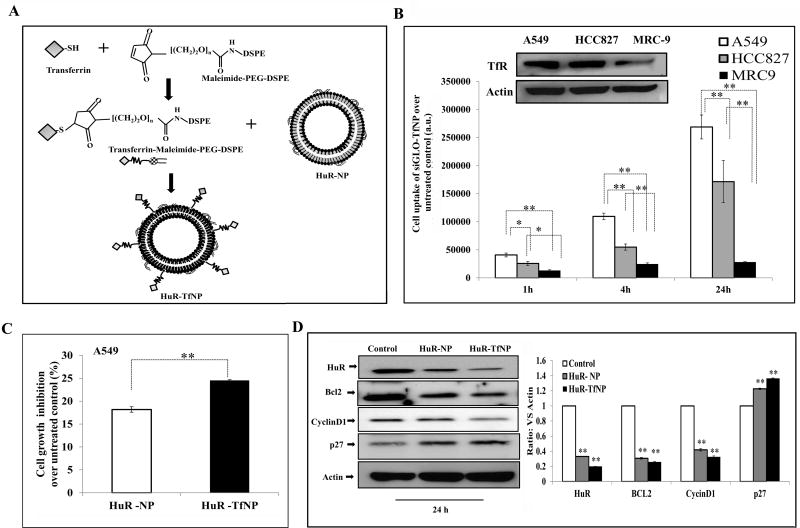
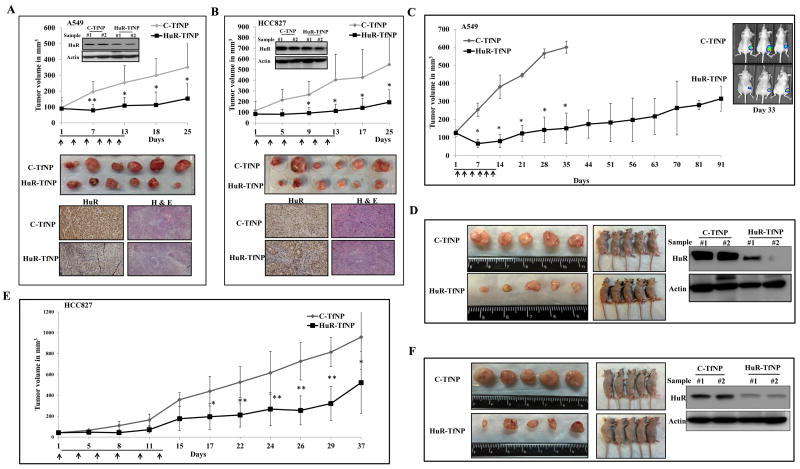
Figure 1. Tumor-targeted HuR silencing suppresses lung tumor cell growth.
A. Schematic illustration of HuR-siRNA-loaded, Tf-modified liposome (HuR-TfNP) preparation. B. Assessment of transfection efficiency of fluorescently-labeled siRNA (siGLO-) TfNP in lung cancer cell lines (A549, HCC827) and normal lung fibroblasts (MRC-9) showed a time-dependent increase in uptake of the NPs by tumor cells compared to normal cells. Inset shows TfR expression is higher in tumor compared to normal cells. C. Cell growth inhibition induced by HuR-TfNP (targeted delivery) compared with HuR-NP (non- targeted delivery) 24 h after treatment of A549 cells. D. Western blot evaluation of HuR and HuR-regulated proteins 24 h after HuR-TfNP or HuR-NP treatment in A549 cells. The quantification of protein bands (Test protein/Actin ratio) is shown as a bar graph at the bottom. * p<0.05, ** p<0.001.
The particle size showed a gradual increase from 180.54 ± 9.6 nm (empty NP) to 313.43 ± 6.5 nm (HuR-TfNP) due to siRNA loading and surface modification with Tf ligand [Table 1]. The siRNA was well condensed by TfNP, resulting in mobility retardation, as shown in the gel electrophoretogram [Supplementary Fig. S1(b)]. The siRNA encapsulation efficiency expressed as percent (% EE) was calculated using the following equation: . The siRNA EE was found to be 80.07 ± 0.35% for HuR-TfNP.
The in vitro release profiles of siRNA-Tf-NP were investigated in three different media conditions such as acetate buffer solution (ABS; pH 5.5), tissue culture medium (pH 7.4), and 10% serum containing culture medium (pH 7.4) at 37 °C over a period of 24 h [Supplementary Fig. S1(c)]. A burst siRNA release pattern was observed for the first 6 h (~25–40% siRNA released) in culture medium and 10% serum containing culture medium (pH 7.4). The observed siRNA release characteristics are typical of a lipid nanoparticle delivery system, and concur with previous studies [39, 40]. However, the siRNA release rate was higher when siRNA-TfNP was incubated in culture medium alone (~46% siRNA released) compared to 10% serum containing medium (~27% siRNA released). The siRNA release was noticeably slower after 6 h until the measurement at 24 h, indicating a sustained release pattern. However, at low pH (5.5), siRNA release was rapid, reaching 64% in 6 h and 93% in 24 h. This result is consistent with our previous report that show enhanced siRNA release rate at low pH (5.5) [24].
In vitro evaluation of siGLO-TfNP
Since the purpose of ligand (Tf) modification of NP was to render specificity in cellular uptake [27], and to achieve enhanced siRNA delivery efficiency in cancer cells, we investigated the cell uptake mechanism of Tf-NP. In one of our studies using desferrioxamine (DFO), an iron chelator, TfR (known as CD71) expression was upregulated in A549 lung cancer cells that were subjected to siGLO-TfNP treatment [Supplementary Fig. S3 (a); upper panel]. We observed a clear difference in fluorescence intensity of TfNP-delivered siGLO in DFO-treated and untreated A549 cells [Supplementary Fig. S3 (a)]. The increased fluorescence intensity in the DFO-treated group indicates enhanced uptake of siGLO-TfNP compared with cells that were not pre-treated with DFO.
To further understand the specificity of TfNP, we conducted a separate study in which human transferrin (HTf) and TfNP were allowed to compete against each other [27], in interacting with TfR-expressing A549 cells. The results showed that HTf (1 µg/well) significantly inhibited the uptake of siGLO-TfNP, as measured by fluorescence intensity, at all time points (1 h, 4 h, 24 h) [Supplementary Fig. S3(b)]. To confirm the TfR-based specificity of TfNP, A549 cells were pretreated with human transferrin receptor-antibody (TfRAb). Significant reduction in siGLO-TfNP fluorescence intensity was observed at 1 h and 4 h with the TfRAb-added group, indicating receptor-mediated siGLO-TfNP uptake is abrogated in presence of TfRAb [Supplementary Fig. S3(c)]. Further, the involvement of a TfR-based endocytosis mechanism in TfNP uptake was demonstrated by incubating the cells at 4 °C or 37 °C while administering siGLO-TfNP treatment. Receptor-mediated endocytosis is known to be affected by low temperature [41]. The results showed a marked decrease, especially at 4 h, in cell uptake of siGLO-TfNP at 4 °C compared with 37 °C, suggesting that the predominant mechanism of cell uptake was via receptor-mediated endocytosis [Supplementary Fig. S3(d)].
Having made it clear that the TfNP internalization depends on TfR expression levels in the target cell, we wanted to verify whether TfR expression levels contribute to TfNP’s selectivity towards lung cancer cells over normal lung fibroblast cells. We found that both A549 and HCC827 tumor cells expressed significantly higher levels of TfR than did normal MRC-9 cells (Fig. 1B; inset). As expected, the cell uptake response varied among cancer cell lines and normal cells at 1 h, 4 h, and 24 h treatment, with siGLO-TfNP uptake corresponding to their TfR expression levels [Fig. 1B]. In A549 cells, the uptake of siGLO-TfNP gradually increased as measured by fluorescence intensity (a.u.) from 40758.6 (1 h) to 109446.6 (2 h), and later to 268930.6 (24 h; p<0.01), compared with untreated control. Though HCC827 cells also showed a similar trend in cell uptake of siGLO-TfNP, the fluorescence intensity values at all time points were significantly less than A549 cells. Importantly, MRC-9 cells showed negligible cell uptake of siGLO-TfNP at all time points and was significantly less than the cancer cell lines studied (p<0.05).
Efficiency of TfNP versus NP in siHuR delivery in Tf-overexpressing lung cancer cells
Since the transfection efficiency of siGLO-TfNP (targeted) over untreated control was confirmed by cell uptake experiments, we replaced siGLO with siRNA against HuR to test it in A549 cells. The HuR gene silencing effect induced by HuR-NP (non-targeted) and HuR-TfNP in A549 cells was compared by analyzing the HuR and associated protein expressions and by cell growth inhibition. As shown in Fig. 1C, growth inhibition induced by HuR-TfNP was 24.5% compared with 18.1% for HuR-NP for a 24 h treatment period (p <0.01), demonstrating the superior efficacy of HuR-TfNP over HuR-NP.
Western blot analysis showed a difference in HuR expression between HuR-NP-treated (HuR/Actin = 0.33) and HuR-TfNP-treated (HuR/Actin = 0.19; p <0.01) groups [Fig. 1D]. This finding suggests that the TfNP facilitated enhanced and more efficient knockdown of HuR compared with NP. The data also showed the effect of HuR knockdown in HuR-regulated proteins, such as BCl2, Cyclin D1, and P27. HuR stabilizes BCl2 mRNA, and thereby the BCl2 family of apoptotic regulatory proteins, by binding to its 3’ untranslated region [42]. Knockdown of HuR deregulates BCl2, reducing its expression (Fig. 1D). Previous studies have shown that downregulation of HuR resulted in reduced expression of cyclins, which are key cell cycle proteins [12, 15, 43, 44]. The reduction of HuR-regulated cyclin D1 as a consequence of HuR knockdown has been established by our studies [Fig. 1D]. siRNA-mediated knockdown is reported to induce endogenous levels of p27 protein, which is a cyclin-dependent kinase inhibitor [45,46]. Consistent with these reports, we observed that p27 was upregulated when HuR was knocked down using either HuR-NP or HuR-TfNP, while the latter significantly increased p27 expression [Fig. 1D]. Based on the above results, we used TfNP as the targeted nanodelivery system for siHuR in later experiments.
The possibility that the observed knockdown of HuR in HuR-NP- or HuR-TfNP-treatment was non-specific was eliminated by testing a control scrambled siRNA and a siRNA used for initial HuR screening (HuRsi21). As shown in Supplementary Figure S4, treatment of A549 cells with NP containing control siRNA (C-NP) or siRNA used for HuR screen (HuRsi21-NP) did not reduce HuR protein expression compared to untreated control cells. In contrast, treatment with HuRsi4 siRNA contained in NP greatly reduced HuR expression in A549 cells. These results show that HuRsi4 siRNA-mediated inhibitory effect on HuR is specific and that inclusion of any siRNA did not produce a similar inhibitory activity on HuR. Based on these results we chose HuRsi4siRNA hereafter referred to as “HuRsiRNA” and encapsulated in TfNP (HuR-TfNP) for conducting the in vitro and in vivo studies and the results of which are described herein.
The target specificity of HuR-TfNP was confined to Tf-expressing cancer cells
After achieving specific targeting and efficiency in HuR gene knockdown, we compared the efficiency of HuR-TfNP in A549 and HCC827 lung cancer cells with varied levels of TfR expression and MRC-9 lung fibroblasts with negligible TfR expression. The downregulation of HuR and corresponding cell growth patterns were analyzed for 24 h and 48 h after HuR-TfNP or C-TfNP (control siRNA [-TfNP]) treatments and compared with untreated controls[Fig. 2A–B]. At both 24 h and 48 h, HuR-TfNP significantly suppressed the expression of HuR mRNA [Supplementary Fig. S5], and protein [Fig. 2A; Supplementary Figs. S6 and S7] accompanied by cell growth inhibition [Fig. 2B; p <0.01] in A549 and HCC827 cells compared with C-TfNP. In HCC827 cells, an unexpected increase in HuR-mRNA expression in C-TfNP-treated cells was observed at 48 h after treatment compared to untreated control cells. The unexpected increase in HuR-mRNA at the present time is not clear and needs further investigation. Nevertheless, the outcome of neither the study nor the conclusions drawn is impacted. Interestingly, HuR-TfNP treatment resulted in negligible cell growth inhibition and minimal reduction in HuR and HuR-regulated target proteins in MRC-9 cells [Fig. 2A–B; Supplementary Fig. S8]. This is probably due to low TfR expression and inefficient HuR-TfNP uptake by MRC-9 cells.
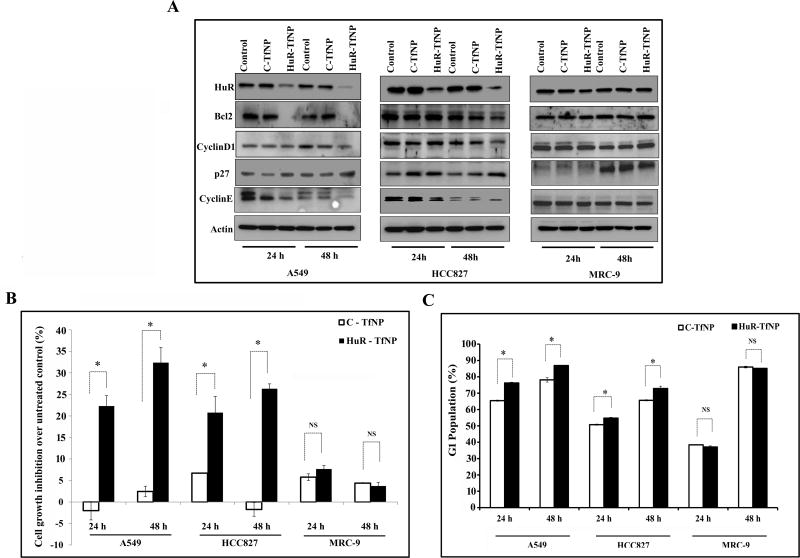
Figure 2. HuR knockdown reduces HuR-target proteins in tumor cells but not normal cells.
A. HuR and HuR-regulated protein expressions in A549, HCC827, and MRC-9 cells treated with C-TfNP or HuR-TFNP at 24 h and 48 h after treatment. Among A549 and HCC 827 groups, HuR-TfNP treatment resulted in better knockdown of HuR protein expression in A549, whereas in MRC-9 when compared to both A549 and HCC827 cancer cells HuR-TfNP induced negligible HuR protein inhibition. HuR knockdown also altered the expression of downstream proteins like BCl2, cyclin D1, P27, and cyclin E in tumor cells but not in normal cells. B. Successful knockdown of HuR resulted in significant growth inhibition in lung cancer cells but not in normal cells, at both 24 h and 48 h after treatment. C. HuR-TfNP induces G1 phase arrest in A549 and HCC827 cells, but not in MRC-9 cells. * p<0.05, “NS” – not significant.
Western blot analysis revealed the impact of HuR knockdown in HuR-associated markers, such as BCl2, Cyclin D1 and E, and p27 [Fig. 2A, Supplementary Fig. S6–S8]. Reduced expression of BCl2 and Cyclin D1 and E were observed in A549 cells at 24 h and 48 h, consistent with HuR knockdown, whereas p27 levels were upregulated. The after-effect of HuR knockdown in HuR-associated markers was also consistent in HCC827 cells, although some observed variations in Cyclin D1 and BCl2 expression at 24 h may be due to cell type effects [Fig. 2A]. As with the poor HuR knockdown in MRC-9 cells, HuR-regulated proteins were less affected at both time points.
Fig. 2B shows the percentage difference in cell growth inhibition over untreated A549, HCC827, and MRC-9 cells upon treatment with HuR-TfNP or C-TfNP. At 24 h, significantly greater growth inhibition was observed in HuR-TfNP-treated A549 cells (22.2 ± 2.3%) than in C-TfNP-treated cells (−2.1 ±. 2.15%; p<0.05). At 48 h, this difference was greater (HuR-TfNP, 32.3 ± 3.5%; C-TfNP, 2.4 ± 1.2%; p<0.05), indicating increased cell growth inhibition over time. A similar trend was observed in HCC827 cells, in which the cell growth in the HuR-TfNP-treated group (20.7 ± 3.7%) was significantly inhibited in comparison with the C-TfNP-treated group (6.7 ± 0.01%; p<0.05) at 24 h. At 48 h, this difference was greater in HCC827 cells (HuR-TfNP, 26.3 ± 1.1%; C-TfNP, −1.7 ± 1.5%). Conversely, no significant difference was observed between C-TfNP- and HuR-TfNP-treated MRC-9 cells at either 24 h or 48 h, indicating less efficient targeted HuR-TfNP-based cell growth inhibition. Hence, Tf-modified NPs carrying siHuR showed selectivity towards TfR-overexpressing lung cancer cells, leading to effective gene knockdown and cell growth inhibition.
HuR-TfNP induces G1 cell cycle arrest and apoptosis in lung cancer cells
The progression of the cell cycle in A549, HCC827, and MRC-9 cells after treatment with either HuR-TfNP or C-TfNP for 24 h and 48 h was evaluated. Compared with cells transfected with C-TfNP, a large increase in the G1 population was observed in HuR-TfNP-treated A549 and HCC827 cells at 24 h and 48 h, indicating a pronounced cell cycle arrest at the G1 phase [Fig. 2C; p <0.05]. Interestingly, no significant cell growth arrest at the G1 phase was observed in MRC-9 cells upon HuR-TfNP treatment compared to C-TfNP treatment [Fig. 2C]. Hence, the possible underlying mechanism of HuR-TfNP- induced cell growth inhibition in lung cancer cells occurs via G1 cell cycle arrest.
One question that arises is whether siRNA-targeted HuR knockdown induces only growth arrest or subsequently proceeds to apoptotic cell death. To answer this question we examined for caspase-9 and PARP by western blotting, and for Annexin V positive cells by flow cytometry respectively. A marked increase in cleaved caspase-9 and PARP was observed in HuR-TfNP-treated A549 cells compared to C-TfNP-treated and unteated control cells [Supplementary Fig. S9a]. Similarly, Annexin V assay showed a significant increase in apopotic cells in HuR-TfNP-treated cells compared to C-TfNP-treated and unteated control cells [Supplementary Fig. S9b; p<0.001]. Our results show that HuR-TfNP treatment produces growth arrest that culminates in apoptotic cell death.
HuR-TfNP inhibits cancer cell migration and invasion
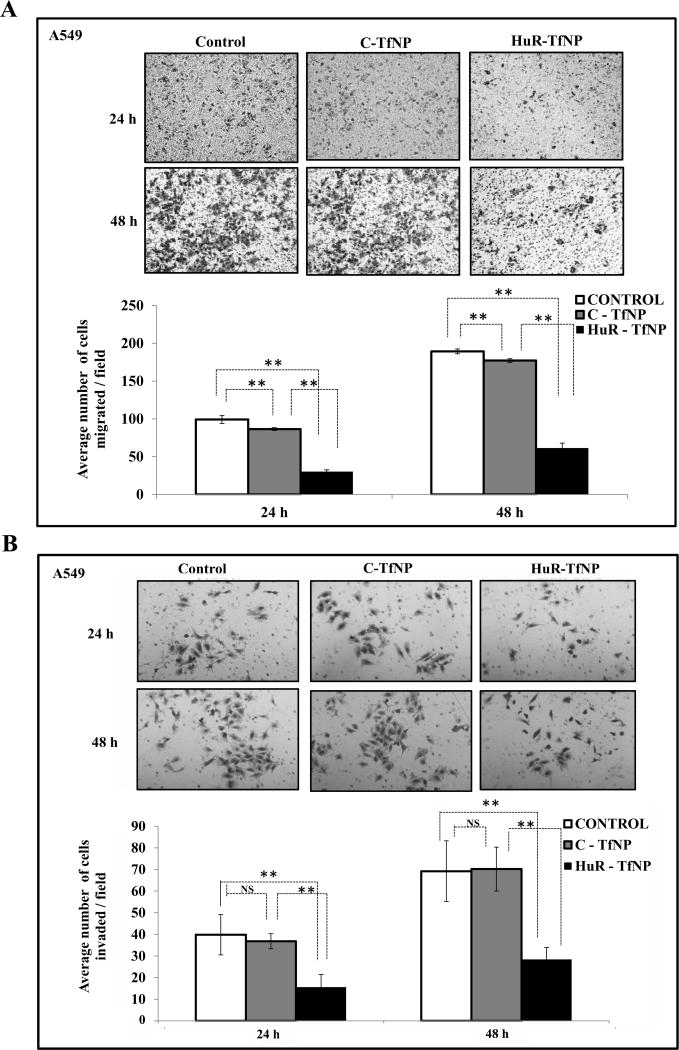
Since we observed that the effectiveness of HuR-TfNP was more prominent in A549 cells, we then evaluated the ability of HuR-TfNP to control migration and invasion in this cell line. Fig. 3A shows the microscopic images from the transwell migration assay; the number of cells migrated per field is represented by bar diagram. The migration rate of A549 cells in HuR-TfNP was significantly reduced compared with C-TfNP and untreated control groups at 24 h and 48 h [p <0.001]. HuR-TfNP treatment also resulted in poor invasion of A549 cells, as evident from the microscopy images, compared with C-TfNP and untreated controls [Fig. 3B; p <0.001]. Significantly fewer cells invaded in the HuR-TfNP group, as shown in the bar diagram, at both 24 h and 48 h [p <0.001].
Figure 3. HuR-TfNP inhibits tumor cell migration and invasion.
Representative images of A, migration and B, invasion assay for A549 tumor cells are shown for C-TfNP and HuR-TfNP treatment groups. Cells that did not receive any treatment served as controls. Quantification of migration and invasion assays expressed by cell counting of respective assays is also presented. * p<0.05, ** p<0.001, “NS” – not significant.
In vivo biodistribution and quantification of siRNA carrying TfNP
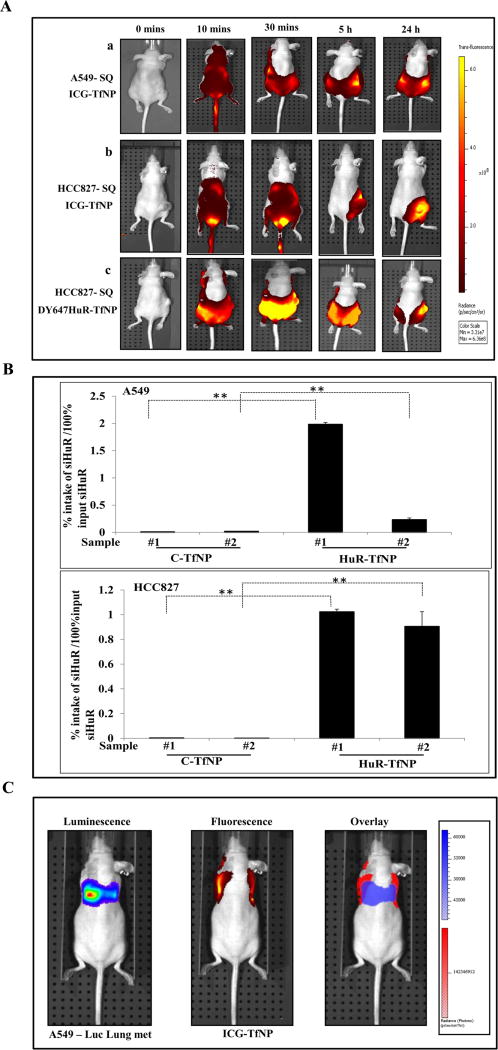
Having demonstrated the in vitro efficacy of HuR-TfNP, our next step was to study the distribution behavior of TfNP in vivo. The biodistribution of TfNP was monitored in subcutaneous A549 and HCC827 tumor models with ICG-TfNP. Fig. 4A shows the IVIS® spectrum live images from tumor-bearing mice that were intravenously (i.v.) injected with ICG-TfNP and monitored for 24 h after injection. Ten minutes after ICG-TfNP injection, the NIR signal was observed throughout the body in mice with both the tumor models. Over time, the fluorescence intensity was confined to the tumor site and the intensity of the signal was diminished throughout the body. Interestingly, the fluorescent signal persisted in the tumors even 24 h after injection. I.V. injection of free ICG did not show preferential tumor accumulation at any time points [Supplementary Fig. S10]. Fluorescent signals obtained over 5 h in tumor-bearing mice suggest the rapid clearance of free ICG. However, intravenous injection of ICG-TfNP into healthy non-tumor-bearing mice showed a prolonged signal for up to 24 h, and the accumulation was mostly confined to the abdominal region, where reticulo-endothelial organs are located. This finding indicates that TfNP could delay the process of ICG clearance in the circulation and, if no target tissues with high TfR expression such as TfR expressing cancer cells are present, ICG-TfNP undergoes normal bio-clearance from the body.
Figure 4. In vivo biodistribution studies.
A. IVIS® Spectrum live images of (a) A549 and (b) HCC827 tumor bearing mice obtained at different time intervals after i.v. injection of Indocyanin green (ICG)-TfNP showed accumulation in the tumor by 24 h. (c) I.v. administration of fluorescent HuR siRNA loaded nanoparticle (DY-647HuR-TfNP) showed NP accumulation in the tumor by 24 h. White circle indicates the location of the tumor on the lower right flank. B. Graphical representation of siRNA accumulation in tumor tissues of A549 and HCC827 mice models as measured by Digital Droplet PCR at 24 h after treatment. Sample # 1 and # 2 represent tumor tissue from two mice of each treatment group analyzed. C. Individual luminescent (A549 luc), fluorescent (Dy647HuR-TfNP), and overlaid images showing localization of DY647HuR siRNA in A549 lung tumors. * p<0.05, ** p<0.001.
Further, we investigated the distribution of fluorescently-labeled siRNA upon in vivo administration in mice bearing HCC827 tumors [Fig. 4A (c)]. I.V. injection of Dy647HuR-TfNP into HCC827 subcutaneous tumor-bearing animals demonstrated accumulation and localization of the particles in the tumor site over time with maximum accumulation as evidenced by highest fluorescence signal occurring at 24 h after injection.
Next we determined the percent intake of siRNA by the tumor after i.v. administration of HuR-TfNP in mice bearing subcutaneous A549 and HCC827 tumors by TaqMan assay using digital droplet PCR (ddPCR). HuR siRNA was detected in tumor tissues harvested from the HuR-TfNP-treated animals, while tumor tissues from C-TfNP-treated animals produced negligible signals for control siRNA, confirming the specificity of siHuR detection. Notably, individual tumor samples accumulated 1.98% and 0.23% of administrated doses of siHuR respectively in A549, and 1.02% and 0.90% respectively in HCC827 [Fig. 4B; p <0.0001]. Although siRNA levels detected among the HuR-TfNP-treated samples varied, nevertheless the levels were significantly higher compared to C-TfNP. Hence, it is evident that HuR-TfNP was capable of successfully delivering its siRNA payload to the target tissue in vivo.
Our next step was to understand the fate of TfNP in a mouse model of metastatic lung tumors. ICG-TfNP was administered i.v. into mice bearing A549-luc tumors, and we monitored the accumulation at tumor metastatic site. Fig. 4C shows the overlaid image of luciferase-expressing mouse lung metastases and ICG (red fluorescence), suggesting the accumulation of ICG-TfNP in A549 lung-metastases over time. These results show that our TfNP targets both subcutaneous and metastatic lung tumors when administered intravenously and thus HuR-TfNP could be a therapeutic for lung cancer treatment.
Efficacy of HuR-TfNP in mouse lung tumor xenografts
Although i.v. administration of HuR-TfNP resulted in its accumulation at the tumor site, we wanted to examine the efficacy of our formulation in subcutaneous lung tumor models upon direct intratumoral administration. Fig. 5 A & B show the tumor growth patterns in animals bearing subcutaneous A549 and HCC827 tumors for a period of 25 days from the day of first treatment with either C-TfNP or HuR-TfNP. Mice receiving C-TfNP treatments showed uncontrolled tumor growth compared with the significantly reduced tumor volumes observed in HuR-TfNP-treated mice bearing A549 tumors [Fig. 5A]. The HuR-TfNP treated tumors, which were harvested after measurement on the 25th day, showed a clear reduction (56% inhibition) in tumor volumes (153.7 mm3; p<0.05) over C-TfNP-treated tumors (351.6 mm3). Western blotting and immunohistochemistry studies showed reduction in the HuR expression that correlated with reduced tumor volume in HuR-TfNP-treated tumors compared to C-TfNP treated tumors [Fig. 5A].
Figure 5. HuR-TfNP treatment suppresses lung tumor growth inhibition in vivo.
A, A549 and, B. HCC827 subcutaneous tumor-bearing mice were treated intratumorally with C-TfNP or HuR-TfNP for 6 doses over two weeks of time period and tumor growth measured. Inset shows HuR expression in tumors after two treatments. Arrows denote treatments. In vivo tumor growth inhibition profile, representative tumors harvested from mice sacrificed on final day of experiment, immunohistochemistry, and Hematoxylin & eosin stained tumor tissue sections from sacrificed mice on final day of the experiment are shown. C. Mice bearing subcutaneous A549-luc tumors were treated i.v. with C-TfNP or HuR-TfNP for 6 doses over two weeks of time period and tumor growth measured by calipers and by bioluminescent imaging. A significant inhibition in tumor growth was observed in HuR-TfNP-treated mice compared to C-TfNP treatment. D. Representative gross tumor size harvested from C-TfNP- and HuR-TfNP-treated mice sacrificed on final day of experiment, and western blotting for HuR in two tumor samples (# 1 and # 2) representative of each treatment group analyzed after two treatments is shown. E. Mice bearing subcutaneous HCC287 tumors were treated i.v. with -TfNP or HuR-TfNP for 6 doses over two weeks of time period. A significant inhibition in tumor growth was observed in HuR-TfNP-treated mice compared to C-TfNP treatment. F. Representative tumors harvested from C-TfNP and HuR-TfNP-treated mice sacrificed on final day of experiment, and western blotting for HuR in tumors harvested from treated are shown. Arrows denote treatments. * p<0.05, ** p<0.001.
Likewise, the reductions in HCC827 tumor volumes were also highly pronounced upon HuR-TfNP treatment. The average tumor volumes at the termination of the study were 548.0 mm3 for C-TfNP-treated tumors versus 195.1 mm3 (64% inhibition; p <0.05) for HuR-TfNP-treated tumors [Fig. 5B]. Tumor growth was clearly inhibited upon intratumoral treatment, indicating the efficacy of HuR-TfNP. Moreover, the HuR protein expression pattern in the isolated A549 [Fig. 5A] and HCC827 [Fig. 5B] tumor tissues from mice treated with HuR-TfNP showed a marked reduction after two treatments compared with mice treated with C-TfNP. This finding suggests that the antitumor activity was caused by HuR knockdown in vivo.
Since it can be argued that intratumoral treatments are not clinically relevant for lung cancer treatment, we next assessed the delivery efficiency and the therapeutic effect of targeted HuR-TfNP upon i.v. administration in mice bearing A549-luc SQ tumors. As shown in Fig. 5C, tumor growth was significantly inhibited upon HuR-TfNP treatment (p < 0.05). The difference in tumor volumes in the C-TfNP- and HuR-TfNP-treatment groups was observed as early as the 7th day of measurement (73% inhibition). The tumors in the C-TfNP-treated group grew uncontrollably, while a significant slower growth pattern was observed in the HuR-TfNP-treated group. On day 35, the tumor volume in the C-TfNP-treated group reached 600 mm3, and all of the mice in that group were euthanized and the tumors were isolated and displayed as shown in Fig. 5D. The tumor volume in the HuR-TfNP-treated group was significantly less and exhibited 75% growth inhibition compared to C-TfNP treatment on day 35. Also shown are representative mice from each treatment group from which the tumors are harvested. Interestingly, we observed that the tumor progression in the HuR-TfNP-treated group was significantly delayed and the tumor volume equivalent to C-TfNP-treated group was not reached till day 91 which time the study was terminated [Fig. 5D]. The tumor volume in HuR-TfNP-treated group on day 91 was only 315.4 mm3.
In parallel to measuring tumor volume by calipers, the efficacy of HuR-TfNP treatment was also assessed by bioluminescence imaging at select time points (day 1, 15 and 33) in the subcutaneous A549-luc tumor-bearing mice and compared to C-TfNP treatment [Supplementary Fig. S11]. The average radiance emitted from tumors in each group were averaged and plotted against the days of measurement. A significant reduction in the average radiance was observed in HuR-TfNP-treated mice on days 15 (2.6 × 104 p/sec/cm2/sr) and 33 (1.14 × 104 p/sec/cm2/sr) compared to the radiance in C-TfNP-treated mice on day 15 (1.89 × 105 p/sec/cm2/sr) and day 33 (3.72 × 105 p/sec/cm2/sr) [Supplementary Fig. S11; p< 0.05]. Bioluminescent imaging was discontinued after day 33 as the control animals were euthanized on day 35 [Fig. 5C].
To further investigate molecular changes indicative of HuR-TfNP-mediated anti-tumor efficacy, tumors harvested at the end of the study were subjected to molecular analysis. Western blotting and RT-PCR analysis of the tumor samples revealed a reduction in the protein and mRNA expression of HuR and HuR-regulated BCL2 whereas P27 showed an upregulation in its expression in HuR-TfNP-treated tumors compared to C-TfNP-treated tumors [Fig. 5D; Supplementary Fig. S12; p <0.05]. Furthermore, HuR-TfNP-treated tumors showed increased activation of caspase-9 and PARP compared to C-TfNP-treated tumors [Supplementary Fig. S12].
A similar study conducted with mice bearing subcutaneous HCC827 tumors demonstrated the efficiency of i.v. administered HuR-TfNP in targeted antitumor activity, mediated by siHuR gene knockdown. The i.v. administration of HuR-TfNP resulted in sluggish tumor growth compared with the active growth observed in C-TfNP-treated tumors [Fig. 5E]. The tumor volumes were 522.5 mm3 for the HuR-TfNP-treated group and 958.9 mm3 for the C-TfNP-treated group (45% inhibition; p <0.05) on the 37th day of measurement. Due to enhanced tumor burden in the control groups, we discontinued the study per the IACUC recommended guidelines. Compared with A549 tumors, the tumor inhibition in HCC827 tumors was slow, possibly because of the aggressive nature of HCC827 growth and relatively low Tf receptor levels. Nevertheless, the HCC827 tumor growth was significantly inhibited with HuR-TfNP treatment, compared with the large tumors observed in the C-TfNP-treated group [Fig. 5F]. The western blot image shows the HuR knockdown obtained in representative HuR-TfNP-treated tumors compared with C-TfNP treated tumors [Fig. 5F]
Immunohistochemical analysis of HuR revealed that the HuR expression was reduced in the subcutaneous A549 and HCC827 tumors from animals that received HuR-TfNP treatment [Supplementary Fig. S13 & S14]. These results are consistent with western blotting and RT-PCR results from the same tumor samples [Fig. 5A and B]. To determine the mechanism involved in the efficacy of HuR-TfNP treatment, the harvested tumors were analyzed for Ki67 and CD31 expression. Ki67 immunohistochemistry can be used to assess the proliferative index of the tumor [47]. In the present study, Ki67-positive cells were significantly higher in C-TfNP–treated tumors (A549 and HCC827) than in the HuR-TfNP-treated tumors [Supplementary Figs. S13 & S14]. These observations suggest that HuR expression is closely associated with cellular proliferation. It confirms our earlier data that inhibition of HuR expression abrogates the tumor proliferation rate. A rapidly growing tumor often expresses CD31, a microvessel density (MVD) marker, which is detectable by immunohistochemistry [29, 48]. We detected significant decrease in CD31 positive microvessels in tissue sections from HuR-TfNP-treated tumors compared with C-TfNP-treated tumors in both A549 and HCC827 mice models [Supplementary Figs. S13 & S14]. Since tumor vasculature is characterized by increased number of tumor vessels, the CD31 immunohistochemistry data suggests less active vasculature in HuR-TfNP-treated groups, specifying the neovascularization inhibitory activity of HuR-TfNP in A549 and HCC827 tumors. Finally, a significant increase in apoptotic cell death as determined by TUNEL staining was also observed in HuR-TfNP-treated A549 tumors that were collected at the end of the study [Supplementary Fig. S13; p<0.05]. Although, we did not perform TUNEL staining in HCC827 tumor tissues we speculate that increased apoptotic cell death also occurs in HuR-TfNP-treated tumors resulting in delayed tumor growth.
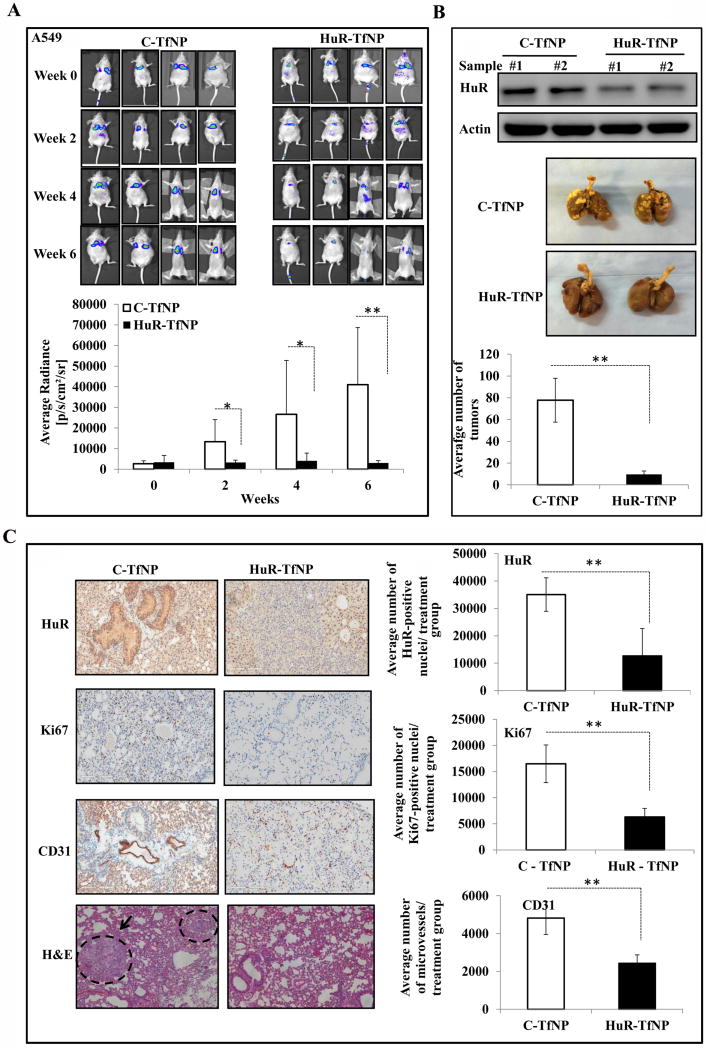
Systemic delivery of HuR-TfNP inhibits experimental lung metastasis in vivo
In order to investigate the antitumor activity of HuR-TfNP on lung metastasis, we monitored the tumor growth in mice bearing A549-luc lung metastasis following HuR-TfNP treatment. As shown in Fig. 6A, the decay of bioluminescence signals from HuR-TfNP-treated A549-luc lung metastases were distinctly visible, especially at the 4th and 6th weeks, indicating a significantly reduced tumor burden compared with C-TfNP-treated groups (93% inhibition at week 6; p < 0.05). The average radiance of the HuR-TfNP-treated groups was reduced or unchanged, compared with the enhanced radiance observed in the C-TfNP-treated groups. Analysis of 6th week images showed an average radiance of 41007.7 (p/sec/cm2/sr) for C-TfNP, which is a 13-fold increase over HuR-TfNP-treated tumor signals of 3025 (p/sec/cm2/sr; p <0.0001). As expected, there was less HuR protein expression in the excised lung tissue from the HuR-TfNP-treated group than in lung tissue from the C-TfNP-treated group [Fig. 6B]. As shown in Fig. 6B, the metastatic nodules in HuR-TfNP-treated lungs (9.57; 88% inhibition) were significantly reduced, whereas tumor nodules occupied a major portion of the C-TfNP treated lungs (77.7; p <0.0001). IHC analysis of the lung lobes revealed a significant, up to threefold reduction in the HuR protein levels in HuR-TfNP-treated groups compared with C-TfNP-treated groups. [Fig. 6C; p <0.0001].
Figure 6. HuR-TfNP treatment suppresses experimental lung metastasis.
Mice bearing A549-luc lung tumors were treated i.v. with C-TfNP or HuR-TfNP for a total of six treatments. Mice were imaged at regular intervals to monitor and measure tumor growth. A. Representative bioluminescence images showing inhibition of experimental lung metastasis in HuR-TfNP-treated mice compared to C-TfNP-treated mice. Bar graph represents the quantitation of bioluminescence intensity as Average Radiance [p/s/cm²/sr]. The gradual decrease in the bioluminescence signals in HuR-TfNP compared to C-TfNP groups suggests significant reduction in tumor growth over the period of treatment. B. Western blot image shows reduced HuR expression in HuR-TfNP-treated A549 lung tumors (upper panel). Sample # 1 and # 2 are representative samples for each treatment group analyzed at end of the study. Lung tumor nodules of a representative animal from each treatment group are also shown (middle panel). Animals were euthanized after six weeks; lungs were inflated with bovine solution and the number of extrapulmonary tumor nodules counted (lower panel). A significant reduction in the number of lung nodules were observed in HuR-TfNP-treated animals compared to control. C. Images represent the IHC staining of lung sections for HuR, CD31, and Ki67. Hematoxylin and eosin (H&E) stain showing the tissue pathology with dashed circle marking tumors in the lung. Bar graph represents the quantitation of IHC stains using. * p<0.05, ** p<0.001.
Correspondingly, there was a two-fold reduction in MVD (indicated by CD31 expression) in sections of lung lobes from HuR-TfNP-treated animals (p <0.0001). Similarly, Ki-67-positive cells were reduced three-fold in HuR-TfNP-treated groups, indicating significantly less proliferation compared with the C-TfNP-treated group (p <0.0001). There were significantly fewer nodules in HuR-TfNP-treated tumors than in C-TfNP-treated tumors, as observed in the H&E stained sections [Fig. 6C]. Thus, systemic delivery of HuRsiRNA using TfNP effectively suppressed HuR expression, and significantly reduced MVD and cell proliferation resulting in inhibition of experimental metastasis.
Discussion
Though the potential of RNAi based gene therapy has been realized over the last decade, the absence of a reliable targeted delivery system limits their efficient use in cancer therapy. Recently, the RNA binding protein HuR overexpressed in NSCLC has been recognized as a potential molecular target for RNAi therapy [11, 12, 14–16]. Since many of the NSCLC tumors are characterized by the overexpression of TfR [4, 5], our goal was to explore the efficacy of a rationally designed liposomal nanocarrier system targeting TfR and specifically deliver HuR siRNA into the tumor cells. Our Tf-NP nanoformulation also helped in understanding the function, fate, biological action and therapeutic effect of HuR siRNA specifically delivered in tumor site.
In the present study the post insertion technique was employed for modifying HuR-NP with Tf ligand [26]. We employed sulfhydryl reactive crosslinking chemistry to synthesize Tf-PEG-DSPE for nanoparticle modification purposes. The target specific cell uptake studies revealed that it is the level of receptor expression which influences the cellular uptake of receptor targeted-nanoparticles [49], and thereby the delivery of its siRNA payload [50]. Increased siRNA delivery by HuR-TfNP in TfR overexpressing cancer cells resulted in the efficient knockdown of HuR and altered the expression levels of its associated proteins [12, 15, 42–46]. Also suppression of HuR using siRNA induced cell cycle arrest in the G1 phase [46]. Multistep processes, such as cell adhesion, migration, and invasion, are involved in cancer metastasis [51], and HuR is known to regulate metastasis-related mRNAs [7, 52]. In the present study, the in vitro anti-cancer efficacy of HuR-TfNP-was demonstrated by growth inhibition, induction of G1 cell-cycle arrest leading to apoptosis, and reduced migration and invasion ability of lung cancer cells. That the observed antitumor activity was specific to HuR silencing was demonstrated by comparing HuR-specific siRNA (HuRsi4) and comparing with control siRNA and a siRNA (HuRsi21) that was initially used for screening HuR inhibitory effects [Supplementary Fig. S4]. Further, it is known that the therapeutic HuR siRNA if not specifically targeted may cause deleterious effects in normal cells too. However, HuR-TfNP caused no significant effect on normal lung tumor fibroblasts, demonstrating safe and selective activity against lung cancer cells.
In vivo biodistribution study demonstrated preferential accumulation of Dy647HuR-TfNP in tumor milieu and this result correlated with ICG-TfNP distribution in tumors. Since neurons and brain capillary endothelial cells in human and mice are known to express TfR in significant levels, one may expect a high accumulation of TfNP in mice brain tissue [53, 54]. However, the accumulation of ICG-TfNP in the brain was minimal and not prolonged as evident in the bio-distribution images obtained [Figure 4, & Supplementary Fig. S10]. The fluorescence from ICG-TfNP faded away from mice head and neck area in less than 30 min after injection, suggesting that the specific accumulation of TfNP in brain area is non-significant. One possibility for negligible TfNP accumulation in the brain is that the TfR expression level in the mice brain is relatively low compared to TfR expression in the tumor resulting in ICG-TfNP fluorescence level in mice brain falling below the detection level of the instrument [Fig. 4 (a) & (b)]. This observation in our study was consistent with a previous report indicating the TfR-targeted nanoparticle accumulation in brain was the lowest among major organs studied, in comparison to the tumor tissues [55]. Another possibility is that the size of the HuR-TfNP (> 200 nm) are large thereby limiting the particles from crossing the blood-brain barrier (BBB) and enter the brain tissue. Since TfR targeting is one of the successful strategies employed to deliver nanoparticle drug carriers across blood brain barrier [56–58], the brain accumulation potential of TfNP if any, will be investigated in future studies.
Systemic delivery of HuR-TfNP showed that it can effectively inhibit the growth of solid tumors of lung origin. The tumor inhibition in subcutaneous models was specific to HuR siRNA treatment, as we accurately detected the presence of HuR siRNA in tumor tissues treated with HuR-TfNP in comparison to C-TfNP treatments by digital droplet PCR technique. The effect of HuR-TfNP treatment in subcutaneous lung tumors via systemic route administration was significant, since tumor volume inhibition was correlated with HuR knockdown. The proliferative nature and active vasculature formation were significantly less in HuR-TfNP treated tumors as analyzed by IHC staining. The mechanism by which HuR-TfNP reduces tumor vasculature is not known and is beyond of the scope of the present study. One possibility is the direct inhibitory activity of HuR-TfNP on vascular endothelial growth factor (VEGF) as a consequence of HuR knockdown. VEGF is a molecular target of HuR [7] However, additional mechanisms for suppressing tumor vasculature may exist that needs to be explored.
Metastatic spread of cancer cells from the primary tumors to the distant organs is a major reason for high mortality among lung cancer patients [59]. To this end the anti-metastatic effect of HuR-TfNP was assessed in a lung metastatic tumor model in mice. Imaging studies revealed preferential accumulation of TfNP in lung metastatic tumor site. Further, HuR-TfNP treatment resulted in efficacious targeted sequence-specific knockdown of HuR and in triggering molecular events that successfully inhibited lung cancer metastasis that represents the clinical relevance of this study. While in the present study we have not investigated the contribution of HuR-TfNP on the chemokine CXCR4 receptor, studies from our laboratory and others have previously shown inhibition of HuR suppresses CXCR4 [12, 60]. Thus, it is possible that the observed inhibition of experimental metastasis could partly be due to CXCR4 inhibition. Should HuR-TfNP treatment contribute to suppression of CXCR4 in vivo and concur with our previous in vitro reports, then combining HuR-TfNP with CXCR4 inhibitors would become a viable therapy for lung cancer [12]. Similarly, conducting combinatorial therapy studies in the future that incorporate chemotherapy or radiation therapy with HuR-TfNP will be of clinical relevance for tackling metastatic disease. Nevertheless, our results convincingly demonstrate that systemic delivery of HuR-TfNP effectively suppresses lung metastasis.
It is important to realize that our studies were conducted in tumor models using immunodeficient mice. It will be of interest to test the efficacy of HuR-TfNP in a tumor model setting that utilizes immunocompetent host to determine the effects of HuR inhibition on host-immune system since HuR has previously been reported to play a role in inflammation [61, 62]. Further, recent reports on the role of PDL1-PD1 in suppressing host immune T-cells begs the question if HuR-TfNP treatment will impact the PDL1-PD1 signaling and alter the host immune system to enhance the anti-tumor immune activity. These questions while of interest are beyond of the scope of the present study but warrant investigation in the future [63, 64]. In conclusion, we have demonstrated HuR-TfNP treatment effectively inhibits lung cancer growth both in vitro and in vivo.
Supplementary Material
Acknowledgments
We thank the Stephenson Cancer Center at the University of Oklahoma Health Sciences Center, Oklahoma City, OK for the use of the Functional Genomics Core, Biostatistics Core, and Small Animal Bioluminescent Imaging Core facilities. Assistance received from the institutional molecular imaging core facility for conducting flow cytometry studies is appreciated. The authors thank Ms. Kathy Kyler at the office of Vice President of Research, OUHSC, for editorial assistance. Rajagopal Ramesh is an Oklahoma TSET Research Scholar and holds the Jim and Christy Everest Endowed Chair in Cancer Developmental Therapeutics.
Financial Support:
The study was supported in part by a grant received from the National Institutes of Health R01 CA167516 (R. Ramesh), an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences (P20 GM103639; A. Munshi, and R. Ramesh) of the National Institutes of Health, and by funds received from the Presbyterian Health Foundation Seed Grant (C5094701 and C5095801; R. Ramesh), Presbyterian Health Foundation Bridge Grant (C5098101; R. Ramesh), Stephenson Cancer Center Seed Grant (R. Ramesh), and Jim and Christy Everest Endowed Chair in Cancer Developmental Therapeutics (R.Ramesh), The University of Oklahoma Health Sciences Center.
Footnotes
Supplementary material is available for the physicochemical characteristics of nanoparticles, cell uptake and ligand optimization studies, mRNA expression analysis, western blots quantification data, imaging data for bio-distribution and anti-tumor efficacy in mouse models, and immunohistochemical (IHC) analysis of tumor tissues.
References
- 1.Chevalier TL. Non-small cell lung cancer: the challenges of the next decade. Front Oncol. 2011;1:29. doi: 10.3389/fonc.2011.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen TM. Ligand-targeted therapeutics in anticancer therapy. Nat Rev Cancer. 2002;2:750–763. doi: 10.1038/nrc903. [DOI] [PubMed] [Google Scholar]
- 3.Bae YH, Park K. Targeted drug delivery to tumors: Myths, reality and possibility. J Control Release. 2011;153:198–205. doi: 10.1016/j.jconrel.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boult J, Roberts K, Brookes MJ, Hughes S, Bury JP, Cross SS, et al. Overexpression of cellular iron import proteins is associated with malignant progression of esophageal adenocarcinoma. Clin Cancer Res. 2008;14:379–387. doi: 10.1158/1078-0432.CCR-07-1054. [DOI] [PubMed] [Google Scholar]
- 5.Whitney JF, Clark JM, Griffin TW, Gautam S, Leslie KO. Transferrin receptor expression in non-small cell lung cancer Histopathologic and clinical correlates. Cancer. 1995;76:20–25. doi: 10.1002/1097-0142(19950701)76:1<20::aid-cncr2820760104>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 6.Filippova N, Yang X, Wang Y, Gillespie GY, Langford C, King PH, et al. The RNA-binding protein HuR promotes glioma growth and treatment resistance. Mol Cancer Res. 2011;9:648–659. doi: 10.1158/1541-7786.MCR-10-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, Zhao W, Guo Y, Zhang B, Xie Q, Xiang D, et al. The expression of RNA-binding protein HuR in non-small cell lung cancer correlates with vascular endothelial growth factor-c expression and lymph node metastasis. Oncology. 2009;76:420–429. doi: 10.1159/000216837. [DOI] [PubMed] [Google Scholar]
- 8.Huang YH, Peng W, Furuuchi N, Gerhart J, Rhodes K, Mukherjee N, et al. Delivery of therapeutics targeting the mRNA-binding protein HuR using 3DNA nanocarriers suppresses ovarian tumor growth. Cancer Res. 2016;76:1549–1559. doi: 10.1158/0008-5472.CAN-15-2073. [DOI] [PubMed] [Google Scholar]
- 9.Dormoy-Raclet V, Ménard I, Clair E, Kurban G, Mazroui R, Di MS, et al. The RNA-binding protein HuR promotes cell migration and cell invasion by stabilizing the beta-actin mRNA in a u-rich-element-dependent manner. Mol Cell Biol. 2007;15:5365–5680. doi: 10.1128/MCB.00113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calaluce R, Gubin MM, Davis JW, Magee JD, Chen J, Kuwano Y, et al. The RNA binding protein HuR differentially regulates unique subsets of mRNAs in estrogen receptor negative and estrogen receptor positive breast cancer. BMC Cancer. 2010;10:126. doi: 10.1186/1471-2407-10-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kakuguchi W, Kitamura T, Kuroshima T, Ishikawa M, Kitagawa Y, Totsuka Y, et al. HuR knockdown changes the oncogenic potential of oral cancer cells. Mol Cancer Res. 2010;8:520–528. doi: 10.1158/1541-7786.MCR-09-0367. [DOI] [PubMed] [Google Scholar]
- 12.Muralidharan R, Panneerselvam J, Chen A, Zhao YD, Munshi A, Ramesh R. HuR-targeted nanotherapy in combination with AMD3100 suppresses CXCR4 expression, cell growth, migration and invasion in lung cancer. Cancer Gene Ther. 2015;22:581–590. doi: 10.1038/cgt.2015.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romeo C, Weber MC, Zarei M, DeCicco D, Chand SN, Lobo AD, et al. HuR contributes to trail resistance by restricting death receptor 4 expression in pancreatic cancer cells. Mol Cancer Res. 2016;14:599–611. doi: 10.1158/1541-7786.MCR-15-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanco FF, Jimbo M, Wulfkuhle J, Gallagher I, Deng J, Enyenihi L, et al. The mRNA-binding protein HuR promotes hypoxia-induced chemoresistance through posttranscriptional regulation of the proto-oncogene pim1 in pancreatic cancer cells. Oncogene. 2016;35:2529–2541. doi: 10.1038/onc.2015.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muralidharan R, Babu A, Basalingappa K, Munshi A, Ramesh R. Targeted disruption of HuR in lung cancer cells results in global knock-down of HuR-regulated oncoproteins, cell-cycle arrest and reduced cell migration. Mol Ther. 2014;22:S44–S44. [Google Scholar]
- 16.Mehta M, Basalingappa K, Griffith JN, Andrade D, Babu A, Amreddy N, et al. HuR silencing elicits oxidative stress and DNA damage and sensitizes human triple-negative breast cancer cells to radiotherapy. Oncotarget. 2016;40:64820–64835. doi: 10.18632/oncotarget.11706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato Y, Note Y, Maeki M, Kaji N, Baba Y, Tokeshi M, et al. Elucidation of the physicochemical properties and potency of siRNA-loaded small-sized lipid nanoparticles for SiRNA delivery. J Control Release. 2016;229:48–57. doi: 10.1016/j.jconrel.2016.03.019. [DOI] [PubMed] [Google Scholar]
- 18.Babu A, Wang Q, Muralidharan R, Shanker M, Munshi A, Ramesh R. Chitosan coated polylactic acid nanoparticle-mediated combinatorial delivery of cisplatin and siRNA/plasmid DNA chemosensitizes cisplatin-resistant human ovarian cancer cells. Mol Pharm. 2014;11:2720–2733. doi: 10.1021/mp500259e. [DOI] [PubMed] [Google Scholar]
- 19.Lee SJ, Yook S, Yhee JY, Yoon HY, Kim MG, Ku SH, et al. Co-delivery of VEGF and Bcl-2 dual-targeted siRNA polymer using a single nanoparticle for synergistic anti-cancer effects in vivo. J Control Release. 2015;220:631–641. doi: 10.1016/j.jconrel.2015.08.032. [DOI] [PubMed] [Google Scholar]
- 20.Gao P, Zhang X, Wang H, Zhang Q, Li H, Li Y, et al. Biocompatible and colloidally stabilized mPEG-PE/calcium phosphate hybrid nanoparticles loaded with siRNAs targeting tumors. Oncotarget. 2016;7:2855–2866. doi: 10.18632/oncotarget.6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee SK, Tung CH. SiRNA nanoparticles for ultra-long gene silencing in vivo. Methods Mol Biol. 2016;1372:113–120. doi: 10.1007/978-1-4939-3148-4_9. [DOI] [PubMed] [Google Scholar]
- 22.Ramesh R. Nanoparticle-mediated gene delivery to the lung. Methods Mol Biol. 2008;433:301–331. doi: 10.1007/978-1-59745-237-3_19. [DOI] [PubMed] [Google Scholar]
- 23.Lu C, Stewart DJ, Lee JJ, Ji L, Ramesh R, Jayachandran G, et al. Phase I clinical trial of systemically administered TUSC2 (Fus1)-nanoparticles mediating functional gene transfer in humans. PLoS One. 2012;7:e34833. doi: 10.1371/journal.pone.0034833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muralidharan R, Babu A, Amreddy N, Basalingappa K, Mehta M, Chen A, et al. Folate receptor-targeted nanoparticle delivery of HuR-RNAi suppresses lung cancer cell proliferation and migration. J Nanobiotechnology. 2016;14:47. doi: 10.1186/s12951-016-0201-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramesh R, Saeki T, Templeton NS, Ji L, Stephens LC, Ito I, et al. Successful treatment of primary and disseminated human lung cancers by systemic delivery of tumor suppressor genes using an improved liposome vector. Mol Ther. 2001;3:337–350. doi: 10.1006/mthe.2001.0266. [DOI] [PubMed] [Google Scholar]
- 26.Allen TM, Sapra P, Moase E. Use of the post-insertion method for the formation of ligand-coupled liposomes. Cell Mol Biol Lett. 2002;7:889–894. [PubMed] [Google Scholar]
- 27.Amreddy N, Muralidharan R, Babu A, Mehta M, Johnson EV, Zhao YD, et al. Tumor-targeted and ph-controlled delivery of doxorubicin using gold nanorods for lung cancer therapy. Int J Nanomedicine. 2015;10:6773–6788. doi: 10.2147/IJN.S93237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yokoyama T, Tam J, Kuroda S, Scott AW, Aaron J, Larson T, et al. EGFR-targeted hybrid plasmonic magnetic nanoparticles synergistically induce autophagy and apoptosis in non-small cell lung cancer cells. PLoS One. 2011;6:e25507. doi: 10.1371/journal.pone.0025507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramesh R, Mhashilkar AM, Tanaka F, Saito Y, Branch CD, Sieger K, et al. Melanoma differentiation-associated gene 7/interleukin (IL)-24 is a novel ligand that regulates angiogenesis via the IL-22 receptor. Cancer Res. 2003;63:5105–5113. [PubMed] [Google Scholar]
- 30.Panneerselvam J, Srivastava A, Muralidharan R, Wang Q, Zheng W, Zhao L, et al. IL-24 modulates the high mobility group (HMG) A1/miR222 /AKT signaling in lung cancer cells. Oncotarget. 2016;7:70247–70263. doi: 10.18632/oncotarget.11838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bustin SA. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol. 2002;29:23–39. doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- 32.Saito Y, Miyahara R, Gopalan B, Litvak A, Inoue S, Shanker M, et al. Selective induction of cell cycle arrest and apoptosis in human prostate cancer cells through adenoviral transfer of the melanoma differentiation-associated −7 (mda-7)/interleukin-24 (IL-24) gene. Cancer Gene Ther. 2005;12:238–247. doi: 10.1038/sj.cgt.7700780. [DOI] [PubMed] [Google Scholar]
- 33.Inoue S, Hartman A, Branch CD, Bucana CD, Bekele BN, Stephens LC, et al. Mda-7 in combination with bevacizumab treatment produces a synergistic and complete inhibitory effect on lung tumor xenograft. Mol Ther. 2007;15:287–294. doi: 10.1038/sj.mt.6300035. [DOI] [PubMed] [Google Scholar]
- 34.Gopalan B, Shanker M, Chada S, Ramesh R. MDA-7/IL-24 suppresses human ovarian carcinoma growth in vitro and in vivo. Mol Cancer. 2007;6:11. doi: 10.1186/1476-4598-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shanker M, Jin J, Branch CD, Miyamoto S, Grimm EA, Roth JA, et al. Tumor suppressor gene-based nanotherapy: from test tube to the clinic. J Drug Deliv. 2011;2011:465845. doi: 10.1155/2011/465845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coutinho de Souza P, Mallory S, Smith N, Saunders D, Li XN, McNall-Knapp RY, et al. Inhibition of pediatric glioblastoma tumor growth by the anti-cancer agent OKN-007 in orthotopic mouse xenografts. PLoS One. 2015;10:e0134276. doi: 10.1371/journal.pone.0134276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Majzoub RN, Chan CL, Ewert KK, Silva BF, Liang KS, Jacovetty EL, et al. Biomaterials. 2014;35:4996–5005. doi: 10.1016/j.biomaterials.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elias DR, Poloukhtine A, Popik V, Tsourkas A. Nanomedicine. 2013;9:194–201. doi: 10.1016/j.nano.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di Paolo D, Brignole C, Pastorino F, Carosio R, Zorzoli A, Rossi M, et al. Neuroblastoma-targeted nanoparticles entrapping siRNA specifically knockdown ALK. Mol Ther. 2011;19:1131–1140. doi: 10.1038/mt.2011.54. 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Arrington L, Boardman D, Davis J, Xu Y, Di FK, et al. The development of an in vitro assay to screen lipid based nanoparticles for siRNA delivery. J Control Release. 2014;28:7–14. doi: 10.1016/j.jconrel.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 41.Goldenthal KL, Pastan I, Willingham MC. Initial steps in receptor-mediated endocytosis. The influence of temperature on the shape and distribution of plasma membrane clathrin-coated pits in cultured mammalian cells. Exp Cell Res. 1984;152:558–564. doi: 10.1016/0014-4827(84)90658-x. [DOI] [PubMed] [Google Scholar]
- 42.Ishimaru D, Ramalingam S, Sengupta TK, Bandyopadhyay S, Dellis S, Tholaikunnel BG, et al. Regulation of Bcl-2 expression by HuR in HL60 leukemia cells and A431 carcinoma cells. Mol Cancer Res. 2009;7:1354–1366. doi: 10.1158/1541-7786.MCR-08-0476. [DOI] [PubMed] [Google Scholar]
- 43.Guo X, Hartley RS. HuR contributes to cyclin E1 deregulation in MCF-7 breast cancer cells. Cancer Res. 2006;66:7948–7956. doi: 10.1158/0008-5472.CAN-05-4362. [DOI] [PubMed] [Google Scholar]
- 44.Kim HH, Gorospe M. Phosphorylated HuR shuttles in cycles. Cell Cycle. 2008;7:3124–3126. doi: 10.4161/cc.7.20.6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morosétti R, Kawamata N, Gombart AF, Miller CW, Hatta Y, Hirama T, et al. Alterations of the p27kip1 gene in non-hodgkin's lymphomas and adult T-cell leukemia/lymphoma. Blood. 1995;86:1924–1930. [PubMed] [Google Scholar]
- 46.Kullmann M, Gopfert U, Siewe B, Hengst L. Elav/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR. Genes Dev. 2002;16:3087–3099. doi: 10.1101/gad.248902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jalava P, Kuopio T, Juntti-Patinen L, Kotkansalo T, Kronqvist P, Collan Y. Ki67 immunohistochemistry: a valuable marker in prognostication but with a risk of misclassification: proliferation subgroups formed based on ki67 immunoreactivity and standardized mitotic index. Histopathology. 2006;48:674–682. doi: 10.1111/j.1365-2559.2006.02402.x. [DOI] [PubMed] [Google Scholar]
- 48.Wang D, Stockard CR, Harkins L, Lott P, Salih C, Yuan K, et al. Immunohistochemistry in the evaluation of neovascularization in tumor xenografts. Biotech Histochem. 2008;83:179–189. doi: 10.1080/10520290802451085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daniels TR, Bernabeu E, Rodríguez JA, Patel S, Kozman M, Chiappetta DA, et al. The transferrin receptor and the targeted delivery of therapeutic agents against cancer. Biochim Biophys Acta. 2012;3:291–317. doi: 10.1016/j.bbagen.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yhee JY, Lee SJ, Lee S, Song S, Min HS, Kang SW, et al. Tumor-targeting transferrin nanoparticles for systemic polymerized SiRNA delivery in tumor-bearing mice. Bioconjug Chem. 2013;24:1850–1860. doi: 10.1021/bc400226b. [DOI] [PubMed] [Google Scholar]
- 51.Martin TA, Ye L, Sanders AJ, Lane J, Jiang WG. Madame Curie Bioscience Database [Internet] Jandial Red. Landes Bioscience; Austin: 2000. Cancer invasion and metastasis: molecular and cellular perspective. [Google Scholar]
- 52.Miyata Y, Watanabe S, Sagara Y, Mitsunari K, Matsuo T, Ohba K, et al. High expression of HuR in cytoplasm, but not nuclei, is associated with malignant aggressiveness and prognosis in bladder cancer. PLoS One. 2013;8:e59095. doi: 10.1371/journal.pone.0059095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rothenberger S, Food MR, Gabathuler R, Kennard ML, Yamada T, Yasuhara O, et al. Coincident expression and distribution of melanotransferrin and transferrin receptor in human brain capillary endothelium. Brain Res. 1996;712:117–121. doi: 10.1016/0006-8993(96)88505-2. [DOI] [PubMed] [Google Scholar]
- 54.Zumbrennen-Bullough KB, Becker L, Garrett L, Hölter SM, Calzada-Wack J, Mossbrugger I, et al. Abnormal brain iron metabolism in Irp2 deficient mice is associated with mild neurological and behavioral impairments. PLoS One. 2014;9:e98072. doi: 10.1371/journal.pone.0098072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang K, Zhang Y, Wang J, Yuan A, Sun M, Wu J, et al. Self-assembled IR780-loaded transferrin nanoparticles as an imaging, targeting and PDT/PTT agent for cancer therapy. Sci Rep. 2016;6:27421. doi: 10.1038/srep27421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Masserini M. Nanoparticles for Brain Drug Delivery. ISRN Biochem. 2013;2013:238428. doi: 10.1155/2013/238428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dixit S, Novak T, Miller K, Zhu Y, Kenney ME, Broome AM. Transferrin receptor-targeted theranostic gold nanoparticles for photosensitizer delivery in brain tumors. Nanoscale. 2015;7:1782–1790. doi: 10.1039/c4nr04853a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cabezón I, Manich G, Martín-Venegas R, Camins A, Pelegrí C, Vilaplana J. Trafficking of gold nanoparticles coated with the 8d3 anti-transferrin receptor antibody at the mouse blood-brain barrier. Mol Pharm. 2015;12:4137–4145. doi: 10.1021/acs.molpharmaceut.5b00597. [DOI] [PubMed] [Google Scholar]
- 59.D’Antonio C, Passaro A, Gori B, Signore ED, Migliorino MR, Ricciardi S, et al. Bone and brain metastasis in lung cancer: recent advances in therapeutic strategies. Ther Adv Med Oncol. 2014;6:101–114. doi: 10.1177/1758834014521110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Al-Souhibani N, Al-Ghamdi M, Al-Ahmadi W, Khabar KS. Posttranscriptional control of the chemokine receptor CXCR4 expression in cancer cells. Carcinogenesis. 2014;35:1983–1992. doi: 10.1093/carcin/bgu080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rhee WJ, Nia C-W, Zhenga Z, Chang K, Joa H, Bao G. HuR regulates the expression of stress-sensitive genes and mediates inflammatory response in human umbilical vein endothelial cells. Proc Natl Acad Sci USA. 2010;107:6858–6863. doi: 10.1073/pnas.1000444107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Katsanou V, Papadaki O, Milatos S, Blackshear PJ, Anderson P, Kollias G, et al. HuR as a negative posttranscriptional modulator in inflammation. Mol Cell. 2005;19:777–789. doi: 10.1016/j.molcel.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 63.Sgambato A, Casaluce F, Sacco PC, Palazzolo G, Maione P, Rossi A. Anti PD-1 and PDL-1 immunotherapy in the treatment of advanced non-small cell lung cancer (NSCLC): a review on toxicity profile and its management. Curr Drug Saf. 2016;11:62–68. doi: 10.2174/1574886311207040289. [DOI] [PubMed] [Google Scholar]
- 64.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nature. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.