Abstract
Tyrosine kinase inhibitors (TKI’s) are currently the drug of choice for management of chronic myeloid leukemia. Imatinib is the most commonly used first line TKI in India. Mutations leading to resistance to imatinib are the most common cause for imatinib failure. We studied pattern of kinase domain mutations in 40 patients of CML who either lost their response or did not achieve it in defined timepoints. Loss of molecular response was the most common indication for asking mutation analysis. Sixteen patients were found to have detectable mutations. M351T was the most common tyrosine kinase mutation followed by Y253H and H396R. Two patients had 2 mutations simultaneously. M351T is the most common mutation in our patient population.
Keywords: Tyrosine kinase inhibitors, Imatinib, Kinase domain mutations, Molecular response
Introduction
Chronic myeloid leukemia is the most common chronic leukemia in India [1]. The pathogenetic abnormality in CML is a well characterized reciprocal translocation between chromosome 9 and 22, t(9:22)(q34;q11), giving rise to shortened chromosome 22 known as Philadelphia chromosome. During this translocation, abelson gene on chromosome 9 comes adjacent to breakpoint cluster region on chromosome 22. This results in BCR–ABL fusion gene which encodes for a 210 kd BCR–ABL oncoprotein which has tyrosine kinase activity responsible for continued signals for proliferation and survival leading to CML [2].
Imatinib prevents binding of ATP molecule on the BCR–ABL protein in a competitive manner. This results in inhibition of substrate phosphorylation and interruption in BCR–ABL signaling. Various downstream signaling pathways like PI3K–AKT, RAS-GDP and STAT are inhibited. Imatinib also shows affinity for platelet derived growth factor and c kit tyrosine kinases [3].
Tyrosine kinase inhibitor, imatinib was approved by US Food and Drug administration for CML in 2002 as a molecularly targeted treatment [4]. This drug changed the landscape of treatment in CML. Survival of CML patients improved drastically with median survival of 8–9 years in chronic phase (CP) and 4.9 years in Accelerated Phase (AP) [5]. This lead to reconsidering the role of stem cell transplantation in management of CML. However the initial enthusiasm is waning with recognition of resistance as well as intolerance to imatinib. Studies in CML patients on imatinib show that approximately 33% of patients with CML treated with imatinib do not achieve a complete cytogenetic response (CCyR), while others have drug resistance or cannot tolerate drug-related toxicities [6]. Six year follow up data of IRIS trial showed that 14% patients discontinued imatinib due to unsatisfactory therapeutic effect [7].
A significant majority of patients have progressive disease from the beginning of therapy or later on in due course on imatinib suggesting resistance. Most common modes of resistance are overexpression of BCR–ABL and tyrosine kinase domain mutations. Almost more than 100 mutations have been reported in literature. These mutations are variably sensitive to other first line TKI’s dasatinib or nilotinib [8]. Mutation analysis thus helps in deciding further treatment in CML. In case of overexpression of BCR–ABL higher doses of imatinib 600–800 may be effective. Mutation profile has been reported from different part of the world [9]. Patterns of receptor mutations may very among different populations in the world. Few centers in India have reported the kinase domain mutation patterns [1, 12, 13]. M351T has been reported as the most common mutation from a centre in India [13]. Tyrosine kinase has various domains like P-loop, SH-2, SH-3 and A-loop domains. Type of mutation and the domain involved also has prognostic significance. Mutations occurring in P-loop and specific mutations like T351I have relatively poor prognosis. There is an urgent and unmet need to identify the kinase domain mutation in the local population with CML on imatinib. This study aims at studying BCR–ABL kinase domain mutations in our patients with CML.
Aims and Objectives
The aim of this study was to evaluate the pattern of kinase domain mutations in CML patients having inadequate response or resistance to imatinib.
Materials and Methods
Out of total 751 patients of CML, 46 (6.1%) patients having poor response or progressive disease on imatinib were selected in this observational study. Patients were followed initially every 2 weeks for 1 month and then once a month regularly and importance of compliance was reinforced at every visit. Imatinib resistance mutation analysis was done using nested RT-PCR to detect the BCR–ABL fusion transcript and ABL kinase domain region. Kinase domain mutations were evaluated using the fluorescent nucleotide sequencing technique followed by bioinformatics tool, multiple nucleotide sequencing alignment. Indications of mutation analysis were based on recent European LeukemiaNet guidelines [14]. Loss of hematological response (HR), cytogenetic response (CyR) or molecular response or delay in attaining defined landmarks were indications for asking imatinib resistance mutation analysis. Hematological response, cytogenetic and molecular responses at different time points were defined as per ELN 2013 guidelines for chronic myeloid leukemia [14]. Poor response was defined as not achieving hematological, cytogenetic or molecular response at defined time points by ELN. Progressive disease was defined as loss of achieved responses defined by ELN.
Results
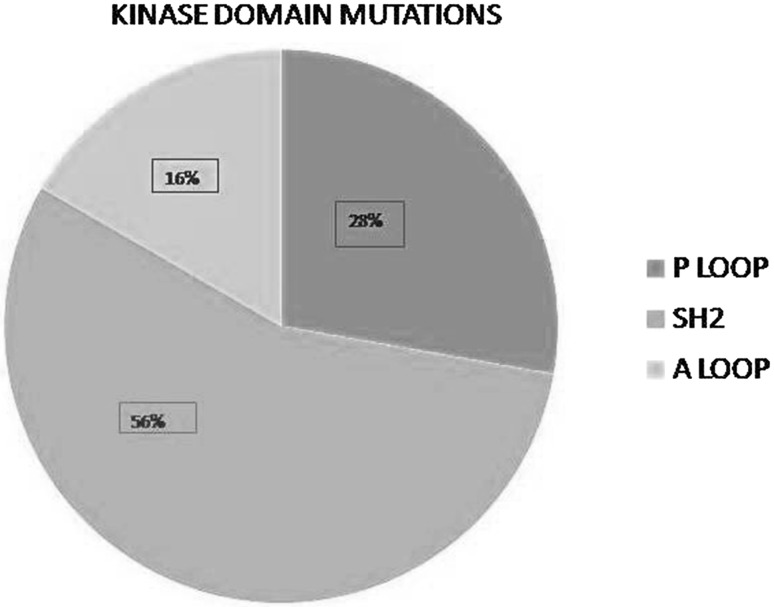
Forty-six patients with imatinib resistance with mean age of 36.8 (15–72) years were included in this study. Male to female ratio was 1.3:1. Median Sokal score was 0.88 (range 0.60–1.66). Thirty-three (71.7%) patients had intermediate or high risk Sokal score and 14 (28.2%) had low risk Sokal score. Mutation analysis was asked after a median of 4.4 (1–12) years of initiation of imatinib. Imatinib was used in standard doses of 400 mg per day. Forty-five patients were in chronic phase and one had accelerated phase at the time of mutation analysis. No patient with blast crises was included. Out of 46 patients, 29 (63%) were upfront started on imatinib and 17 (37%) received hydroxyurea initially later changed to imatinib. Mutation analysis could be done in 40 patients. Mutation analysis was asked at loss of Hematological response (HR), Cytogenetic response (CyR), and Molecular Response (MR) in 7 (17.5%), 13 (32.5%), and 20 (50%) patients respectively. Kinase domain mutations were detected in 16 (40%) patients. Two patients were having 2 detectable mutations so total detectable mutations were 18. Table 1 shows various mutations and their frequency in our patients. Out of 18 mutations detected, 5 mutations were p loop mutations, 10 SH2 mutations and 3 A loop mutations (Fig. 1). Five patients were found to have mutations (Y253H in 3 patients and F359V in 2 patients) which involve amino acids directly involved in imatinib binding. None of our patients had T315I mutation which is considered to be resistant to all first line tyrosine kinase inhibitors.
Table 1.
Showing different types of kinase domain mutations detected in our patients
| Kinase domain mutations | Number of patients (n = 16) |
|---|---|
| M244V | 1 |
| N231S | 1 |
| Y253H | 3 |
| M351T | 7 |
| F359V | 2 |
| E355G | 1 |
| H396R | 3 |
Fig. 1.
Showing different mutation sites in tyrosine kinase domain and their relative frequency in our patients
Among patients negative for mutations 9, 10, 2 and 3 patients were changed to imatinib 600 OD, nilotinib 400 bid, dasatinib 100 OD and no change respectively. Thus most patients either received higher doses of imatinib or nilotinib. Patients with kinase domain mutations were changed to nilotinib 400 BD, dasatinib 100 OD and imatinib 600 OD in 8, 5, and 3 patients respectively based on sensitivity of mutation detected and affordability.
Molecular response data at 12 month was available for 8 out of 16 patients. Two patients achieved major molecular responses (MMR), 5 patients achieved <MMR and 1 did not achieve any molecular response. Table 2 shows summary of therapy given post mutation analysis and response to same. Four deaths occurred in patients without kinase domain mutations with an overall median survival of 6.5 months (3–18 months). Four deaths occurred in patients with kinase domain mutations having median survival of 9 months (3–22 months). These patients had M351T, F359V and M351T, E355G and N231S mutations. Three patients had SH2 domain mutations and one patient had p-loop mutation. Survival of the patient with p loop mutation was only 3 months. Table 3 shows the outcome details with different mutations and different second line therapies.
Table 2.
Showing summary of therapy given post mutation analysis and response to change of TKI
| Drug | Dose | Number of patients | Number of patients with response data available at 12 months | Molecular response at 12 months |
|---|---|---|---|---|
| Nilotinib | 400 mg BD | 8 | 4 | MMR = 1 |
| <MMR = 2 | ||||
| No MR = 1 | ||||
| Dasatinib | 100 mg OD | 5 | 3 | MMR = 1 |
| <MMR = 2 | ||||
| Imatinib | 600 mg OD | 3 | 1 | <MMR |
MR molecular response, MMR major molecular response
Table 3.
Outcome details with different common mutations and different second line therapies in our patients
| Mutation | Number of patients | Second line therapy | Mean follow up (in months) after initiation of second line therapy | Molecular response at last follow up | Overall survival (%) |
|---|---|---|---|---|---|
| M351Ta | 7 | Nilotinib = 5 | <MMR-2 | ||
| Dasatinib = 1 | 15.4 (3–29) | No data-5 | 57 | ||
| Imatinib = 1 | |||||
| Y253H | 3 | Nilotinib-2 | 24.3 (8–44) | MMR-2 | 100 |
| Dasatinib-1 | <MMR-1 | ||||
| H396R | 3 | Nilotinib-1 | 24 (15–33) | <MMR-2 | 100 |
| Dasatinib-1 | No data-1 | ||||
| Imatinib-1 |
aOne patient had 2 mutations (M351T and F395V)
Discussion
Imatinib was a landmark discovery in the field of molecularly targeted therapy which changed the landscape of therapeutics in CML. Off late resistance to imatinib was detected in good number of patients leading to inadequate or no response. Tyrosine kinase activity of BCR–ABL transcript is the major driving force in CML. Imatinib acts through binding at ATP binding site and inhibiting kinase activity. Mutation in various domains (p-loop, A-loop, SH-2 and SH3 domains) of tyrosine kinase prevents adequate contact and binding of imatinib to its site on ABL. Different mutations have different IC50 values based on which tyrosine kinase inhibitors are selected against them. Table 4 shows some common mutations and their IC50 values for different TKI’S (fold increase from wild type) [9].
Table 4.
Showing some common mutations and their IC50 values and fold increase with respect to the IC50 value of wild type reported in literature [9]
| Mutation | Imatinib | Nilotinib | Dasatinib |
|---|---|---|---|
| Wild type | 1 | 1 | 1 |
| M351T | 1.8 | 0.4 | 0.9 |
| T315I | >25 | >150 | >250 |
| H396R | 3.9 | 3.1 | 1.6 |
| F359V | 7.0 | 13.5 | 2.8 |
| M244V | 7.7 | 2.9 | 1.6 |
| Y253H | >25 | 35 | 1.6 |
| E255K | 20 | 15 | 7 |
| E255V | >25 | 33.1 | 13.8 |
Our study results show that only 40% of the patients had detectable mutations in ABL kinase domain, the most common being M351T. Seven different mutations were detected in 16 patients and none of our patients was found to have T315I, which has been documented to be the most frequent mutation in world literature. M351T mutation is sensitive to higher doses of imatinib, nilotinib as well as dasatinib. It has a clinical significance as most of our patients can’t afford nilotinib and dasatinib.
Imatinib resistant mutations were analyzed in 125 Malaysian patients by Elias et al. [10]. Twenty-eight (22.4%) patients were found to have a total of 15 different kinase domain mutations, T351I being the most predominant one. This study also stressed that mutations present in different part of the BCR–ABL kinase cause variable resistance to Imatinib.
Soverini et al. [11] studied contribution of ABL kinase domain mutations in imatinib resistance in different subsets of CML patients. This study showed 43% of patients having mutations and E255V/K was the most common mutation followed by Y253H/F.
The importance of presence of these mutations in CML patients on imatinib without resistance has been studied by Branford S et al. This study found that 0% of early chronic phase and 27% of late chronic phase patients were found to have mutation even without presence of clinical resistance to imatinib. M351T mutation was the most common and patients with p loop mutation did worse [12].
Babu [13] reported mutation analysis in 101 South Indian patients of CML who did not achieve milestones or had a loss of response. Only 27 patients (27%) had mutation detected, M351T, T315I being most common in 4 patients each and F359I in 3 patients. Prakash et al. [14] reported analysis of mutations in 25 patients with CML on Imatinib at the lime of loss of response to the drug in Asian-Indian patients. Twenty-two patients were in chronic phase and median age was 40 years. The analysis revealed no mutations in 11 patients, M351T in 4 patients, G250E in 3 patients. Srivastava [15] has mentioned Indian experience of Imatinib resistance and mutations. Total of 41.53% patients were found to have mutations and 4.9% exhibited more than one mutation. T315I was the most common mutation. A trend towards preferential association of p-loop and drug binding domain mutation was found on analyzing the domain localization of the mutations.
Our data shows M351T as the most common mutation in our patients. T315I or E255V/K has been reported from rest of world as the most common mutation. Surprisingly we did not get any patient with T315I mutation. This may be partly due to less number of patients or geographical variation in mutation frequency as most of the Indian data is from south India. We suggest a study with more number of patients at different hematology centers in north India to have more robust data on mutational frequency.
Compliance with Ethical Standards
Conflict of interest
All authors declared no conflict of interest.
Ethical Approval
All procedures performed in this study were in accordance with the ethical standards of the institution and with the 1964 Helsinki’s Declaration.
Informed Consent Statement
Informed consent was obtained for all individuals participating in this study.
References
- 1.Bansal S, Prabhash K, Parikh P. Chronic myeloid leukemia data from India. Indian J Med Paediatr Oncol. 2013;34(3):154–158. doi: 10.4103/0971-5851.123711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee SJ. Chronic myelogenous leukaemia. Br J Haematol. 2000;111:993–1009. doi: 10.1046/j.1365-2141.2000.02216.x. [DOI] [PubMed] [Google Scholar]
- 3.Druker B, Lydon N. Lessons learned from the development of an Abl tyrosine kinase inhibitor for chronic myelogenous leukemia. J Clin Invest. 2000;105:3–7. doi: 10.1172/JCI9083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen MH, Williams G, Johnson JR, et al. Approval summary for imatinib mesylate capsules in the treatment of chronic myelogenous leukemia. Clin Cancer Res. 2002;8:935–942. [PubMed] [Google Scholar]
- 5.Kantarjian H, O’Brien S, Jabbour E, et al. Improved survival in chronic myeloid leukemia since the introduction of imatinib therapy: a single institution historical experience. Blood. 2012;119(9):1981–1987. doi: 10.1182/blood-2011-08-358135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kantarjian H, Hochhaus A, Saglio G, Souza C, Flinn I, Stenke L, et al. Nilotinib versus imatinib for the treatment of patients with newly diagnosed chronic phase, Philadelphia chromosome-positive, chronic myeloid leukaemia: 24-month minimum follow-up of the phase 3 randomised ENESTnd trial. Lancet Oncol. 2011;12:841–851. doi: 10.1016/S1470-2045(11)70201-7. [DOI] [PubMed] [Google Scholar]
- 7.Hochhaus A, O’Brien S, Guilhot F, Druker B, Branford S, Foroni L, et al. Six-year follow-up of patients receiving imatinib for the first-line treatment of chronic myeloid leukemia. Leukemia. 2009;23:1054–1061. doi: 10.1038/leu.2009.38. [DOI] [PubMed] [Google Scholar]
- 8.Bhamidipati PK, Kantarjian H, Cortes J, et al. Management of imatinib resistant patients with chronic myeloid leukemia. Ther Adv Hematol. 2013;4(2):103–117. doi: 10.1177/2040620712468289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soverini S, Hochhaus A, Nicolini FE, Franz G, Lange T, Saglio G, et al. BCR–ABL kinase domain mutation analysis in chronic myeloid leukemia patients treated with tyrosine kinase inhibitors: recommendations from an expert panel on behalf of European leukemiaNet. Blood. 2011;118(5):1208–1215. doi: 10.1182/blood-2010-12-326405. [DOI] [PubMed] [Google Scholar]
- 10.Elias MH, Baba AA, Azian H, et al. BCR–ABL kinase domain mutations, including 2 novel mutations in imatinib resistant Malaysian chronic myeloid leukemia patients—frequency and clinical outcome. Leuk Res. 2014;38(4):454–549. doi: 10.1016/j.leukres.2013.12.025. [DOI] [PubMed] [Google Scholar]
- 11.Soverini S, Colarossi S, Gnani A, Rosti G, Castagnetti F, Poerio A, et al. Contribution of ABL kinase domain mutations in imatinib resistance in different subsets of Philadelphia positive patients: by the GIMEMA working party on Chronic Myeloid Leukemia. Clin Cancer Res. 2006;12:7374–7379. doi: 10.1158/1078-0432.CCR-06-1516. [DOI] [PubMed] [Google Scholar]
- 12.Branford S, Rudzki Z, Walsh S, Parkinson I, Grigg A, Szer J, et al. Detection of BCR–ABL mutations in patients with CML treated with imatinib is virtually always accompanied by clinical resistance, and mutations in the ATP phosphate binding loop(p-loop) are associated with poor prognosis. Blood. 2003;102:276–283. doi: 10.1182/blood-2002-09-2896. [DOI] [PubMed] [Google Scholar]
- 13.Babu G. Report of patients with chronic myeloid leukemia Kidwai memorial institute of oncology Bangalore over 15 years. Indian J Med Paediatr Oncol. 2013;34(3):196–198. doi: 10.4103/0971-5851.123736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prabhash K, Vora T, Chadyalpatil NS, et al. Patterns of imatinib resistance mutation analysis in chronic myeloid leukemia patients on imatinib at the time of loss of response to the drug in Asian Indian subjects. J Clin Oncol. 2009;27:7079. [Google Scholar]
- 15.Srivastava S, Dutta S. Imatinib mesylate resistance and mutations: an Indian experience. Indian J Med Paediatr Oncol. 2013;34(3):213–220. doi: 10.4103/0971-5851.123748. [DOI] [PMC free article] [PubMed] [Google Scholar]