Abstract
Potassium channels are responsible for the selective permeation of K+ ions across cell membranes. K+ ions permeate in single file through the selectivity filter, a narrow pore lined by backbone carbonyls that compose 4 K+ binding sites. Here, we report 2D IR spectra of a semisynthetic KcsA channel with site-specific 13C18O isotope labels in the selectivity filter. The ultrafast time-resolution of 2D IR spectroscopy provides an instantaneous snapshot of the multi-ion configurations and structural distributions that occur spontaneously in the filter. Two elongated features are resolved, revealing the statistical weighting of two structural conformations. The spectra are reproduced by MD simulations of structures with water separating two K+ ions in the binding sites, ruling out configurations with ions occupying adjacent sites.
Main Text
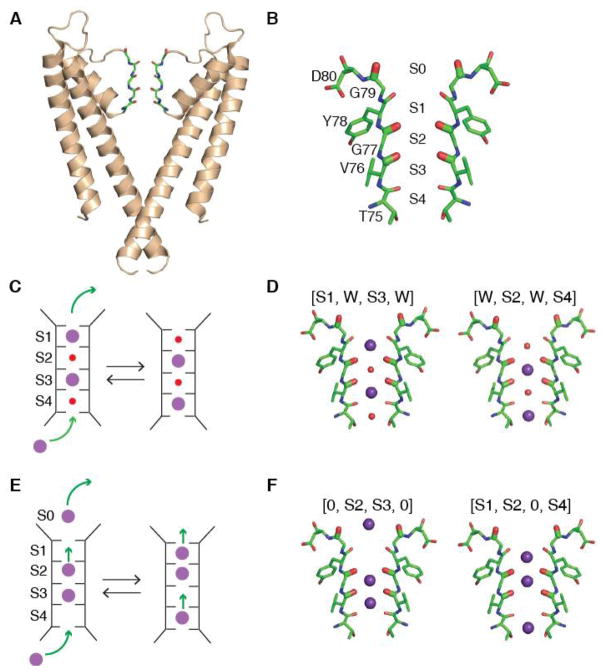
Ion channels that mediate the flux of K+ are responsible for determining the resting membrane potential and for the repolarization phase of action potentials of excitable cells (1). Ion permeation through all K+ channels is characterized by high selectivity and throughput approaching the diffusion limit. These permeation properties are defined by the selectivity filter, a highly conserved structural element consisting of four K+ binding sites (S1–S4, labeled extracellular to intracellular, Fig. 1B). During translocation, K+ coordination is carried out by main chain carbonyl oxygens (plus a threonine side chain in S4) (2, 3).
Figure 1. Overview of Ion Permeation Mechanisms of KcsA.
(A) X-ray crystal structure of KcsA (PDB ID 1K4C) with two of the four protein subunits shown. (B) The selectivity filter of KcsA is lined with the carbonyl groups (oxygen in red) of four amino acids (Thr75, Val76, Gly77, and Tyr78). Amino acids that were 13C18O isotope labeled for the experiment are shown bolded (Val76, Gly77, and Gly79). The binding sites (S1 to S4) span the selectivity filter. (C) Knock-on mechanism for potassium ion permeation through the channel. In this mechanism, K+ (purple), alternate with water molecules (red) and collectively move through the filter when a new K+ enters the filter. (D) The two most prominent ion-binding configurations for the knock-on mechanism used to simulate the 2D IR spectra: (left) [S1,W,S3,W] and (right) [W,S2,W,S4]. (E) The hard-knock model for ion conduction. When a K+ ion enters the S4 binding site, strong electrostatic repulsions simultaneously translocate two ions upward. No water is involved in this mechanism. (F) The two ion configurations used to simulate the 2D IR spectra: (left) [0,S2,S3,0] and (right) [S1,S2,0,S4]. Simulations for 97% of the ion configurations proposed by the hard-knock mechanism are shown in the SM, Fig. S7.
The mechanism of ion permeation through the selectivity filter has been investigated using diverse experimental techniques such as radiotracer flux assays, single channel electrophysiological measurements, X-ray crystallography and computational studies (3–11). These experiments are broadly supportive of a mechanism commonly referred to as “knock–on” permeation (4). This mechanism posits that the narrow selectivity filter is simultaneously occupied by two K+ ions in single file, either at sites S1 and S3 or at sites S2 and S4, with intervening water molecules (Fig. 1C, D) (9–13). The approach of a third K+ ion from the intracellular side of the channel would advance the single file of alternating ions and water to the extracellular side, resulting in the net translocation of one K+ ion and one water molecule across the membrane. However, the experimental data is not sufficiently conclusive to rule out alternative models for the permeation mechanism (14–16). Of these, a recently proposed “hard-knock” permeation model suggests a very different mechanism. It postulates that the conductance of K+ channels arises from direct, short-range ion-ion collisions inside the filter, so that a pair of adjacent K+ ions occupy the selectivity filter at adjacent sites S2 and S3 with no intervening water molecules [0,S2,S3, 0] (17). In this model, permeation would take place when a third K+ ion enters the S4 binding site [0,S2,S3,S4], colliding directly with the S2/S3 ion pair to create the ion configuration [S1,S2,0,S4]. Eventually, S1 would leave the filter and the K+ at S4 transitions to S3, starting over again (17). Here, we have used two-dimensional infrared (2D IR) spectroscopy and protein semisynthesis to test these permeation models, providing new experimental data on the ion and water configurations in the selectivity filter of the KcsA channel.
Atomic bond vibrations are sensitive to their electrostatic environment, and so the frequencies are dictated by nearby ions and water as well as the structure and dynamics of the protein itself (18). The inherent time-resolution of 2D IR spectroscopy is a few picoseconds, which implies that a 2D IR spectrum provides a nearly instantaneous snapshot of the dynamic channel and the content of the pore (19). In essence, it is a weighted average of the ion configurations and protein structural distributions (20). For the 2D IR experiments, we used protein semisynthesis to selectively isotope label the ion binding sites in the selectivity filter. Using the native chemical ligation reaction, we assembled the KcsA protein from a synthetic peptide (residues 70–81) that encompasses the selectivity filter and two recombinant peptides corresponding to the rest of the protein. (21, 22). We introduced 13C18O labels on the backbone carbonyls of Val76, Gly77, and Gly79 by using the appropriately labeled residues during synthesis of the peptide fragment (Fig. 1B). The 13C18O labels cause a 65 cm−1 shift of the carbonyl frequency (amide I vibrations) (23), enabling us to spectroscopically isolate and probe the selectivity filter. Gly77 and Val76 probe the S1, S2, and S3 binding sites. These are good sites for labeling as these sites have different ion occupancies according to the two models. Labeling Gly77 and Val76 together makes the spectra particularly sensitive to ion configurations because of vibrational coupling between these two residues (24).
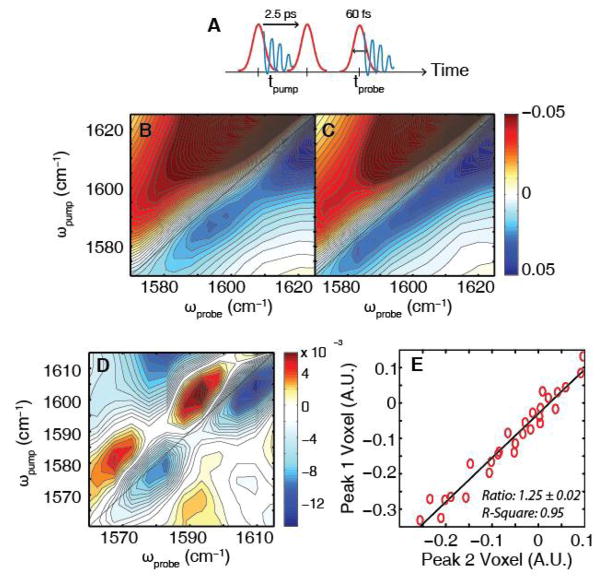
Shown in Fig. 2C is the 2D IR spectrum of labeled KcsA. This spectrum was collected using the pulse sequence shown in Fig. 2A. This approach is analogous to that used in 2D NMR spectroscopy, but probing vibrational modes. This pulse sequence produces an infrared signal that oscillates as a function of the time-delay between the pump pulses and after the probe pulse, respectively. The signal is Fourier transformed along these two time-axes to give a 2D IR spectrum that correlates the two frequency axes. The spectrum in Fig. 2C is shown in the region that the 13C18O labeled carbonyls absorb (full spectra in Supplementary Material (SM)). Acidic side chains also absorb in this region, creating a background (25, 26). To remove the background, we also measured a KcsA sample without isotope labels under identical conditions. The intensity of the unlabeled KcsA spectrum is about 50% lower in this region, which is approximately what we would expect based on the ratio of labels to acidic side chains.
Figure 2. Experimental 2D IR spectra of unlabeled and 13C18O-labeled KcsA at high potassium concentrations.
(A) Pulse sequence for 2D IR spectrum with illustrated vibrational coherences (blue). Fourier transform of the coherence during tprobe to give the ωprobe axis while the time between the pump pulses, tpump, is computationally Fourier transformed to give the ωpump axis. (B) 2D IR spectrum of unlabeled KcsA showing the absorption of side chains in the region where the isotope labeled features are expected to appear. (C) 2D IR spectrum of labeled KcsA shows a 50% increase in intensity in this region, consistent with absorption by the 13C18O labeled residues. (D) Subtraction of the unlabeled spectrum from the labeled spectrum yields two pairs of peaks. The bleaches (blue) appear at (ωpump,ωprobe )= (1603 cm−1, 1609 cm−1) and (1580 cm−1, 1582 cm−1), respectively. (E) For every unique combination of difference spectra, the intensities of the 3x3 pixel array centered at the pixels for (ωpump, ωprobe) = (1603 cm−1, 1609 cm−1) (peak1) and (1580 cm−1, 1582 cm−1) (peak 2) were integrated, respectively. The slope of the fit yielded an intensity ratio of 1.25 ± 0.02 with an R-square of 0.95.
Fig. 2D shows the difference spectrum generated by subtracting Fig. 2B and 2C. It contains two pairs of oppositely-signed peaks, with blue, negative peaks corresponding to the 0→1 vibrational transition and red, positive peaks corresponding to the 1→2 vibrational transition. For each isotope label, our pulse sequence will create a pair of oppositely-signed peaks, with the negative peak on the diagonal. Thus, the difference spectrum resolves two vibrational features, which we refer to by their frequencies along the y-axis, ωpump = 1603 cm−1 and 1580 cm−1. The frequencies reflect the magnitude of the electric fields inside the filter as well as the coupling between residues set by the backbone structure (27, 28). The two pairs of peaks have very different frequencies, with a peak separation of 23 cm−1 along ωpump, indicating that there is either a single protein structure or ion configuration that has distinct electrostatics at the separate binding sites or a weighted average of two conformations with very different electrostatics. A statistical analysis is presented in the SM that address reproducibility and statistical significance. A portion of this analysis is shown in Fig. 2E, which plots the intensity of the 1603 and 1580 cm−1 peaks for every combination of foreground and background spectra that we collected. If the experimental peaks were not reproducible, then this plot would be uncorrelated. Instead, a strong correlation is observed with a slope of 1.25 ± 0.02, which equals the relative intensities of the two peaks. As we show below, the relative intensity gives the statistical weighting of ion channel structures.
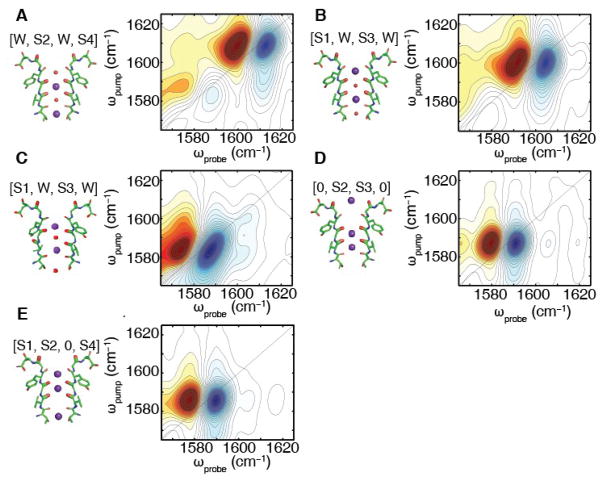
To test if these experimental features can be explained by either of the two permeation models described above, we conducted molecular dynamics (MD) simulations and computed 2D IR spectra for all relevant ion configurations (see SM for Methods). Shown in Fig. 3A and 3B are 2D IR spectra calculated for each of the two multi-ion configurations predicted to occur during ion conduction according to the knock-on model, [W,S2,W,S4] and [S1,W,S3,W]. The spectrum of the [W,S2,W,S4] ion configuration has an intense pair of peaks at 1608 cm−1. A similar spectrum is predicted for the [S1,W,S3,W] ion binding configuration, albeit centered at 1600 cm−1. Simulations of the individual residues reveals that Val76, which is coordinating the S2 K+ ion, is mostly responsible for the frequency and the spectral intensity we observe in this ion binding configuration (see SM). MD simulations also predict that the Val76 carbonyl is often flipped outwards (16, 29), away from the pore (see Fig. 3C) when the ions are in the [S1,W,S3,W] configuration. Simulations of that structure predict a pair of peaks that are much lower in frequency, absorbing at 1583 cm−1. The flipped state is not predicted to exist for the [W,S2,W,S4] ion configuration (29).
Figure 3. Calculated 2D IR spectra to test proposed ion configurations of the KcsA selectivity filter.
The calculated 2D IR spectra for the (A) [W,S2,W,S4], (B) [S1,W,S3,W], (C) [S1,W,S3,W] with a single Val76 flip, (D) [0,S2,S3,0], and (E) [S1,S2,0,S4] K+ binding configurations of the labeled KcsA filter from MD simulations using a K+ charge profile described in (35). In (A), we have the spectrum for the [W,S2,W,S4] ion binding configuration in which all of the Val76 residues point into the filter. In this case, we observe peaks at (ωpump, ωprobe) = (1608 cm−1, 1612 cm−1). (B) For the [S1,W,S3,W] configuration, we see a similar spectrum with (ωpump, ωprobe) = (1600 cm−1, 1604 cm−1). (C) For the [S1,W,S3,W] ion configuration of the Val76-flipped state, we get a pair of inhomogeneous peaks at (ωpump, ωprobe) = (1583 cm−1, 1587 cm−1). Fig. 3(D) and (E) are the calculated 2D IR spectra of the [0,S2,S3,0] and [S1,S2,0,S4] binding configurations, respectively. In (E), we observe a single homogenous peak at (ωpump, ωprobe) = (1587 cm−1, 1591 cm−1), which is similar to the homogeneous peak we observe in (F) at (wpump, Wpwbe) (1585 ern·′, 1589 ern−1. The spectra have been normalized for visualization purposes.
Shown in Fig. 3C and 3D are the calculated 2D IR spectra for two of the ion configurations of the new hard-knock model [0,S2,S3,0] and [S1,S2,0,S4]. Both ion configurations predict nearly the same 2D IR spectrum, with frequencies of 1587 and 1585 cm−1. Ions in the channel tend to shift carbonyls to lower frequencies because of their charge, which is why this model predicts low frequencies for both ion configurations. The hard-knock model also contains other ion configurations in its mechanism, whose computed spectra are given in the SM (Fig. S7). The predicted spectra for the ion configuration corresponding to the knock-on and the hard-knock models are very different from one another in both peak frequency and 2D lineshape.
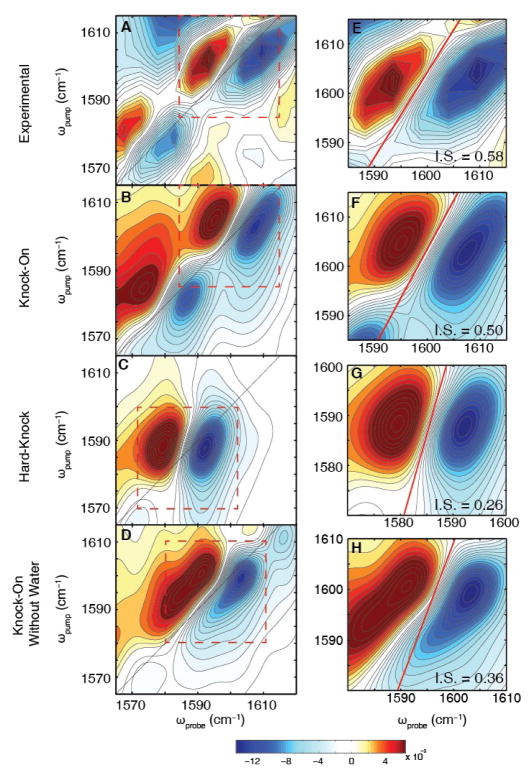
For comparison to experiment, the simulated spectra of the structures in the knock-on model ([W,S2,W,S4]: [S1,W,S3,W]:flipped[S1,W,S3,W]) were added at a ratio of 0.3:0.3:0.4 to match the relative intensities in the experimental spectra (the experimental peaks have a ratio of 1.25 ± 0.02 from Fig. 2E). The agreement between the knock-on model and experiment is good (Fig. 4B vs. 4A), especially considering that the only adjustable parameters in these calculations are the relative ratios. We also added together the 2D spectra for 97% of the ion configurations predicted from the hard-knock model (Fig. 3D, 3E and S7), which gives the spectrum in Fig. 4C (17). The agreement to experiment is poor; the simulations only predict one set of peaks instead of two. There is no linear combination of hard-knock ion configurations or structures that can match the experiment. We have also explored whether the ion configurations of the hard-knock model occur, albeit less frequently along with the configurations for the knock-on states. To do so, we added the spectra of the hard-knock configurations to the knock-on states (Fig. S12). We estimate that there cannot be more than 15% of the states in the hard-knock configuration, as increasing the population beyond this limitation decreases the frequency separation and changes the 2D lineshapes of the peaks. Therefore, our data are consistent with ion distributions in the filter predicted by the classic knock-on model, but not the hard-knock model proposed more recently.
Figure 4. Comparison of 2D IR experimental spectrum to the simulated spectra of the knock-on models (with and without water) and the hard-knock model.
(A) Experimental difference spectrum shown in Fig. 2C. (B) Simulated 2D IR spectrum for the knock-on model with water generated by a weighted average of the [W,S2,W,S4], [S1,W,S3,W], and Val76 flipped [S1,W,S3,W] ion configurations in a ratio of 0.3:0.3:0.4, respectively. The simulated spectrum agrees well with peak positions, 2D lineshapes, and intensity ratio. (C) Simulated 2D IR spectrum for the hard-knock model from a weighted average of [0,S2,S3,0] and [S1,S2,0,S4] along with other hard-knock states (see SM). (D) Simulated 2D IR spectrum of the knock-on model with water removed with the same ratios of states used to match experiment (Fig. 4B). No linear combination of these states without water (see Fig. S14) reproduces the experimental spectrum. Therefore, water must be present within the filter. (E–H) Zoom-ins of the high frequency peak of each spectra. The nodal line slopes are illustrated by red lines with values of the inverse slope (IS).
An analysis of the 2D lineshapes corroborates our conclusions and provides additional insights. The calculated 2D lineshapes are elongated for the knock-on model and rounder for the hard-knock model (Fig. 4B and D, respectively). 2D lineshapes are elongated along the diagonal when there is a heterogeneous subset of frequencies—and accordingly, structures—each with its own set of electric fields. In essence, the 2D lineshapes measure the structural distribution of the selectivity filter for each ion configuration (30). Shown in Fig. 4E–H (red lines) are nodal line slopes that quantify the inhomogeneity of the 2D lineshapes. The experiment and simulations of the knock-on model both have inverse slopes of about 0.6, whereas the hard-knock model has an inverse slope of about 0.3 (see Table S3). To test the effect of water on the knock-on spectra, we reran the molecular dynamics simulations without water in the selectivity filter (Fig. 4D). The simulations predict that the high frequency peak bifurcates because the [0,S2,0,S4] ion binding configuration blueshifts 4 cm−1 (see Fig. S13) and the inverse slope is too small. There are no linear combinations of the knock-on states without water that can reproduce the experimental spectrum in Fig. 4B. Thus, water must be present in the channel in order to reproduce the experimental 2D lineshape. It follows that during ion conduction, one water molecule is transported for each K+ ion, in accord with streaming potential (9, 12, 13).
Analyzing the angles of the isotope labeled carbonyls inside the selectivity filter, we found that removing the water from the knock-on model reduces their angular distribution by as much as 36% (see Fig. S11). The hard-knock model, which also lacks water, has a similarly narrow angular distribution. Without the water, the high positive charge inside the filter causes a more rigid protein conformation and less dynamic K+ ions, similar to experiments on K+ binding to model peptides (24).
Since the temporal resolution of 2D IR spectroscopy is orders-of-magnitude shorter than the timescales for ion permeation, we can determine from the 2D IR spectra the relative populations of each of these ion binding states. The experimental peaks have a ratio of 1.25 ± 0.02 from Fig. 2E. It follows that the relative equilibrium population of the [W,S2,W,S4], [S1,W,S3,W], and Val76-flipped [S1,W,S3,W] ion configurations required to account for differences in lineshape and transition dipoles strengths can be determined to be 30%, 30% and 40%, respectively. The [W,S2,W,S4] and [S1,W,S3,W] ion configurations are equally populated, to within 5% of each other, which is in agreement with the ratio of states found in X-Ray diffraction studies (6, 9). Thus, the Val76-flipped [S1,W,S3,W] ion configurations accounts for roughly 40% of the states at equilibrium. This flipped state has been reported in MD simulations (29), but has not been observed with X-ray crystallography.
The ultrafast time resolution of 2D IR spectroscopy provides an opportunity to test short-lived configurations extracted from MD simulations. Because the 2D IR time-resolution of 1–2 ps is faster than almost all protein dynamics, the spectrum reports on the instantaneous distribution of ions, water, and protein structures. As a result, 2D IR spectroscopy provides a means to test protein conformations observed in crystal structures and to empirically refine simulations to effectively reweight configurations. Energy differences on the order of kBT are below the accuracy of current force fields (31), and so simulations generated from different force fields could lead to different conclusions (31). Rather than test individual hypotheses, one might also simulate 2D IR spectra generated from Markov state models to analyze entire trajectories (32, 33). Essential for this study was the ability to selectively introduce 13C18O isotope labels into the ion binding sites in the filter. Advances in the semisynthesis methodology or alternatively, the use of nonsense suppression approaches have widely expanded the range of membrane proteins that can be selectively labeled and therefore investigated using 2D IR spectroscopy (34). We anticipate that the combination of these new labeling methods, molecular dynamics, and 2D IR spectroscopy will be an important addition to the toolkit presently used to investigate the functional mechanisms that operate in membrane proteins.
Supplementary Material
Acknowledgments
This research was supported by US National Institutes of Health grants NIH DK79895 (H.T.K., A.L.S., S.D.M., M.M., and M.T.Z), the University of Wisconsin Foundation (J.K.C. and J.L.S.), NIH R01-GM062342 (H.L. and B.R.), NIH GM087546 (K.M., A.W.A., and F.V.) NIH R01- GM057846 (E.P. and H.R) and the Membrane Protein Structural Dynamics Consortium U54 GM087519. F.V. and A.W.A. would like to thank members of the Xiao group for advice on chemical synthesis. M.T.Z. is an owner of PhaseTech Spectroscopy, Inc. Additional materials, methods, and supplementary text detailing the data reported in this paper can be found in the Supplementary Online Materials.
Footnotes
References and Notes
- 1.Hille B. Ion Channels of Excitable Membranes. Sinauer Associates, Inc; Sunderland, Massachusetts: 2001. p. 814. [Google Scholar]
- 2.Heginbotham L, Abramson T, MacKinnon R. A functional connection between the pores of distantly related ion channels as revealed by mutant K+ channels. Science. 1992;258:1152–1155. doi: 10.1126/science.1279807. [DOI] [PubMed] [Google Scholar]
- 3.Doyle DA, et al. The Structure of the Potassium Channel: Molecular Basis of K+ Conduction and Selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 4.Hodgkin AL, Keynes RD. The Potassium Permeability of a Giant Nerve Fibre. J Physiol. 1955;128:61–88. doi: 10.1113/jphysiol.1955.sp005291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uysal S, et al. Crystal structure of full-length KcsA in its closed conformation. Proc Natl Acad Sci USA. 2009;106:6644–6649. doi: 10.1073/pnas.0810663106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morais-Cabral JH, Zhou Y, MacKinnon R. Energetic optimization of ion conduction rate by the K+ selectivity filter. Nature. 2001;414:37–42. doi: 10.1038/35102000. [DOI] [PubMed] [Google Scholar]
- 7.Thompson AN, et al. Mechanism of potassium-channel selectivity revealed by Na(+) and Li(+) binding sites within the KcsA pore. Nat Struct Mol Biol. 2009;16:1317–1324. doi: 10.1038/nsmb.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noskov SY, Berneche S, Roux B. Control of ion selectivity in potassium channels by electrostatic and dynamic properties of carbonyl ligands. Nature. 2004;431:830–834. doi: 10.1038/nature02943. [DOI] [PubMed] [Google Scholar]
- 9.Zhou Y, MacKinnon R. The Occupancy of Ions in the K+ Selectivity Filter: Charge Balance and Coupling of Ion Binding to a Protein Conformational Change Underlie High Conduction Rates. J Mol Biol. 2003;333:965–975. doi: 10.1016/j.jmb.2003.09.022. [DOI] [PubMed] [Google Scholar]
- 10.Berneche S, Roux B. Energetics of ion conduction through the K+ channel. Nature. 2001;414:73–77. doi: 10.1038/35102067. [DOI] [PubMed] [Google Scholar]
- 11.Berneche S, Roux B. A microscopic view of ion conduction through the K+ channel. Proc Natl Acad Sci USA. 2003;100:8644–8648. doi: 10.1073/pnas.1431750100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alcayaga C, Cecchi X, Alvarez O, Latorre R. Streaming potential measurements in Ca2+-activated K+ channels from skeletal and smooth muscle. Coupling of ion and water fluxes. Biophys J. 1989;55:367–371. doi: 10.1016/S0006-3495(89)82814-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwamoto M, Oiki S. Counting ion and water molecules in a streaming file through the open- filter structure of the K channel. J Neurosci. 2011;31:12180–12188. doi: 10.1523/JNEUROSCI.1377-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aqvist J, Luzhkov V. Ion permeation mechanism of the potassium channel. Nature. 2000;404:881–884. doi: 10.1038/35009114. [DOI] [PubMed] [Google Scholar]
- 15.Furini S, Domene C. Atypical mechanism of conduction in potassium channels. Proc Natl Acad Sci USA. 2009;106:16074–16077. doi: 10.1073/pnas.0903226106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fowler PW, Abad E, Beckstein O, Sansom MS. Energetics of Multi-Ion Conduction Pathways in Potassium Ion Channels. J Chem Theory Comput. 2013;9:5176–5189. doi: 10.1021/ct4005933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kopfer DA, et al. Ion permeation in K(+) channels occurs by direct Coulomb knock-on. Science. 2014;346:352–355. doi: 10.1126/science.1254840. [DOI] [PubMed] [Google Scholar]
- 18.Fried SD, Bagchi S, Boxer SG. Measuring electrostatic fields in both hydrogen-bonding and non-hydrogen-bonding environments using carbonyl vibrational probes. J Am Chem Soc. 2013;135:11181–11192. doi: 10.1021/ja403917z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghosh A, Qiu J, DeGrado WF, Hochstrasser RM. Tidal surge in the M2 proton channel, sensed by 2D IR spectroscopy. Proc Natl Acad Sci USA. 2011;108:6115–6120. doi: 10.1073/pnas.1103027108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stevenson P, et al. Visualizing KcsA Conformational Changes upon Ion Binding by Infrared Spectroscopy and Atomistic Modeling. J Phys Chem B. 2015;119:5824–5831. doi: 10.1021/acs.jpcb.5b02223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valiyaveetil FI, MacKinnon R, Muir TW. Semisynthesis and Folding of the Potassium Channel KcsA. J Am Chem Soc. 2002;124:9113–9120. doi: 10.1021/ja0266722. [DOI] [PubMed] [Google Scholar]
- 22.Komarov AG, Linn KM, Devereaux JJ, Valiyaveetil FI. Modular strategy for the semisynthesis of a K+ channel: investigating interactions of the pore helix. ACS Chem Biol. 2009;4:1029–1038. doi: 10.1021/cb900210r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torres J, Kukol A, Goodman JM, Arkin IT. Site-specific examination of secondary structure and orientation determination in membrane proteins: The peptidic (13)C=(18)O group as a novel infrared probe. Biopolymers. 2001;59:396–401. doi: 10.1002/1097-0282(200111)59:6<396::AID-BIP1044>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 24.Ganim Z, Tokmakoff A, Vaziri A. Vibrational excitons in ionophores: experimental probes for quantum coherence-assisted ion transport and selectivity in ion channels. New J Phys. 2011;13:113030. [Google Scholar]
- 25.Barth A. The infrared absorption of amino acid side chains. Prog Biophys Mol Biol. 2000;74:141–173. doi: 10.1016/s0079-6107(00)00021-3. [DOI] [PubMed] [Google Scholar]
- 26.Barth A, Zscherp C. What vibrations tell about proteins. Q Rev Biophys. 2002;35:369–430. doi: 10.1017/s0033583502003815. [DOI] [PubMed] [Google Scholar]
- 27.Kim H, Cho M. Infrared Probes for Studying the Structure and Dynamics of Biomolecules. Chem Rev. 2013;113:5817–5847. doi: 10.1021/cr3005185. [DOI] [PubMed] [Google Scholar]
- 28.Kim YS, Hochstrasser RM. Applications of 2D IR spectroscopy to peptides, proteins, and hydrogen-bond dynamics. J Phys Chem B. 2009;113:8231–8251. doi: 10.1021/jp8113978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berneche S, Roux B. A gate in the selectivity filter of potassium channels. Structure. 2005;13:591–600. doi: 10.1016/j.str.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 30.Hamm P, Zanni MT. Concepts and Methods of 2D Infrared Spectroscopy. Cambridge University Press; New York: 2011. p. 286. [Google Scholar]
- 31.Li H, et al. Representation of Ion-Protein Interactions Using the Drude Polarizable Force-Field. J Phys Chem B. 2015;119:9401–9416. doi: 10.1021/jp510560k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baiz CR, et al. A molecular interpretation of 2D IR protein folding experiments with Markov state models. Biophys J. 2014;106:1359–1370. doi: 10.1016/j.bpj.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwantes CR, Pande VS. Improvements in Markov State Model Construction Reveal Many Non-Native Interactions in the Folding of NTL9. J Chem Theory Comput. 2013;9:2000–2009. doi: 10.1021/ct300878a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leisle L, Valiyaveetil F, Mehl RA, Ahern CA. In: Novel Chemical Tools to Study Ion Channel Biology. Ahern C, Pless S, editors. Vol. 869. Springer; New York: 2015. pp. 119–151. chap. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kraszewski S, Boiteux C, Ramseyer C, Girardet C. Determination of the charge profile in the KcsA selectivity filter using ab initio calculations and molecular dynamics simulations. Phys Chem Chem Phys. 2009;11:8606–8613. doi: 10.1039/b905991a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.