Abstract
Specific macromolecular transport systems, ion channels and pumps, provide the pathways to facilitate and control the passage of ions across the lipid membrane. Ion channels provide energetically favorable passage for ions to diffuse rapidly and passively according to their electrochemical potential. Selective ion channels are essential for the excitability of biological membranes: the action potential is a transient phenomenon that reflects the rapid opening and closing of voltage-dependent Na+-selective and K+-selective channels. One of the most critical functional aspects of K+ channels is their ability to remain highly selective for K+ over Na+ while allowing high-throughput ion conduction at a rate close to the diffusion limit. Permeation through the K+ channel selectivity filter is believed to proceed as a “knock-on” mechanism in which 2–3 K+ ions interspersed by water molecules move in single file. Permeation through the comparatively wider and less selective Na+ channels also proceeds via a loosely coupled knock-on mechanism, although the ions do not need to be fully dehydrated. While simple structural concepts are often invoked to rationalize the mechanism of ion selectivity, a deeper analysis shows that subtle effects play an important role in these flexible dynamical structures.
INTRODUCTION
Specific macromolecular transport systems, ion channels and pumps, provide the pathways to facilitate and control the passage of ions across the lipid membrane. Without these macromolecular systems, the lipid membrane would present a prohibitively high-energy barrier to the passage of any ion. Ion channels provide energetically favorable passage for ions to diffuse rapidly and passively according to their electrochemical potential. Ion pumps exploit ATP hydrolysis to establish and maintain a concentration gradient across the cell membrane by moving ions uphill against their electrochemical potential. Biological membrane channels and pumps can both be very selective for specific ions. What is unique about ion channels is their remarkably high rate of transport. The rate of transport of ions through one open channel is often on the order of several millions per second, whereas the maximum turnover of one ATP-driven ion pump or a co-transport protein is often less than a few hundreds per second.
As established long ago by Hodgkin and Huxley (Hodgkin and Huxley, 1952), voltage-activated selective ion channels are absolutely essential for the generation and propagation of action potentials in electrically excitable cells. By virtue of the long-range nature of the Coulombic interactions, the changes in the membrane potential generated by the selective movements of ions across the cell membrane constitutes a unified communication system that insures the synchronization of specific molecular processes taking place at distances far larger than typical short range intermolecular forces. At the simplest level, the word “selective” indicates that ions of the “correct” type can permeate through these channels more easily (i.e., at a faster rate) than ions of the “wrong” type. Selective membrane permeability is a key ingredient supporting the concept of membrane potential: a potential difference (traditionally called the Nernst potential) appears spontaneously between two ionic solutions of different concentrations separated by a semipermeable membrane (Nernst, 1889). Due to the high basal K+ permeability, the resting Nernst potential of living cells is normally dominated by the high and low concentration of K+ ions inside and outside the cell, respectively. The action potential that underlies the propagation of the nerve impulse reflects the rapid opening and closing of Na+- and K+-selective channels in response to changes in the transmembrane potential. After a stimulus, the (very fast) opening of voltage-activated Na+ (Nav) channels causes a depolarization of the membrane. While the Nav channels are transiently open, the membrane Nernst potential is briefly dominated by the low and high concentration of Na+ ions inside and outside the cell, respectively. The transient membrane depolarization subsequently triggers the (slower) opening of the voltage-activated K+ (Kv) channels, which then repolarize the membrane toward the resting potential (Lacroix et al., 2013). One of the most critical functional aspects of Kv channels is their ability to remain highly selective for K+ over Na+ while allowing high-throughput ion conduction at a rate close to the diffusion limit (Bezanilla and Armstrong, 1972; Eisenman, 1973; Hille, 1973). In comparison, Nav channels, which serve as a trigger to initiate the action potential with their fast gating kinetics, do not need to be as highly selective as Kv channels. Thus, the difference in selectivity is consistent with the respective functional contribution of these channels to the generation of the action potential. The large conductance and high selectivity of K+ channels, in particular, is absolutely critical to ensure rapid return to the resting potential after membrane depolarization.
The relation of structure to function of ion channels is of central interest for physiologists and considerable efforts are dedicated to the characterization of ion permeation in molecular terms. The determination of the three-dimensional atomic structure of several ion channels in the last decades represents an extraordinary opportunity for understanding ion permeation in biological membrane channels at the molecular level. Leaning on the availability of molecular structures at atomic resolution, computational studies offer a powerful avenue to better understand ion permeation and selectivity. The approach called “molecular dynamics” (MD) is of particular importance (Smith and Roux, 2013). It consists of constructing detailed atomic models of the macromolecular system and, having described the microscopic forces with a potential function, using Newton’s classical equation, F=MA, to literally “simulate” the dynamical motions of all the atoms as a function of time. The calculated MD trajectory, though an approximation to the real world, provides detailed information about the time course of the atomic motions, which is impossible to access experimentally. The scope of MD simulations can be greatly expanded by using free energy perturbation and potential of mean force approaches (Chipot, 2008). Nevertheless, even though computational studies offer a great complement to the information from X-ray crystallography and functional measurements, it is nonetheless important to keep in mind that the results can be sensitive to the inaccuracies in the force field used to generate MD simulations (Jensen et al., 2013). Over the years there have been numerous studies of the K+ channel (Åqvist and Luzhkov, 2000; Bernèche and Roux, 2001; Noskov et al., 2004; Roux, 2005; Furini and Domene, 2009; Thompson et al., 2009; Egwolf and Roux, 2010; Kim and Allen, 2011; Kopfer et al., 2014; Medovoy et al., 2016), and more recently of Na+ channels (Chakrabarti et al., 2013; Ulmschneider et al., 2013; Bagneris et al., 2015; Ing and Pomes, 2016; Naylor et al., 2016).
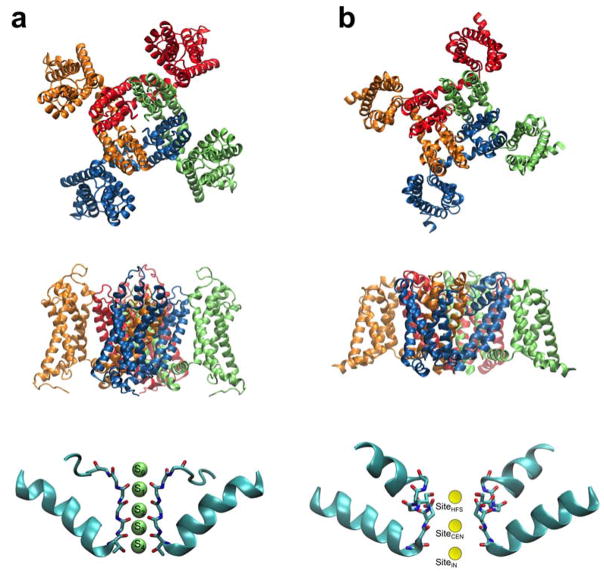
A first view of the overall molecular architecture of K+ channels was provided by the X-ray structures of the bacterial KcsA channel (Doyle et al., 1998; Zhou et al., 2001). Because of the high sequence similarity with eukaryotic channels, it was expected that the structure would be of relevance for the entire K+ channel family, a fact that was confirmed when the structure of the Kv1.2 channel was determined (Long et al., 2005; Long et al., 2007). The general tetrameric architecture of a Kv channel is shown in Figure 1a. The pore domain, surrounded by four voltage sensing domain (VSD), comprises four identical subunits of two transmembrane (TM) α-helices separated by a long reentrant loop disposed symmetrically around common axis corresponding to the ion conducting pore. The reentrant loop includes a pore helix making an angle of approximately 45° with the pore axis. The narrowest region of the pore (Figure 1a, bottom), formed by the backbone carbonyl oxygens of a highly conserved sequence TTVGYGD, is believed to act as the selectivity filter for K+ ions (Heginbotham, 1992; Heginbotham et al., 1994). In comparison, no atomic structure of mammalian Nav channel is available yet. But, as shown in Figure 1b, their overall topology is expected to be generally similar to that of Kv channels according to X-ray structures of a number of bacterial Nav channels (Payandeh et al., 2011; McCusker et al., 2012; Payandeh et al., 2012; Zhang et al., 2012). The pore domain comprises four identical subunits of two TM α-helices separated by a long reentrant loop disposed around common axis corresponding to the ion conducting pore. A noticeable difference with the pore domain of Kv channels is the reentrant loop, which includes two short α-helices (Figure 1b, bottom). The selectivity filter of these channels is also quite different. As shown in Figure 1a (bottom), five specific cation binding sites, called S0 to S4, are disposed along the narrow selectivity filter of K+ channels (Zhou et al., 2001). In contrast, the selectivity filter of bacterial Nav channels is lined with amino acid side chains (Figure 1b, bottom); a few ion bindings sites have been identified (Payandeh et al., 2011), though the situation remains uncertain due to the limited information (Naylor et al., 2016). While studies of these Nav bacterial channel structures have been very informative, one striking difference with mammalian Nav channels is that the latter are constituted by four distinct domains from a single polypeptide chain assembled into an asymmetric pore (Catterall, 2014; Catterall and Zheng, 2015). Importantly, an asymmetric ring of four amino acids of sequence “DEKA” clearly responsible for maintaining Na+ selectivity was identified through site-directed mutagenesis experiments on the mammalian Nav channel (Heinemann, 1992). Such structural differences between Nav channels from prokaryotes (homotetramers) and eukaryotes (asymmetric, four-domain proteins) suggest that our understanding of the molecular mechanism of permeation and selectivity in these channels remain uncertain (Finol-Urdaneta et al., 2014; Catterall and Zheng, 2015).
Figure 1.
Overall structure of voltage-gated K+ (Kv) and Na+ (Nav) channels. The Kv channel (a) is the mammalian Kv1.2/Kv2.1 chimera structure protein data base (PDB) entry: 2R9R (Long et al., 2007). The Nav channel (b) is the bacterial NavAb structure PDB entry 3RVY (Payandeh et al., 2011). At the top is a view of the tetrameric channels seen from the extracellular side. In the middle is a side view of the channels. At the bottom is a close-up view of the selectivity filter. In the Kv channel (a), the narrow selectivity filter of K+ channels comprises 5 distinct binding sites (S0 to S4) where the permeating K+ ions are coordinated by backbone carbonyl oxygens. In the Nav channel (b), the selectivity filter is relatively wide, allowing the passage of partially hydrated ions. A few binding sites have also been identified (Payandeh et al., 2011).
Conceptually, the permeation process of K+ ions through the K+ channel is believed to proceed predominantly as a multi-ion “knock-on” mechanism, in which the single-file translocation of 2–3 K+ ions interspersed by water molecules through the five ion binding sites, S0 to S4 (Figure 1a) disposed along the narrow selectivity filter (Bernèche and Roux, 2001, 2003). For example, considering the single file of water and ions distributed over the five binding sites S0 to S4, the approach of a third K+ ion from the intracellular side of the channel would advance the single file of alternating ions and water (W) to the extracellular side, K+ + W/K+/W/K+/W → K+/W/K+/W/K+ → W/K+/W/K+/W + K+, resulting in the net translocation of one K+ ion and one water molecule across the membrane. While this single file knock-on mechanism of ion conduction is consistent with streaming potential measurements indicating that about one water molecule is transported on average for each permeating K+ ion (Alcayaga et al., 1989), proposed alternative permeation mechanisms (Kopfer et al., 2014) could not be definitively ruled out due to the lack of experimental data. The issue was resolved using ultrafast time-resolution of 2D IR spectroscopy to detect the instantaneous multi-ion configurations in the narrow pore, revealing the predominance of single file structures with water molecules separating neighboring K+ ions (Kratochvil et al., 2016). Analysis of ions dynamics in simulations of bacterial Na+ channels indicates that a loosely coupled knock-on mechanism, with alternating occupancy of the channel by 2 and 3 ions, is prevalent (Chakrabarti et al., 2013; Finol-Urdaneta et al., 2014; Ing and Pomes, 2016). However, because the narrowest part of the pore is significantly wider than that of K+ channels, permeating ions remain at least partly hydrated (Figure 1b, bottom). In the following, we will focus mainly on the selectivity of Kv channels, which is most important for cellular excitability.
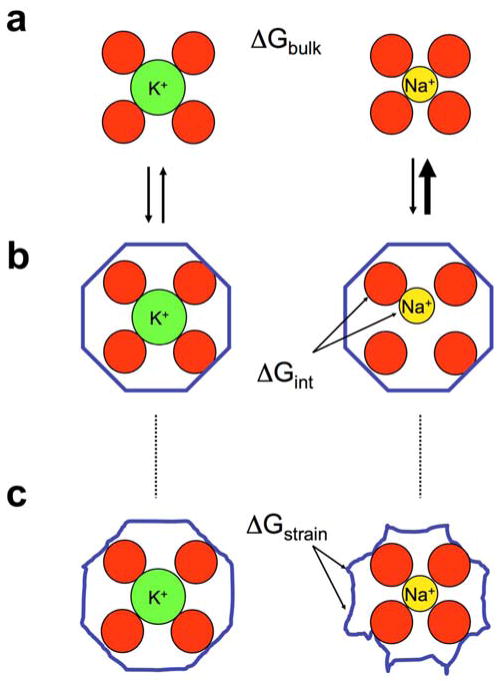
One unambiguous observation based on the crystallographic X-ray structures of K+ channels is that the K+ ions must become essentially dehydrated to permeate through the narrow selectivity filter (Zhou et al., 2001). In trying to understand the basis of selectivity of K+ channels, considerations of ion hydration is, thus, of paramount importance. Small alkali ions are very strongly hydrated in the bulk phase; the hydration free energy of Na+ and K+ is on the order of −98 kcal/mol and −80 kcal/mol, respectively (Friedman and Krishnan, 1973). To be “recognized” by a channel, an ion must shed at least part of its first hydration shell in the bound state. This implies that the free energy cost for dehydration must be compensated by interactions gained in the binding site for an ion to favorably occupy some location in the pore. Selectivity for K+ over Na+ arises when the difference in free energies of those ions in the pore departs from the corresponding difference in the bulk solution. It follows that ion selectivity of binding is fundamentally governed by differences in relative free energies, and that the problem of ion selectivity can be stated from the point of view of thermodynamic equilibrium as,
| (1) |
The difference of 18 kcal/mol in the hydration free energies of Na+ and K+ ions, corresponding to ΔGbulk, sets the fundamental “baseline” for the K+ or Na+ selectivity of any biological ion channels.
On the basis of the X-ray structure, the high selectivity for K+ over Na+ has traditionally been explained by pointing out that the carbonyl ligands along the selectivity filter provide a coordination structure perfectly adapted to K+ ions, but that Na+ ions are too small (the Pauling radius of Na+ is smaller than that of K+ by 0.38 Å) to fit well in the sites (Doyle et al., 1998; Hille, 1999; Zhou et al., 2001). This intuitively appealing explanation of K+ selectivity is the concept of the “snug-fit” proposed in the early 1970’s (Bezanilla and Armstrong, 1972). Similar concepts are invoked in host-guest chemistry (Dietrich, 1985), in which a host molecule with a (pre-organized) cavity binds an ion of the appropriate radius. As depicted in Figure 2a and 2b, the snug-fit mechanism posits that the binding site is, for structural reasons, rigidly constrained in an optimal geometry so that a dehydrated K+ fits with proper coordination but that Na+ is too small and is, thus, poorly coordinated by the host. Selectivity then comes about from the difference in the interaction of the ion with the coordinating ligands compared to the hydration free energy. The concept of snug-fit as pictured in Figure 2b is, however, obviously an idealization. In reality, channels are flexible macromolecules and may be able to structurally deform and adapt (to some extent) to a bound ion. Numerous computational studies based on atomic models highlighted the fluctuating and dynamical “liquid-like” nature of the selectivity filter of K+ channels (Guidoni et al., 1999; Allen, 2000; Åqvist and Luzhkov, 2000; Bernèche and Roux, 2000, 2001; Biggin et al., 2001; Luzhkov and Åqvist, 2001; Shrivastava et al., 2002; Noskov et al., 2004), suggesting that the origin of ion selectivity could be more complex (for review see (Roux et al., 2011)). When there is flexibility, the net difference in the direct ion-ligands interaction energy may exactly balance the difference in hydration free energy for both K+ and Na+ and yet, selectivity may be preserved as long as there is a sufficient build-up of unfavorable energy to deform and adapt the host for a given ion. This relates to the classical concept of strain energy invoked in host-guest chemistry (Dietrich, 1985). At first sight, the contribution from strain energy may seem somewhat counterintuitive because both ions appear well coordinated in the bound complexes (Figure 2c). For example, a visual examination of a crystallographic structure with a bound K+ would be unable to reveal if one is dealing with the situation of Figure 2b (left) or 2c (left). The concept of strain energy in host-guest chemistry is traditionally associated with structural distortions of the host (e.g., involving bonds, angles, and dihedrals). The implication is that size selectivity would be expected to vanish in the limit of a flexible host without sufficient structural stiffness. Quantitatively, the effective elastic restoring forces associated with the molecular stiffness are inversely proportional to the magnitude of the atomic thermal fluctuations (Allen et al., 2004; Roux et al., 2011). However, it can be shown that factors other than architectural deformations such as the ligand-ligand interactions can make important contributions also to the strain energy in a highly flexible site. This issue was clarified through a formal statistical mechanical analysis dissecting the contribution from the coordinating ligands from the rest of the protein (Yu et al., 2010). In the limit where a binding site is structurally very stiff and unable to adapt to a smaller or larger ion, ion selectivity is controlled by the ion-ligand interactions according to the familiar “snug-fit” cavity size mechanism of host-guest chemistry. But in the limit where a binding site is fairly flexible, the ligands behave as a confined liquid-like microdroplet that is free to fluctuate and adapt to ions of different sizes, and the relative binding free energy is then controlled by the interplay of ion-ligand and ligand-ligand interactions (Yu et al., 2010). This analysis explains how, in flexible binding sites, ion selectivity is ultimately controlled by the number and the physico-chemical properties of the ion-coordinating ligands (Noskov and Roux, 2006, 2007; Dixit et al., 2009; Roux, 2010; Yu et al., 2010). These concepts have also been helpful in explaining the selectivity of ion binding sites in transporters (Caplan et al., 2008; Larsson et al., 2010; Zhao and Noskov, 2011; Zhao et al., 2012) and the Na,K ATPase pump (Yu et al., 2011; Rui et al., 2016).
Figure 2.
Schematic illustration of fundamental concepts in thermodynamic ion binding selectivity. In (a), the K+ and Na+ ions are pictured in bulk solution with their first hydration shell. The difference in hydration free energy ΔGbulk between these two cations is ~18 kcal/mol. Binding to a rigid host (b) with a cavity size matching precisely a K+ ion (left) does not provide a favorable environment for the smaller Na+ (right). In this case, selectivity arises from the poor coordination interaction free energy ΔGint between the ion and its rigid host. This is the classical snug-fit mechanism of host-guest chemistry. However, selectivity may also be achieved by a flexible host (c) that is able to deform and adapt to both K+ and Na+ ion if there is a sufficient build-up of strain energy ΔGstrain (illustrated as a deformation of the blue molecular scaffolding). Reproduced with permission from (Noskov and Roux, 2007).
Up until now, our discussion has mainly focused on thermodynamic factors affecting ion selectivity. The last few years have seen an emergence of exciting ideas about the selectivity of K+ channels that departs from a view that strictly relies on thermodynamic equilibrium. Nimigean and Allen have emphasized the kinetic aspects of selectivity based on studies of Na+ blocks in the bacterial KcsA channel (Thompson et al., 2009; Kim and Allen, 2011; Nimigean and Allen, 2011). Moreover, results from a number of studies have highlighted the remarkable importance of the multi-ion single file on the selectivity of K+ channels. By examining the properties of MthK (Ye et al., 2010) and NaK (Derebe et al., 2011; Sauer et al., 2011) mutants, Jiang and co-workers showed that the channel becomes selective only if four consecutive binding sites exist along the narrow selectivity filter. This has culminated more recently with studies of two engineered mutants of the NaK channel, referred to as “NaK2K” and “NaK2CNG” (Liu and Lockless, 2013; Sauer et al., 2013). According to single-channel recordings, the NaK2K construct is K+-selective and the NaK2CNG construct is non-selective. Remarkably, despite being non-selective in ion permeation, the NaK2CNG filter display equilibrium binding preference for K+ over Na+, as indicated by measurements with isothermal titration calorimetry (ITC) and concentration-dependent ion replacement within the filter observed through crystallographic titration experiments. The K+-selective channels bind two or more K+ ions in the narrow filter, whereas the non-selective channels bind fewer ions. Based on the crystallographic titration experiments, the NaK2K construct has two high-affinity K+ sites while the NaK2CNG construct has only one K+-selective site. These experiments show that both K+-selective and non-selective channels select K+ over Na+ ions at equilibrium, implying that equilibrium binding at one site is insufficient to yield selective ion conduction (Liu and Lockless, 2013; Sauer et al., 2013). The data indicate that having multiple K+ ions bound simultaneously is required for selective K+ conduction, and that a reduction in the number of bound K+ ions destroys the multi-ion selectivity mechanism utilized by K+ channels. The implication is that the multi-ion character of the permeation process must, somehow, be a critical element for establishing the selective ion conductive through K+-channels. Under physiological conditions corresponding to high K+ concentration, it is expected that the permeation of one isolated Na+ “defect” would be “flanked” by two K+ ions along the narrow pore. The presence of the K+ ions on both side of the defect, perhaps, contributes to “chaperon” the structure of the selectivity filter into its conductive and selective conformation. Therefore, because of its highly concerted nature, ion conduction through the narrow selectivity filter of K+ channels is a multi-ion process that involves the evolution of several dynamical variables that are tightly coupled. Interesting, a computational study showed that when one allows the two outer ions in the single file K+/W/K+/W/K+, or K+/W/Na+/W/K+, to sample a Boltzmann-weighted average of their available positions then the central site S2 is not selective for K+ or Na+ (Medovoy et al., 2016). This is truly an unexpected result, demonstrating that selectivity arises as the result of a multi-ion process in this system. The selectivity for K+ over Na+ in the KcsA channel is a feature that emerges only as the result of the multi-ion process when the central ion crosses the site S2–an observation that is lacking in a simple analysis of the free energy of the central ion with the other at equilibrium in their binding sites.
The above discussion was mainly focused on the selectivity displayed by a K+ channel that is in its conductive open state. In fact, the molecular determinants of selectivity in K+ channels are sometimes entangled with many of the complex factors responsible for the overall conformational plasticity and relative stability of the conductive and non-conductive form of the filter (Callahan and Korn, 1994; Korn and Ikeda, 1995; Liu et al., 1996; Starkus et al., 1997; Kiss et al., 1999; Cheng et al., 2011; McCoy and Nimigean, 2012; Hoshi and Armstrong, 2013). For instance, the filter of some K+ channels appears to be unable to remain in its conductive conformation when exposed to an ionic solution with few or no K+ ions. For example, the selectivity filter of the KcsA channel rapidly converts to a non-conductive state under such conditions because K+ ions are needed to maintain the conductive state of the channel, giving rise to some kind of conformationally-driven super-selectivity, a complex phenomenon that must be distinguished from the more traditional notion of conductive-state selectivity (Alam and Jiang, 2011; Roux et al., 2011).
CONCLUSION
Ion channels are large transmembrane proteins that serve to provide a pathway for ions to diffuse at a high rate across the cell membrane according to their electrochemical potential. The action potential of excitable cells reflects the rapid opening and closing of voltage-activated Na+- and K+-selective channels in response to changes in the transmembrane potential (Hodgkin and Huxley, 1952). In the last decades, our understanding of ion channels has greatly advanced through the determination of three-dimensional structure by X-ray crystallography (Doyle et al., 1998; Zhou et al., 2001; Payandeh et al., 2011) and the utilization of computational methods to simulate the dynamical motions of the atoms as a function of time (Roux, 2005; Ing and Pomes, 2016). Architecturally, Na+ and K+ ion channels comprise a narrow pore region called the selectivity filter where permeating ions are either completely or partially dehydrated (Zhou et al., 2001; Payandeh et al., 2011). Issues of hydration free energy are thus of fundamental importance to understand ion selectivity. The permeation process of K+ ions through the K+ channel is believed to proceed predominantly as a multi-ion “knock-on” mechanism, in which the single-file translocation of K+ ions diffuses along the narrow selectivity filter (Kratochvil et al., 2016). A similar mechanism has been proposed for the permeation through bacterial Na+ channels (Ing and Pomes, 2016). Theoretical and computational analysis has led to a better understanding of the role of conformational flexibility and structural strain energy on ion selectivity (Noskov et al., 2004; Yu et al., 2010). Additional kinetic factors related to the multi-ion nature of the pore appear to be also important for the selectivity of K+ channels (Medovoy et al., 2016). Lastly, the selectivity of the latter appears to be also entangled with the conformational plasticity and relative stability of the conductive and non-conductive form of the pore (Roux et al., 2011).
Summary points.
Ion channels provide energetically favorable passage for ions to diffuse rapidly and passively according to their electrochemical potential.
Selective ion channels are essential for the excitability of biological membranes: the action potential is a transient phenomenon that reflects the rapid opening and closing of voltage-dependent Na+-selective and K+-selective channels.
Voltage-dependent Na+-selective and K+-selective channels share a similar overall topology
Rapid permeation through the K+ channel selectivity filter proceeds as a “knock-on” mechanism in which 2–3 K+ ions interspersed by water molecules move in single file.
Computational analysis shows that complex effects play an important role in setting the selectivity of K+ channels
Acknowledgments
This work was supported by the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health (NIH) through grant R01-GM062342. The author is grateful to Jing Li for his help.
References
- Alam A, Jiang Y. Structural studies of ion selectivity in tetrameric cation channels. J Gen Physiol. 2011;137:397–403. doi: 10.1085/jgp.201010546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcayaga C, Cecchi X, Alvarez O, Latorre R. Streaming potential measurements in Ca2+-activated K+ channels from skeletal and smooth muscle. Coupling of ion and water fluxes. Biophys J. 1989;55:367–371. doi: 10.1016/S0006-3495(89)82814-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TW, Andersen OS, Roux B. On the importance of atomic fluctuations, protein flexibility, and solvent in ion permeation. Journal of General Physiology. 2004;124:679–690. doi: 10.1085/jgp.200409111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TW, Bliznyuk A, Rendell AP, Kyucak S, Chung SH. The potassium channel: Structure, selectivity and diffusion. J Chem Phys. 2000;112:8191–8204. [Google Scholar]
- Åqvist J, V, Luzhkov B. Ion permeation mechanism of the potassium channel. Nature. 2000;404:881–884. doi: 10.1038/35009114. [DOI] [PubMed] [Google Scholar]
- Bagneris C, Naylor CE, McCusker EC, Wallace BA. Structural model of the open-closed-inactivated cycle of prokaryotic voltage-gated sodium channels. The Journal of general physiology. 2015;145:5–16. doi: 10.1085/jgp.201411242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernèche S, Roux B. Molecular dynamics of the KcsA K+ channel in a bilayer membrane. Biophysical Journal. 2000;78:2900–2917. doi: 10.1016/S0006-3495(00)76831-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernèche S, Roux B. Energetics of ion conduction through the K+ channel. Nature. 2001;414:73–77. doi: 10.1038/35102067. [DOI] [PubMed] [Google Scholar]
- Bernèche S, Roux B. A microscopic view of ion conduction through the K+ channel. Proc Natl Acad Sci USA. 2003;100:8644–8648. doi: 10.1073/pnas.1431750100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla F, Armstrong CM. Negative conductance caused by entry of sodium and cesium ions into the potassium channels of squid axons. J Gen Physiol. 1972;60:588–608. doi: 10.1085/jgp.60.5.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin PC, Smith GR, Shrivastava I, Choe S, Sansom MS. Potassium and sodium ions in a potassium channel studied by molecular dynamics simulations. Biochim Biophys Acta. 2001;1510:1–9. doi: 10.1016/s0005-2736(00)00345-x. [DOI] [PubMed] [Google Scholar]
- Callahan MJ, Korn SJ. Permeation of Na+ through a delayed rectifier K+ channel in chick dorsal root ganglion neurons. The Journal of general physiology. 1994;104:747–771. doi: 10.1085/jgp.104.4.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan DA, Subbotina JO, Noskov SY. Molecular mechanism of ion-ion and ion-substrate coupling in the Na+-dependent leucine transporter LeuT. Biophysical journal. 2008;95:4613–4621. doi: 10.1529/biophysj.108.139741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall WA. Structure and function of voltage-gated sodium channels at atomic resolution. Exp Physiol. 2014;99:35–51. doi: 10.1113/expphysiol.2013.071969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall WA, Zheng N. Deciphering voltage-gated Na(+) and Ca(2+) channels by studying prokaryotic ancestors. Trends in biochemical sciences. 2015;40:526–534. doi: 10.1016/j.tibs.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti N, Ing C, Payandeh J, Zheng N, Catterall WA, Pomes R. Catalysis of Na+ permeation in the bacterial sodium channel Na(V)Ab. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:11331–11336. doi: 10.1073/pnas.1309452110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng WW, McCoy JG, Thompson AN, Nichols CG, Nimigean CM. Mechanism for selectivity-inactivation coupling in KcsA potassium channels. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5272–5277. doi: 10.1073/pnas.1014186108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipot C. Free energy calculations applied to membrane proteins. Methods in molecular biology. 2008;443:121–144. doi: 10.1007/978-1-59745-177-2_7. [DOI] [PubMed] [Google Scholar]
- Derebe MG, Sauer DB, Zeng W, Alam A, Shi N, Jiang Y. Tuning the ion selectivity of tetrameric cation channels by changing the number of ion binding sites. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:598–602. doi: 10.1073/pnas.1013636108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich B. Coordination chemistry of alkali and alkaline-earth cations with macrocyclic ligands. J Chem Edu. 1985;62:954–964. [Google Scholar]
- Dixit PD, Merchant S, Asthagiri D. Ion selectivity in the KcsA potassium channel from the perspective of the ion binding site. Biophysical journal. 2009;96:2138–2145. doi: 10.1016/j.bpj.2008.12.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- Egwolf B, Roux B. Ion selectivity of the KcsA channel: a perspective from multi-ion free energy landscapes. J Mol Biol. 2010;401:831–842. doi: 10.1016/j.jmb.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenman G, Krasne S. Further considerations on the ion selectivity of carrier molecules and membranes. IV International Biophysics Congress Symposium on Membrane Structure and Function.1973. [Google Scholar]
- Finol-Urdaneta RK, Wang Y, Al-Sabi A, Zhao C, Noskov SY, French RJ. Sodium channel selectivity and conduction: prokaryotes have devised their own molecular strategy. J Gen Physiol. 2014;143:157–171. doi: 10.1085/jgp.201311037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman HL, Krishnan CV. Water : A Comprehensive Treatise. Vol. 3. Plenum Press; New York: 1973. [Google Scholar]
- Furini S, Domene C. Atypical mechanism of conduction in potassium channels. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:16074–16077. doi: 10.1073/pnas.0903226106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidoni L, Torre V, Carloni P. Potassium and sodium binding to the outer mouth of the K+ channel. Biochemistry. 1999;38:8599–8604. doi: 10.1021/bi990540c. [DOI] [PubMed] [Google Scholar]
- Heginbotham L, Abramson T, MacKinnon R. A functional connection between the pores of distantly related ion channels as revealed by mutant K+ channels. Science. 1992;258:1152. doi: 10.1126/science.1279807. [DOI] [PubMed] [Google Scholar]
- Heginbotham L, Lu Z, Abramson T, Mackinnon R. Mutations in the K+ channel signature sequence. Biophysical journal. 1994;66:1061–1067. doi: 10.1016/S0006-3495(94)80887-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann SH, Terlau H, Stuhmer W, Imoto K, Numa S. Calcium channel characteristics conferred on the sodium channel by single mutations. Nature. 1992;356:441–443. doi: 10.1038/356441a0. [DOI] [PubMed] [Google Scholar]
- Hille B. Potassium channels in myelinated nerve-selective permeability to small cations. J Gen Physiol. 1973;61:599. doi: 10.1085/jgp.61.6.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille B, Armstrong CM, MacKinnon R. Ion channels:From idea to reality. Nature Medicine. 1999;5:1105–1109. doi: 10.1038/13415. [DOI] [PubMed] [Google Scholar]
- Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation of nerve. J Physiol (Lond) 1952;117:500–544. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshi T, Armstrong CM. C-type inactivation of voltage-gated K+ channels: Pore constriction or dilation? The Journal of general physiology. 2013;141:151–160. doi: 10.1085/jgp.201210888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ing C, Pomes R. Simulation Studies of Ion Permeation and Selectivity in Voltage-Gated Sodium Channels. Curr Top Membr. 2016;78:215–260. doi: 10.1016/bs.ctm.2016.07.005. [DOI] [PubMed] [Google Scholar]
- Jensen MO, Jogini V, Eastwood MP, Shaw DE. Atomic-level simulation of current-voltage relationships in single-file ion channels. The Journal of general physiology. 2013;141:619–632. doi: 10.1085/jgp.201210820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Allen TW. On the selective ion binding hypothesis for potassium channels. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:17963–17968. doi: 10.1073/pnas.1110735108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss L, LoTurco J, Korn SJ. Contribution of the selectivity filter to inactivation in potassium channels. Biophysical journal. 1999;76:253–263. doi: 10.1016/S0006-3495(99)77194-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopfer DA, Song C, Gruene T, Sheldrick GM, Zachariae U, de Groot BL. Ion permeation in K(+) channels occurs by direct Coulomb knock-on. Science. 2014;346:352–355. doi: 10.1126/science.1254840. [DOI] [PubMed] [Google Scholar]
- Korn SJ, Ikeda SR. Permeation selectivity by competition in a delayed rectifier potassium channel. Science. 1995;269:410–412. doi: 10.1126/science.7618108. [DOI] [PubMed] [Google Scholar]
- Kratochvil HT, Carr JK, Matulef K, Annen AW, Li H, Maj M, Ostmeyer J, Serrano AL, Raghuraman H, Moran SD, Skinner JL, Perozo E, Roux B, Valiyaveetil FI, Zanni MT. Instantaneous ion configurations in the K+ ion channel selectivity filter revealed by 2D IR spectroscopy. Science. 2016;353:1040–1044. doi: 10.1126/science.aag1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix JJ, Campos FV, Frezza L, Bezanilla F. Molecular bases for the asynchronous activation of sodium and potassium channels required for nerve impulse generation. Neuron. 2013;79:651–657. doi: 10.1016/j.neuron.2013.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson HP, Wang X, Lev B, Baconguis I, Caplan DA, Vyleta NP, Koch HP, Diez-Sampedro A, Noskov SY. Evidence for a third sodium-binding site in glutamate transporters suggests an ion/substrate coupling model. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13912–13917. doi: 10.1073/pnas.1006289107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Lockless SW. Equilibrium selectivity alone does not create K(+)-selective ion conduction in K(+) channels. Nature communications. 2013;4:2746. doi: 10.1038/ncomms3746. [DOI] [PubMed] [Google Scholar]
- Liu Y, Jurman ME, Yellen G. Dynamic rearrangement of the outer mouth of a K+ channel during gating. Neuron. 1996;16:859–867. doi: 10.1016/s0896-6273(00)80106-3. [DOI] [PubMed] [Google Scholar]
- Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- Luzhkov VB, Åqvist J. K(+)/Na(+) selectivity of the KcsA potassium channel from microscopic free energy perturbation calculations. Biochim Biophys Acta. 2001;1548:194–202. doi: 10.1016/s0167-4838(01)00213-8. [DOI] [PubMed] [Google Scholar]
- McCoy JG, Nimigean CM. Structural correlates of selectivity and inactivation in potassium channels. Biochim Biophys Acta. 2012;1818:272–285. doi: 10.1016/j.bbamem.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker EC, Bagneris C, Naylor CE, Cole AR, D’Avanzo N, Nichols CG, Wallace BA. Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing. Nat Commun. 2012;3:1102. doi: 10.1038/ncomms2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medovoy D, Perozo E, Roux B. Multi-ion free energy landscapes underscore the microscopic mechanism of ion selectivity in the KcsA channel. Biochim Biophys Acta. 2016;1858:1722–1732. doi: 10.1016/j.bbamem.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor CE, Bagneris C, DeCaen PG, Sula A, Scaglione A, Clapham DE, Wallace BA. Molecular basis of ion permeability in a voltage-gated sodium channel. The EMBO journal. 2016;35:820–830. doi: 10.15252/embj.201593285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nernst W. Die elektromotorische Wirksamkeit der Ionen. Z Phys Chem. 1889;4:129–181. [Google Scholar]
- Nimigean CM, Allen TW. Perspectives: Origins of ion selectivity in potassium channels from the perspective of channel block. The Journal of general physiology. 2011 doi: 10.1085/jgp.201010551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noskov SY, Bernèche S, Roux B. Control of ion selectivity in potassium channels by electrostatic and dynamic properties of carbonyl ligands. Nature. 2004;431:830–834. doi: 10.1038/nature02943. [DOI] [PubMed] [Google Scholar]
- Noskov SY, Roux B. Ion selectivity in potassium channels. Biophysical Chemistry. 2006;124:279–291. doi: 10.1016/j.bpc.2006.05.033. [DOI] [PubMed] [Google Scholar]
- Noskov SY, Roux B. Importance of hydration and dynamics on the selectivity of the KcsA and NaK channels. J Gen Physiol. 2007;129:135–143. doi: 10.1085/jgp.200609633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payandeh J, Gamal El-Din TM, Scheuer T, Zheng N, Catterall WA. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature. 2012;486:135–139. doi: 10.1038/nature11077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B. Ion conduction and selectivity in K+ channels. Annual Review of Biophysics and Biomolecular Structure. 2005;34:153–171. doi: 10.1146/annurev.biophys.34.040204.144655. [DOI] [PubMed] [Google Scholar]
- Roux B. Exploring the ion selectivity properties of a large number of simplified binding site models. Biophys J. 2010;98:2877–2885. doi: 10.1016/j.bpj.2010.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B, Berneche S, Egwolf B, Lev B, Noskov SY, Rowley CN, Yu H. Ion selectivity in channels and transporters. J Gen Physiol. 2011;137:415–426. doi: 10.1085/jgp.201010577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui H, Artigas P, Roux B. The selectivity of the Na(+)/K(+)-pump is controlled by binding site protonation and self-correcting occlusion. eLife. 2016;5 doi: 10.7554/eLife.16616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer DB, Zeng W, Canty J, Lam Y, Jiang Y. Sodium and potassium competition in potassium-selective and non-selective channels. Nature communications. 2013;4:2721. doi: 10.1038/ncomms3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer DB, Zeng W, Raghunathan S, Jiang Y. Protein interactions central to stabilizing the K+ channel selectivity filter in a four-sited configuration for selective K+ permeation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16634–16639. doi: 10.1073/pnas.1111688108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava IH, Tieleman DP, Biggin PC, Sansom MS. K(+) versus Na(+) ions in a K channel selectivity filter: a simulation study. Biophys J. 2002;83:633–645. doi: 10.1016/s0006-3495(02)75197-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JC, Roux B. Eppur si muove! The 2013 nobel prize in chemistry. Structure. 2013;21:2102–2105. doi: 10.1016/j.str.2013.11.005. [DOI] [PubMed] [Google Scholar]
- Starkus JG, Kuschel L, Rayner MD, Heinemann SH. Ion conduction through C-type inactivated Shaker channels. The Journal of general physiology. 1997;110:539–550. doi: 10.1085/jgp.110.5.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AN, Kim I, Panosian TD, Iverson TM, Allen TW, Nimigean CM. Mechanism of potassium-channel selectivity revealed by Na(+) and Li(+) binding sites within the KcsA pore. Nature structural & molecular biology. 2009;16:1317–1324. doi: 10.1038/nsmb.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmschneider MB, Bagneris C, McCusker EC, Decaen PG, Delling M, Clapham DE, Ulmschneider JP, Wallace BA. Molecular dynamics of ion transport through the open conformation of a bacterial voltage-gated sodium channel. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:6364–6369. doi: 10.1073/pnas.1214667110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S, Li Y, Jiang Y. Novel insights into K+ selectivity from high-resolution structures of an open K+ channel pore. Nature structural & molecular biology. 2010;17:1019–1023. doi: 10.1038/nsmb.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Noskov SY, Roux B. Two mechanisms of ion selectivity in protein binding sites. Proc Natl Acad Sci USA. 2010;107:20329–20334. doi: 10.1073/pnas.1007150107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, I, Ratheal M, Artigas P, Roux B. Protonation of key acidic residues is critical for the K-selectivity of the Na/K pump. Nat Struct & Mol Biol. 2011;18:1159–1163. doi: 10.1038/nsmb.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ren W, DeCaen P, Yan C, Tao X, Tang L, Wang J, Hasegawa K, Kumasaka T, He J, Clapham DE, Yan N. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature. 2012;486:130–134. doi: 10.1038/nature11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Noskov SY. The role of local hydration and hydrogen-bonding dynamics in ion and solute release from ion-coupled secondary transporters. Biochemistry. 2011;50:1848–1856. doi: 10.1021/bi101454f. [DOI] [PubMed] [Google Scholar]
- Zhao C, Stolzenberg S, Gracia L, Weinstein H, Noskov S, Shi L. Ion-controlled conformational dynamics in the outward-open transition from an occluded state of LeuT. Biophysical journal. 2012;103:878–888. doi: 10.1016/j.bpj.2012.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 2001;414:43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]