Abstract
RNA helicases, like their DNA-specific counterparts, can function as processive enzymes, unwinding RNA with a defined step size in a unidirectional fashion. Recombinant nuclear DEAD-box protein p68 and its close relative p72 are reported here to function in a similar fashion, though the processivity of both RNA helicases appears to be limited to only a few consecutive catalytic steps. The two proteins resemble each other also with regard to other biochemical properties. We have found that both proteins exhibit an RNA annealing in addition to their helicase activity. By using both these activities the enzymes are able in vitro to catalyse rearrangements of RNA secondary structures that otherwise are too stable to be resolved by their low processive helicase activities. RNA rearrangement proceeds via protein induced formation and subsequent resolution of RNA branch migration structures, whereby the latter step is dependent on ATP hydrolysis. The analysed DEAD-box proteins are reminiscent of certain DNA helicases, for example those found in bacteriophages T4 and T7, that catalyse homologous DNA strand exchange in cooperation with the annealing activity of specific single strand binding proteins.
INTRODUCTION
Regulated rearrangement of structured RNA in macromolecular assemblies is essential during processes such as ribosomal biogenesis or RNA splicing, transport, translation and degradation. Furthermore, the folding of large RNAs is often a multi-step process itself. It includes intermediate conformations which require protein-catalysed alterations on the way to the correct fold (reviewed in 1–4).
The resolution of double-stranded (ds) RNA is catalysed by RNA helicases in a nucleotide triphosphate-dependent manner (reviewed in 5). Most of these proteins possess a variant of a so-called helicase domain characterised by eight blocks of conserved amino acid sequences (6). One of these motifs, the ATP hydrolysis motif (Walker B motif), allows them to be grouped into distinct families such as the DEAD-, DEAH- and DExH-box families. Many DExD/H-box proteins are putative or demonstrated RNA helicases (7,8). Like DNA helicases (reviewed in 9), at least some of them seem to act in a processive and unidirectional fashion with a step size of roughly one half helix turn (10). Processivity reflects the probability that a helicase will perform the next unwinding step rather than dissociate from the substrate and individual RNA and DNA helicases show quantitative differences in this property (10,11). The exact molecular basis of processivity is unknown, but a quantitative correlation between ATP utilization and helicase processivity has been demonstrated (10).
Another feature of several RNA-binding proteins is to enhance the annealing of complementary single-stranded (ss) RNA (12). Such an RNA-folding activity, among other things, has been reported to play an important role in splicing (13,14) and translation (15,16). Some of these proteins contain a common arginine/serine-rich motif (17) though others, like the tumour suppressor p53, do not (18). Interestingly, a DExD-box protein with demonstrated RNA helicase activity, RNA helicase II/Gu, has been reported to contain an extra RNA annealing activity, the function of which is unclear (19). It is also unknown whether the two activities of the enzyme can cooperate under certain conditions, which is conceivable. Thus, it has been speculated that the dissociation of dsRNA structures is functionally coupled to the formation of new structures in the splicing of pre-mRNA, where several DExD/H-box proteins are involved (reviewed in 20,21). Furthermore, a structure mimicking a Holliday junction has been proposed to exist in the newly assembled spliceosome, which may serve to juxtapose the 5′ and 3′ splice sites and to allow essential conformational changes to occur, probably by branch migration (22). Holliday structures are well-characterized intermediates in homologous recombination of DNA molecules. In Escherichia coli, they are induced and processed by RecA, a protein with DNA-dependent ATPase and DNA annealing activity that are both essential for homologous DNA strand exchange (reviewed in 23,24). Moreover, detailed structural analyses of RecA and several DNA and RNA helicases have shown that their active centres have similar folds (25–28), indicating that all helicases may be structural homologues of RecA. Thus, the question arises whether RecA-like recombinational activities are also inherent in some RNA-specific ATPases.
The nuclear DEAD-box protein p68 was identified 20 years ago because of its immunological relations to the SV40 large tumour antigen (29,30). Biochemical studies revealed that both proteins possessed NTP-dependent RNA helicase activities, the cellular functions of which are still unclear (31,32). Recently, another human DEAD-box protein, p72, has been detected which is highly homologous to p68 (33). The two relatives (and possibly additional not yet identified ones) may form a subgroup with similar biochemical properties within the DEAD-box protein family. p68 and p72 were found in a wide range of vertebrate species (33–35 and our own observations), but only one gene, closely related to both, is encoded by the yeast Saccharomyces cerevisiae (36). p68 seems to be required for normal growth and development and its expression appears to be regulated in a complex way (34,37,38).
We report here that p68 and p72 possess RNA annealing activity in addition to a low processive RNA helicase activity. We show that coupling of both activities enables the proteins in vitro to rearrange RNA secondary structures that are otherwise too stable to be resolved by their helicases. Implications of this new activity for the cellular role(s) of the proteins will be discussed.
MATERIALS AND METHODS
Cloning procedures and preparation of recombinant proteins
pDOR6 was constructed by cloning a PCR product (nucleotides 2213–2303, amplified with a sense and an antisense primer containing additional BamHI and HindIII sites) of the E.coli deaD gene (accession number M63288) in the respective site of pGEM-7Zf(-) (Promega). A description of the pGEM derivatives C4, MO1, MO1/2 and CS is given in Scheffner et al. (32). To generate a recombinant p68-baculovirus strain, the coding sequence of the p68-cDNA (nucleotide sequence 1–1849; 39) was subcloned into the baculovirus transfer vector pAc610 (Dianova). A recombinant p72-baculovirus strain was obtained via the recombinant transfer vector pBAKPac-His-p72, which was generated by PCR amplification of the human p72-cDNA (primers located at 1–21 and 1963–1984 and containing additional BamHI and HindIII sites) and cloning of the BamHI–HindIII digested fragment into the transfer vector pBAKPac-His1 (Clontech). The nucleotide sequence of the resulting construct was verified by DNA sequencing and coded for p72 containing a histidine tag at the N-terminus. To generate recombinant baculovirus strains coding for p68 deletion mutants, respective parts of the p68-cDNA (p681–189, an AcyI–SspI fragment; p681–386, an AcyI–EcoRI fragment and p68387–614, an EcoRI–KpnI fragment) were inserted into pAcSG-His NT (Dianova) resulting in respective polypeptides with histidine tags at their N-termini. For isolation of recombinant proteins, Sf9 cells (grown at 27°C in SF900 medium containing 7.5% FCS) were infected with the respective baculoviruses at a m.o.i. of 10 and harvested 40 h post-infection. All subsequent steps were performed at 3°C. Nuclear extracts of wild-type and mutant DEAD-box proteins were prepared as described (31). p68 was immunopurified using monoclonal antibody C10 crosslinked to protein A–Sepharose (Pharmacia). Bound p68 was eluted with the oligopeptide used for immunization (20 mg/ml). The eluant was dialysed against buffer A (25 mM Tris–HCl pH 7.8, 50 mM NaCl, 20% glycerol, 1 mM EDTA, 10 mM mercaptoethanol, 0.1% Triton X-100) and p68 was stored in aliquots at –70°C. p72 was purified from nuclear extracts by affinity chromatography on Ni2+ NTA–cellulose (Qiagen) eluting at 200 mM imidazol and further purified by heparin–Sepharose (1 ml HiTrap column; Pharmacia) and ssDNA–cellulose affinity chromatography, dialysed against buffer A and stored at –70°C. Purified p72 as well as p68 sedimented as a homogeneous peak at about 4–5 S in a sucrose gradient (31). Mutant p68 polypeptides (p681–386, p681–189, p68387–614) were purified from insect cell extracts infected with the respective baculoviruses by affinity chromatography on Ni2+ NTA–cellulose and obtained polypeptides were further purified by ssDNA–cellulose column chromatography. Mutant proteins were recoverd at 200–300 mM NaCl, dialysed against buffer A and stored at –70°C.
RNA
RNA was produced by run-off transcription of the respective linearised pGEM3- (Promega) and pGEM-7Zf(-) derivatives as recommended by the supplier. Where indicated, it was uniformly labelled with [α-32P]CTP (specific activity: 2 × 104 d.p.m./pmol). Partially dsRNAs were prepared by hybridisation of the respective complementary transcripts at a 3-fold excess of unlabelled over labelled strands in 80% formamide, 400 mM NaCl, 1 mM EDTA and 40 mM PIPES pH 6.4 (Table 1). For hybridisation, RNA mixtures were heated at 80°C for 15 min and further incubated at 50°C for 3 h. Hybrid RNAs were isolated after electrophoretic separation in non-denaturing polyacrylamide gels (in 0.5× TBE) by elution from respective crushed gel slices in 300 mM sodium acetate, 5 mM EDTA pH 5.3 at room temperature. RNA concentrations are expressed in terms of moles of nucleotides. The nucleotide sequences of all RNAs used were of random choice and were not found to have an influence on p68 or p72 activities.
Table 1. Transcripts used for preparation of hybrid RNAs.
| Hybrid RNA |
Strand |
Strand length (nt) |
Polymerasea |
Vector |
Digested with |
| 17 bp RNA |
I |
194 |
SP6 |
pGEM-C4 |
PvuII |
| |
IIb |
130 |
T7 |
pGEM-MO1 |
HindIII |
| 25 bp RNA |
I |
103 |
T7 |
pGEM-Cs– |
RsaI |
| |
IIb |
107 |
SP6 |
pGEM-Cs– |
HaeII |
| 41 bp RNA |
Ib |
130 |
T7 |
pGEM-MO1 |
HindIII |
| |
II |
85 |
SP6 |
pGEM-MO1/2 |
EcoRI |
| 5′-RNA |
Ib |
89 |
T7 |
pGEM-MO1/2 |
HindIII |
| |
II |
35 |
SP6 |
pGEM-MO1/2 |
XbaI |
| 51 bp RNA |
Ib |
130 |
T7 |
pGEM-MO1 |
HindIII |
| |
II |
127 |
SP6 |
pGEM-MO1/2 |
PvuII |
| 44 bp RNA |
Ib |
314 |
T7 |
pGEM-3 |
NheI |
| |
II |
84 |
SP6 |
pGEM-MO1/2 |
EcoRI |
| 72 bp RNA |
Ib |
314 |
T7 |
pGEM-3 |
NheI |
| |
II |
158 |
SP6 |
pGEM-3 |
HaeII |
| 76 bp RNA |
I |
195 |
T7 |
pDOR6 |
BamHI |
| IIb | 233 | SP6 | pGEM-7Zf(-) | HaeII | |
| 123 bp RNA | I | 323 | T7 | pGEM-7Zf(-) | HaeII |
| IIb | 233 | SP6 | pGEM-7Zf(-) | HaeII |
aThe two RNA strands of each hybrid were transcribed with SP6 or T7 polymerase from indicated DNA vectors, digested with given restriction enzymes.
bThese strands were synthesised in the presence of [α-32P]CTP.
Assays
RNA-binding activity was determined by the retention of 32P-RNA (synthesized by run-off transcription of the PvuII-restricted plasmid pGEM-C4 with SP6 polymerase) (Table 1) on nitrocellulose filters as previously described (31). Reaction mixtures (40 µl) contained indicated amounts of protein, 25 nM RNA, 80 mM NaCl, 1 mM EDTA, 1.5 mM DTT and 40 mM Tris–HCl pH 7.5 and were incubated for 30 min on ice.
RNA helicase activity was assayed as decribed previously (32). In brief, reactions were performed in 40 µl volumes containing 12.5 nM RNA, 0–100 mM NaCl, 5 mM MgCl2, 4 mM ATP, 1.5 mM DTT, 30 mM Tris–HCl pH 7.5, 30 µg/ml RNase-free BSA and 0.5 U/µl RNAsin. After incubation at 37°C for indicated times, reactions were stopped by addition of 0.1 vol of 3% SDS, 150 mM EDTA and analysed by SDS–PAGE and autoradiography. Assay conditions for RNA–RNA annealing activity and analysis of reaction products were exactly as descibed for RNA unwinding, except that Mg2+-ATP was not essential and was omitted. The partially complementary RNA strands were used at a 1:3 molar ratio with 12.5 nM of the labelled strand (130 nt in length) and 36.6 nM of the unlabelled strand (127 nt in length). Strand exchange reaction conditions also corresponded to those of RNA helicase reactions. A 1:3 ratio (with respect to RNA molecules) of dsRNA to homologous third strand was used in all assays. For strand exchange reactions with the 17 bp RNA as the substrate, the third strand used was obtained by transcription of pGEM-MO1/2 (digested with PvuII) with SP6 polymerase, for strand exchange reactions with the 44 bp RNA substrate, the third strand used was obtained by transcription of pGEM-3 (digested with HaeII) with SP6 polymerase and for strand exchange reactions with the 76 bp RNA substrate, the third strand used was obtained by transcription of pGEM-7Zf(-) (digested with HaeII) with T7 polymerase. Autoradiographic signals of labelled products of the RNA helicase and annealing reactions were quantified with a Molecular Dynamics densitometer.
RESULTS
The p68 subfamily of RNA helicases
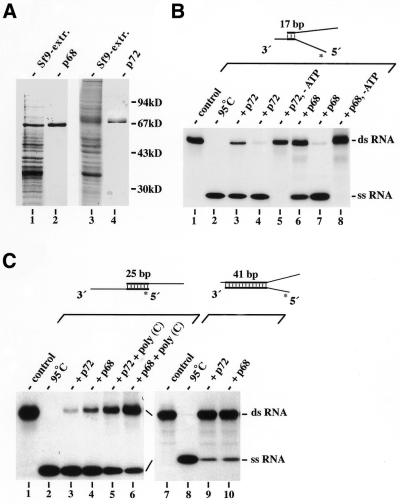
p68 (31,40) and p72 (33) have been shown to be RNA-dependent ATPases, whereas RNA helicase activity so far could only be demonstrated for p68 (31). We were interested in a comparative biochemical analysis of this subgroup of DEAD-box proteins and first subjected the purified recombinant proteins expressed in the baculovirus insect cell system (Fig. 1A) to RNA helicase activity analysis monitored by gel-shift electrophoresis (Fig. 1B and C). As expected, recombinant p72 as well as p68 showed an ATPase-dependent RNA helicase activity with a partial dsRNA as a substrate containing a small duplex (17 bp in length, Fig. 1B). Further experiments revealed that a ss 3′ overhang of the RNA substrate is essential and sufficient for unwinding with both proteins (left panel of Fig. 1C); dsRNA with only a ss 5′ overhang was not unwound (data not shown). The two helicases only differed slightly with regard to the dependence on ionic strength and the acceptance of nucleotide triphosphates other than ATP as an energy source (data not shown). Respective RNAs with a duplex length of 25 bp (Fig. 1C) or 32 bp (data not shown) were also unwound though with decreased efficiencies (defined as the final fraction of unwound substrate RNA), whereas RNA duplexes of ≥41 bp (Figs 1C and 5A, lanes 3 and 4) were hardly separated into their single strands or not at all, respectively. The inability to separate longer duplexes (>40 bp) most probably results from premature dissociation during the course of reaction followed by immediate reannealing of partially unwound strands before another enzyme molecule is bound. (Note that the helicase assay used does not give a signal until the duplex is completely unwound.) The RNA helicase NPH-II, as well as some DNA helicases (10), has been shown to act with a step size of roughly one half helix turn (41). If this is also the case with p68 and p72, then both enzymes need to perform six to seven continuous unwinding steps in order to separate the strands of a 30–35 bp duplex RNA and therefore must be regarded to be processive helicases that nevertheless have a relative high tendency to release the substrate. In good agreement with this conclusion, p68 and p72 also unwound the 25 bp RNA substrate (though with decreased efficiency) under single-turnover conditions with respect to the RNA substrate, obtained by adding a 500-fold excess of non-specific ‘trap RNA’ to prevent the helicases from reassociation with duplex once they fall off during the course of reaction (Fig. 1C, lanes 5 and 6).
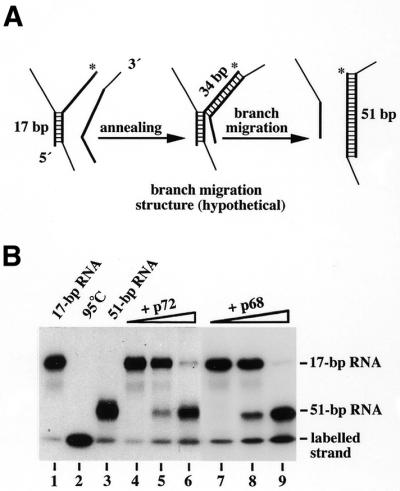
Figure 1.
RNA helicase activity analysis of recombinant p68 and p72. (A) SDS–PAGE (10%) analysis of crude extracts (20 µg each, lanes 1 and 3) and purified p68 and p72 (0.5 µg each, lanes 2 and 4, respectively) isolated from Sf9 cells infected with the respective recombinant baculoviruses. The gel was stained with Coomassie blue. Quantitative determination with a Bioanalyser (Agilent Technologies) using the Protein 200 LabChip Kit revealed 95 and 92% homogeneity of purified p68 and p72, respectively. (B) RNA helicase activity of p72 and p68. Helicase reactions were run with a 17 bp substrate RNA (12.5 nM) plus 0.7 nM (lane 3) or 1.25 nM (lanes 4 and 5) p72, or 1.25 nM (lane 6) or 2.5 nM (lanes 7 and 8) p68 for 15 min at 37°C. Control reactions performed without protein (control, lane 1) or without ATP (lanes 5 and 8) are also shown. One reaction mixture was heat denatured at 95°C before gel electrophoresis (lane 2). (C) Restriction of the unwinding on small dsRNA regions suggests low processivity of the helicases. Helicase reactions were run without protein (lanes 1 and 7) or with p72 (1.25 nM; lanes 3, 5 and 9) or p68 (2.5 nM; lanes 4, 6 and 10) and a 25 (lanes 1–6) or 41 bp (lanes 7–10) substrate (12.5 nM each) as described in (B). Reactions in lanes 5 and 6 additionally contained 6.25 µM of poly(C) as a trap RNA that could be replaced with the same effect by in vitro transcribed, unspecific RNA. RNA helicase activity analysis in (B) and (C) was monitored by gel-shift electrophoresis and autoradiography. A diagram of the substrates used is given on top of each part with an asterisk marking the labelled strand. Note that the upper strand of the 41 bp RNA is identical to that of the 17 bp RNA.
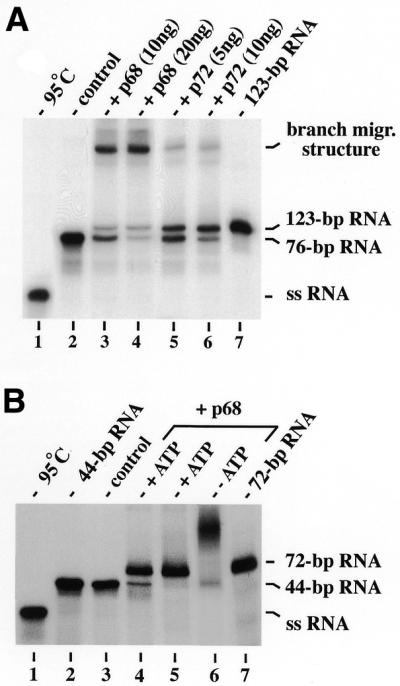
Figure 5.
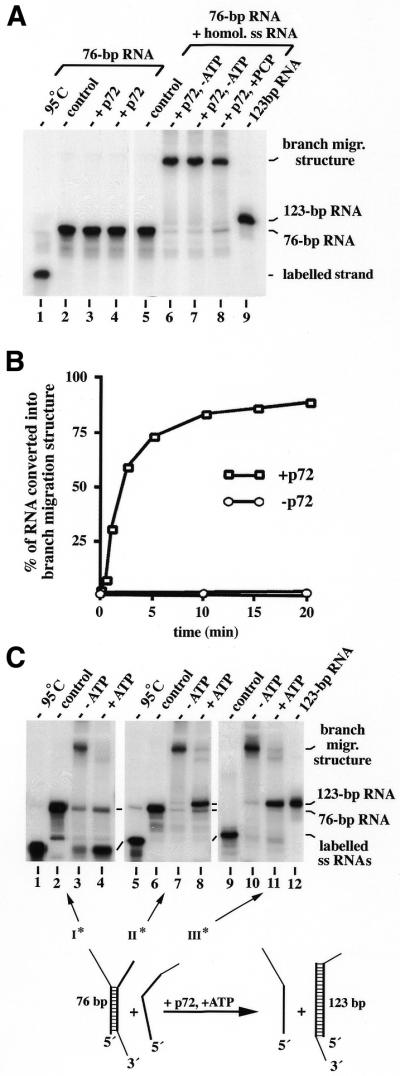
ATP-dependent RNA branch migration catalysed by p72. (A) Formation of a stable RNA branch migration structure in the absence of ATP. To the left (lanes 1–4), RNA helicase reactions (see Fig. 1C) with the partially ds 76 bp RNA as a substrate [with one strand, strand II of the design in (C), being 32P-labelled] are shown. Probes were incubated either without protein (lane 2) or with 2 nM (lane 3) or 4 nM (lane 4) p72 at 37°C for 30 min. To the right (lanes 5–8), the same labelled 76 bp RNA (12.5 nM) plus the non-labelled homologous ssRNA [strand III of the design in (C); 37.5 nM] were used as a substrate in strand exchange reactions without ATP (lanes 5–7) or with PCP (4 mM, lane 8), incubated without protein (lane 5) or at 1 (lane 6) or 2 nM (lanes 7 and 8) p72 for 30 min at 37°C. For comparison, the 123 bp RNA prepared by hybridisation of the respective RNA strands [strands II and III of the design in (C)] in 80% formamide at 50°C are shown (lane 9). (B) Time course of p72-catalysed and protein-independent formation of the stable RNA branch migration structure. Strand exchange reactions without ATP were performed in the presence (2 nM; for 0.5, 1, 2, 5, 10, 15 and 20 min) or absence of p72 (0, 10 and 20 min) exactly as described in (A). Autoradiographic signals of labelled RNA in branch migration structures were quantified with a densitometer (Molecular Dynamics). (C) ATP-dependent branch migration catalysed by p72. As a substrate, we used the same RNAs as in lanes 5–8 of (A) with a different strand being 32P-labelled in each column (strand I, lanes 1–4; strand II, lanes 5–8; strand III, lanes 9–11; see also the design of the reaction at the bottom). Strand exchange reactions were performed with p72 (2 nM) in the absence of ATP for 15 min exactly as described above [lanes 5–8 in (A)] and samples were either analysed directly thereafter (lanes 3, 7 and 10) or after an additional incubation in the presence of ATP (4 mM) for 15 min at 37°C (lanes 4, 8 and 11). Some probes were heat denatured instead of incubated (lanes 1 and 5) or were incubated without protein (lanes 2, 6 and 9). A design of the RNA strand exchange reaction is shown at the bottom with the individual strands marked by roman numbers, and arrows with asterisks indicating the labelled strand in respective reactions. The formation of joint molecules and RNA strand exchange activity was monitored by gel-shift electrophoresis in (A), (B) and (C).
RNA renaturation activity
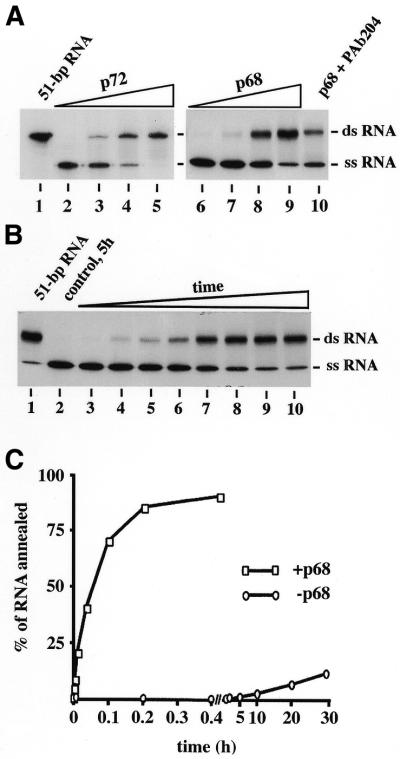
At increasing p68 or p72 concentrations, RNA of high molecular weight, which was sensitive to heat denaturation, was obtained in the respective helicase assays (data not shown). Therefore, we assumed that these DEAD-box proteins may also have the capacity to enhance the annealing of complementary strands. The annealing activity may predominate then at over-saturating protein concentrations, leading to cross hybridisation products. In fact, in separate annealing assays each protein was found to increase the renaturation rate of complementary RNA strands on dependence of protein concentrations. (Fig. 2; note that in the annealing assay complementary RNA stretches were chosen that were >40 bp, and thus cannot be re-unwound by the helicase activity). The optimal annealing conditions were found to be similar to those for unwinding, but Mg2+-ATP was not essential. When the rates of p68- (Fig. 2C) or p72-catalysed annealing (data not shown) were compared with those of uncatalysed hybridisation, a factor of rate enhancement of 1600 was obtained. The helicase, ATPase and ATP-independent annealing activities showed identical profiles during sucrose gradient sedimentation, excluding that the annealing activity is being performed by a contaminating protein (data not shown). Furthermore, binding of monoclonal antibody Pab 204 to p68 (which binds to amino acids 506–523) (30) partially inhibited the annealing activity (Fig. 2A, lane 10) whereas monoclonal C10 (which binds to amino acids 600–614) (38) had no effect (data not shown).
Figure 2.
p68- and p72-catalysed RNA annealing. A mixture of two RNA chains (127 and 130 bp long, the latter 32P-labelled) with a 51 bp complementary region was used as substrate. RNA annealing activity was monitored by gel-shift electrophoresis and autoradiography. (A) Dependence of RNA annealing on protein concentrations. Annealing reactions were incubated at increasing concentrations of the indicated proteins (lanes 2 and 6, no protein; lanes 3 and 7, 0.4 nM; lane 4 and 8, 2 nM; lanes 5, 9 and 10, 4 nM; of p72 or p68, respectively) at 37°C for 30 min. The reaction shown in lane 10 additionally contained 1 µg of PAb 204. (B) Dependence of RNA annealing on time. The autoradiogramm illustrates the annealing of RNA complementary strands by p68 (3 nM). Reactions were run for 0 (lane 3), 0.2 (lane 4), 0.5 (lane 5), 1 (lane 6), 2 (lane 7), 5 (lane 8), 10 (lane 9) and 20 min (lane 10). A control reaction run without protein for 5 h is also shown (lane 2). Products of the annealing reactions were run in parallel with the hybrid RNA obtained by hybridisation of the two chains in 80% formamide at 50°C [51 bp RNA, lane 1 in (A) and (B)]. (C) Kinetic plots of p68-dependent and -independent RNA annealing.
RNA-binding proteins may induce renaturation of complementary RNA strands by assisting their finding through simultaneous binding to several RNA molecules (16). Because p68 and p72 were not found to form oligomeric complexes (H.Stahl, unpublished results), we analysed one of them, p68, for occurrence of multiple RNA-binding sites within the polypeptide chain by mutant studies. We found that purified p68 deletion mutants containing either amino acids 1–189 (p681–189), 1–386 (p681–386) or 387–614 (p68387–614) bound to a test RNA with an efficiency similar to that of the wild-type protein in a nitrocellulose filter binding assay (Fig. 3A). However, only mutant p681–386 exhibited a significant RNA annealing activity, whereas that of the other mutants was strongly reduced (Fig. 3B). Mutant p681–386 seems to contain, therefore, more than one RNA-binding site encoded by small arginine stretches as well as arginine–lysine–lysine (RGG) repeats present in this part of p68 (as well as of p72) and this may provide a structural basis for its annealing activity. As was expected, neither mutant showed RNA unwinding activity (data not shown) (42).
Figure 3.
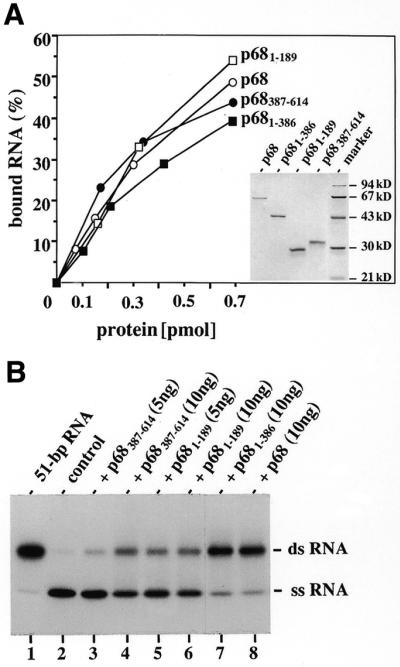
Biochemical activities of p68 deletion mutants. (A) RNA-binding activity. Isolated deletion mutants p681–189, p681–386 and p68387–614 were analysed for binding by the retention of 32P-RNA on nitrocellulose filters. Reaction mixtures contained 1 pmol RNA plus the indicated amounts of protein and were incubated for 30 min on ice before filtration. The insert shows SDS–PAGE (12%) analysis of the isolated proteins (0.5 µg each) visualised by Coomassie blue staining. (B) RNA-annealing activity. Annealing activity assays with the indicated amounts of respective proteins were performed exactly as described in (A).
Formation of joint molecules and catalysis of branch migration
Because p68 and p72 can mediate RNA annealing and unwinding with similar efficiencies under physiological conditions, a rearrangement of structured RNA could be envisaged to be catalysed by the two activities. In DNA strand exchange reactions some proteins (e.g. Rad51) (43) require an overhanging complementary end on the dsDNA to initiate the reaction. We therefore used similar RNA substrates (Fig. 4A) to test whether p68 and p72 could (i) catalyse the formation of joint RNA molecules and (ii) resolve such structures which are stable under physiological conditions by catalysing branch migration through their homologous ds part in an ATP-dependent manner.
Figure 4.
ATP-independent RNA strand exchange. (A) Design of substrate RNAs (with an asterisk marking the labelled strand) and postulated course of the strand exchange reaction. Strand exchange was performed as an intermolecular process between a partially dsRNA and ssRNA, but may also proceed intramoleculary in a similar way. Thick lines indicate homologous regions. Note that the branch migration structure, implicated as a reaction intermediate, is unstable due to the small length (17 bp) of the homologous ds part. (B) ATP-independent strand exchange catalysed by p68 and p72. Reaction mixtures contained the partially ds 17 bp RNA (12.5 nM) plus the respective homologous ssRNA (37.5 nM) and were incubated at 37°C for 30 min at increasing concentrations of p68 or p72 (lanes 4 and 7, no protein; lanes 5 and 8, 2 nM; lanes 6 and 9, 4 nM). For comparison, the 32P-labelled RNA strand (lane 2) and the 17 (lane 1) and 51 bp (lane 3) RNAs prepared by hybridisation of the corresponding RNA strands in 80% formamide at 50°C are shown.
Branch migration structures are very unstable under reassociation conditions (Tm – 25°C), and thus the displacement of small DNA or RNA strands (up to 35 nt in length) by branch migration proceeds spontaneously at 37°C (44). Consequently, RNA strand exchange concerning such short homologous duplexes can be promoted under physiological conditions by proteins with an RNA annealing activity, like the tumour suppressor p53 or eIF-4B, in an ATP-independent manner (16,18). We found that p72 and p68 as well as the deletion mutant, that retained the annealing activity (p681–386; data not shown), also mediated strand exchange in the absence of ATP, when RNA substrates with corresponding short homologous duplexes were used (Fig. 4B); the other p68 mutants were inactive (data not shown). In the absence of protein, no strand exchange was observed under the reaction conditions and the data strongly confirm that the two DEAD-box proteins can catalyse the formation of joint RNA molecules most probably by their renaturation activity.
In homologous recombination in E.coli, branch migration can be catalysed by specialized DNA helicases (reviewed in 23). It has been suggested that the mechanism of branch migration might involve the transient unwinding and rewinding of dsDNA (reviewed in 9). Interestingly, the E.coli RuvB DNA helicase, which has been shown to catalyse branch migration, works less processively in isolated unwinding reactions and it has been shown that a mutant RuvB with a damaged DNA helicase activity can still catalyse branch migration (45). The low processive helicase activity of RuvB is paralleled in p68 and p72 and this may point to an ATP-dependent function of p68 and p72 in RNA rearrangement processes, possibly also involving protein-catalysed RNA branch migration. To test this hypothesis, we used RNA substrates, suitable to form branch migration structures that are stable under the strand exchange reaction conditions and need to be resolved enzymatically, like that shown in Figure 5C with a homologous 76 bp ds region. First, we confirmed (in the absence of the homologous third strand) that the input 76 bp RNA could not be unwound by the p72 (Fig. 5A, lanes 3 and 4) or the p68 (data not shown) RNA helicase. Next, we tested whether a stable branch migration structure was formed from the input RNAs in the strand exchange assay. Indeed, p72 (Fig. 5A) and p68 (Fig. 6A and data not shown) induced the formation of a specific larger RNA, most likely the expected branch migration structure, in the absence of ATP or in the presence of a non-hydrolysable ATP analogue. To confirm that all three strands were present within this larger RNA, we radioactively labelled either one of the three input RNA strands and, in each case, the same RNA product was obtained (Fig. 5C). Furthermore, after isolation from the gel, each product was found to yield the respective labelled free single strand by heat denaturation (95°C for 5 min). The partially ds nature of the branch migration structure was confirmed by resistance of the base paired parts of the first and the third strand against S1 nuclease digestion (data not shown). The branch migration structure appeared to be stable; in particular, no dissociation by strand displacement into the final 123 bp hybrid RNA was observed after incubation under the strand exchange conditions (37°C) for several hours. In a more detailed analysis, it showed the same thermal stability as the branch migration structure formed from the same RNAs under hybridisation conditions. Structures were stable at 37°C and dissociated after 10 min at 60°C, which confirms that RNA-, like DNA-branch migration structures, dissociate very rapidly by strand displacement under reassociation conditions (data not shown) (44). When the rate of formation of the branch migration structure was determined (Fig. 5B), it was found to be in the same range as that of p68- and p72-catalysed annealing, confirming the hypothesis that the annealing activity of these proteins is responsible.
Figure 6.
p68-catalysed branch migration. (A) Strand exchange reaction with an RNA substrate containing a 76 bp homologous region. Reactions were performed with the same labelled substrate used in lanes 5–8 of Figure 5A, but with ATP (4 mM) being present during the whole 30 min incubation time either without protein (lane 2) or at 1.5 nM p68 (lane 3), 3 nM p68 (lane 4), 1 nM p72 (lane 5) or 2 nM p72 (lane 6). One probe without protein was denatured at 95°C (lane 1) and the 123 bp RNA formed under hybridisation conditions was run in parallel (lane 7). (B) Strand exchange reaction with an RNA substrate containing a 44 bp homologous region. As a substrate a 44 bp partially dsRNA plus a respective homologous single strand was used, the structure of which was otherwise similar to that of the 76 bp substrate (Materials and Methods). RNA strand exchange activity analysis was performed as described above at 1.5 (lane 4) or 3 nM (lanes 5 and 6) p68 with or without ATP as indicated.
Next, we addressed whether the branch migration structure formed in the absence of ATP can be resolved by p68 or p72 after the addition of Mg2+-ATP. We found that each protein was able to convert the branch migration structure into a product that comigrated with a 123 bp hybrid RNA formed under hybridisation conditions from the two strands that contain the homologous region (RNA strands II and III in Fig. 5C). Furthermore, when the non-homologous strand of the 76 bp RNA (strand I in Fig. 5C) carried the radioactive label, its dissociation from the branch migration structure could be traced in the strand exchange assay (Fig. 5C, lanes 2–4) and when the third strand was labelled, it became part of the 123 bp RNA (Fig. 5C, lanes 9–11). p72- and p68-catalysed branch migration was dependent on protein concentrations and was also observed when Mg2+-ATP was added to the reaction prior to the proteins (Fig. 6A). As can be seen from Figure 6A, p68 was less active than p72 in the resolution of the 76 bp branch migration structure at saturating protein levels, which may reflect differences in their processive mode of action that became not obvious in the helicase reactions (Fig. 1C). Indeed, when we reduced the length of the homologous ds part in the branch migration structure to 44 bp, the efficiency of the p68-catalysed strand exchange reaction strongly increased though it was strictly dependent on the hydrolysis of ATP in all cases (Fig. 6B). This may indicate that protein-catalysed RNA branch migration, like RNA unwinding, is also performed with a defined step size and that p68 dissociates from branch migration structures after each step with higher probability than p72. However, the matter needs to be analysed in more detail by kinetic experiments.
DISCUSSION
We have shown here that p72 is an ATP-dependent RNA helicase with properties similar to that of p68. RNA unwinding with both enzymes proceeds in a defined 3′ to 5′ direction with respect to the essential ss 3′ overhang. Strand separation was also observed under conditions that prevent the helicases from restarting the reaction once they fall off during the course of unwinding. Therefore, the analysed RNA helicases seem to act in a processive manner, though the size of duplexes that can be separated efficiently was <40 bp. In contrast to the DExH RNA helicase NPH-II from vaccinia virus II that can unwind >100 bp (10), the processivity of p68 and p72 must be regarded as low, allowing them to only perform six to seven continuous unwinding steps, provided that their step sizes equals those of other RNA and DNA helicases (10,41,46). Our studies add support to the notion that processivity varies with the RNA helicase used and seems to be an important characteristic with respect to function. Notably, the prototype of DEAD-box proteins, eIF4A, acts in a non-processive manner and dissociates from the substrate after each unwinding step (46). Consequently, eIF4A can only separate very small RNA duplexes (up to ∼12 bp) in a process composed of ATP-dependent unwinding of 5–6 bp (equivalent to approximately one half helix turn) and thermal melting of the rest (46).
RNA-binding and unwinding by DExD/H-box proteins (with the exception of E.coli DbpA that specifically interacts with 23 S ribosomal RNA) (47) apparently shows no substrate specificity in vitro and this raises the question of the mechanisms or factors that direct the roles of these enzymes in vivo. RNA helicases may be restricted to defined cellular compartments by specific interaction with anchoring proteins (48) and indeed, p68 and p72 seem to be bound to subnuclear structures from which they can only be extracted under stringent conditions (31). Another way to prevent inopportune resolutions of at least more stable RNA secondary structures may be to limit the processivity of RNA helicases, forcing them to prematurely dissociate from their substrates (and allowing the partially unwound strands to reanneal). Consistent with this idea, the processivity of p68 and p72 is intrinsically low, though it may be upregulated in vivo by factors that control the accessibility of their NTP binding sites (46).
We have shown here that recombinant p68 and p72 have an annealing in addition to the RNA helicase activity and that both proteins can couple the unwinding of existing RNA secondary structures to the rearrangement of new ones in an in vitro assay. Though their in vivo functions are not known, this could be a means in the cell to compensate for the low processivity of these RNA helicases in processes that demand the resolution of longer RNA duplexes. Protein-induced annealing seems to require more than one nucleic acid binding site within a given enzyme (e.g. RecA) (49), which could be provided by multiple subunits of an oligomeric complex. Though dimerisation or hexamer formation is common to several DNA and RNA helicases (reviewed in 50), p68 and p72 seem to function as monomers in agreement with our mutant studies that show that the p68 polypeptide has multiple RNA-binding sites, which are regarded as a prerequisite for monomeric helicases. On the other hand, we cannot exclude that oligomerisation is finally induced by contact with the substrate as may be the case with the RNA helicase from hepatitis C virus (51). The annealing activity may also explain the previously reported p68 inhibition of DNA unwinding through several DNA helicases (31).
With their biochemical activities, p68 and p72 resemble the RecA family proteins that, among others, have a DNA-dependent ATPase, a DNA annealing activity and catalyse branch migration (reviewed in 23). Furthermore, DNA branch migration is also catalysed by replicative DNA helicases, such as those encoded by the the bacteriophages T4 and T7, which in complex with an annealing activity mediate DNA strand exchange (52–54). The mechanistic manner of how p68 and p72 form branch migration structures and drive branch migration through a duplex in an ATP-dependent manner remains to be elucidated. However, the observed RNA strand exchange does not simply result from an isolated action of their unwinding and annealing activities, but rather appears to reflect an extra enzymatic quality. This can be concluded from the apparent higher processivity observed in the branch migration reaction as compared with unwinding (Fig. 6A). The input partially dsRNA of the used branch migration substrates contains two forks, of which only that involved in branch migration is essential (H.Stahl, unpublished results). The helicase that associates with the fork of the branch migration structure may simultaneously bind the third strand and continuous contact during unwinding may then enable its concomitant annealing, which in turn may enhance the processivity of the helicase.
p68 genes from yeast to man contain a large intron near the 3′ end, the position of which is conserved (36,38). The presence of this intron has been shown to regulate gene expression negatively by providing a negative feedback loop (37,38). The helicase activity of p68 is essential for this autoregulation and it is possible that p68 alters the secondary structure of its pre-mRNA by the strand exchange activity in a way that interferes with splicing. Furthermore, the detection of p68 in spliceosomes (55 and our own observations) may point to a role of this protein (and possibly also of p72) in post-transcriptional regulation of the expression of other genes as well.
Finally, p68 has also been shown in our laboratory to unwind RNA–DNA hybrids, if the RNA strand contains the essential ss 3′ overhang (H.Stahl, unpublished results). This may be relevant for the function of p68 present in the highly purified 5-methylcytosine–DNA glycosylase complex isolated from developing chicken embryos (56). RNA rich in CpGs is also present in this complex and it was shown that it serves as a guide for the demethylation complex to hemimethylated sites (57). The molecular mechanism of this process is unknown, but an involvement of RNA–DNA hybrid structures (R-loops), being built up and/or resolved by strand exchange reactions, is conceivable. In support of this notion, it was shown that RecA can catalyse assimilation of complementary RNA into a homologous region of a DNA duplex. In E.coli, this R-loop formation by RNA strand exchange is likely to be required for recombination-dependent DNA replication (58,59).
Acknowledgments
ACKNOWLEDGEMENTS
We thank K.Hilgert for excellent technical assistance and H.Uhlmann-Schiffler for helpful discussions and critical reading of the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (SFB 399) and the Fonds der Chemie.
References
- 1.Herschlag D. (1995) RNA chaperones and the RNA folding problem. J. Biol. Chem., 270, 20871–20874. [DOI] [PubMed] [Google Scholar]
- 2.Pan J., Thirumalai,D. and Woodson,S.A. (1997) Folding of RNA involves parallel pathways. J. Mol. Biol., 273, 7–13. [DOI] [PubMed] [Google Scholar]
- 3.Weeks K.M. (1997) Protein-facilitated RNA folding. Curr. Opin. Struct. Biol., 7, 336–342. [DOI] [PubMed] [Google Scholar]
- 4.Clodi E., Semrad,K. and Schroeder,R. (1999) Assaying RNA chaperone activity in vivo using a novel RNA folding trap. EMBO J., 18, 3776–3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lüking A., Stahl,U. and Schmidt,U. (1998) The protein family of RNA helicases. Crit. Rev. Biochem. Mol. Biol., 33, 259–296. [DOI] [PubMed] [Google Scholar]
- 6.Linder P., Lasko,P.F., Ashburner,M., Leroy,P., Nielsen,P.J., Nishi,K., Schnier,J. and Slonimski,P.P. (1989) Birth of the D-E-A-D box. Nature, 337, 121–122. [DOI] [PubMed] [Google Scholar]
- 7.de la Cruz J., Kressler,D. and Linder,P. (1999) Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci., 24, 192–198. [DOI] [PubMed] [Google Scholar]
- 8.Linder P. and Daugeron,M.C. (2000b) Are DEAD-box proteins becoming respectable helicases? Nat. Struct. Biol., 7, 97–99. [DOI] [PubMed] [Google Scholar]
- 9.West S.C. (1997) Processing of recombination intermediates by the RuvABC proteins. Annu. Rev. Genet., 31, 213–244. [DOI] [PubMed] [Google Scholar]
- 10.Jankowsky E., Gross,C.H., Shuman,S. and Pyle,A.M. (2000) The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature, 403, 447–451. [DOI] [PubMed] [Google Scholar]
- 11.Lohman T.M. and Bjornson,K.P. (1996) Mechanisms of helicase-catalysed DNA unwinding. Annu. Rev. Biochem., 65, 169–214. [DOI] [PubMed] [Google Scholar]
- 12.Portman D.S. and Dreyfuss,G. (1994) RNA annealing activities in HeLa nuclei. EMBO J., 13, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee C.G., Zamore,P.D., Green,M.R. and Hurwitz,J. (1993) RNA annealing activity is intrinsically associated with U2AF. J. Biol. Chem., 268, 13472–13478. [PubMed] [Google Scholar]
- 14.Raghunathan P.L. and Guthrie,C. (1998) A spliceosomal recycling factor that reanneals U4 and U6 small nuclear ribonucleoprotein particles. Science, 279, 857–860. [DOI] [PubMed] [Google Scholar]
- 15.Altmann M., Wittmer,B., Methot,N., Sonenberg,N. and Trachsel,H. (1995) The Saccharomyces cerevisiae translation initiation factor Tif3 and its mammalian homologue, eIF-4B, have RNA annealing activity. EMBO J., 14, 3820–3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niederberger N., Trachsel,H. and Altmann,M. (1998) The RNA recognition motif of yeast translation initiation factor Tif3/eIF4B is required but not sufficient for RNA strand-exchange and translational activity. RNA, 4, 1259–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burd C.G. and Dreyfuss,G. (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science, 265, 615–621. [DOI] [PubMed] [Google Scholar]
- 18.Oberosler P., Hloch,P., Ramsperger,U. and Stahl,H. (1993) p53-catalysed annealing of complementary single-stranded nucleic acids. EMBO J., 12, 2389–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valdez B.C., Henning,D., Perumal,K. and Busch,H. (1997) RNA-unwinding and RNA-folding activities of RNA helicase II/Gu — two activities in separate domains of the same protein. Eur. J. Biochem., 15, 800–807. [DOI] [PubMed] [Google Scholar]
- 20.Staley J.P. and Guthrie,C. (1998) Mechanical devices of the spliceosome: motors, clocks, springs and things. Cell, 92, 315–326. [DOI] [PubMed] [Google Scholar]
- 21.Murray H.L. and Jarrel,K.A. (1999) Flipping the switch to an active spliceosome. Cell, 96, 599–602. [DOI] [PubMed] [Google Scholar]
- 22.Steitz J.A. (1992) Splicing takes a holliday. Science, 257, 888–889. [DOI] [PubMed] [Google Scholar]
- 23.Bianco P.R., Tracy,R.B. and Kowalczykowski,S.C. (1998) DNA strand exchange proteins: a biochemical and physical comparison. Front. Biosci., 3, 570–603. [DOI] [PubMed] [Google Scholar]
- 24.Shinohara A. and Ogawa,T. (1999) Rad51/RecA protein families and the associated proteins in eukaryotes. Mutat. Res., 435, 13–21. [DOI] [PubMed] [Google Scholar]
- 25.Egelman E.H. (1998) Bacterial helicases. J. Struct. Biol., 124, 123–128. [DOI] [PubMed] [Google Scholar]
- 26.Johnson E.R. and McKay,D.B. (1999) Crystallographic structure of the amino terminal domain of yeast initiation factor 4A, a representative DEAD-box RNA helicase. RNA, 5, 1526–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benz J., Trachsel,H. and Baumann,U. (1999) Crystal structure of the ATPase domain of translation initiation factor 4A from Saccharomyces cerevisiae—the prototype of the DEAD box protein family. Structure Fold. Des., 7, 671–679. [DOI] [PubMed] [Google Scholar]
- 28.Subramanya H.S., Bird,L.E., Brannigan,J.A. and Wigley,D.B. (1996) Crystal structure of a DExx box DNA helicase. Nature, 384, 379–383. [DOI] [PubMed] [Google Scholar]
- 29.Lane D.P. and Hoeffler,W.K. (1980) SV40 large T shares an antigenic determinant with a cellular protein of molecular weight 68,000. Nature, 288, 167–170. [DOI] [PubMed] [Google Scholar]
- 30.Ford M.J., Anton,I.A. and Lane,D.P. (1988) Nuclear protein with sequence homology to translation initiation factor eIF-4A. Nature, 332, 736–738. [DOI] [PubMed] [Google Scholar]
- 31.Hirling H., Scheffner,M., Restle,T. and Stahl,H. (1989) RNA helicase activity associated with the human p68 protein. Nature, 339, 562–564. [DOI] [PubMed] [Google Scholar]
- 32.Scheffner M., Knippers,R. and Stahl,H. (1989) RNA unwinding activity of the SV40 large T antigen. Cell, 57, 955–963. [DOI] [PubMed] [Google Scholar]
- 33.Lamm G.M., Nicol,S.M., Fuller-Pace,F.V. and Lamond,A.I. (1996) p72: a human nuclear DEAD box protein highly related to p68. Nucleic Acids Res., 24, 3739–3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stevenson R.J., Hamilton,S.J., MacCallum,D.E., Hall,P. and Fuller-Pace,F.V. (1998) Expression of the ‘dead box’ RNA helicase p68 is developmentally and growth regulated and correlates with organ differentiation/maturation in the fetus. J. Pathol., 184, 351–359. [DOI] [PubMed] [Google Scholar]
- 35.Ip F.C., Chung,S.S., Fu,W.Y. and Ip,N.Y. (2000) Developmental and tissue-specific expression of DEAD box protein p72. Neuroreport, 11, 457–462. [DOI] [PubMed] [Google Scholar]
- 36.Iggo R.D., Jamieson,D.J., MacNeill,S.A., Southgate,J., McPheat,J. and Lane,D.P. (1991) p68 RNA helicase: identification of a nucleolar form and cloning of related genes containing a conserved intron in yeasts. Mol. Cell Biol., 11, 1326–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barta I. and Iggo,R. (1995) Autoregulation of expression of the yeast Dbp2p ‘DEAD-box’ protein is mediated by sequences in the conserved DBP2 intron. EMBO J., 14, 3800–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roessler O.G., Hloch,P., Schuetz,N., Weitzenegger,T. and Stahl,H. (2000) Structure and expression of the human p68 RNA helicase gene. Nucleic Acids Res., 28, 932–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hloch P., Schiedner,G. and Stahl,H. (1990) Complete cDNA sequence of the human p68 protein. Nucleic Acids Res., 18, 3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iggo R.D. and Lane,D.P. (1989) Nuclear protein p68 is an RNA-dependent ATPase. EMBO J., 8, 1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ali J.A. and Lohman,T.M. (1997) Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science, 275, 377–380. [DOI] [PubMed] [Google Scholar]
- 42.Pause A. and Sonenberg,N. (1992) Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J., 11, 2643–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Namsaraev E. and Berg,P. (1997) Characterization of strand exchange activity of yeast Rad51 protein. Mol. Cell Biol., 17, 5359–5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsieh P., Camerini-Otero,C.S. and Camerini-Otero,R.D. (1990) Pairing of homologous DNA sequences by proteins: evidence for three-stranded DNA. Genes Dev., 4, 1951–1963. [DOI] [PubMed] [Google Scholar]
- 45.George H., Mezard,C., Stasiak,A. and West,S.C. (1999) Helicase-defective RuvB(D113E) promotes RuvAB-mediated branch migration in vitro. J. Mol. Biol., 293, 505–519. [DOI] [PubMed] [Google Scholar]
- 46.Rogers G.W.,Jr, Richter,N.J. and Merrick,W.C. (1999) Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J. Biol. Chem., 274, 12236–12244. [DOI] [PubMed] [Google Scholar]
- 47.Pugh G.E., Nicol,S.M and Fuller-Pace,F.V. (1999) Interaction of the Escherichia coli DEAD box protein DbpA with 23 S ribosomal RNA. J. Mol. Biol., 292, 771–778. [DOI] [PubMed] [Google Scholar]
- 48.Nicol S.M., Causevic,M., Prescott,A.R. and Fuller-Pace,F.V. (2000) The nuclear DEAD box RNA helicase p68 interacts with the nucleolar protein fibrillarin and colocalizes specifically in nascent nucleoli during telophase. Exp. Cell Res., 257, 272–280. [DOI] [PubMed] [Google Scholar]
- 49.Kurumizaka H., Ikawa,S., Sarai,A. and Shibata,T. (1999) The mutant RecA proteins, RecAR243Q and RecAK245N, exhibit defective DNA binding in homologous pairing. Arch. Biochem. Biophys., 365, 83–91. [DOI] [PubMed] [Google Scholar]
- 50.Soultanas P. and Wigley,D.B. (2000) DNA helicases: inching forward. Curr. Opin. Struct. Biol., 10, 124–128. [DOI] [PubMed] [Google Scholar]
- 51.Levin M.K. and Patel,S.S. (1999) The helicase from hepatitis C virus is active as an oligomer. J. Biol. Chem., 274, 31839–31846. [DOI] [PubMed] [Google Scholar]
- 52.Salinas, F. and Kodadek,T. (1995) Phage T4 homologous strand exchange: a DNA helicase, not the strand transferase, drives polar branch migration. Cell, 82, 111–119. [DOI] [PubMed] [Google Scholar]
- 53.Kong D. and Richardson,C.C. (1996) Single-stranded DNA binding protein and DNA helicase of bacteriophage T7 mediate homologous DNA strand exchange. EMBO J., 15, 2010–2019. [PMC free article] [PubMed] [Google Scholar]
- 54.Kong, D., Nossal,N.G. and Richardson,C.C. (1997) Role of the bacteriophage T7 and T4 single-stranded DNA-binding proteins in the formation of joint molecules and DNA helicase-catalyzed polar branch migration. J. Biol. Chem., 272, 8380–8387. [DOI] [PubMed] [Google Scholar]
- 55.Neubauer G., King,A., Rappsilber,J., Calvio,C., Watson,M., Ajuh,P., Sleeman,J., Lamond,A. and Mann,M. (1998) Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat. Genet., 20, 46–50. [DOI] [PubMed] [Google Scholar]
- 56.Jost J.P., Schwarz,S., Hess,D., Angliker,H., Fuller-Pace,F.V., Stahl,H., Thiry,S. and Siegmann,M. (1999) A chicken embryo protein related to the mammalian DEAD box protein p68 is tightly associated with the highly purified protein–RNA complex of 5-MeC–DNA glycosylase. Nucleic Acids Res., 27, 3245–3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jost J.P., Fremont,M., Siegmann,M. and Hofsteenge,J. (1997) The RNA moiety of chick embryo 5-methylcytosine–DNA glycosylase targets DNA demethylation. Nucleic Acids Res., 25, 4545–4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kasahara M., Clikeman,J.A., Bates,D.B. and Kogoma,T. (2000) RecA protein-dependent R-loop formation in vitro. Genes Dev., 14, 360–365. [PMC free article] [PubMed] [Google Scholar]
- 59.Zaitsev E.N. and Kowalczykowski,S.C. (2000) A novel pairing process promoted by Escherichia coli RecA protein: inverse DNA and RNA strand exchange. Genes Dev., 14, 740–749. [PMC free article] [PubMed] [Google Scholar]