Abstract
Aspergillus is a genus of ubiquitous fungi that are pathologically & therapeutically important. Aspergillus Secondary Metabolites Database (A2MDB) is a curated compendium of information on Aspergillus & its secondary metabolome. A2MDB catalogs 807 unique non-redundantsecondary metabolites derived from 675 Aspergillus species. A2MDB has a compilation of 100 cellular targets of secondary metabolites, 44 secondary metabolic pathways, 150 electron and light microscopy images of various Aspergillus species. A phylogenetic representation of over 2500 strains has been provided. A2MDB presents a detailed chemical information of secondary metabolites and their mycotoxins. Molecular docking models of metabolite-target protein interactions have been put together. A2MDB also has epidemiological data representing Aspergillosis and global occurrence of Aspergillus species. Furthermore a novel classification of Aspergillosis along with 370 case reports with images, were made available. For each metabolite catalogued, external links to related databases have been provided. All this data is available on A2MDB, launched through Indian Institute of Chemical Technology, Hyderabad, India, as an open resource http://www.iictindia.org/A2MDB. We believe A2MDB is of practical relevance to the scientific community that is in pursuit of novel therapeutics.
Introduction
The initial description and nomenclature of Aspergillus was credited to Micheli (1729), Haller (1768) and Fries (1832) who proposed a generic name for a group of fungi sharing similar morphological characteristics1. Microscopic examination reveals a spore bearing structure conidiophore, having aspergillum-like morphology. Aspergillus species thrive as endophytes2, saprophytes3, parasites4 and also as human pathogens5–7. Aspergillus species cause localized and systemic human conditions termed as Aspergilloses8, 9. They are also responsible for diseases in agricultural crops10 and domestic animals10.
Fungi are categorized and classified based on morphology and molecular genetics. Morphological characterization of various Aspergillus spp. requires a catalog of microscopic images and genetic examination of clinical strains needs the sequence information of ITS regions. Aspergillus species although ubiquitous are more frequently observed in immuno-compromised individuals upon inhalation of conidia. Most usual complications are lung and cutaneous infections. Over the last decade there has been an increase in Aspergillosis reports, an emerging infectious disease that can be fatal. There is a need for better understanding of Aspergillosis and a resource containing a collection of case reports and a fresh classification is very much necessary, as information is scattered all over the literature.
Aspergillus spp. produces a wide range of structurally heterogeneous secondary metabolites. that are of considerable interest to the scientific research community. Fungi of this genus produce important secondary metabolites that have industrial importance11, 12 and therapeutic significance like antibiotics13 and lovastatins14. As numerous natural products are being identified each day, a plethora of compounds still await discovery and a database can act as platform for their collection and annotation. Due to the need for Aspergillus centric metabolome repository, we have developed an open, user friendly resource; A2MDB that has experimental metabolomic data, catalogued and annotated with literature information. A2MDB provides an easy access to unbiased, comprehensive information about Aspergillosis, Aspergillus species, their secondary metabolites and cellular targets, molecular docking of metabolites–target interaction, secondary metabolic pathways, ITS based phylogeny and microscopic morphology. A2MDB also provides latest classification of Aspergillosis and collection of 370 case reports with over 70 reported variants of Aspergillosis. In the future more number of species, metabolite and molecular target data along with Aspergiloosis case reports will be included as and when additional information becomes available.
A2MDB Database Development
Data Mining
Articles containing the search term Aspergillus (42189 articles as on 12/08/2016) and secondary metabolites (13770 articles as on 12/08/2016) were screened for cataloguing secondary metabolites from genus Aspergillus and this was collected with NCBI taxonomy ID and Mycobank ID.
Microscopy images of Aspergillus species, Aspergillosis case reports and Secondary metabolite biosynthetic pathways were searched and collected from PubMed.
ITS sequences of 7715 different Aspergillus species were collected from NCBI database for the primary analysis, out of which 2580 non-redundant sequences were from unique strains and species. The criterion for selection of a sequence was the availability of complete “18S-ITS1–5.8S-ITS2-28S” sequence. The non-redundant data was considered for the phylogenetic analysis.
Multiple sequence alignment was performed using MAFFT 715 and tree was constructed using UPGMA method. “iTOL” web interface was used for tree viewing and editing purpose16.
All secondary metabolites archived in A2MDB have been linked to Public chemical compound databases17, PubChem18, ChemSpider19, TOXNET20, ChEBI21 and Chemical abstracts22. Classification of the 805 secondary metabolites has been done according to IUPAC nomenclature established and verified using NCBI-MeSH23.
Tertiary structures of proteins described as cellular targets for the secondary metabolites were retrieved from Protein Data Bank (PDB). Protein Structures were optimized using Discovery Studio 4.5 Client24. The structures of secondary metabolites were downloaded from PubChem and optimized in Autodock25 and docked using AutoDock Vina26. The probable 3D models were generated using PyMOL27.
A2MDB database technology: A2MDB database runs on MicroSoft.NET technology. ASP.NET and C#.net technologies have been used to build the dynamic web interface. C#, a server side scripting language, provides interface and assists in fetching data. ASP.NET Web pages function as HTML pages at run time. JavaScript was applied to ASP web pages for generating faster output with less stress on the server. A2MDB uses custom-designed lookup tables that ensure rapid responses to search queries. The relational architecture of A2MDB ensures data integrity and expandability, scope of the database. SQL server 2008 was used to facilitate back-end database support for storing the data and Asp.net as front end is used for fetching the data. Database has been provided a refined customized search functionality and search capability especially for the Aspergillus species, metabolites and docking images.
Epidemiology: In order to visualize the global distribution of Aspergillus along with the Aspergillosis disease location, data mining was done from PubMed and based on the literature a global map was created using a R-package “rworldmap” (version 1.3–6)28 and spotting of the regions were done in Adobe Illustrator CC 2015, to showcase the incidence of Aspergillus spp. and Aspergillosis.
Results and Discussion
A2MDB is a database that is organized around the central entity, Aspergillus genus focusing primarily on its Secondary Metabolites and their biological interactions with the basic goal of understanding metabolic pathways in Aspergillus spp. and Aspergillosis (Fig. 1). A2MDB is a one of a kind resource that provides access to unique secondary metabolites produced by Aspergillus species. A2MDB is an efficient, non-redundant, user-friendly resource for viewing, sorting and extracting information. Each set of data is connected to every other set of data so that every possible aspect related to species, metabolite, metabolic pathways and cellular targets available so far, is brought together and can be downloaded.
Figure 1.
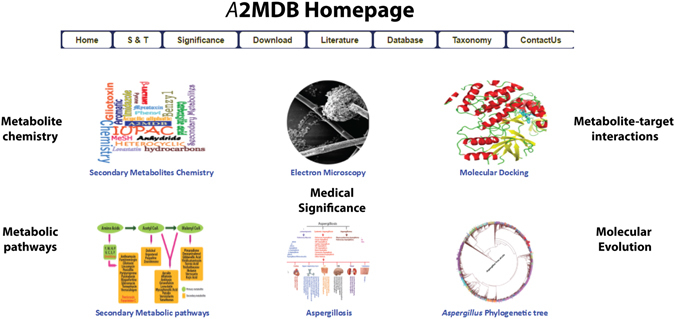
A screenshot of A2MDB showing multiple features in the homepage. The figure shows how clicking on various icons can navigate to the required option. Main page has icons for secondary metabolites chemistry, biological targets, Secondary metabolic pathways, microscopy images, molecular docking, Aspergillosis and phylogenetic analysis, apart from buttons which when clicked, explain science and significance of the project, database pages where one can surf and download data, taxonomy and contact us pages. The database has been officially launched through CSIR-IICT, India webpages.
A2MDB data retrieval, quality check and annotation
A2MDB is a curated secondary database to showcase metabolome of a group of fungi belonging to the genus Aspergillus. Information contained in it is, data related to Aspergillus ITS sequences, metabolites and their targets, electron microscopic images, information on Aspergillosis are based purely on text mining from PubMed and other authentic primary databases17, 18, 29.
Data entry in A2MDB follows a set protocol that enables (1) Identification of a new of species/strain with NCBI taxonomy ID, search and identification of any secondary metabolites with their chemical identifiers, (2) Collection of its genomic ITS data (3) search and identification of a metabolic target (4) metabolite-target interaction modeling (5) Search for a pathway (6) Search for microscopy information and further Aspergillosis components. In the Data curation of A2MDB, all the data has been verified by a curator besides the contributor and an additional scientist.
The quality of the data coming from a publication has already been peer reviewed and hence without bias full reference of the publication has been given for each and every piece of gathered data that has been provided. For quality check during curation of the data, \we generally follow a three step strategy.
Data collection: - The text mining is done only in the authenticated primary database and those data having valid reference are collected for entry in the A2MDB after being verified by 2 qualified scientists.
Pre-entry review: - In this step we follow the simple duplicate data entry validation system. For this purpose two independent files are produced for each dataset and these are compared to check for any discrepancy by two different scientists. After removing discrepancy and redundancy a final dataset is generated for entry to the database.
Post-entry review: - After the final tables were prepared, we check for any missing data and then a random sample of data is audited against the entered data to find manual errors.
The newer data coming in either by our group or externally will be entered after the three step cycle.
The data retrieval from primary databases although performed manually, was carried out with a systematic approach that dramatically reduced the work load in manual reviewing. We used the boolean and field operators in PubMed search30 to make our raw dataset as accurate as possible. Screening or pre-processing of the literature dataset was based on standard text mining procedure and after the data was collected it was annotated by scientists and uploaded and verified before making the database public.
Searching and browsing A2MDB
A2MDB web application allows users to search or browse for Aspergillus Species resources by either specifying a search string or by choosing different optional filters to species name, metabolites, biological function, cellular target, PMID and Tax ID links. Search results are usually displayed as a tabular format and some browsable elements as PDF format. All the data available through A2MDB is downloadable and the content is available as tables as well as PDF files. A2MDB application also allows users to view all results on the same page at one time. The table consists of columns showing, for each resource, its species name, metabolites, biological function, cellular target, PMID and NCBI Taxonomy ID. In addition to providing comprehensive data, each metabolite and cellular target also contains hyperlinks to other authentic databases (PubChem, PubMed), references, digital images and applets for viewing directly from the primary database. Docking images are shown as image objects that show details of docking upon clicking the object.
Metabolite information and classification
A2MDB has a collection of about 675 Aspergillus species, linked to a Taxonomy ID in NCBI database. 581 species have been annotated with links to Mycobank. 807 unique metabolites isolated from 324 species of this genus are incorporated so far with the objective of providing complete chemical and biological target related information. Nearly 25 species were identified for their variety of secondary metabolies produced. In an attempt to give complete chemical information, 523 of these secondary metabolites were connected to chemical databases like PubChem, ChemSpider, ChEBI, TOXNET. Among the secondary metabolites 213 were isolates from A. flavus, A. fumigatus, A. oryzae and A. niger of which 90 were from A. niger alone (Fig. 2A). This culmination among the highly enriched ones, is because these species are most studied owing to their etiological importance of some of the species.
Figure 2.
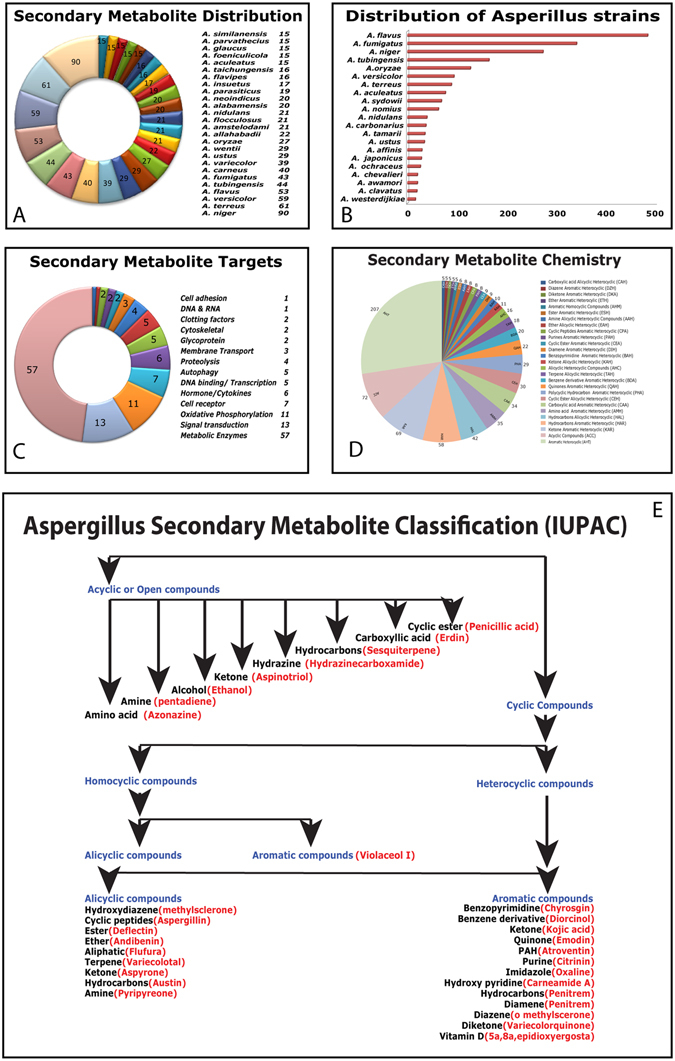
Analysis of the data collected for assimilation in to A2MDB: (A) pie chart representing the number of metabolites identified from major Aspergillus species, (B) bar graph shows the clinical distribution of biologically, medically and important strains of major Aspergillus species, (C) gene ontology based classification of cellular targets based on molecular function, (D) Distribution of secondary metabolites based on their chemical property, (E) IUPAC based classification of secondary metabolites in to various chemical compounds shown with examples under each category.
Out of the 807 metabolites identified, majority of the secondary metabolites have been identified from around 25 Aspergillus species (Fig. 2A). 60 different Mycotoxins were identified (Supplementary Table 1) from 35 species of Aspergillus that pose a considerable amount of threat to veterinary and human health31. Aflatoxins produced by Aspergillus flavus and Aspergillus parasiticus have been known to be carcinogenic and hepatotoxic in nature. Aflatoxin B1 the most toxic mycotoxin can penetrate through the skin32. Ochratoxin, contaminant of cereals, is primarily produced by Aspergillus ochraceus 33 causes liver damage, enterititis, immunosuppression, teratogenesis, nephrotoxicity and renal tumors31 Citrinin is a polyketide mycotoxin, produced by Aspergillus candidus and Aspergillus carneus shows nephrotoxic, hepatotoxic and cyototoxic behaviour. Another lethal mycotoxin sterigmatocystin is mainly found in dairy products, is a possible carcinogen mainly produced by Aspergillus nidulans and Aspergillus versicolor. Patulin is a mycotoxin produced by Aspergillus giganteus usually associated with spoilage of apple and grapes. causes cerebral hemorrhage. Within each species, several biologically important strains; were reported that are either pathogenic, or that are industrially or biologically important (Fig. 2B). A vast majority of strains that are clinically, biolocically and industrially important were identified to be from 18 species (Fig. 2B) which not only shows the infectious nature of these species in producing toxins but also of their industrial importance by the nature of the important secondary metabolites produced. We observed that majority of these secondary metabolites had been found to be targeting metabolic enzymes in order to manipulate cellular machinery (Fig. 2C). These metabolites have been linked to a unique registry number provided by Chemical Abstracts Service (CAS) of the American Chemical Society22 and classified into categories based on IUPAC nomenclature (Fig. 2D,E). Whole genome sequencing of Aspergillus species has pointed out to a varying diversity of the enzymes involved in the secondary metabolism and a range of novel compounds remain elusive and uncharacterized34. Analysis is ongoing to identify secondary metabolism gene clusters in over 250 species of Aspergillus.
Secondary metabolic pathways
Primary metabolism is well studied in fungi and well documented in many databases and secondary metabolites and metabolic pathways in fungi are underrepresented in databases. Precursors derived from the primary metabolic pathways are siphoned into secondary metabolic pathways to synthesize compounds that have unusual structures and biological properties (Figs 2D and 3A). There is an increasing need to understand secondary metabolism to exploit these organisms, and control the production of potential drugs and toxins. In A2MDB we provide 44 secondary metabolic pathway illustrations directly taken from the literature with references. 133 biological targets for 135 of these metabolites have been identified from the literature so far. We have also furnished external links to 1578 metabolic pathways from KEGG database that are Aspergillus specific (1447 primary and 131 secondary metabolic pathways). We have also provided information about anti-fungal compounds and their bioactivity on Aspergillus species, with relevant information from ChEMBL35.
Figure 3.
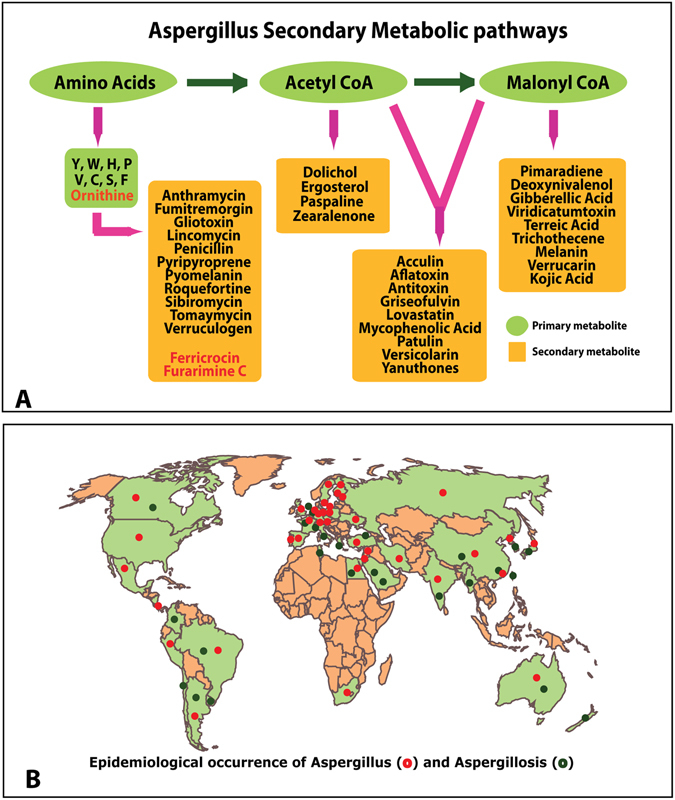
(A) A representation of metabolic pathways where in the primary metabolic intermediates serve as precursors for secondary metabolite production in Aspergillus, (B) A qualitative epidemiological distribution map was created using a R-package “rworldmap” (version 1.3–6) to showcase the incidence of Aspergillus spp. and Aspergillosis.
Epidemiology
As Aspergillus produces small hydrophobic conidia that easily disperse into air and can survive in the drastic environmental condition, distribution of Aspergillus species is found to be ubiquitous across different ecological niche3. The geographical map was provided that provides clear correlation of the occurrence of the pathogen and disease (Fig. 3B).
Molecular modelling of secondary metabolite - target interactions were provided to further validate their likely metabolite-target interactions with PDB structures based on docking energies and binding affinities (Fig. 4). Further studies are being carried out to understand their structure activity relationships.
Figure 4.
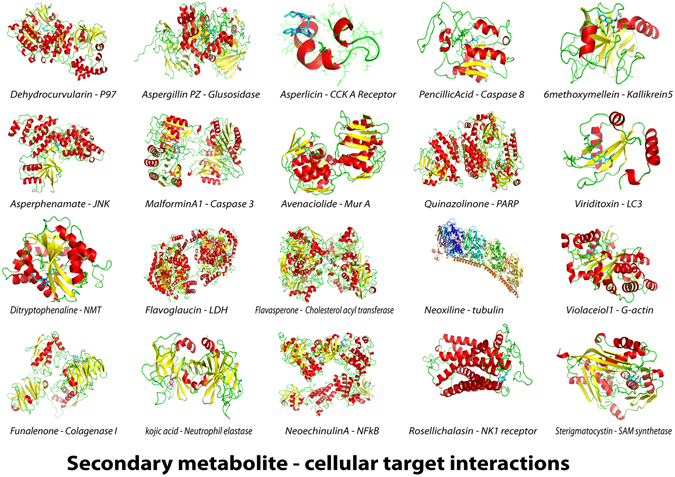
Molecular docking models representing secondary metabolite and cellular target interactions as reported in literature. Docking was performed using Auto Dock Vena. The protein structure was from PDB, metabolite structure from one of the chemical databases as explained in the text and binding energies are also represented.
Molecular and Microscopic examination
In A2MDB we provide 223 Microscopy images of Aspergillus spp.; a collection of literature derived electron and light microscopy images is provided with links to original published articles. Morphological information by means of optical and electron microscopy images as well as molecular phylogenetic information by means of ITS sequences as reported earlier was provided.
Aspergillosis classification and Case studies
Aspergillosis caused by various Aspergillus spp. has been classified broadly in to 5 different categories and over 70 different types (Fig. 5) that fall under 5 primary categories (invasive aspergillosis, pneumomycosis, systemic aspergillosis, aspergilloma and allergic aspergillosis. Aspergillosis in humans and animals has been mentioned in depth using case reports and CT scan images wherever available from 370 PubMed articles each one available for download with article identifier.
Figure 5.
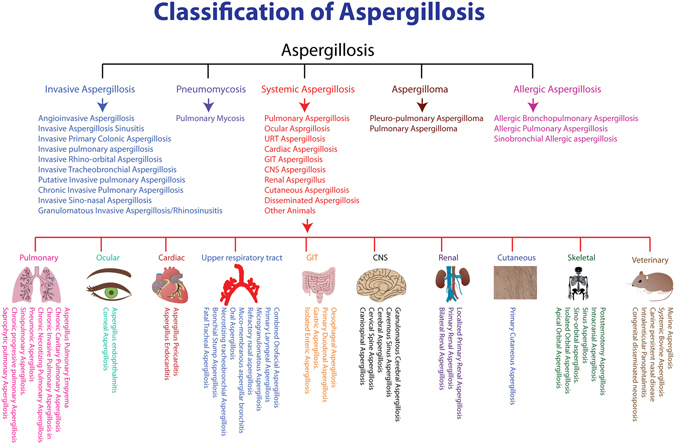
A detailed classification of Aspergillosis with case reports and related images from the literature available so far from humans and animals with DOI literature references. All the organ systems have been drawn using the software Edraw (Version 8.6) trial version.
Taxonomical information
A phylogenetic tree of life based on 2580 ITS sequences, selected from 175 unique strains and species of Aspergillus (Fig. 6) was represented. A significant number of Aspergillus species can be associated with 11 different sexual stages (teleomorphs); Emericella, Eurotium, Fennellia, Neosartorya, Petromyces, Dichotomomyces, Chaetosartorya, Cristaspora, Penicilliopsis, Sclerocleista and Phialosimplex that have now come under the umbrella of Aspergillus after the decision of “one name one fungi” by International Code of Nomenclature. U.S. Department of Energy, Joint Genome Institute (DOE-JGI) has about 270 Aspergillus species/strains/teleomorphs in their genome sequencing list, out of which nearly 90 have been sequenced and the others in either sequencing pipeline or in review as part of 1000 fungal genome project.
Figure 6.
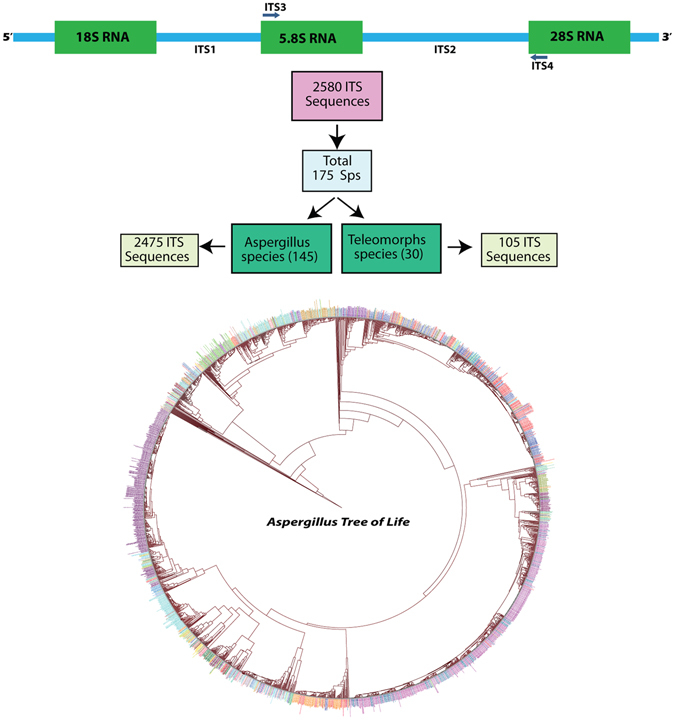
Phylogenetic tree of life for 2850 Aspergillus strains using ITS sequences. All the sequences are unique, various strains of Aspergillus belong mainly to 175 species.
Comparison with other fungal and metabolic databases
A2MDB is a non-redundant comprehensive compilation of Aspergillus specific secondary metabolism information. There are few databases that provide species specific information and even fewer on secondary metabolism and secondary metabolites of biological importance. While databases like PHI-base36, EHFPI37, catalogue genetic information to aide pathogenicity studies, ITS238, metaMicrobesOnline39, Mycobank29 provide for phylogenetic and taxonomic information. Primary metabolic pathways in eukaryotes are well annotated by databases like KEGG40, Reactome41 and HMDB42, while Metacyc, Biocyc and Humancyc are pathway genome databases43, 44. Metacyc provides a list of natural compounds but is not organism specific. AntiSMASH45 and SMURF46 are web applications that predict bacterial and fungal secondary metabolic gene clusters45. Databases like YEASTRACT47, PomBase48, NetwoRx49 are specific to Saccharomyces species, but not to secondary metabolite/metabolism information. ‘Aspergillus and Aspergillosis’ resource provides secondary metabolite information from Aspergillus species, but redundancy exists and sources remain undefined with major focus on aspergillosis50 and is currently merged with Central Aspergillus Data REpository (CADRE)51. CADRE is a repository of genomic data of Aspergillus species51. Aspergillus Genome Database (AspGD) is a genetic information resource specific to Aspergillus spp.52. However, A2MDB is a unique database that provides comprehensive secondary metabolite information related to vast number of Aspergillus species with resourceful evidence based information pertaining to Aspergillosis, Aspergillus specific anti-fungal compounds, ATCC Aspergillus collection, secondary metabolic pathways, phylogeny and other related databases. We believe that A2MDB will find a global niche with its unique content.
Future Directions
A2MDB has documented 807 secondary metabolites and almost 500 of these have chemical information from established chemical databases and we are trying to provide hyperlinks for the rest of the compounds to well established chemical and metabolite databases. Search for new metabolites is being actively done from the 351 species, that do not have prior reports on secondary metabolite isolation.
A2MDB has 370 case reports with images from 70 different types of Aspergillosis that was classified afresh. We are also trying to find information about Aspergillus causing diseases in plants.
Since some of the species are endophytes we are trying to identify and categorize Aspergillus species as endophytes and relationship with their host plants.
More species are being added along with their electron and light microscopy images as and when they become available.
Addition of transformation products of Aspergillus and derivatives of its secondary metabolites is under progress as well as their pathway annotation.
Molecular docking studies are being attempted using QSAR that might result in discovery of new therapeutic compounds.
Analysis is ongoing on identification of secondary metabolism gene clusters in all of the genome sequenced Aspergillus species.
Identification of primary metabolite that becomes the parent compound for the secondary metabolite is also being carried out.
Conclusions
A2MDB is a unique non-redundant resource of secondary metabolites and pathways information dedicated to Aspergillus species and Aspergillosis. A2MDB is regularly updated by database administrators and scientists. We welcome community participation in depositing and sharing the data. A2MDB is available for free without any registration. We believe A2MDB will be of immense importance to mycologists as well as scientists looking for important natural products obtained from Aspergillus species.
Data Accessibility
All the data being reported in this manuscript that has been collected, curated and deposited has been made publicly available directly through our database in the database download options (http://www.iictindia.org/a2mdb).
Electronic supplementary material
Acknowledgements
VVR would like to thank to Ministry of Earth Sciences, New Delhi, India (MoES/36/OOIS/Extra/18/2013) for financial support towards Research Associateship. MR would like to thank Department of Science and Technology, India for the DST Inspire fellowship (GAP-0347). SG thanks Council of Scientific & Industrial Research (CSIR), India for the support received in the form of Ph. D studentship (P811101). We thank Sri A. Radhakrishna, CSIR-IICT computer centre for technical help regarding SQL Server database management. We also thank the Director, IICT for granting us permission to launch A2MDB database through CSIR-IICT webpages.
Author Contributions
Research design: R.A., V.V., N.B.; Data mining: V.V., N.B., M.R., S.G., P.C.; Data analysis and annotation: N.B., R.A.; Molecular Docking: K.R.Y., V.V.; Phylogeny: N.B.; Database development: K.R.Y., P.R., S.P.G.; Contribution of tools: V.L.N.S., U.S.N.M.; Manuscript preparation: R.A., V.V., N.B.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Varahalarao Vadlapudi, Nabajyoti Borah and Kanaka Raju Yellusani contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-07436-w
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Suryanarayana Murty Upadhyayula, Email: director@niperguwahati.ac.in.
Ramars Amanchy, Email: ramars@iict.res.in.
References
- 1.Samson RA, et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Studies in mycology. 2014;78:141–173. doi: 10.1016/j.simyco.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.You YH, Kwak TW, Kang SM, Lee MC, Kim JG. Aspergillus clavatus Y2H0002 as a New Endophytic Fungal Strain Producing Gibberellins Isolated from Nymphoides pe ltata in Fresh Water. Mycobiology. 2015;43:87–91. doi: 10.5941/MYCO.2015.43.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dagenais TR, Keller NP. Pathogenesis of Aspergillus fumigatus in Invasive Aspergillosis. Clinical microbiology reviews. 2009;22:447–465. doi: 10.1128/CMR.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ogawa A, et al. Pseudodeflectusin, a novel isochroman derivative from Aspergillus pseudodeflectus a parasite of the sea weed, Sargassum fusiform, as a selective human cancer cytotoxin. Bioorganic & medicinal chemistry letters. 2004;14:3539–3543. doi: 10.1016/j.bmcl.2004.04.050. [DOI] [PubMed] [Google Scholar]
- 5.Hedayati MT, Pasqualotto AC, Warn PA, Bowyer P, Denning DW. Aspergillus flavus: human pathogen, allergen and mycotoxin producer. Microbiology. 2007;153:1677–1692. doi: 10.1099/mic.0.2007/007641-0. [DOI] [PubMed] [Google Scholar]
- 6.Behnsen J, et al. The opportunistic human pathogenic fungus Aspergillus fumigatus evades the host complement system. Infection and immunity. 2008;76:820–827. doi: 10.1128/IAI.01037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paulussen C, et al. Ecology of aspergillosis: insights into the pathogenic potency of Aspergillus fumigatus and some other Aspergillus species. Microbial biotechnology. 2016 doi: 10.1111/1751-7915.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sivak-Callcott JA, et al. Localised invasive sino-orbital aspergillosis: characteristic features. The British journal of ophthalmology. 2004;88:681–687. doi: 10.1136/bjo.2003.021725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aleksenko A, Gyasi R. Disseminated invasive aspergillosis. Ghana medical journal. 2006;40:69–72. doi: 10.4314/gmj.v40i2.36021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perrone G, et al. Biodiversity of Aspergillus species in some important agricultural products. Studies in mycology. 2007;59:53–66. doi: 10.3114/sim.2007.59.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoon J, Kikuma T, Maruyama J, Kitamoto K. Enhanced production of bovine chymosin by autophagy deficiency in the filamentous fungus Aspergillus oryzae. PloS one. 2013;8:e62512. doi: 10.1371/journal.pone.0062512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh BK, et al. Kojic Acid Peptide: A New Compound with Anti-Tyrosinase Potential. Annals of dermatology. 2016;28:555–561. doi: 10.5021/ad.2016.28.5.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Then Bergh K, Brakhage AA. Regulation of the Aspergillus nidulans penicillin biosynthesis gene acvA (pcbAB) by amino acids: implication for involvement of transcription factor PACC. Applied and environmental microbiology. 1998;64:843–849. doi: 10.1128/aem.64.3.843-849.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu Y, Wu P, Xue J, Wei X, Li H. Versicorin, a new lovastatin analogue from the fungus Aspergillus versicolor SC0156. Natural product research. 2015;29:1363–1368. doi: 10.1080/14786419.2015.1026342. [DOI] [PubMed] [Google Scholar]
- 15.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular biology and evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic acids research. 2011;39:W475–478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams AJ. Public chemical compound databases. Current opinion in drug discovery & development. 2008;11:393–404. [PubMed] [Google Scholar]
- 18.Kim S, et al. PubChem Substance and Compound databases. Nucleic acids research. 2016;44:D1202–1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Editorial: ChemSpider–a tool for Natural Products research. Natural product reports32, 1163–1164, doi:10.1039/c5np90022k (2015). [DOI] [PubMed]
- 20.Wexler P. TOXNET: an evolving web resource for toxicology and environmental health information. Toxicology. 2001;157:3–10. doi: 10.1016/S0300-483X(00)00337-1. [DOI] [PubMed] [Google Scholar]
- 21.Hastings J, et al. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic acids research. 2016;44:D1214–1219. doi: 10.1093/nar/gkv1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buntrock RE. Chemical registries–in the fourth decade of service. Journal of chemical information and computer sciences. 2001;41:259–263. doi: 10.1021/ci000109q. [DOI] [PubMed] [Google Scholar]
- 23.Sartor MA, et al. Metab2MeSH: annotating compounds with medical subject headings. Bioinformatics. 2012;28:1408–1410. doi: 10.1093/bioinformatics/bts156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brooks BR, et al. CHARMM: the biomolecular simulation program. Journal of computational chemistry. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forli S, et al. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nature protocols. 2016;11:905–919. doi: 10.1038/nprot.2016.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of computational chemistry. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeLano WL. Unraveling hot spots in binding interfaces: progress and challenges. Current opinion in structural biology. 2002;12:14–20. doi: 10.1016/S0959-440X(02)00283-X. [DOI] [PubMed] [Google Scholar]
- 28.R-Core-Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/ (2016).
- 29.Robert V, et al. MycoBank gearing up for new horizons. IMA fungus. 2013;4:371–379. doi: 10.5598/imafungus.2013.04.02.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ebbert JO, Dupras DM, Erwin PJ. Searching the medical literature using PubMed: a tutorial. Mayo Clinic proceedings. 2003;78:87–91. doi: 10.4065/78.1.87. [DOI] [PubMed] [Google Scholar]
- 31.Brase S, Encinas A, Keck J, Nising CF. Chemistry and biology of mycotoxins and related fungal metabolites. Chemical reviews. 2009;109:3903–3990. doi: 10.1021/cr050001f. [DOI] [PubMed] [Google Scholar]
- 32.Patten RC. Aflatoxins and disease. The American journal of tropical medicine and hygiene. 1981;30:422–425. doi: 10.4269/ajtmh.1981.30.422. [DOI] [PubMed] [Google Scholar]
- 33.Kolakowski B, O’Rourke SM, Bietlot HP, Kurz K, Aweryn B. Ochratoxin A Concentrations in a Variety of Grain-Based and Non-Grain-Based Foods on the Canadian Retail Market from 2009 to 2014. Journal of food protection. 2016;79:2143–2159. doi: 10.4315/0362-028X.JFP-16-051. [DOI] [PubMed] [Google Scholar]
- 34.Craney A, Ahmed S, Nodwell J. Towards a new science of secondary metabolism. The Journal of antibiotics. 2013;66:387–400. doi: 10.1038/ja.2013.25. [DOI] [PubMed] [Google Scholar]
- 35.Gaulton, A. et al. The ChEMBL database in 2017. Nucleic acids research, doi:10.1093/nar/gkw1074 (2016). [DOI] [PMC free article] [PubMed]
- 36.Urban M, et al. The Pathogen-Host Interactions database (PHI-base): additions and future developments. Nucleic acids research. 2015;43:D645–655. doi: 10.1093/nar/gku1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y, et al. EHFPI: a database and analysis resource of essential host factors for pathogenic infection. Nucleic acids research. 2015;43:D946–955. doi: 10.1093/nar/gku1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koetschan C, et al. The ITS2 Database III–sequences and structures for phylogeny. Nucleic acids research. 2010;38:D275–279. doi: 10.1093/nar/gkp966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chivian D, Dehal PS, Keller K, Arkin AP. MetaMicrobesOnline: phylogenomic analysis of microbial communities. Nucleic acids research. 2013;41:D648–654. doi: 10.1093/nar/gks1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanehisa M, et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic acids research. 2014;42:D199–205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fabregat A, et al. The Reactome pathway Knowledgebase. Nucleic acids research. 2016;44:D481–487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wishart DS, et al. HMDB 3.0–The Human Metabolome Database in 2013. Nucleic acids research. 2013;41:D801–807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caspi R, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic acids research. 2016;44:D471–480. doi: 10.1093/nar/gkv1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romero P, et al. Computational prediction of human metabolic pathways from the complete human genome. Genome biology. 2005;6:R2. doi: 10.1186/gb-2004-6-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Medema MH, et al. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic acids research. 2011;39:W339–346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khaldi N, et al. SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal genetics and biology: FG & B. 2010;47:736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teixeira MC, et al. The YEASTRACT database: an upgraded information system for the analysis of gene and genomic transcription regulation in Saccharomyces cerevisiae. Nucleic acids research. 2014;42:D161–166. doi: 10.1093/nar/gkt1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McDowall MD, et al. PomBase 2015: updates to the fission yeast database. Nucleic acids research. 2015;43:D656–661. doi: 10.1093/nar/gku1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fortney K, et al. NetwoRx: connecting drugs to networks and phenotypes in Saccharomyces cerevisiae. Nucleic acids research. 2013;41:D720–727. doi: 10.1093/nar/gks1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mabey Gilsenan JE, et al. Aspergillus genomes and the Aspergillus cloud. Nucleic acids research. 2009;37:D509–514. doi: 10.1093/nar/gkn876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mabey Gilsenan J, Cooley J, Bowyer P. CADRE: the Central Aspergillus Data REpository 2012. Nucleic acids research. 2012;40:D660–666. doi: 10.1093/nar/gkr971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cerqueira GC, et al. The Aspergillus Genome Database: multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic acids research. 2014;42:D705–710. doi: 10.1093/nar/gkt1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data being reported in this manuscript that has been collected, curated and deposited has been made publicly available directly through our database in the database download options (http://www.iictindia.org/a2mdb).


