Abstract
Necrotic enteritis (NE) caused by Clostridium perfringens is one of the most detrimental infectious diseases in poultry. This study examined the effect of blends of essential oils (BEOs) (25% thymol and 25% carvacrol) on NE and bacterial dynamics and functions in chicks challenged with C. perfringens. Chicks were assigned to a Control diet and BEOs diet (Control diet + 120 mg/kg BEOs), were challenged with C. perfringens from days 14 to 20 and were killed on day 21 for assessment. Supplementation with BEOs decreased the mortality, alleviated gut lesions, and decreased the virulence factors of pathogenic bacteria (VF 0073-ClpE, VF0124-LPS, and VF0350-BSH). Lack of supplementation also changed the nutrient and immunological dynamics of host microbiota in responding to C. perfringens infection. Adding BEOs changed the host ileum microbial population by increasing the numbers of Lactobacillus crispatus and Lactobacillus agilis, and decreasing Lactobacillus salivarius and Lactobacillus johnsonii. The functional roles of these changing host bacterial populations coupled with the putative reduced pathogenicity of C. perfringens by BEOs contributed to the reduction in gut lesions and mortality in infected chickens. It suggests that dietary supplementation with BEOs could significantly reduce the impact of NE caused by C. perfringens on broilers.
Introduction
Necrotic enteritis (NE) is one of the most detrimental infectious diseases in poultry. Productivity losses and treatment costs as a result of NE are estimated to total about 6 billion US dollars globally1. The etiological agent of NE is C. perfringens, a Gram-positive anaerobic spore-forming bacterium2, 3. Clostridium is a genus of Gram-positive members of the domain Bacteria 4 comprised mainly of rod-shaped, anaerobic, endospore forming, and non-sulfur-reducing bacteria. The genus Clostridium is grouped in the phylum Firmicutes, the class Clostridia, the order Clostridales and the family Clostridiaceae.
C. perfringens grows extremely rapidly, with a generation time of 8–10 min, and growth is accompanied by abundant gas production5. C. perfringens strains are classified into types A-E depending on the production of major toxins (α-, β-, ε-, and ι-toxins)6. Other studies have discovered some novel toxins produced by C. perfringens such as NetB7 and Tpel8. C. perfringens is unique not only in terms of the variety and number of toxins it produces, but also in terms of their toxicity and lethal activity. It is speculated that the pathogenesis of C. perfringens infection can be broken down into several stages including colonization of the site of disease, multiplication, acquisition of nutrients to allow further multiplication, evasion of host defenses, damages to the host, and transmission of toxins9.
In our previous studies, C. perfringens challenge increased the intestinal populations of C. perfringens and Escherichia subgroup strains10–12 and led to damage in the intestinal mucosa. The C. perfringens challenge also resulted in a significant increase in bacterial translocation10, 12 and induced strong inflammatory response in the birds10–13, which may have inhibited nutrient digestion and absorption14, subsequently suppressing chicken growth3, 10, 11, 14, 15.
The antibacterial properties of essential oils (EOs) have long been recognized and widely tested in vitro against a wide range of pathogenic bacteria, including both Gram positive and Gram negative bacteria16, 17. Essential oils have been reported to improve intestinal integrity and strengthen the mucosal barrier18, 19, improve cellular and humoral immunity20, 21, and modulate the immunity related gene expression of chickens22. Thymol and carvacrol are major components of commonly used EOs, such as thyme and oregano oils23. In rodents, thymol and carvacrol have been reported to inhibit pro-inflammatory cytokines, decrease the inflammatory cell recruitment and alleviate the oxidative damage24, 25. Our recent study suggested that the supplemental BEOs product (a mixture of thymol and carvacrol) could decrease tumor necrotic factor (TNF)-α gene expression and increase interleukin (IL)-4 gene expression in the spleen of the broilers injected with lipopolysaccharides (LPS)11, and could alleviate intestinal injury by improving intestinal integrity and modulating immune responses in the C. perfringens-challenged broiler chickens12. However, the underlying mechanisms linking dietary inclusion of essential oil mixture to immune response has yet to be elucidated.
However, in our previous study we showed that BEOs were beneficial for the Lactobacillus strains in the caecum of broilers, but had no influence on the abundance of C. perfringens in the C. perfringens-challenged broilers11. The traditional identification methods of bacteria have their limitations. However, 16 S rRNA analysis can identify the species represented in a habitat and detects those that cannot be cultivated by conventional techniques26.
Metagenomics analyses have been employed in a range of studies to assess the distribution of bacterial membership and function of gut microbes and this has proven to be a powerful tool for understanding the factors that shape microbial communities, due to both the informative and predictive potential of metagenomic data27. In this study, we used metagenomics to analyze the distribution of bacterial species and functions in the ileum microbiota of chicken fed the blends of essential oils (BEOs) (25% thymol and 25% carvacol) and challenged with C. perfringes. We also explored the mechanism of BEOs products’ effect on the pathogenicity of C. perfringes.
Results
Mortality and intestinal lesion scores of broiler chickens
The C. perfringens challenged broilers whose diet was supplemented with BEOs had almost a 5-fold reduction (4 vs 20%) in mortality compared to their control counterparts that did not receive the BEOs-supplementation (Fig. 1). The BEOs supplemented birds also had significant reduction in lesion scores (P < 0.01) compared to their control counterparts (Fig. 2).
Figure 1.

Effects of challenge and supplemented BEOs on the mortality of broiler chickens during d 14–21 (Ctrl: C. perfringens challenge; BEOs: C. perfringens challenge and supplemental blends of essential oils 120 mg/kg).
Figure 2.

Effects of challenge and supplemented BEOs on the intestinal lesion scores of broiler chickens (Ctrl: C. perfringens challenge; BEOs: C. perfringens challenge and supplemental blends of essential oils 120 mg/kg).
Variation in ileum microbiome
The number of bacterial operational taxonomic units (OTUs) and sample richness and diversity are shown in Table S1. There were no significant differences in total tags, taxon tags and OTUs between treatments. Good’s coverage index was the same (0.998) between the two treatments, which also suggested a high coverage. Birds in Ctrl and BEOs shared 216 OTUs, and there were 23 OTUs unique to the Ctrl group and 24 OTUs unique to the BEOs group (Figure S1). Alpha and beta diversity were inspected using all the metrics available in the QIIME package. There were no significant differences in the Chao 1 index and ACE between groups. However, BEOs supplementation significantly decreased the Shannon and Simpson diversity indices compare to the control (P < 0.05).
The relative abundance (%) of taxa of ileal bacteria is shown in Fig. 3. Based on the phylum level (Fig. 3A), Firmicutes is the most dominant microbiota in the control group, followed by Cyanobacteria and Proteobacteria. Dietary supplementation with BEOs increased the relative taxa abundance of Firmicutes to 93.5% (compared to 68.1% in the control group), and decreased the relative taxa abundance of Cyanobacteria and Proteobacteria (Figure S2A).
Figure 3.
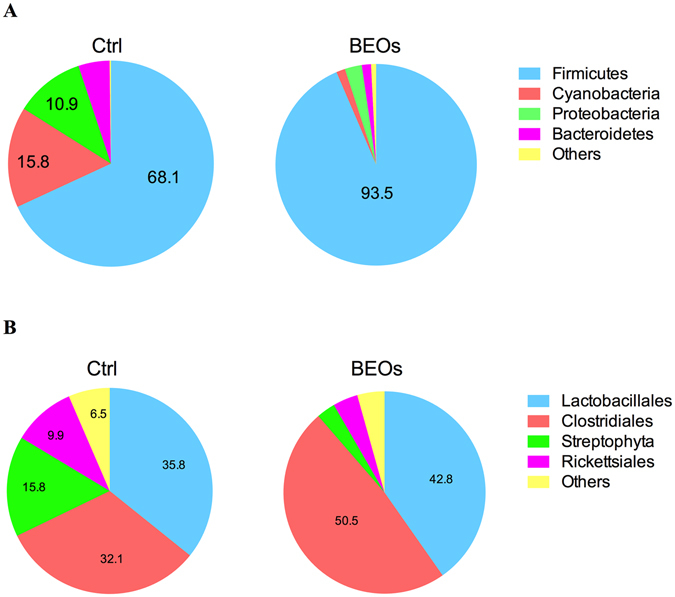
Compositions of the ileal microbiota of the broilers. Relative taxa abundance (%) of ileal bacteria of broilers at phylum level (A) and order taxonomic level (B) (Ctrl: C. perfringens challenge; BEOs: C. perfringens challenge and supplemental blends of essential oils 120 mg/kg).
Based on taxonomic order (Fig. 3B), the relative taxa abundance in the control treatment of Lactobacillales was 35.8% and of Clostridiales was 32.1%. However, supplemental BEOs increased the relative taxa abundance of Clostridiales and Lactobacillales to 50.5% and 42.8%, respectively. In addition, BEOs decreased the relative taxa abundance of Streptophyta and Rickettsiales. The relative taxa abundance (%) of ileal bacteria on the family level is shown in Figure S2B. The relative taxa abundance of ileal bacteria at genus level is shown in Figure S2C, and at the species level is shown in Fig. 4. L. salivarius, L. crispatus, and L. johnsonii were the dominant microbial strains found in the control group. BEOs supplementation significantly decreased the relative abundance of L. salivarius and L. johnsonii, and increased the abundance of L. crispatus, L. agilis and E. coli (P < 0.05).
Figure 4.
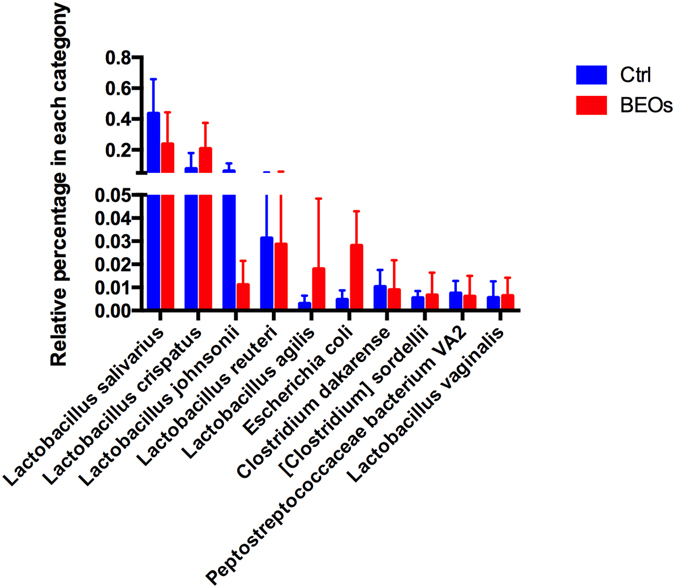
Relative abundance of different annotated species in ileal microbiota of broilers (Top 10)(Ctrl: C. perfringens challenge; BEOs: C. perfringens challenge and supplemental blends of essential oils 120 mg/kg).
According to PCA analysis, the microbial communities of the control and the BEOs groups were clustered into two groups (Fig. 5A,B). The LEfSE test detecting the differences in relative abundance of bacterial taxa across samples indicated that, at the phylum level, Cyanobacteria and Proteobacteria were significantly enriched in control samples, while the phylum Firmicutes was significantly enriched in BEOs samples (LDA >2, P < 0.05)(Fig. 6).
Figure 5.
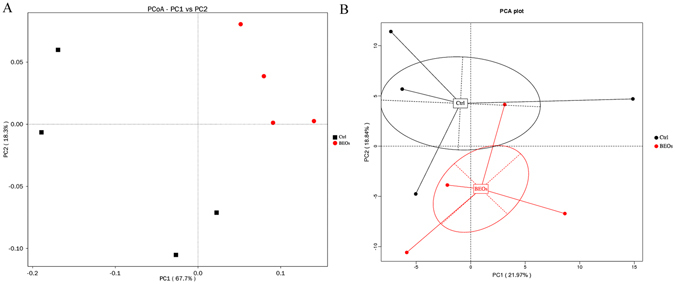
Comparison of the compositions of the ileal microbiota of the broilers. A Principal coordinate analysis (PCoA) based on the weighted unifrac diatance of 16S rRNA of ileal bacteria of broilers (A). Principal component analysis (PCA) of microbiota community by Bray-Curtis distance (B). The circles were drawn around microbiota from the same treatment.
Figure 6.
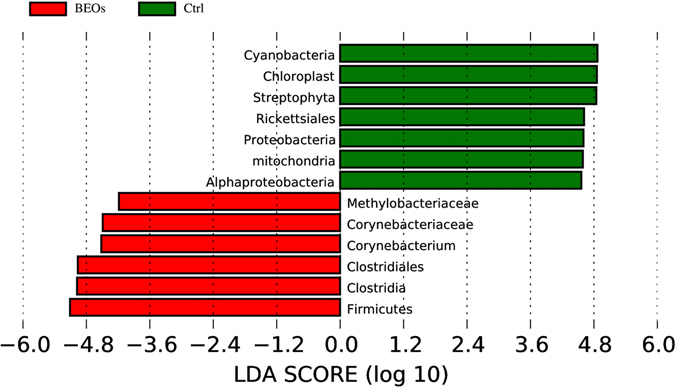
Phylum and genus differentially represented between BEOs and Ctrl samples identified by linear discriminant analysis coupled with effect size (LEfSe) (LDA >2, P < 0.05). (Ctrl: (green): C. perfringens challenge; BEOs (red): C. perfringens challenge and supplemental blends of essential oils 120 mg/kg).
Variation of metagenomes between treatments
By applying the KEGG orthologues (KO) and eggNog orthologue group markers, we assessed the potential microbial functional roles in the gut microbiota of each treatment. In general, the supplemental BEOs significantly decreased the representation of KEGG categories of 6 main processes, especially taxa involved in the KEGG of metabolism, genetic information processing and environmental information processing (Fig. 7A). The eggNOG orthologues showed that supplemental BEOs could decrease the replication, recombination and repair processes, and the translation, ribosomal structure and biogenesis related genes significantly (Fig. 7B). In the control treatment, carbohydrate metabolism and amino acid metabolism were notable. Carbohydrate transport and metabolism and amino acid transport and metabolism were significantly enriched EKGG pathways (P < 0.05) (Fig. 7B). The relative abundance of genes encoded at KEGG level 2 pathways showed that carbohydrate metabolism, amino acid metabolism, translation, and membrane transport were the most predominant activities among the microbiota (Fig. 8). The gut microbiota in the control treatment was functionally characterized and showed enrichment in biosynthesis of amino acids, including the biosynthesis of glycine and lysine. We also observed that the gut microbiota of the control group was rich in many processes related to immune activities such as immune system, cell growth and death (Fig. 8).
Figure 7.
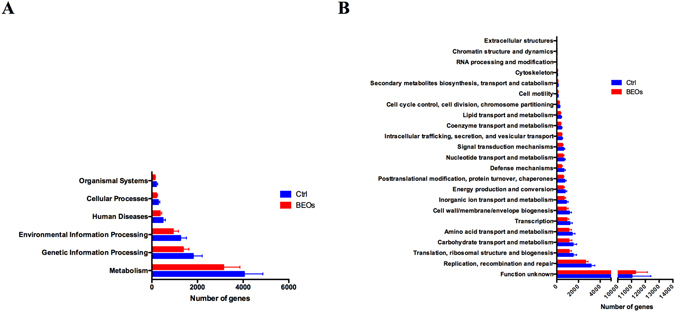
The different functions of the ileal microbiota of the broilers. Statistics of the number of annotated genes at KEGG meatabolic pathway level one (A) and eggNOG level one (B). (Ctrl: (blue): C. perfringens challenge; BEOs (red): C. perfringens challenge and supplemental BEOs 120 mg/kg).
Figure 8.
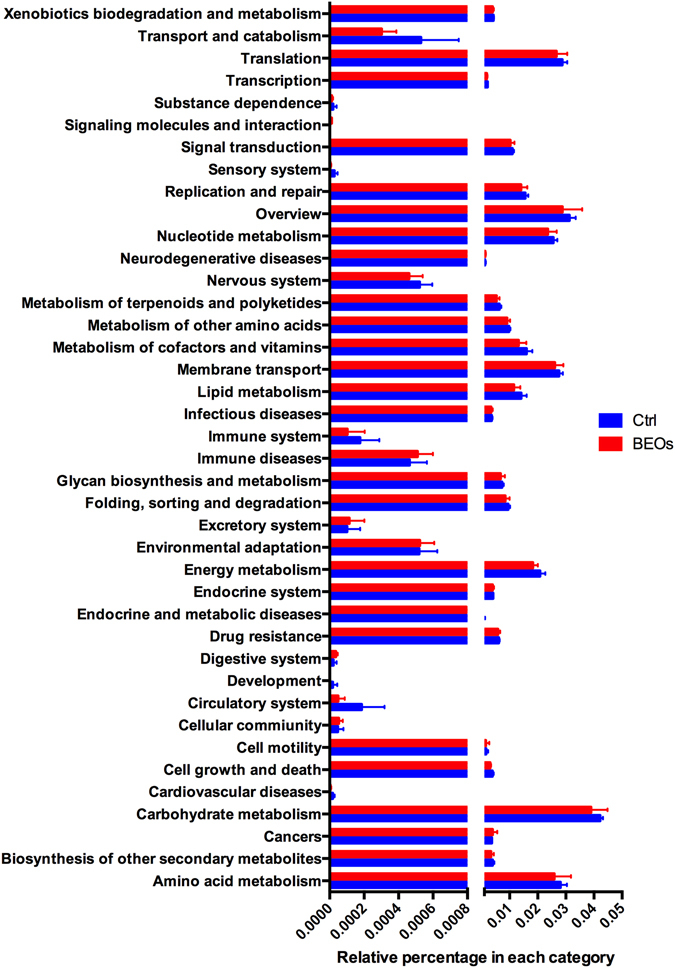
Comparison of the gene pathways of the ileum microbiota of broilers annotated genes at KEGG pathways at level two (Ctrl: (blue): C. perfringens challenge; BEOs (red): C. perfringens challenge and supplemental blends of essential oils 120 mg/kg).
The KEGG pathways analysis at level 3 showed in an enrichment of the PI3K-AKT signaling pathway in the BEOs group. To explore the variation of metagenomes in the immune system pathways, we analyzed the relative abundances of KEGG pathways in ileum microbiota annotated by KEGG pathways at level three. We found that five markers of the control-enriched KEGG orthologues were related to the immune system, including antigen processing and presentation, Nod-like receptor signaling pathway, leukocyte transendothelial migration, platelet activation and chemokine signaling pathway (Fig. 9). The control group exhibited increased relative abundance of the immune pathways compared to the BEOs group. We also analyzed the virulence factors of ileal bacteria (Fig. 10). Supplemental BEOs decreased the abundance of genes encoding factors VF 0073-ClpE, VF0124-LPS, and VF0350-BSH compared with the non-supplemented control.
Figure 9.
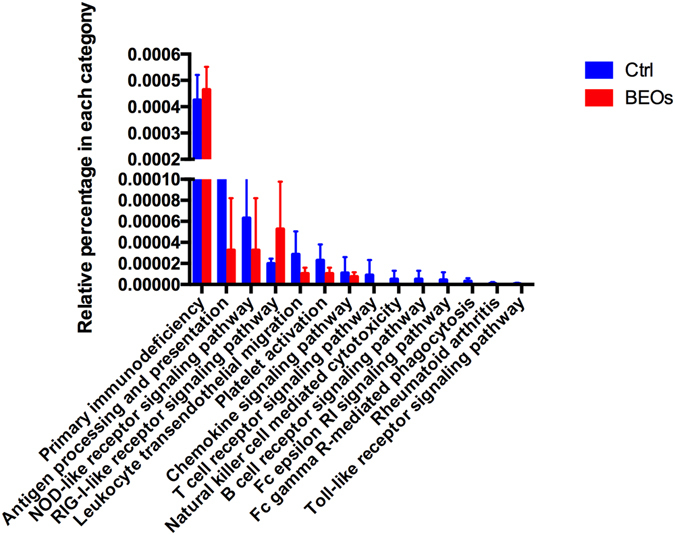
Comparison of immune system pathways in ileum microbiota annotated by KEGG pathways at level three (Ctrl: (blue): C. perfringens challenge; BEOs: (red): C. perfringens challenge and supplemental blends of essential oils 120 mg/kg).
Figure 10.
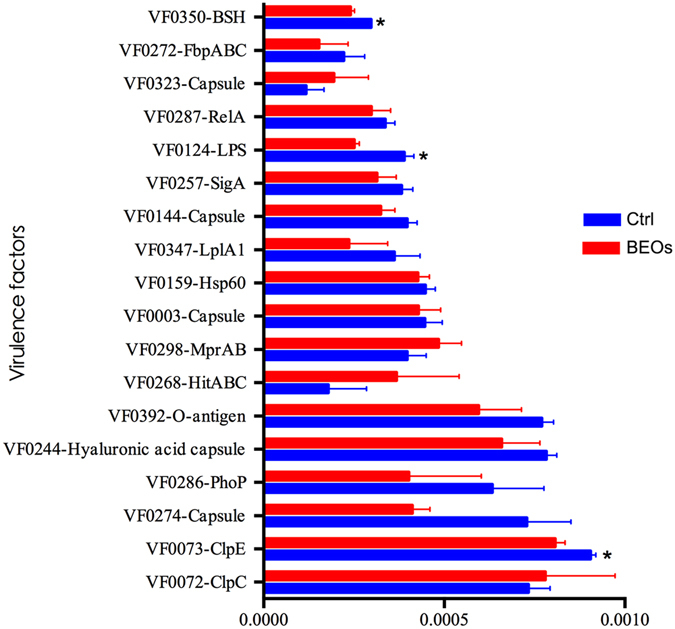
Comparison of relative abundance of annotated genes at VFDB in ileum microbiota of broilers (Ctrl: (blue): C. perfringens challenge; BEOs (red): C. perfringens challenge and supplemental blends of essential oils 120 mg/kg) * Means significantly diffference (P < 0.05).
Discussion
C. perfringens infection in poultry has been demonstrated to increase mortality and increase gut lesions. In the current study, we have shown that inclusion of thymol and carvacrol in the diet prior to C. perfringens infection can significantly reduce both mortality and gut lesions. These data are consistent with the study of Du et al.11. Possibly, the presence of thymol and carvacrol modulate the pathogenicity of C. perfringens. Bacterial virulence factors can be divided into several groups on the basis of the mechanism of virulence and function. Gram-positive bacteria are naturally surrounded by a thick cell wall that has a low permeability to the surrounding environment, while in Gram-negative bacteria the major outer membrane glycolipid, lipopolysaccharide (LPS), can protect against complement-mediated lysis. LPS activates the host complement pathway and is a potent inducer of inflammation28. The antibacterial properties of BEOs have been well recognized and widely tested in-vitro against a wide range of pathogenic bacteria, including both Gram-positive and Gram-negative bacteria11, 29. The mode of antibacterial action of BEOs consists of degradation of the cell wall, damage to cytoplasmic membrane, damage to membrane proteins, leakage of cell contents, coagulation of cytoplasm, and depletion of the proton motive force30. Qiu et al.31 showed that essential oil could inhibit pathogenicity of S. aureus to secrete a number of virulence factors. In the current study, supplementing the poultry diet with BEOs prior to C. perfringens infection led to significant reductions in VF0073-ClpE, VF0124-LPS, and VF0350-BSH compared to controls. VF 0073-ClpE is an ATP-dependent protease that plays a role posttranslational modification, protein turnover, and chaperones. VF0124-LPS is an endotoxin that functions in cell wall/membrane/envelope biogenesis. VF0350-BSH is a bile salt hydrolase that regulates cell membrane and envelope biogenesis. Thus, reduction in gut lesions due to be due to essential oil’s effect on mediating the pathogenicity of C. perfringes in the gut reducing VF 0073-ClpE, VF0124-LPS, and VF0350-BSH.
C. perfringens is model species for genetic studies because of its tolerance of oxygen, high growth rate, and ability to lend itself to genetic manipulation32. Shimizu found many virulence-associated genes in the C. perfringens genome33. Five putative haemolysin genes were identified, based on their similarity to haemolysins previously described in other bacterial species. Enterotoxins are the best understood toxins among all the C. perfringens toxins34. They interact with epithelial cell tight junction proteins, leading to diarrhea and intestinal cramping caused by leakage of water and ions35. The newly-discovered toxin NetB, which makes holes in cell membranes that cause leakage of the contents and destroy the cell, is now considered as the essential factor that initiates disease9, while Tpel can enhance the virulence of the toxin gene netB-containing strains36, 37. C. perfringens lacks genes for the biosynthesis of many amino acids33. To obtain the required nutrients, C. perfringens must secrete toxins and enzymes which act synergistically to degrade the mucus barrier in the host and then allow for rapid uptake of the nutrients into the bacterial cells, which is critical for bacterial survival in the host38. Therefore virulence and metabolism are closely linked in C. perfringens and are directed by a complex regulatory network39. It is, therefore, expected that the virulence of C. perfringes and the metabolism of the ileum microbiota would be modulated by thymol and carvacrol, reducing intestinal damage and subsequently reducing mortality. Challenging broiler chickens with C. perfringens resulted in an increase in C. perfringens in the gut and decreased the claudin-1 and occludin gene expression, disrupted the tight junctions between epithelial cells10–12, increased the mucosal sIgA levels, and enhanced the expression of TLR2 and IL-1β genes in the ileum12 thereby contributing to the damaging of the intestinal epithelium and to higher mortality10–12. We did not observe any increase in the abundance of C. Perfringens in the gut of the BEOs group, suggesting that BEOs supplementation directly or indirectly limited the growth of the pathogen. Dietary BEOs supplementation down-regulated TLR2 and TNF-α gene expressions and up-regulated mRNA expression of occluding, suggesting that the BEOs supplement had protective effects against C. perfringens due to its modulation of intestinal integrity and immunity12.
The microbiota contributes to the development and maintenance of the intestinal epithelial barrier, development of the immune system and competition with pathogenic microorganisms40. The microbiota affects the host immune system through multiple factors, which include microbial components and their metabolites. In the current study we found five of the control-enriched KEGG orthologue markers were related to immune system, including genes related to antigen processing and presentation, Nod-like receptor signaling, leukocyte transendothelial migration, platelet activation and chemokine signaling. Many pathways in NE are specifically known to have heterogenous effects when activated in different cell types. In epithelial cells, autophagy pathways play a key role in bacterial clearance, and the same autophagy genes affect the ability of cells to secrete IL-1β in host defense41. We also observed that supplemental BEOs could enrich the ATG1 gene, which can regulate autophagy. Moreover, IL-1β can act through both innate lymphoid cells and CD4 T cells to stimulate IL-17 and IL-22 secretion and induce intestinal inflammation42. Schirmer43 reported that Streptococcus was associated with changes in IL-1β. TLRs is one of several proteins that the host uses to recognize44. Many studies have shown that the commensal bacteria appear to be important in suppressing inflammatory responses and promoting immunological tolerance through TLRs45. TLR ligands stimulate DCs to express inducible iNOS. The gaseous nitric oxide produced by iNOS then induces the expression of B cell activating factor46. Interferon (IFN)- γ and TNF-α are inflammatory cytokines induced by TLRs which can act as precipitating factors for IBD by modifying tight junction function in intestinal epithelial cells and increasing epithelial barrier leakage47. NOD1 recognizes intestinal commensal and pathogenic bacteria and plays critical roles in the regulation, activation, and organization of both local and systemic innate and adaptive immune responses40. The greate relative abundance of NOD-like receptor signaling showed in the control group might suggest that C. perfringens challenge can induce the innate and adaptive immune responses. On the other hand, supplemental BEOs could relieve the immune responses caused by C. perfringens. Impaired NOD function has been implicated in a potentially distinct subtype of microbial imbalances (REF). Clearly, infection with C. perfringens in chickens affects the immune system, and allocation of resources to elicit immune response could potentially affect growth. Additionally, carbohydrate and amino acids metabolism pathways were enriched in the control group. The pathway analysis also suggests, increased nutrient metabolism of the host microbiota in the control group when infected with C. perfringes. However, dietary supplementation with BEOs could potentially modulate the C. perfringes numbers in the gut to possibly limit C. perfringens’ dependency on the host microbiota’s nutrient resources.
We found that a diversity indicator (Simpson index) was significantly higher for the control treatment than the BEOs treatment. The BEOs group also had increased relative taxa abundance of Clostridiales and Lactobacillales, which was in concordance with studies11, 12, indicating that dietary supplementation with essential oil benefits Lactobacillus. The current study found that supplementing the poultry diet with BEOs prior to C. perfringens infection led to an increased abundance of L. crispatus, L. agilis and E. coli and decreased abundance of L. salivarius and L. johnsonii. L. crispatus is a rod-shaped species of the genus Lactobacillus and is a hydrogen peroxide-producing beneficial microbial species that plays a crucial role in protecting the host from infection48. L. crispatus also induces NF-jB activation in epithelial cells and do not elicit expression of innate immunity mediators IL-8, IL-1b, IL-1a and TNF-α49. L. agilis is a facultatively heterofermentative bacteria. Genome analysis shows that this bacterium encodes several enzymes that participate in carbohydrate transport and metabolism and two enzymes (acetyl-CoA acetyltransferase and carboxylesterase type B) involved in lipid transport and metabolism50. A biosurfactant produced by a L. agilis strain exhibited considerable anti-adhesive activity against S. aureus, as well as antimicrobial activity against S. aureus, S. agalactiae and P. aeruginosa 51.
L. salivarius and L. johnsonii were the first and second most dominant microbiota in the control group. Supplemental BEOs reduced L. salivarius number, and greatly decreased the abundance of L. johnsonii. Similar to L. agilis, L. salivarius is a bacterium that has the ability to re-establish proper microbial balance by the formation of lactate and propionate, and to stimulate butyrate-producing bacteria to produce butyrate in the chicken cecum52. L. salivarius may suppress the pro-inflammatory cytokines and further suppress bacterial overgrowth in the small intestine leading to a reduction in bacterial translocation. L. salivarius reduces the interleukin (IL)-17-producing T cells [T helper 17 (Th17)] cell fraction, and increases the regulatory T cell fraction and anti-inflammatory IL-10 levels in serum53. L. johnsonii is one of the many species typically found in human and animal gastrointestinal tract as a part of the normal commensal microbiota. Pasciak et al.53 showed that the glycoconjugate extract from L. johnsonii cell wall acts as antigens and may represent new inflammatory bowel disease diagnostic biomarkers.
Conclusion
Infecting chickens with C. perfringens has been shown to increase intestinal lesions and mortality. Supplementing the diet with thymol and carvacrol prior to infection significantly reduces intestinal lesions and mortality. The essential oil appears to limit bacterial growth and modulate the pathogenicity of the bacteria in the gut. C. perfringens infection changes the nutrient metabolism of the host microbiota and elicits inflammatory responses, but addition of thymol and carvacrol to the diet changes the host ileum microbial population dynamics to increase the abundance of L. crispatus and L. agilis, and decrease L. salivarius and L. johnsonii. The functional roles of these host bacteria coupled with the possible reduced pathogenicity of C. perfringens by BEOs treatment provide clues to the underlying mechanisms by which gut lesions and mortality are significantly reduced in chickens whose diet are supplemented with BEOs prior to C. perfringes infection.
Materials and Methods
All animal work was approved by the China Agricultural University Animal Care and Use Committee (permit number SYXK20130013). All experiments used in this study were performed in accordance with protocols, approved guidelines, and regulations.
Chemicals and bacterial strain
The commercial BEOs product used contained 25% thymol and 25% carvacrol as active components, 37% silicon dioxide as a caking inhibitor, and 13% glycerides as stabilizing agents (Novus International Inc. (St Charles, MO, USA). The chicken C. perfringens field strains (CVCC2027 and CVCC2030) were obtained from the China Veterinary Culture Collection Center (Beijing, China).
Animals and diet
A total of 112 1d-old male broiler chicks (Arbor Aces) were assigned to 2 treatments supplemented with 0 and 120 mg/kg BEOs (7 replicates and 8 chickens per replicate). At 15 to 21 days of age, broilers were challenged with C. perfringens (No BEOs supplement and pathogen challenge, CTRL; supplemental BEOs and pathogen challenged, BEOs). The trial was finished at 28 d of age. The diets were formulated to meet or exceed the feeding standards of China (NY/T 2004) for broilers. The treatment diet was supplemented at 120 mg BEOs per kg of feed.
Pathogen challenge
All C. perfringens challenges were conducted as originally developed by Dahiya et al.54. The particular strain used, CVCC2030, was a type A field strain, isolated from a clinical case of NE in chickens and did not carry the NetB gene, as determined by polymerase chain reaction (PCR). Briefly, the bacterium was cultured anaerobically on tryptose-sulphite-cycloserine agar base at 37 °C for 18 h, aseptically inoculated into cooked meat medium and incubated anaerobically at 37 °C overnight. All birds were orally gavaged in the crop once per day with 1.0 mL of actively growing C. perfringens culture (1.0 × 108 cfu/mL) from days 15 to 21.
Chicken management
Chickens were reared in cages, and had access to feed and water ad libitum. The room temperature was maintained at 34 °C for the first week and then reduced by 3 °C each week until reaching 22 °C. The lighting schedule was 23 h light and 1 h dark throughout the experiment. In addition, the chickens were vaccinated against Newcastle disease virus (NDV) and Infectious Bronchitis Virus on days 7 and 21, respectively, and against bursa disease virus according to the routine immunization program.
Sampling, tissue collection and performance
Mortality and culling were recorded daily for each pen and were used for determining the mortality rate. On day 21, one bird per replicate was randomly selected and killed by administration of sodium pentobarbital (30 mg/kg body weight). Genomic DNA was isolated from 200 mg of digesta from ileum using a commercial kit (QIAamp® Fast DNA Stool Mini Kit, Qiagen Inc., Germany). Extracted DNA was stored at −20 °C until analysis.
16S rRNA gene sequencing
The microbial 16 S rRNA gene was amplified with indexed and adaptor-linked universal primers (341F: ACTCCTACGGGAGGCAGCAG, 806R:GGACTACHVGGGTWTCTAAT) targeting the V3-4 region, purified with QIAquick PCR Purification Kit (QIAGEN), and quantified by Qubit 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, USA) to pool at equal concentrations. Amplicon libraries were sequenced on Illumina HiSeq. 2500 PE250 platform (Illumina, San Diego, US) for paired-end reads of 300 bp. The paired-end reads were assembled into longer tags and quality-filtered to remove tags with length of <220 nt, average quality score of <20, and tags containing >3 ambiguous bases by PANDAseq. After discarding the singletons, the high-quality tags were clustered into operational taxonomic units (OTUs) using Uparse in QIIME software (Uparse v7.0.1001, http://drive5.com/uparse/) with a similarity threshold of 97%. The OTUs were obtained from Mothur, and were sorted from most to least abundant. Sequence abundance values within each OTU were normalized for comparisons of V3 OTU abundance between samples. The OTUs were further subjected to the taxonomy-based analysis by the RDP algorithm using the Greengenes database (http://greengenes.lbl.gov). Alpha diversity (Shannon) and beta diversity (weighted UniFrac, principal coordinate analysis (PCoA)) were analyzed using QIIME (Version 1.7.0. Linear discriminant analysis (LDA) effect size (LEfSe) analyses were performed with the LEfSe tool (http://huttenhower.sph.harvard.edu/lefse/). Standard curves for RT-PCR were prepared using DNA extracted from pure cultures to produce a high concentration of the target DNA by normal PCR amplification. Primer sequences designed on the basis of 16 S rRNA sequences were used in previous studies55–57.
Metagenomic Sequencing
DNA library construction was performed following the manufacturer’s instructions and libraries were sequenced by Illumina Hiseq. 4000. High quality reads were obtained by filtering low quality reads, adapter contamination, or DNA contamination (Gallus gallus, Triticum aestivum and Glycine max) from the Illumina raw data. We used SOAPdenovo58 and MetaGeneMark59 to perform de novo assembly and gene prediction, respectively, with the high quality reads. All predicted genes were aligned by CD-HIT (identity >95% and coverage >90%)60 to get the non-redundant gene catalogue. To obtain the relative gene abundance for each gene, the high quality reads from each sample were aligned against the gene catalogue by SOAP2 (identity >95%). We aligned the gene catalogue against the NCBI-nr database by DIAMOND61 and performed taxonomic binning by assigning genes in the NCBI taxonomy using the LCA algorithm62, 63. We aligned putative amino acid sequences from the gene catalogue against VFDB, CAZy, eggNOG and KEGG databases (release 59.0) using BLASTP (e-value ≤ 1e-5). Samples were clustered and visualized by PCA implemented in “ade4” package in R software. Functional predictions were categorized into KEGG pathways and statistical analysis was performed using STAMP v 2.0
Statistical Analysis
Differences were analyzed by the Mann-Whitney U test (GraphPad Prism, version 6.01). LEfSe analysis uses the Kruskal-Wallis rank sum test to detect significantly different abundances and performs LDA scores to estimate the effect size (threshold: ≥ 2). A P-value ≤ 0.05 with a q-value (false discovery rate) less than 0.05 in 16S rRNA gene sequence analysis and metagenomic analysis was considered significantly (q < 1, trend).
Accession codes
The raw sequences of this study have been deposited in the Sequence Read Archive (accession number: SRR5483007, SRR5483006, SRR5483005, SRR5483004, SRR5483003, SRR5483002).
Electronic supplementary material
Acknowledgements
This research was supported by the earmarked fund for China Agricultural Research Systems (CARS-42).
Author Contributions
Conceived and designed the experiments: Y.G. Performed the experiments: E.D., Y.W., J.G. Analyzed the data: D.Y. Interpretation of the data: D.Y. and S.E.A. Contributed to the writing of the manuscript: S.E.A., J.Y.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Dafei Yin, Encun Du and Jianmin Yuan contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-07420-4
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wade, B. & Keyburn, A. The true cost of necrotic enteritis. http://www.poultryworld.net/Meat/Articles/2015/10/The-true-cost-of-necrotic-enteritis-2699819W/#comments (2016) (Date of access:09/10/2015).
- 2.Van Immerseel F, et al. Clostridium perfringens in poultry: An emerging threat for animal and public health. Avian Pathol. 2004;33:537–549. doi: 10.1080/03079450400013162. [DOI] [PubMed] [Google Scholar]
- 3.Van Immerseel F, et al. Rethinking our understanding of the pathogenesis of necrotic enteritis in chickens. Trends Microbiol. 2009;17:32–36. doi: 10.1016/j.tim.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Stackebrandt, E. & Rainey, F. A. Phylogenetic relationships. In The Clostridia - molecular biology and pathogenesis (ed. Rood, J. I., McClane, B. A., Songer, G. & Titball. R. W.) Chapter 17 (1997).
- 5.Bryant, A. E. & Stevens, L. S. The Pathogenesis of Gas Gangrene. Academic Press. San Diego. 186–187 (1997).
- 6.Songer JG, Meer RR. Genotyping of Clostridium perfringens bypolymerase chain reaction is a useful adjunct to diagnosis of clostridial enteric disease in animals. Anaerobe. 1996;2:197–203. doi: 10.1006/anae.1996.0027. [DOI] [Google Scholar]
- 7.Keyburn AL, et al. NetB, a new toxin that is associated with avian necrotic enteritis caused by Clostridium perfringens. PLoS Pathog. 2008;4:e26. doi: 10.1371/journal.ppat.0040026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amimoto K, Noro T, Oishi E, Shimizu M. A novel toxin homologous to large clostridial cytotoxins found in culture supernatant of Clostridium perfringens type C. Microbiology. 2007;4:1198–1206. doi: 10.1099/mic.0.2006/002287-0. [DOI] [PubMed] [Google Scholar]
- 9.Prescott JF, Parreira VR, Mehdizadeh GI, Lepp D, Gong J. The pathogenesis of necrotic enteritis in chickens: what we know and what we need to know: a review. Avian Pathol. 2016;45:288–294. doi: 10.1080/03079457.2016.1139688. [DOI] [PubMed] [Google Scholar]
- 10.Liu D, et al. Exogenous lysozyme influences Clostridium perfringens colonization and intestinal barrier function in broiler chickens. Avian Pathol. 2010;39:17–24. doi: 10.1080/03079450903447404. [DOI] [PubMed] [Google Scholar]
- 11.Du E, et al. In vitro antibacterial activity of thymol and carvacrol and their effects on broiler chickens challenged with Clostridium perfringens. J. Anim. Sci. Biotech. 2015;6:58–71. doi: 10.1186/s40104-015-0055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du E, et al. Effects of thymol and carvacrol supplementation on intestinal integrity and immune responses of broiler chickens challenged with Clostridium perfringens. J. Anim. Sci. Biotechnol. 2016;7:19–29. doi: 10.1186/s40104-016-0079-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collier CT, et al. Coccidia-induced mucogenesis promotes the onset of necrotic enteritis by supporting Clostridium perfringens growth. Vet. Immun. Immunopathol. 2008;122:104–115. doi: 10.1016/j.vetimm.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 14.Dahiya JP, et al. Potential strategies for controlling necrotic enteritis in broiler chickens in post-antibiotic era. Anim. Feed Sci. Technol. 2006;129:60–88. doi: 10.1016/j.anifeedsci.2005.12.003. [DOI] [Google Scholar]
- 15.Guo S, et al. Inflammatory responses to a Clostridium perfringens type a strain and alpha-toxin in primary intestinal epithelial cells of chicken embryos. Avian Pathol. 2015;44:81–91. doi: 10.1080/03079457.2015.1005573. [DOI] [PubMed] [Google Scholar]
- 16.Windisch W, et al. Use of phytogenic products as feed additives for swine and poultry. J. Anim. Sci. 2008;86:E140–E148. doi: 10.2527/jas.2007-0459. [DOI] [PubMed] [Google Scholar]
- 17.Brenes A, Roura E. Essential oils in poultry nutrition: Main effects and modes of action. Anim. Feed Sci. Technol. 2010;158:1–14. doi: 10.1016/j.anifeedsci.2010.03.007. [DOI] [Google Scholar]
- 18.Placha. I, et al. Effect of thyme essential oil and selenium on intestine integrity and antioxidant status of broilers. Br. Poult. Sci. 2014;55:105–114. doi: 10.1080/00071668.2013.873772. [DOI] [PubMed] [Google Scholar]
- 19.Wlodarska M, et al. Phytonutrient diet supplementation promotes beneficial clostridia species and intestinal mucus secretion resulting in protection against enteric infection. Sci. Rep. 2015;5:9253. doi: 10.1038/srep09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee SH, et al. Effects of dietary supplementation with phytonutrients on vaccine-stimulated immunity against infection with eimeria tenella. Vet. Parasitol. 2011;181:97–105. doi: 10.1016/j.vetpar.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 21.Awaad MHH, Elmenawey M, Ahmed KA. Effect of a specific combination of carvacrol, cinnamaldehyde, and capsicum oleoresin on the growth performance, carcass quality and gut integrity of broiler chickens. Vet. World. 2014;7:284–290. doi: 10.14202/vetworld.2014.284-290. [DOI] [Google Scholar]
- 22.Kim DK, et al. High-throughput gene expression analysis of intestinal intraepithelial lymphocytes after oral feeding of carvacrol, cinnamaldehyde, or capsicum oleoresin. Poult. Sci. 2010;89:68–81. doi: 10.3382/ps.2009-00275. [DOI] [PubMed] [Google Scholar]
- 23.Bassole IH, Juliani HR. Essential oils in combination and their antimicrobial properties. Molecules. 2012;17:3989–4006. doi: 10.3390/molecules17043989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riella KR, et al. Anti-inflammatory and cicatrizing activities of thymol, a monoterpene of the essential oil from Lippia gracilis, in rodents. J. Ethnopharmacol. 2012;143:656–663. doi: 10.1016/j.jep.2012.07.028. [DOI] [PubMed] [Google Scholar]
- 25.Guimaraes AG, et al. Carvacrol attenuates mechanical hypernociception and inflammatory response. Naunyn-Schmiedebergs Arc. Pharmacol. 2012;385:253–263. doi: 10.1007/s00210-011-0715-x. [DOI] [PubMed] [Google Scholar]
- 26.Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waite DW, Taylor MW. Characterizing the avian gut microbiota: membership, driving influences, and potential function. Front Microbiol. 2014;5:223. doi: 10.3389/fmicb.2014.00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu H, Wang AH, Jennings MP. Discovery of virulence factors of pathogenic bacteria. Curr. Opin. Chem. Biol. 2008;12:1–9. doi: 10.1016/j.cbpa.2008.01.023. [DOI] [PubMed] [Google Scholar]
- 29.Si W, et al. Antimicrobial activity of essential oils and structurally related synthetic food additives towards Clostridium perfringens. J. Appl. Microbiol. 2009;106:213–220. doi: 10.1111/j.1365-2672.2008.03994.x. [DOI] [PubMed] [Google Scholar]
- 30.Burt S. Essential oils: their antibacterial properties and potential applications in foods-a review. Int. J. Food Microbiol. 2004;94:223–253. doi: 10.1016/j.ijfoodmicro.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 31.Qiu J, et al. Subinhibitory concentrations of Perilla Oil affect the expression of secreted virulence factor genes in Staphylococcus aureus. PLoS ONE. 2011;6:e16160. doi: 10.1371/journal.pone.0016160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rood JI. Virulence genes of Clostridium perfringens. Annu. Rev. Microbiol. 1998;52:333–60. doi: 10.1146/annurev.micro.52.1.333. [DOI] [PubMed] [Google Scholar]
- 33.Shimizu T, et al. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. USA. 2002;99:996–1001. doi: 10.1073/pnas.022493799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Granum PE. Clostridium perfringens toxins involved in food poisoning. Inter. J. Food Microbiol. 1990;10:101–112. doi: 10.1016/0168-1605(90)90059-E. [DOI] [PubMed] [Google Scholar]
- 35.McClane BA. The complex interactions between Clostridium perfringens enterotoxin and epithelial tight junctions. Toxicon. 2001;39:1781–1791. doi: 10.1016/S0041-0101(01)00164-7. [DOI] [PubMed] [Google Scholar]
- 36.Chalmers G, et al. Multilocus sequence typing analysis of Clostridium perfringens isolates from necrotic enteritis outbreaks in broiler chicken populations. J. Clin. Microbiol. 2008;46:3957–3964. doi: 10.1128/JCM.01548-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coursodon CF, Glock RD, Moore KL, Cooper KK, Songer JG. TpeL-producing strains of Clostridium perfringens type A are highly virulent for broiler chicks. Anaerobe. 2012;18:117–121. doi: 10.1016/j.anaerobe.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 38.Ohtani K, Shimizu T. Regulation of Toxin Production in Clostridium perfringens. Toxins. 2016;8:207. doi: 10.3390/toxins8070207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohtani K, Shimizu T. Regulation of toxin gene expression in Clostridium perfringens. Res. Microbiol. 2015;166:280–289. doi: 10.1016/j.resmic.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 40.Honda K, Littman DR. The microbiome in infectious disease and inflammation. Immunology. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lassen KG, et al. Atg16l1 t300a variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense. Proc. Nat. Acad. Sci. USA. 2014;111:7741–7747. doi: 10.1073/pnas.1407001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coccia M, et al. IL-1beta mediates chronic intestinal inflammation by promoting the accumulation of IL-17A secreting innate lymphoid cells and CD4 (+) Th17 cells. J. Exp. Med. 2012;209:1595–1609. doi: 10.1084/jem.20111453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schirmer M, et al. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell. 2016;167:1125–1136. doi: 10.1016/j.cell.2016.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Round JL, et al. The toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tezuka H, et al. Regulation of iga production by naturally occurring tnf/inos-producing dendritic cells. Nature. 2007;448:929–933. doi: 10.1038/nature06033. [DOI] [PubMed] [Google Scholar]
- 47.Turner JR. Intestinal mucosal barrier function in health and disease. Nat. Rev. Immunol. 2009;9:799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 48.Shipitsyna. E, et al. Composition of the vaginal microbiota in women of reproductive age-sensitive and specific molecular diagnosis of bacterial vaginosis is possible? PLoS ONE. 2013;8:e60670. doi: 10.1371/journal.pone.0060670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abramov V, et al. Melnikov probiotic properties of Lactobacillus crispatus 2,029: Homeostatic interaction with cervicovaginal epithelial cells and antagonistic activity to genitourinary pathogens. Probiotics Antimicro. Proteins. 2014;6:165–176. doi: 10.1007/s12602-014-9164-4. [DOI] [PubMed] [Google Scholar]
- 50.Drissi, F., Labas, N., Merhej, V. & Raoult, D. Draft Genome Sequence of the Lactobacillus agilis Strain Marseille. Genome Announc. 3, e00840–15 (2015). [DOI] [PMC free article] [PubMed]
- 51.Gudina EJ, Fernandes EC, Teixeira JA, Rodrigues LR. Antimicrobial and anti-adhesive activities of cellbound biosurfactant from Lactobacillus agilis CCUG31450. RSC Adv. 2015;5:90960–90968. doi: 10.1039/C5RA11659G. [DOI] [Google Scholar]
- 52.Meimandipour A, et al. Selected microbial groups and short-chain fatty acids profile in a simulated chicken cecum supplemented with two strains of Lactobacillus. Poult. Sci. 2010;89:470–476. doi: 10.3382/ps.2009-00495. [DOI] [PubMed] [Google Scholar]
- 53.Liu X, et al. Lactobacillus salivarius isolated from patients with rheumatoid arthritis suppresses collagen-induced arthritis and increases treg frequency in mice. J. Interferon Cytokine Res. 2016;36:706–712. doi: 10.1089/jir.2016.0057. [DOI] [PubMed] [Google Scholar]
- 54.Pasciak M, Orska SG, Jawiarczyk N, Gamian A. Lactobacillus johnsonii glycolipids, their structure and immunoreactivity with sera from inflammatory bowel disease patients. Microb. Biotechnol. 2016;0:000–000. doi: 10.1111/1751-7915.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dahiya JP, Hoehler D, Wilkie DC, Van Kessel AG, Drew MD. Dietary glycine concentration affects intestinal Clostridium perfringens and Lactobacilli populations in broiler chickens. Poult. Sci. 2005;84:1875–1885. doi: 10.1093/ps/84.12.1875. [DOI] [PubMed] [Google Scholar]
- 56.Deplancke B, et al. Selective growth of mucolytic bacteria including Clostridium perfringens in a neonatal piglet model of total parenteral nutrition. Am. J. Clin. Nutr. 2002;76:1117–1125. doi: 10.1093/ajcn/76.5.1117. [DOI] [PubMed] [Google Scholar]
- 57.Malinen E, Kassinen A, Rinttila T, Palva A. Comparison of real-time PCR with SYBR Green I or 5’-nuclease assays and dot-blot hybridization with rDNAtargeted oligonucleotide probes in quantification of selected faecal bacteria. Microbiology. 2003;149:269–277. doi: 10.1099/mic.0.25975-0. [DOI] [PubMed] [Google Scholar]
- 58.Steed H, et al. Bacterial translocation in cirrhosis is not caused by an abnormal small bowel gut microbiota. FEMS Immunol. Med. Mic. 2011;63:346–354. doi: 10.1111/j.1574-695X.2011.00857.x. [DOI] [PubMed] [Google Scholar]
- 59.Luo R, Liu B, Xie Y, Li Z, Huang W. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu W, Lomsadze A, Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010;38:e132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 63.Huson DH, Mitra S, Ruscheweyh H, Weber N, Schuster SC. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


