Abstract
Activation of the immune system needs to be tightly regulated to provide protection against infections and, at the same time, to prevent excessive inflammation to limit collateral damage to the host. This tight regulation includes regulating the activation of TLRs, which are key players in the recognition of invading microbes. A group of short cationic antimicrobial peptides, called cathelicidins, have previously been shown to modulate TLR activation by synthetic or purified TLR ligands and may play an important role in the regulation of inflammation during infections. However, little is known about how these cathelicidins affect TLR activation in the context of complete and viable bacteria. In this article, we show that chicken cathelicidin-2 kills Escherichia coli in an immunogenically silent fashion. Our results show that chicken cathelicidin-2 kills E. coli by permeabilizing the bacterial inner membrane and subsequently binds the outer membrane–derived lipoproteins and LPS to inhibit TLR2 and TLR4 activation, respectively. In addition, other cathelicidins, including human, mouse, pig, and dog cathelicidins, which lack antimicrobial activity under cell culture conditions, only inhibit macrophage activation by nonviable E. coli. In total, this study shows that cathelicidins do not affect immune activation by viable bacteria and only inhibit inflammation when bacterial viability is lost. Therefore, cathelicidins provide a novel mechanism by which the immune system can discriminate between viable and nonviable Gram-negative bacteria to tune the immune response, thereby limiting collateral damage to the host and the risk for sepsis.
Introduction
Toll-like receptors are crucial in the initiation of an immune response against invading pathogens. They are expressed by numerous cell types, including macrophages and epithelial cells, and activation of TLRs leads to immune activation, which includes the production of cytokines and chemokines (1, 2). These chemokines promote the recruitment of additional immune cells, including neutrophils, which are subsequently activated by the proinflammatory environment at the site of infection and further boost the inflammatory response (3). Although this positive feedback is useful to elicit a rapid inflammatory response during infections, it also needs to be tightly controlled to prevent excessive inflammation, which can lead to tissue damage and sepsis. To prevent this, various regulatory mechanisms are in place to keep inflammation in check (3, 4).
Cathelicidins are short host-derived cationic peptides with an important role in the protection against infections (5–7). The main source of cathelicidins in vivo is neutrophils, which store high concentrations of cathelicidin in their specific granules (8), which can be released upon neutrophil activation (9–12). In addition, epithelial cells can produce cathelicidins at mucosal surfaces (13, 14), such as the lung or the gut, whereas keratinocytes produce cathelicidin in the skin (15). Cathelicidin concentrations in vivo can range from 0.2 μM in plasma (8, 16) to 1–3 μM at epithelial surfaces (17) or in sweat (18) and to 4–6 μM in ascites fluid and saliva (16). In extreme conditions, such as psoriatic lesions, >300 μM of cathelicidin can be detected (19). The importance of cathelicidin in the protection against infections has been demonstrated in mouse models, where loss of cathelicidin expression increases the susceptibility to Escherichia coli infections (6, 7).
Many cathelicidin functions have been described over the years that can play a role in their protective effects during infections (20, 21). First, cathelicidins are well known for their antimicrobial activity, although this activity has been questioned, because many cathelicidins show a reduced antimicrobial activity when tested under more physiological conditions (22, 23). Second, cathelicidins have been shown to regulate activation of a wide variety of TLRs, including many mammalian TLRs, which can play an important role in the modulation of the inflammatory response during infections (24–29). For instance, cathelicidins, such as human LL-37 and chicken cathelicidin-2 (CATH-2), have been shown to inhibit LPS-induced TLR4 activation and to enhance DNA-induced TLR9 or TLR21 activation (23, 28). Nevertheless, these effects have only been tested in the context of specific purified or synthetic TLR ligands, and it is not known whether cathelicidins can still exert these effects in the context of complete and viable bacteria.
In this study, we aimed to identify the effect of cathelicidins on macrophage activation in the context of complete and viable bacteria. Our results show that CATH-2 can kill E. coli under physiological conditions in an immunogenically silent fashion (i.e., CATH-2 kills E. coli and, at the same time, prevents immune activation by the killed bacteria). This silent killing depends on two events: CATH-2 first kills E. coli by permeabilizing the bacterial inner membrane (IM) and subsequently binds the outer membrane (OM)–derived lipoproteins and LPS to inhibit TLR2 and TLR4 activation, respectively. In addition, we show that, although many cathelicidins, including human LL-37, lack antimicrobial activity under physiological cell culture conditions, they retain the ability to inhibit macrophage activation by nonviable E. coli. Together, these results demonstrate a novel cathelicidin function: cathelicidins can dampen inflammation during infections by Gram-negative bacteria once the bacterial threat has been neutralized and, thereby, can limit unnecessary inflammation and prevent tissue damage and sepsis.
Materials and Methods
Reagents
CATH-2 and LL-37 were synthesized by Fmoc-chemistry at CPC Scientific (Sunnyvale, CA). All other peptides (see later in the Results section) were synthesized by Fmoc-chemistry at the Academic Centre for Dentistry Amsterdam (Amsterdam, the Netherlands). Pam2CSK4, Pam3CSK4, E. coli LPS O111:B4, recombinant Salmonella typhimurium flagellin, polyinosinic-polycytidylic acid, CL264, and ODN-2006 were obtained from InvivoGen (Toulouse, France). Human TNF-α was obtained from Miltenyi Biotec (Bergisch Gladbach, Germany). Gentamicin was obtained from Sigma-Aldrich (St. Louis, MO).
Cell culture
J774.A1 cells (30) were a kind gift of Prof. J. van Putten (Utrecht University, Utrecht, the Netherlands). J774.A1 cells were cultured in DMEM (Thermo Fisher Scientific, Waltham, MA) supplemented with 10% FCS (Bodinco, Alkmaar, the Netherlands) at 37°C, 5.0% CO2. HEK–Blue–human TLR (hTLR) cell lines expressing specific TLRs, together with a NF-κB–SEAP reporter gene, were obtained from InvivoGen and cultured according to the manufacturer’s protocol. HEK-293 cells overexpressing hTLR5 and harboring a pNIFTY NF-κB luciferase reporter plasmid were cultured in DMEM supplemented with 10% FCS, 10 μg/ml blasticidin, and 250 μg/ml zeocin (both from InvivoGen). Chicken PBMCs were obtained from healthy adult chickens. Blood was diluted in PBS, and blood cells were separated by Ficoll density gradient centrifugation. PBMCs were collected at the interphase and subsequently washed with RPMI 1640 medium. Bone marrow cells were obtained by flushing the femur and tibia of C57BL/6J mice. All mice were kept under the guidelines and approval of the animal ethical committee of Utrecht University and had free access to food and water. For differentiation of bone marrow–derived macrophages (BMDMs), bone marrow cells were cultured in RPMI 1640 supplemented with 10% FCS, 1% Pen/Strep, and 20 ng/ml recombinant murine M-CSF (PeproTech, Rocky Hill, NJ) for 3 d, after which cells were washed with RPMI 1640 and incubated for another 3 d in RPMI 1640 supplemented with 10% FCS and 20 ng/ml M-CSF.
Bacterial culture
E. coli O78 and S. enteritidis phage type 13a were obtained from Zoetis Animal Health (Kalamazoo, MI). E. coli K12 MC4100 was a kind gift from Dr. Luirink (Vrije Universiteit Amsterdam, Amsterdam, the Netherlands), and E. coli ATCC 25922 was obtained from the American Type Culture Collection (Manassas, VA). All bacteria were cultured in Luria Broth (BioTRADING Benelux, Mijdrecht, the Netherlands). Prior to use, bacteria were grown to log-phase, centrifuged at 1200 × g for 10 min at 4°C, and diluted in RPMI 1640 or DMEM to the correct bacterial density.
Macrophage stimulation
A total of 7.5 × 104 J774.A1 cells per well was seeded in a 96-well plate and allowed to adhere overnight. For stimulation, bacteria were diluted to the appropriate density and mixed with cathelicidins at the indicated concentrations, after which the mixtures were used for cell stimulation. To compare different types of killing, E. coli was heat killed (70°C, 30 min), gentamicin killed (37°C, 30 min, 250 μg/ml gentamicin), CATH-2 killed (37°C, 30 min, 5 μM) or left untreated (4°C, 30 min). To determine TNF-α release, macrophages were stimulated for 2 h, after which supernatants were harvested for ELISA. To measure the release of other cytokines, macrophages were stimulated for 2 h with bacteria in the presence or absence of CATH-2, after which the cells were washed twice and incubated for an additional 22 h in DMEM + 10% FCS + 250 μg gentamicin in the absence of any stimuli.
ELISA
ELISA DuoSet kits for mouse TNF-α, IL-1β, IL-6, IL-10, RANTES, IP-10, and IFN-β were obtained from R&D Systems (Minneapolis, MN). ELISAs were performed following the manufacturer’s protocol, and samples were diluted in 1% BSA (Sigma-Aldrich) in PBS (pH 7.4). Measurements were performed with a FLUOstar Omega microplate reader and analyzed with MARS data analysis software (both from BMG Labtech, Ortenberg, Germany). OD450 measurements were corrected by subtracting OD570 measurements.
Colony count assay
Colony count assays were performed by incubating bacteria with the indicated final concentrations of cathelicidins in 20 μl of DMEM or RPMI 1640 supplemented with 10% FCS at 37°C for 2 h. After incubation, samples were diluted with 180 μl of PBS, followed by spread-plating 10-fold dilutions in PBS on Tryptone Soy Agar plates (Oxoid, Hampshire, U.K.). After overnight incubation at 37°C, CFU were counted (detection limit = 102 CFU/ml).
Quantitative PCR
For quantitative PCR (qPCR) experiments, chicken PBMCs were stimulated with 5 × 105 CFU/ml E. coli O78 in the presence or absence of 5 μM CATH-2 for 2 h at 41°C, 5% CO2. After the stimulation, cells were centrifuged at 400 × g for 8 min. Lysis of the cell pellet and RNA isolation were performed using a High Pure RNA Tissue Kit (Roche, Basel, Switzerland). RNA was converted to cDNA using an iScript cDNA Synthesis Kit (Bio-Rad, Veenendaal, the Netherlands). qPCR was performed using primers, probes (see later in the Results section), and IQ Supermix (Bio-Rad), and plates were run in a CFX Connect Real-Time PCR Detection System with CFX Manager 3.0 software (both from Bio-Rad). Quantification cycle values were corrected for PCR efficiency and housekeeping gene expression of GAPDH. When no signal was detected after 40 cycles, a quantification cycle value of 40 was given. Unstimulated samples were set to 1.
Transmission electron microscopy
For transmission electron microscopy (TEM), E. coli O78 was grown to log phase and diluted to 108 E. coli O78 per milliliter in DMEM. Bacteria were left untreated, incubated at 70°C, or incubated with 40 μM CATH-2, 40 μM LL-37, or 250 μg/ml gentamicin at 37°C for 0.5 or 2.5 h. Mixtures were fixed with 2% glutaraldehyde (Polysciences, Eppelheim, Germany), 5 mM CaCl2, and 10 mM MgCl2 (both from Merck, Darmstadt, Germany) in 0.1 M sodium cacodylate buffer (Sigma-Aldrich) (pH 7.4) overnight at 4°C. After washing (three times for 10 min each) in sodium cacodylate buffer, bacteria were embedded in 2% low-melting point agarose v/v (Sigma-Aldrich) and postfixed with 4% osmium tetroxide (Electron Microscopy Sciences, Hatfield, PA) and 1.5% K4Fe(CN)6-3H2O (Merck) in distilled water for 2 h at 4°C. Bacteria were rinsed with distilled water (five times for 10 min each) and incubated in 0.5% uranyl acetate (Electron Microscopy Sciences) for 1 h at 4°C. After washing (three times for 10 min each) with distilled water, samples were embedded in Epon, and ultrathin sections (50 nm) of each block were prepared using a Leica UCT ultramicrotome (Leica, Vienna, Austria). Finally, sections were stained with uranyl acetate and lead citrate using a Leica AC20 system (Leica). Electron microscopy was performed with an FEI Tecnai 12 electron microscope (FEI, Eindhoven, the Netherlands) at 80 kV.
HEK-TLR assay
Stimulation of HEK-TLR cells containing the NF-κB SEAP reporter gene with 105 CFU/ml heat-killed E. coli O78 was performed over 18 h at 37°C, 5% CO2 in the presence or absence of 5 μM CATH-2 or LL-37. After incubation, NF-κB activity was determined by measuring SEAP activity with QUANTI-Blue (InvivoGen). Stimulation of HEK–hTLR5–luciferase cells was performed over 6 h, after which NF-κB activity was determined by measuring luciferase activity with Bright-Glo (Promega, Fitchburg, WI).
Flow cytometry
For flow cytometry, 106 CFU/ml E. coli O78 was incubated with the indicated concentrations of FITC–CATH-2 or FITC–LL-37 for 2 h at 37°C in DMEM + 10% FCS. After incubation, bacteria were centrifuged at 1200 × g for 10 min and fixed in 4% PFA (Sigma-Aldrich) in 0.1 M phosphate buffer (Na2HPO4 + KH2PO4) (pH 7.4). Bacteria were washed and resuspended in PBS for measurement on a FACSCalibur (BD Biosciences, San Jose, CA). Data were analyzed with Flow Jo software (TreeStar, Ashland, OR)
Confocal microscopy
For confocal microscopy, 106 CFU/ml live or heat-killed E. coli O78 was incubated with FITC–CATH-2 or FITC–LL-37 for 2 h at 37°C in DMEM + 10% FCS. After incubation, bacteria were washed, resuspended in PBS, pipetted onto a microscopy slide, air-dried, heat-fixed, and mounted in FluorSave (Merck Millipore). Confocal imaging was performed on a Leica SPE-II DMI4000 microscope with LAS-AF software (Leica, Wetzlar, Germany) using a 100× HCX PLAN APO Oil CS objective. Brightness and contrast were adjusted equally for all images and on entire images using ImageJ (National Institutes of Health, Bethesda, MD).
IM permeabilization
Live, heat-killed, or gentamicin-killed E. coli O78 were incubated with the indicated concentrations of CATH-2 or LL-37 for 30 min at 37°C in DMEM + 10% FCS. After incubation, bacteria were centrifuged at 1200 × g for 10 min at 4°C and washed with PBS. Subsequently, bacteria were resuspended in PBS with 2 μM SYTOX Green Nucleic Acid Stain (Thermo Fisher Scientific) and transferred to a black 96-well plate. After 5 min, fluorescence was determined using a FLUOstar Omega microplate reader and analyzed with MARS data analysis software (both from BMG Labtech).
Isothermal titration calorimetry
Isothermal titration calorimetry (ITC) was performed using a Low Volume NANO ITC (TA Instruments-Waters, New Castle, DE). The 50-μl syringe was filled with 400 μM CATH-2 or LL-37 for titration into 190 μl of 50 μM LPS O111:B4 or 165 μM Pam3CSK4. All components were diluted in PBS (6.04 mM Na2HPO4, 1.10 mM KH2PO4, 103.45 mM NaCl, 2.0 mM KCl, 0.37 mM MgCl2, 0.68 mM CaCl2) (Thermo Fisher Scientific). Titrations were incremental, with 2-μl injections at 300-s intervals. Experiments were performed at 37°C. Data were analyzed with NanoAnalyze software (TA Instruments-Waters).
Statistical analysis and graphics
Statistical analysis was performed using Prism 7 software (GraphPad, La Jolla, CA) and IBM SPSS Statistics 20 (IBM, Armonk, NY). Graphics were designed using Prism 7 software (GraphPad), Adobe InDesign, and Adobe Illustrator (both from Adobe Systems, San Jose, CA).
Results
CATH-2 kills E. coli and S. enteritidis in an immunogenically silent fashion
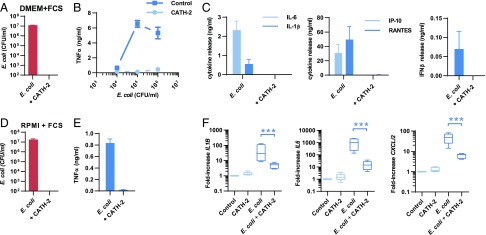
Chicken CATH-2 has previously been shown to be a potent antimicrobial peptide with TLR modulatory activity (28, 31). To investigate the role of CATH-2 on E. coli–induced macrophage activation, we first performed a colony count assay to determine whether CATH-2 is capable of killing E. coli under cell culture conditions. After incubation of 106 CFU/ml E. coli O78 with 5 μM CATH-2 in DMEM + 10% FCS, we observed that CATH-2 retains its antimicrobial activity under cell culture conditions (Fig. 1A). Next, to determine whether CATH-2 affects macrophage activation by E. coli, J774.A1 macrophages were incubated with E. coli alone or E. coli in combination with CATH-2, after which cytokine release was determined. Macrophage stimulation with E. coli alone resulted in rapid TNF-α release that could be detected after 2 h of stimulation for E. coli concentrations ranging from 104 to 106 CFU/ml (Fig. 1B). However, stimulation of macrophages for 2 h with E. coli in the presence of 5 μM CATH-2 resulted in inhibition of this TNF-α release. To determine whether the release of other cytokines that are produced at later time points was also affected by CATH-2, macrophages were stimulated with 106 CFU/ml E. coli in the presence or absence of CATH-2 for 2 h, after which the cells were washed and incubated for another 22 h in normal cell culture media containing 250 μg/ml gentamicin in the absence of stimuli. This showed that, similar to the inhibition of TNF-α after 2 h, CATH-2 inhibited E. coli–induced IL-6, IL-1β, RANTES, IP-10, and IFN-β release after 24 h as well (Fig. 1C). In addition, CATH-2 was able to kill E. coli 25922 and E. coli K12 in DMEM + 10% FCS (Supplemental Fig. 1A), and it inhibited the E. coli K12– and E. coli 25922–induced TNF-α release by macrophages (Supplemental Fig. 1B). Furthermore, CATH-2 killed S. enteritidis in DMEM + 10% FCS (Supplemental Fig. 1C), and it inhibited S. enteritidis–induced macrophage activation, as determined by TNF-α (2 h), IL-6 (24 h), and IL-1β (24 h) release (Supplemental Fig. 1D). This showed that the observed inhibition of macrophage activation by CATH-2 is not specific for E. coli alone. To determine whether CATH-2 could inhibit the activation of primary cells by E. coli, murine BMDMs were stimulated with 106 CFU/ml E. coli O78 in combination with 5 μM CATH-2 in RPMI 1640 + 10% FCS. Similar to the results using DMEM + 10% FCS, CATH-2 also killed E. coli in RPMI 1640 + 10% FCS (Fig. 1D) and was able to prevent E. coli–induced TNF-α release by BMDMs (Fig. 1E). Finally, because CATH-2 is a chicken-derived peptide, we tested whether it could also prevent E. coli–induced activation of chicken PBMCs. To this end, chicken PBMCs were stimulated with 106 CFU/ml E. coli O78 in the presence or absence of 5 μM CATH-2 in RPMI 1640 + 10% FCS for 2 h, after which cytokine gene expression was determined by qPCR (Table I). This showed that, similar to the results with murine cells, CATH-2 inhibited E. coli–induced CXCLi2, IL6, and IL1B gene expression in these chicken-derived cells (Fig. 1F). Together, these results indicate that CATH-2 kills E. coli and S. enteritidis in an immunogenically silent fashion and prevents E. coli–induced cytokine responses in a nonspecies-specific manner.
FIGURE 1.
CATH-2 inhibits E. coli–induced macrophage activation. (A) Colony count assay of 106 CFU/ml E. coli O78 + 5 μM CATH-2 in DMEM + 10% FCS after 2 h of incubation (n = 3). Error bars show SEM. (B) TNF-α release, as determined by ELISA, by J774.A1 macrophages incubated for 2 h with 104–106 CFU/ml E. coli O78 in the presence or absence of 5 μM CATH-2 (n = 3). Error bars show SEM. (C) Stimulation of J774.A1 macrophages for 2 h with 106 CFU/ml E. coli O78 in the presence or absence of 5 μM CATH-2, after which cells were washed with PBS and incubated for 22 h with 250 μg/ml gentamicin in the absence of other stimuli. After incubation, IL-6, IL-1β, IP-10, RANTES, and IFN-β were determined by ELISA (n = 3). Error bars show SEM. (D) Colony count assay of 106 CFU/ml E. coli O78 + 5 μM CATH-2 in RPMI 1640 + 10% FCS after 2 h of incubation (n = 3). Error bars show SEM. (E) TNF-α release, as determined by ELISA, by BMDMs incubated for 2 h with 106 CFU/ml E. coli O78 in the presence or absence of 5 μM CATH-2 (n = 3). Error bars show SEM. (F) Chicken PBMCs were left untreated or were incubated with 5 μM CATH-2 alone or with 5 × 105 CFU/ml E. coli O78 in the presence or absence of 5 μM CATH-2. After 2 h, RNA was isolated, and IL1B, IL6, and CXCLi2 gene expression was determined by qPCR. Statistical analysis was performed by repeated-measures ANOVA on log-transformed data, followed by the Bonferroni post hoc test. Box plots show the median, and whiskers represent minimal and maximal values. n = 5. ***p < 0.001.
Table I. Primers and probes for qPCR on chicken genes.
| Forward (5′–3′) | Reverse (5′–3′) | Probe (5′–3′) | |
|---|---|---|---|
| IL1B | GCTCTACATGTCGTGTGTGATGAG | TGTCGATGTCCCGCATGA | CCACACTGCAGCTGGAGGAAGCC |
| IL6 | GTCGAGTCTCTGTGCTAC | GTCTGGGATGACCACTTC | ACGATCCGGCAGATGGTGA |
| CXCLi2 | GCCCTCCTCCTGGTTTCA | CGCAGCTCATTCCCCATCT | TGCTCTGTCGCAAGGTAGGACGCTG |
| GAPDH | GCCGTCCTCTCTGGCAAAG | TGTAAACCATGTAGTTCAGATCGATGA | AGTGGTGGCCATCAATGATCCC |
Direct interaction between E. coli and CATH-2 is required to inhibit macrophage activation and leads to E. coli membrane disruption
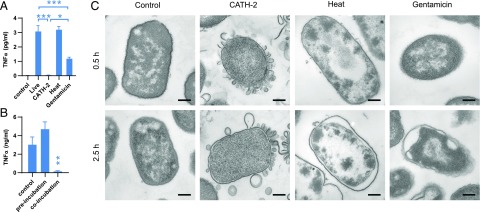
Next, we aimed to determine how CATH-2 inhibits macrophage activation. To determine whether the loss of bacterial viability itself would affect macrophage activation, E. coli was killed by CATH-2 (5 μM, 30 min), heat (70°C, 30 min) or gentamicin (250 μg/ml, 30 min), after which macrophages were stimulated, and TNF-α release was determined. This showed that although CATH-2–killed E. coli were unable to induce a TNF-α response, heat-killed and gentamicin-killed bacteria did induce TNF-α release (Fig. 2A). This indicates that loss of bacterial viability itself did not prevent E. coli from inducing macrophage activation. Next, to determine whether CATH-2–mediated inhibition requires direct interaction between CATH-2 and the bacteria, J774.A1 macrophages were preincubated with CATH-2 for 2 h, washed, and subsequently incubated with E. coli or were left untreated for the first 2 h and then incubated with E. coli and CATH-2 simultaneously. This showed that only incubation with E. coli and CATH-2 simultaneously resulted in inhibition of macrophage activation and indicates the importance of direct interaction between CATH-2 and E. coli for this inhibition (Fig. 2B). To visualize what happens to E. coli when it is killed by CATH-2, TEM was performed. This showed that CATH-2 had a membrane-disruptive effect on E. coli and induced the release of membrane fragments (Fig. 2C). In contrast, although killing by heat or gentamicin did affect the bacterial membrane, this appeared to primarily involve detachment of the OM from the IM; no fragmentation, as induced by CATH-2, was observed. These results indicate that CATH-2 kills E. coli through disruption of bacterial membrane integrity and that the interaction between CATH-2 and E. coli is required for the inhibition of E. coli–induced macrophage activation.
FIGURE 2.
Only CATH-2–mediated killing inhibits macrophage activation. (A) Stimulation of J774.A1 macrophages for 2 h with 106 CFU/ml E. coli, that were alive, CATH-2 killed (5 μM, 30 min), heat killed (70°C, 30 min), or gentamicin killed (250 μg/ml, 30 min), after which TNF-α release was determined by ELISA (n = 4). Error bars show SEM. *p < 0.05, ***p < 0.001, repeated-measures ANOVA, followed by the Bonferroni post hoc test. (B) J774.A1 macrophages were preincubated with 5 μM CATH-2 for 2 h, washed, and incubated with 106 CFU/ml E. coli for 2 h or were left untreated for 2 h, washed, and incubated for 2 h with 106 CFU/ml E. coli O78 and 5 μM CATH-2 simultaneously. Supernatants were used to determine TNF-α release by ELISA (n = 5). Error bars show SEM. **p < 0.01 versus control, repeated-measures ANOVA, followed by the Dunnett post hoc test. (C) TEM images of 108 CFU/ml E. coli O78 in DMEM that were left untreated or were treated with 40 μM CATH-2, heat (70°C), or gentamicin (250 μg/ml) for 0.5 or 2.5 h. Images are representative of two independent experiments. Scale bars, 200 nm.
CATH-2 binds E. coli and permeabilizes the bacterial IM before it neutralizes E. coli immunogenicity
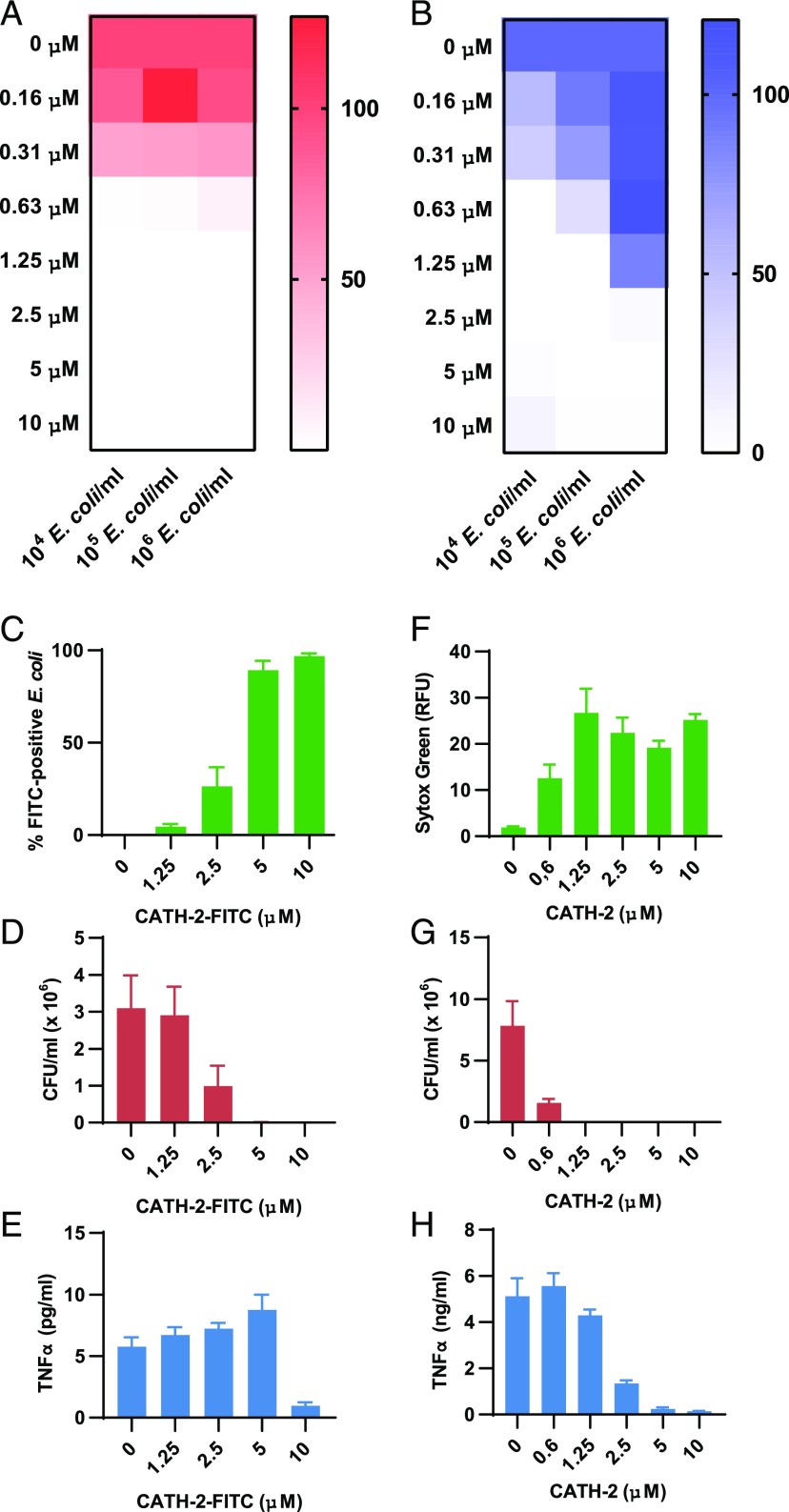
To better understand whether CATH-2 uses one mechanism to kill E. coli and inhibit macrophage activation or whether these effects are the results of two distinct events, different ratios of CATH-2 and E. coli were mixed and used to determine bacterial viability, as well as to stimulate macrophages and determine TNF-α release. These results show that 0.6 μM CATH-2 appears to be the threshold at which most E. coli (91–99%) gets killed (for concentrations between 104 and 106 CFU/ml) (Fig. 3A). However, inhibition of TNF-α release was only observed at 0.6 μM for the lowest E. coli concentration (104 CFU/ml), whereas 105 CFU/ml E. coli required 1.25 μM CATH-2 and 106 CFU/ml E. coli required 2.5 μM CATH-2 to obtain an inhibition of TNF-α release > 90% (Fig. 3B). Interestingly, when 106 CFU/ml E. coli were heat killed prior to macrophage stimulation, CATH-2 already showed significant inhibition at 0.6 μM (Supplemental Fig. 2A). This suggests that bacterial killing and inhibition of macrophage activation are caused by two distinct mechanisms. In addition, these results show that, when bacteria are killed by a method other than CATH-2, CATH-2 can still inhibit E. coli–induced macrophage activation. Next, to determine when CATH-2 and E. coli interact, binding of a fluorescently labeled CATH-2 peptide (FITC–CATH-2) to E. coli was assessed by flow cytometry. Binding of FITC–CATH-2 to E. coli correlates with its antimicrobial activity, with 90% of all E. coli bound by FITC–CATH-2 and killed at 5 μM (Fig. 3C, 3D, Supplemental Fig. 2B). However, inhibition of macrophage activation was not yet observed at 5 μM and required a higher concentration (10 μM) of FITC–CATH-2 (Fig. 3E). Next, we examined the effect of CATH-2 on E. coli IM integrity by SYTOX Green staining. Permeabilization of the IM by CATH-2 started at 0.6 μM and was maximal at 1.25 μM (Fig. 3F), which corresponds to the concentration of CATH-2 needed to kill E. coli (Fig. 3G). However, inhibition of TNF-α release was only observed at concentrations of CATH-2 > 1.25 μM (Fig. 3H), which showed that CATH-2 penetrates the bacterial membrane prior to exerting its inhibitory effect. Together, these results show that CATH-2 cannot sustain interaction with E. coli before it reaches antimicrobial concentrations. In addition, it indicates that the peptide targets the IM to kill the bacteria before it is able to prevent E. coli–induced macrophage activation.
FIGURE 3.
Antimicrobial activity and inhibition of E. coli–induced macrophage activation are two distinct effects of CATH-2. Different ratios of CATH-2 (0–10 μM) and E. coli O78 (104–106 CFU/ml) were mixed in DMEM + 10% FCS and used for a colony count assay (A) or for a 2-h stimulation of J774.A1 macrophages, after which TNF-α release was determined by ELISA (B). CFU counts and TNF-α release at 0 μM CATH-2 were set to 100% for each E. coli concentration (n = 3). A total of 106 CFU/ml E. coli O78 was mixed with CATH-2 (0–10 μM), followed by flow cytometric analysis of the percentage of FITC–CATH-2+ E. coli (C), a colony count assay (D), and stimulation of J774.A1 macrophages to determine TNF-α secretion (E) (n ≥ 3). Error bars show SEM. A total of 106 CFU/ml E. coli O78 was mixed with CATH-2 (0–10 μM), followed by analysis of IM permeabilization by assessment of SYTOX Green fluorescence (F), a colony count assay (G), and stimulation of J774.A1 macrophages to determine TNF-α secretion (H) (n ≥ 3). Error bars show SEM.
CATH-2 neutralizes OM-derived lipoproteins and LPS to inhibit E. coli–induced TLR2 and TLR4 activation
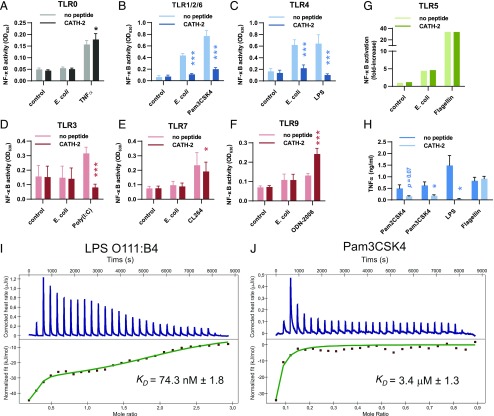
HEK-TLR cell lines were used to determine which bacterial factors are targeted by CATH-2 to inhibit E. coli–induced macrophage activation. These cell lines specifically overexpress different TLRs, together with an NF-κB reporter gene. First, TLR0 cells, which express the NF-κB reporter gene but no TLRs, were stimulated with live E. coli or CATH-2–killed E. coli. This showed that live E. coli, but not CATH-2–killed E. coli, can activate NF-κB in a TLR-independent manner (Supplemental Fig. 3). Therefore, to further study which TLRs are activated by E. coli and to study whether activation of these TLRs is inhibited by CATH-2, we used heat-killed E. coli. This ensured that no TLR-independent activation was induced (Fig. 4A), whereas CATH-2 previously showed that it can still inhibit macrophage activation in the context of heat-killed bacteria (Supplemental Fig. 2).
FIGURE 4.
Inhibition of E. coli–induced TLR2 and TLR4 activation by CATH-2. (A–F) HEK-TLR cells overexpressing no TLR (TLR0), TLR1, 2, and 6 (TLR1/2/6), TLR3, TLR4, TLR7, or TLR9, as well as a SEAP reporter gene, were stimulated with 5 × 104 CFU/ml heat-killed E. coli O78 or TNF-α (50 ng/ml), Pam3CSK4 (5 ng/ml), LPS (0.5 ng/ml), Poly(I:C) (250 ng/ml), CL264 (250 ng/ml), or ODN-2006 (50 nM) in the presence or absence of 5 μM CATH-2. After 18 h, the supernatant was used to determine NF-κB activation through QUANTI-Blue analysis (n ≥ 3). Error bars show SEM. *p < 0.05, ***p < 0.001, two-way repeated-measures ANOVA with Bonferroni post hoc test. (G) HEK–TLR5–luciferase cells were stimulated with 5 × 104 CFU/ml heat-killed E. coli O78 or flagellin (10 ng/ml) in the presence or absence of 5 μM CATH-2. NF-κB activation was determined after 6 h by analyzing luciferase activity by Bright-Glo. Results are representative of three independent experiments. (H) J774.A1 cells were stimulated with Pam2CSK4 (10 pg/ml), Pam3CSK4 (10 ng/ml), LPS (10 ng/ml), or flagellin (1 μg/ml) in the presence or absence of 5 μM CATH-2 for 2 h, after which TNF-α secretion was determined by ELISA (n ≥ 3). Error bars show SEM. *p < 0.05, paired t test. Analysis of ITC titration of CATH-2 into LPS O111:B4 (I) or Pam3CSK4 (J) solution. Images are representative for n = 2. The KD value shown is the mean calculated KD for n = 2 ± SEM.
Stimulation of HEK-TLR cells by heat-killed E. coli showed that E. coli can activate cells expressing TLR1, TLR2 and TLR6, TLR4, or TLR5, but it cannot activate cells expressing TLR3, TLR7, or TLR9 (Fig. 4A–G). Stimulation of these cells with heat-killed E. coli in the presence of CATH-2 showed that CATH-2 could inhibit TLR1/2/6 and TLR4 activation but not TLR5 activation. In addition, NF-κB activation by Pam3CSK4 and LPS, the specific TLR ligands for TLR1/2/6-expressing and TLR4-expressing cells, respectively, was also inhibited by CATH-2 (Fig. 4B, 4C), whereas TLR5 activation by flagellin was unaffected (Fig. 4G). Furthermore, TNF-α release from J774.A1 cells stimulated with LPS or the lipoprotein Pam2CSK4 or Pam3CSK4, but not flagellin, was also inhibited in the presence of CATH-2 (Fig. 4H). Finally, because CATH-2 inhibition required direct interaction with the bacteria, we used ITC to determine whether CATH-2 could directly interact with TLR2 and TLR4 ligands. This showed that CATH-2 directly interacts with LPS (KD = 74.3 nM) (Fig. 4I) and also directly interacts with Pam3CSK4 (KD = 3.4 μM) (Fig. 4J). Together, these results demonstrate that CATH-2 inhibits E. coli–induced macrophage activation by neutralizing lipoproteins and LPS from the bacterial OM, which prevents the activation of TLR2 and TLR4, respectively.
Cathelicidins from various species are capable of inhibiting macrophage activation by nonviable E. coli
Although CATH-2 potently kills E. coli and inhibits macrophage activation, it is unclear whether this is a conserved function of cathelicidins. To test whether this is the case, 12 cathelicidins from six species were examined for their antimicrobial activity, as well as their ability to inhibit E. coli–induced macrophage activation (Table II ). From the 12 cathelicidins, only chicken CATH-2 and porcine PMAP-36 showed strong antimicrobial activity under cell culture conditions (Fig. 5A). In addition, only these two peptides could inhibit live E. coli–induced macrophage activation (Fig. 5B). However, when bacteria were killed first by gentamicin, 5 of 12 cathelicidins significantly inhibited macrophage activation (Fig. 5C). These cathelicidins include chicken CATH-2, human LL-37, mouse CRAMP, dog K9, and porcine PMAP-36. Furthermore, when bacteria were heat killed, these five cathelicidins, as well as chicken CATH-1 and -3 and equine eCATH-2, were all able to significantly inhibit macrophage activation (Fig. 5D). These results demonstrate that various cathelicidins have the ability to inhibit macrophage activation only in the context of nonviable E. coli. This again indicated that the antimicrobial activity of cathelicidins and their ability to inhibit macrophage activation are two distinct functions. To obtain some insight into which peptide regions in CATH-2 are important for the antimicrobial and inhibitory activity, different truncated CATH-2 peptides were synthesized (Table III). Truncation of the last 5 aa (C1-21*) from the C terminus did not abrogate the antimicrobial activity of CATH-2 and also did not affect the inhibition of macrophage activation by live and heat-killed E. coli (Supplemental Fig. 4A, 4B). However, a 3 aa N-terminal truncation strongly inhibited the antimicrobial activity of the peptide and resulted in a loss of inhibitory activity in the context of live bacteria. Nevertheless, significant inhibition of heat-killed bacteria was still observed. Further N-terminal truncation reduced the inhibitory activity, which was completely lost when the first 7 aa were omitted. These results further support the idea that the antimicrobial and immune-inhibitory functions of CATH-2 depend on two distinct mechanisms.
Table II. Cathelicidin peptide sequences.
| Peptide | Sequence |
|---|---|
| LL-37 | LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES |
| CRAMP | GLLRKGGEKIGEKLKKIGQKIKNFFQKLVPQPEQ |
| K9CATH | RLKELITTGGQKIGEKIRRIGQRIKDFFKNLQPREEKS |
| chCATH-1 | RVKRVWPLVIRTVIAGYNLYRAIKKK |
| chCATH-2 | RFGRFLRKIRRFRPKVTITIQGSARF-NH2 |
| chCATH-3 | RVKRFWPLVPVAINTVAAGINLYKAIRRK |
| eCATH-1 | KRFGRLAKSFLRMRILLPRRKILLAS |
| eCATH-2 | KRRHWFPLSFQEFLEQLRRFRDQLPFP |
| eCATH-3 | KRFHSVGSLIQRHQQMIRDKSEATRHGIRIITRPKLLLAS |
| PMAP-23 | RIIDLLWRVRRPQKPKFVTVWVR |
| PMAP-36 | Ac-GRFRRLRKKTRKRLKKIGKVLKWIPPIVGSIPLGCG |
| PR-39 | RRRPRPPYLPRPRPPPFFPPRLPPRIPPGFPPRFPPRFP |
FIGURE 5.
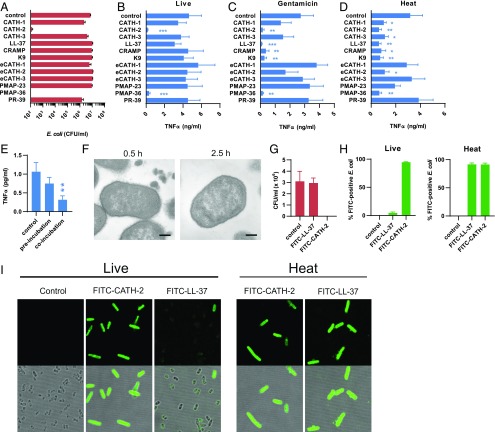
Inhibition of E. coli–induced macrophage activation by cathelicidin is partially conserved between different species. (A) Colony count assay of 106 CFU/ml E. coli O78 + 5 μM of the indicated cathelicidins in DMEM + 10% FCS after 2 h of incubation (n = 4). Error bars show SEM. Stimulation of J774.A1 macrophages for 2 h with 106 CFU/ml live E. coli O78 (B), 106 CFU/ml gentamicin-killed E. coli O78 (C), or 106 CFU/ml heat-killed E. coli O78 (D), in the presence or absence of 5 μM of the indicated cathelicidins in DMEM + 10% FCS, after which TNF-α release was determined by ELISA (n = 4). Error bars show SEM. (E) J774.A1 macrophages were incubated with 5 μM LL-37 for 2 h, washed, and incubated with 106 CFU/ml E. coli for 2 h or were left untreated for 2 h, after which cells were washed and incubated for 2 h with 106 CFU/ml E. coli O78 and 5 μM LL-37 simultaneously. Supernatants were subsequently used to determine TNF-α release by ELISA. n = 6. Error bars show SEM. (F) TEM images of 108 CFU/ml E. coli O78 + 40 μM LL-37 in DMEM for 0.5 or 2.5 h. Images are representative of two independent experiments. Scale bars, 200 nm. (G) Colony count assay of 106 CFU/ml E. coli O78 + 5 μM FITC-LL-37 or FITC-CATH-2 in DMEM + 10% FCS after 2 h of incubation (n = 4). Error bars show SEM. (H) A total of 106 CFU/ml live or heat-killed E. coli O78 was incubated for 2 h with 10 μM FITC–LL-37 or FITC–CATH-2 in DMEM + 10% FCS, followed by flow cytometric analysis of the percentage of FITC+ E. coli (n = 4). Error bars show SEM. (I) Confocal microscopy (original magnification ×100) of 106 CFU/ml live or heat-killed E. coli O78 incubated with 10 μM FITC–CATH-2 or FITC–LL-37 for 2 h in DMEM + 10% FCS. Images are representative of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 versus control, repeated-measures ANOVA, followed by the Dunnett post hoc test.
Table III. Truncated CATH-2 sequences.
| Peptide | Sequence |
|---|---|
| C1-26* (CATH-2) | RFGRFLRKIRRFRPKVTITIQGSARF-NH2 |
| C1-21* | RFGRFLRKIRRFRPKVTITIQ-NH2 |
| C5-21* | FLRKIRRFRPKVTITIQ-NH2 |
| C7-21* | RKIRRFRPKVTITIQ-NH2 |
| C8-21* | KIRRFRPKVTITIQ-NH2 |
| C9-21* | IRRFRPKVTITIQ-NH2 |
Human LL-37 and chicken CATH-2 use the same inhibitory mechanism to prevent macrophage activation by nonviable E. coli
Because both antimicrobial CATH-2 and nonantimicrobial LL-37 show similar inhibitory activity against nonviable E. coli, we aimed to determine whether LL-37 would use the same mechanism as CATH-2 to inhibit E. coli–induced macrophage activation. First, to determine whether LL-37 requires direct interaction with the bacteria to inhibit macrophage activation, macrophages were preincubated for 2 h with LL-37, washed, and stimulated for 2 h with heat-killed E. coli or were left untreated for the first 2 h, washed, and incubated for 2 h with LL-37 and heat-killed E. coli simultaneously. This showed that LL-37 also requires interaction with E. coli to inhibit macrophage activation (Fig. 5E). In addition, it was observed that with HEK-TLR cells, LL-37 inhibited TLR2 (Supplemental Fig. 4C) and TLR4 (Supplemental Fig. 4D) activation by heat-killed E. coli but not TLR5 activation (Supplemental Fig. 4E). Furthermore, LL-37 inhibited macrophage activation by the TLR2 ligand Pam3CSK4 and the TLR4 ligand LPS (Supplemental Fig. 4F). Finally, ITC analysis showed that LL-37 directly interacts with LPS (KD = 33.9 nM ± 26.0) (Supplemental Fig. 4G) and Pam3CSK4 (KD = 30.4 μM ± 14.1) (Supplemental Fig. 4H), indicating that CATH-2 and LL-37 use the same inhibitory mechanism to prevent macrophage activation by nonviable E. coli. Interestingly, although LL-37 is unable to kill E. coli or inhibit live E. coli–induced macrophage activation, TEM images of LL-37–treated E. coli did show the release of bacterial fragments from E. coli after 0.5 h of incubation (Fig. 5F). However, this appears to be a transient effect, because no antimicrobial activity was observed and release of fragments was no longer visible after 2.5 h of incubation. In addition, FITC–LL-37 was unable to kill E. coli or interact with live E. coli, as determined by flow cytometry and confocal microscopy (Fig. 5G–I). In contrast, when E. coli was heat killed, FITC–LL-37 and FITC–CATH-2 were able to interact with E. coli (Figs. 5H, 5I, 6) and inhibit the E. coli–induced macrophage activation (Supplemental Fig. 4I). Together, these results show that sustained interaction between cathelicidins and E. coli only occurs when E. coli is killed, either by the cathelicidin itself or in other ways, and that inhibition of TLR2 and TLR4 activation is partially conserved between cathelicidins but only occurs in the context of nonviable bacteria (Fig. 6).
FIGURE 6.
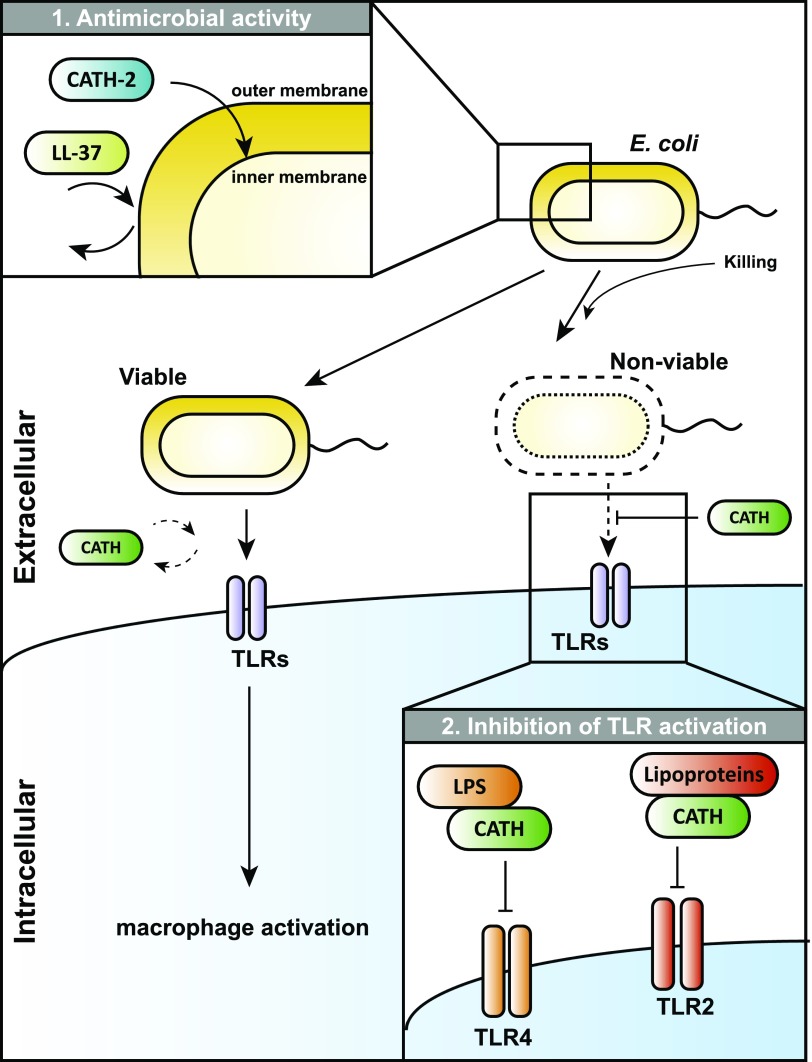
Model for the effects of cathelicidins on E. coli viability and E. coli–induced macrophage activation. 1) Cathelicidins are attracted to the Gram-negative bacterial OM, possibly as the result of an ionic interaction between cationic residues on the cathelicidins and the anionic LPS. Depending on the cathelicidin, this is followed by bacterial killing due to translocation to and permeabilization of the IM or bacterial survival that results from displacement of cathelicidins from the bacterial surface. 2) Upon killing of Gram-negative bacteria by cathelicidins or other antimicrobial mechanisms, cathelicidins can interact with the LPS and lipoproteins from the bacterial OM to prevent activation of TLR4 and TLR2, respectively. In contrast, when bacterial viability remains intact, cathelicidins are unable to inhibit macrophage activation.
Discussion
The immune system needs to induce a balanced immune response during infections to kill invading pathogens, as well as to prevent tissue damage and sepsis by excessive inflammation. Therefore, mechanisms need to be in place to regulate inflammation, depending on the viability of the bacteria and the threat the bacteria pose to the host. In this study, we show how cathelicidins can play a role in this process, by inhibiting TLR2 and TLR4 activation by Gram-negative bacteria upon the loss of bacterial viability. Our results show that CATH-2 can kill E. coli by permeabilizing the bacterial IM, and it subsequently neutralizes lipoproteins and LPS from the bacterial OM to prevent TLR2 and TLR4 activation. In addition, our results show that several other cathelicidins, including human LL-37, have a similar inhibitory effect and inhibit macrophage activation by nonviable E. coli, despite their lack of direct antimicrobial activity under physiological cell culture conditions (Fig. 6).
During an infection, inflammation is induced by activation of pattern-recognition receptors, such as TLRs (1). As a result of the difference in pathogen-associated molecular patterns presented by pathogens, different TLRs are activated by different pathogens. Sensing of E. coli by macrophages largely depends on TLR2 and TLR4, and loss of TLR2 and TLR4 expression in mice renders them more susceptible to E. coli infections (32–35). Although previous studies have shown that various cathelicidins, including CATH-2 and LL-37, can inhibit TLR4 activation by purified LPS (23–25, 36–40), this study shows for the first time, to our knowledge, that cathelicidins inhibit TLR2 and TLR4 activation in the context of complete bacteria and that the inhibition is dependent on the loss of bacterial viability. In addition, our results indicate that the inhibition is dependent on direct interaction of cathelicidins with LPS and lipoproteins from the bacterial OM. Furthermore, because bacteria other than the ones tested in this study also activate TLR2 and/or TLR4 (32, 41–43), the inhibitory effects of cathelicidins observed in this study are most likely applicable to other Gram-negative bacteria as well.
Although the killing of E. coli and inhibition of E. coli–induced macrophage activation by CATH-2 can occur simultaneously, it appears that these two effects are caused by two distinct mechanisms. Our results suggest that CATH-2 first permeabilizes the bacterial IM to kill E. coli, which is followed by disruption of the OM and neutralization of LPS and lipoproteins. Interestingly, in a recent study (44) immuno-electron microscopy showed that CATH-2 can rapidly reach the cytosol of E. coli at antimicrobial concentrations, supporting rapid IM permeabilization. In addition, it was shown that CATH-2 molecules can be observed on the OM fragments released from E. coli upon killing. This further supports our results that CATH-2 directly interacts with the OM-derived LPS and lipoproteins to inhibit TLR4 and TLR2 activation.
Although most cathelicidins tested in this study have previously been described for their direct broad-spectrum antimicrobial activity (31, 45–53), many lacked antimicrobial activity against E. coli in our experiments. This includes human LL-37, which also did not show any interaction with live E. coli, as determined by flow cytometry. Interestingly, even when bacteria were killed with gentamicin, which disrupts the OM but leaves the IM intact (54, 55), LL-37 did not affect the IM integrity (data not shown). This is in contrast to a previous report on the antimicrobial mechanism of LL-37, which shows that LL-37 can kill E. coli through IM permeabilization (56). These discrepancies in antimicrobial activity and lack of IM permeabilizing activity are most likely the result of the presence of serum components and monovalent or divalent cations in the cell culture media, which are known to limit the antimicrobial activity of various antimicrobial peptides, including cathelicidins (13, 22, 23, 57). This serum and cation sensitivity could mean that, in vivo, other antimicrobial mechanisms are needed to kill bacteria, and cathelicidins are instead required to reduce immune activation against the dead bacteria. Cathelicidins have been shown to act in synergy with other host-derived antimicrobial components, such as lysozyme and lactoferrin, which are stored in the same neutrophil granules as cathelicidins (13, 58, 59). Thus, although most cathelicidins have limited antimicrobial activity under cell culture conditions in vitro, their bactericidal activity during infections in vivo might be enhanced by acting in concert with other host-derived antimicrobial components.
Although LL-37 was unable to kill E. coli under the conditions tested in this study, TEM images do show a stress response from E. coli in the presence of LL-37. This stress response appears to be the shedding of membrane fragments to prevent sustained interaction between LL-37 and the bacterial membrane. The release of OM vesicles has been reported to play a role in the defense of E. coli against antimicrobial components and can be used to prevent accumulation of unwanted components in the bacterial membrane (60, 61). This defense mechanism would also explain the lack of binding observed between FITC–LL-37 and E. coli using flow cytometry.
Although various cathelicidins tested in this study were able to inhibit macrophage activation by nonviable E. coli, only CATH-2 and PMAP-36 combined this with potent antimicrobial activity. This dual function makes these peptides nonimmunogenic, or “silent” killers (i.e., they both kill the bacteria and subsequently inhibit immune activation) (62). This could be interesting from a pharmaceutical point of view for the development of novel anti-infective therapies that limit inflammation and sepsis when the bacterial threat has been eliminated. Furthermore, other studies have shown that CATH-2 acts against a wide variety of pathogens, without inducing strong resistance (31, 63), and retains its antimicrobial activity under more physiological conditions, which makes it an interesting template for the development of novel anti-infective therapies against the growing number of antibiotic-resistant bacterial infections.
In conclusion, this study shows for the first time, to our knowledge, the regulatory effects of cathelicidins during macrophage activation by whole bacteria. Our results show how CATH-2 can cause nonimmunogenic or silent killing of Gram-negative bacteria by permeabilizing the bacterial IM and subsequently neutralizing lipoproteins and LPS released from the bacterial OM to prevent TLR2 and TLR4 activation, respectively. In addition, although most other cathelicidins, including LL-37, have limited bactericidal activity under physiological conditions, many of these peptides are able to inhibit macrophage activation by nonviable E. coli. These results describe a novel role for cathelicidins in the discrimination between viable and nonviable bacteria by the immune system by only inhibiting TLR activation when the bacterial threat has been neutralized to prevent excessive inflammation and sepsis.
Supplementary Material
Acknowledgments
We thank George Posthuma and René Scriwanek for assistance with TEM experiments and analysis.
This work was supported by the Immuno Valley Alternatives to Antibiotics Animal Specific Immunomodulatory Antimicrobials 2 program of the Dutch Ministry of Economic Affairs.
The online version of this article contains supplemental material.
- BMDM
- bone marrow–derived macrophage
- CATH-2
- cathelicidin-2
- hTLR
- human TLR
- IM
- inner membrane
- ITC
- isothermal titration calorimetry
- OM
- outer membrane
- qPCR
- quantitative PCR
- TEM
- transmission electron microscopy.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Pandey S., Kawai T., Akira S. 2014. Microbial sensing by Toll-like receptors and intracellular nucleic acid sensors. Cold Spring Harb. Perspect. Biol. 7: a016246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gewirtz A. T., Navas T. A., Lyons S., Godowski P. J., Madara J. L. 2001. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J. Immunol. 167: 1882–1885. [DOI] [PubMed] [Google Scholar]
- 3.Soehnlein O., Lindbom L. 2010. Phagocyte partnership during the onset and resolution of inflammation. Nat. Rev. Immunol. 10: 427–439. [DOI] [PubMed] [Google Scholar]
- 4.Liew F. Y., Xu E. K., Brint, O’Neill L. A. J. 2005. Negative regulation of toll-like receptor-mediated immune responses. Nat. Rev. Immunol. 5: 446–458. [DOI] [PubMed] [Google Scholar]
- 5.Nizet V., Ohtake T., Lauth X., Trowbridge J., Rudisill J., Dorschner R. A., Pestonjamasp V., Piraino J., Huttner K., Gallo R. L. 2001. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature 414: 454–457. [DOI] [PubMed] [Google Scholar]
- 6.Chromek M., Arvidsson I., Karpman D. 2012. The antimicrobial peptide cathelicidin protects mice from Escherichia coli O157:H7-mediated disease. PLoS One 7: e46476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chromek M., Slamová Z., Bergman P., Kovács L., Podracká L., Ehrén I., Hökfelt T., Gudmundsson G. H., Gallo R. L., Agerberth B., Brauner A. 2006. The antimicrobial peptide cathelicidin protects the urinary tract against invasive bacterial infection. Nat. Med. 12: 636–641. [DOI] [PubMed] [Google Scholar]
- 8.Sørensen O., Cowland J. B., Askaa J., Borregaard N. 1997. An ELISA for hCAP-18, the cathelicidin present in human neutrophils and plasma. J. Immunol. Methods 206: 53–59. [DOI] [PubMed] [Google Scholar]
- 9.van Dijk A., Tersteeg-Zijderveld M. H., Tjeerdsma-van Bokhoven J. L., Jansman A. J., Veldhuizen E. J., Haagsman H. P. 2009. Chicken heterophils are recruited to the site of Salmonella infection and release antibacterial mature cathelicidin-2 upon stimulation with LPS. Mol. Immunol. 46: 1517–1526. [DOI] [PubMed] [Google Scholar]
- 10.Jann N. J., Schmaler M., Kristian S. A., Radek K. A., Gallo R. L., Nizet V., Peschel A., Landmann R. 2009. Neutrophil antimicrobial defense against Staphylococcus aureus is mediated by phagolysosomal but not extracellular trap-associated cathelicidin. J. Leukoc. Biol. 86: 1159–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wan M., Sabirsh A., Wetterholm A., Agerberth B., Haeggström J. Z. 2007. Leukotriene B4 triggers release of the cathelicidin LL-37 from human neutrophils: novel lipid-peptide interactions in innate immune responses. FASEB J. 21: 2897–2905. [DOI] [PubMed] [Google Scholar]
- 12.Amulic B., Cazalet C., Hayes G. L., Metzler K. D., Zychlinsky A. 2012. Neutrophil function: from mechanisms to disease. Annu. Rev. Immunol. 30: 459–489. [DOI] [PubMed] [Google Scholar]
- 13.Bals R., Wang X., Zasloff M., Wilson J. M. 1998. The peptide antibiotic LL-37/hCAP-18 is expressed in epithelia of the human lung where it has broad antimicrobial activity at the airway surface. Proc. Natl. Acad. Sci. USA 95: 9541–9546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schauber J., Svanholm C., Termén S., Iffland K., Menzel T., Scheppach W., Melcher R., Agerberth B., Lührs H., Gudmundsson G. H. 2003. Expression of the cathelicidin LL-37 is modulated by short chain fatty acids in colonocytes: relevance of signalling pathways. Gut 52: 735–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorschner R. A., Pestonjamasp V. K., Tamakuwala S., Ohtake T., Rudisill J., Nizet V., Agerberth B., Gudmundsson G. H., Gallo R. L. 2001. Cutaneous injury induces the release of cathelicidin anti-microbial peptides active against group A Streptococcus. J. Invest. Dermatol. 117: 91–97. [DOI] [PubMed] [Google Scholar]
- 16.Byfield F. J., Wen Q., Leszczynska K., Kulakowska A., Namiot Z., Janmey P. A., Bucki R. 2011. Cathelicidin LL-37 peptide regulates endothelial cell stiffness and endothelial barrier permeability. Am. J. Physiol. Cell Physiol. 300: C105–C112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaller-Bals S., Schulze A., Bals R. 2002. Increased levels of antimicrobial peptides in tracheal aspirates of newborn infants during infection. Am. J. Respir. Crit. Care Med. 165: 992–995. [DOI] [PubMed] [Google Scholar]
- 18.López-García B., Lee P. H., Yamasaki K., Gallo R. L. 2005. Anti-fungal activity of cathelicidins and their potential role in Candida albicans skin infection. J. Invest. Dermatol. 125: 108–115. [DOI] [PubMed] [Google Scholar]
- 19.Ong P. Y., Ohtake T., Brandt C., Strickland I., Boguniewicz M., Ganz T., Gallo R. L., Leung D. Y. M. 2002. Endogenous antimicrobial peptides and skin infections in atopic dermatitis. N. Engl. J. Med. 347: 1151–1160. [DOI] [PubMed] [Google Scholar]
- 20.Cuperus T., Coorens M., van Dijk A., Haagsman H. P. 2013. Avian host defense peptides. Dev. Comp. Immunol. 41: 352–369. [DOI] [PubMed] [Google Scholar]
- 21.Vandamme D., Landuyt B., Luyten W., Schoofs L. 2012. A comprehensive summary of LL-37, the factotum human cathelicidin peptide. Cell. Immunol. 280: 22–35. [DOI] [PubMed] [Google Scholar]
- 22.Bowdish D. M., Davidson D. J., Scott M. G., Hancock R. E. 2005. Immunomodulatory activities of small host defense peptides. Antimicrob. Agents Chemother. 49: 1727–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coorens M., Scheenstra M. R., Veldhuizen E. J., Haagsman H. P. 2017. Interspecies cathelicidin comparison reveals divergence in antimicrobial activity, TLR modulation, chemokine induction and regulation of phagocytosis. Sci. Rep. 7: 40874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Dijk A., van Eldik M., Veldhuizen E. J., Tjeerdsma-van Bokhoven H. L., de Zoete M. R., Bikker F. J., Haagsman H. P. 2016. Immunomodulatory and anti-inflammatory activities of chicken cathelicidin-2 derived peptides. PLoS One 11: e0147919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scott M. G., Davidson D. J., Gold M. R., Bowdish D., Hancock R. E. 2002. The human antimicrobial peptide LL-37 is a multifunctional modulator of innate immune responses. J. Immunol. 169: 3883–3891. [DOI] [PubMed] [Google Scholar]
- 26.Pistolic J., Cosseau C., Li Y., Yu J. J., Filewod N. C., Gellatly S., Rehaume L. M., Bowdish D. M., Hancock R. E. 2009. Host defence peptide LL-37 induces IL-6 expression in human bronchial epithelial cells by activation of the NF-kappaB signaling pathway. J. Innate Immun. 1: 254–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nijnik A., Pistolic J., Filewod N. C., Hancock R. E. 2012. Signaling pathways mediating chemokine induction in keratinocytes by cathelicidin LL-37 and flagellin. J. Innate Immun. 4: 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coorens M., van Dijk A., Bikker F., Veldhuizen E. J., Haagsman H. P. 2015. Importance of endosomal Cathelicidin degradation to enhance DNA-induced chicken macrophage activation. J. Immunol. 195: 3970–3977. [DOI] [PubMed] [Google Scholar]
- 29.Lande R., Gregorio J., Facchinetti V., Chatterjee B., Wang Y.-H., Homey B., Cao W., Wang Y.-H., Su B., Nestle F. O., et al. 2007. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature 449: 564–569. [DOI] [PubMed] [Google Scholar]
- 30.Ralph P., Prichard J., Cohn M. 1975. Reticulum cell sarcoma: an effector cell in antibody-dependent cell-mediated immunity. J. Immunol. 114: 898–905. [PubMed] [Google Scholar]
- 31.van Dijk A., Molhoek E. M., Veldhuizen E. J., Bokhoven J. L., Wagendorp E., Bikker F., Haagsman H. P. 2009. Identification of chicken cathelicidin-2 core elements involved in antibacterial and immunomodulatory activities. Mol. Immunol. 46: 2465–2473. [DOI] [PubMed] [Google Scholar]
- 32.Elson G., Dunn-Siegrist I., Daubeuf B., Pugin J. 2007. Contribution of Toll-like receptors to the innate immune response to gram-negative and gram-positive bacteria. Blood 109: 1574–1583. [DOI] [PubMed] [Google Scholar]
- 33.Roger T., Froidevaux C., Le Roy D., Reymond M. K., Chanson A.-L., Mauri D., Burns K., Riederer B. M., Akira S., Calandra T. 2009. Protection from lethal gram-negative bacterial sepsis by targeting Toll-like receptor 4. Proc. Natl. Acad. Sci. USA 106: 2348–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van ’t Veer C., van den Pangaart P. S., Kruijswijk D., Florquin S., de Vos A. F., van der Poll T. 2011. Delineation of the role of Toll-like receptor signaling during peritonitis by a gradually growing pathogenic Escherichia coli. J. Biol. Chem. 286: 36603–36618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zanoni I., Ostuni R., Marek L. R., Barresi S., Barbalat R., Barton G. M., Granucci F., Kagan J. C. 2011. CD14 controls the LPS-induced endocytosis of Toll-like receptor 4. Cell 147: 868–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larrick J. W., Hirata M., Zheng H., Zhong J., Bolin D., Cavaillon J. M., Warren H. S., Wright S. C. 1994. A novel granulocyte-derived peptide with lipopolysaccharide-neutralizing activity. J. Immunol. 152: 231–240. [PubMed] [Google Scholar]
- 37.Mookherjee N., Brown K. L., Bowdish D. M., Doria S., Falsafi R., Hokamp K., Roche F. M., Mu R., Doho G. H., Pistolic J., et al. 2006. Modulation of the TLR-mediated inflammatory response by the endogenous human host defense peptide LL-37. J. Immunol. 176: 2455–2464. [DOI] [PubMed] [Google Scholar]
- 38.Molhoek E. M., den Hertog A. L., de Vries A.-M. B., Nazmi K., Veerman E. C., Hartgers F. C., Yazdanbakhsh M., Bikker F. J., van der Kleij D. 2009. Structure-function relationship of the human antimicrobial peptide LL-37 and LL-37 fragments in the modulation of TLR responses. Biol. Chem. 390: 295–303. [DOI] [PubMed] [Google Scholar]
- 39.Nijnik A., Pistolic J., Wyatt A., Tam S., Hancock R. E. 2009. Human cathelicidin peptide LL-37 modulates the effects of IFN-gamma on APCs. J. Immunol. 183: 5788–5798. [DOI] [PubMed] [Google Scholar]
- 40.Scott A., Weldon S., Buchanan P. J., Schock B., Ernst R. K., McAuley D. F., Tunney M. M., Irwin C. R., Elborn J. S., Taggart C. C. 2011. Evaluation of the ability of LL-37 to neutralise LPS in vitro and ex vivo. PLoS One 6: e26525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bernheiden M., Heinrich J.-M., Minigo G., Schütt C., Stelter F., Freeman M., Golenbock D., Jack R. S. 2001. LBP, CD14, TLR4 and the murine innate immune response to a peritoneal Salmonella infection. J. Endotoxin Res. 7: 447–450. [PubMed] [Google Scholar]
- 42.Faure K., Sawa T., Ajayi T., Fujimoto J., Moriyama K., Shime N., Wiener-Kronish J. P. 2004. TLR4 signaling is essential for survival in acute lung injury induced by virulent Pseudomonas aeruginosa secreting type III secretory toxins. Respir. Res. 5: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiss D. S., Raupach B., Takeda K., Akira S., Zychlinsky A. 2004. Toll-like receptors are temporally involved in host defense. J. Immunol. 172: 4463–4469. [DOI] [PubMed] [Google Scholar]
- 44.Schneider V. A., Coorens M., Ordonez S. R., Tjeerdsma-van Bokhoven J. L., Posthuma G., van Dijk A., Haagsman H. P., Veldhuizen E. J. 2016. Imaging the antimicrobial mechanism(s) of cathelicidin-2. Sci. Rep. 6: 32948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zanetti M., Storici P., Tossi A., Scocchi M., Gennaro R. 1994. Molecular cloning and chemical synthesis of a novel antibacterial peptide derived from pig myeloid cells. J. Biol. Chem. 269: 7855–7858. [PubMed] [Google Scholar]
- 46.Xiao Y., Dai H., Bommineni Y. R., Soulages J. L., Gong Y. X., Prakash O., Zhang G. 2006. Structure-activity relationships of fowlicidin-1, a cathelicidin antimicrobial peptide in chicken. FEBS J. 273: 2581–2593. [DOI] [PubMed] [Google Scholar]
- 47.Skerlavaj B., Scocchi M., Gennaro R., Risso A., Zanetti M. 2001. Structural and functional analysis of horse cathelicidin peptides. Antimicrob. Agents Chemother. 45: 715–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scocchi M., Zelezetsky I., Benincasa M., Gennaro R., Mazzoli A., Tossi A. 2005. Structural aspects and biological properties of the cathelicidin PMAP-36. FEBS J. 272: 4398–4406. [DOI] [PubMed] [Google Scholar]
- 49.Sang Y., Teresa Ortega M., Rune K., Xiau W., Zhang G., Soulages J. L., Lushington G. H., Fang J., Williams T. D., Blecha F., Melgarejo T. 2007. Canine cathelicidin (K9CATH): gene cloning, expression, and biochemical activity of a novel pro-myeloid antimicrobial peptide. Dev. Comp. Immunol. 31: 1278–1296. [DOI] [PubMed] [Google Scholar]
- 50.Gallo R. L., Kim K. J., Bernfield M., Kozak C. A., Zanetti M., Merluzzi L., Gennaro R. 1997. Identification of CRAMP, a cathelin-related antimicrobial peptide expressed in the embryonic and adult mouse. J. Biol. Chem. 272: 13088–13093. [DOI] [PubMed] [Google Scholar]
- 51.Bommineni Y. R., Dai H., Gong Y. X., Soulages J. L., Fernando S. C., Desilva U., Prakash O., Zhang G. 2007. Fowlicidin-3 is an alpha-helical cationic host defense peptide with potent antibacterial and lipopolysaccharide-neutralizing activities. FEBS J. 274: 418–428. [DOI] [PubMed] [Google Scholar]
- 52.Agerberth B., Lee J. Y., Bergman T., Carlquist M., Boman H. G., Mutt V., Jörnvall H. 1991. Amino acid sequence of PR-39. Isolation from pig intestine of a new member of the family of proline-arginine-rich antibacterial peptides. Eur. J. Biochem. 202: 849–854. [DOI] [PubMed] [Google Scholar]
- 53.Agerberth B., Gunne H., Odeberg J., Kogner P., Boman H. G., Gudmundsson G. H. 1995. FALL-39, a putative human peptide antibiotic, is cysteine-free and expressed in bone marrow and testis. Proc. Natl. Acad. Sci. USA 92: 195–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hancock R. E., Farmer S. W., Li Z. S., Poole K. 1991. Interaction of aminoglycosides with the outer membranes and purified lipopolysaccharide and OmpF porin of Escherichia coli. Antimicrob. Agents Chemother. 35: 1309–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mortimer F. C., Mason D. J., Gant V. A. 2000. Flow cytometric monitoring of antibiotic-induced injury in Escherichia coli using cell-impermeant fluorescent probes. Antimicrob. Agents Chemother. 44: 676–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sochacki K. A., Barns K. J., Bucki R., Weisshaar J. C. 2011. Real-time attack on single Escherichia coli cells by the human antimicrobial peptide LL-37. Proc. Natl. Acad. Sci. USA 108: E77–E81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johansson J., Gudmundsson G. H., Rottenberg M. E., Berndt K. D., Agerberth B. 1998. Conformation-dependent antibacterial activity of the naturally occurring human peptide LL-37. J. Biol. Chem. 273: 3718–3724. [DOI] [PubMed] [Google Scholar]
- 58.Mayadas T. N., Cullere X., Lowell C. A. 2014. The multifaceted functions of neutrophils. Annu. Rev. Pathol. 9: 181–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van der Linden D. S., Short D., Dittmann A., Yu P. L. 2009. Synergistic effects of ovine-derived cathelicidins and other antimicrobials against Escherichia coli O157:H7 and Staphylococcus aureus 1056 MRSA. Biotechnol. Lett. 31: 1265–1267. [DOI] [PubMed] [Google Scholar]
- 60.McBroom A. J., Kuehn M. J. 2007. Release of outer membrane vesicles by gram-negative bacteria is a novel envelope stress response. Mol. Microbiol. 63: 545–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Manning A. J., Kuehn M. J. 2011. Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol. 11: 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dupont A., Kaconis Y., Yang I., Albers T., Woltemate S., Heinbockel L., Andersson M., Suerbaum S., Brandenburg K., Hornef M. W. 2015. Intestinal mucus affinity and biological activity of an orally administered antibacterial and anti-inflammatory peptide. Gut 64: 222–232. [DOI] [PubMed] [Google Scholar]
- 63.Veldhuizen E. J. A., Brouwer E. C., Schneider V. A. F., Fluit A. C. 2013. Chicken cathelicidins display antimicrobial activity against multiresistant bacteria without inducing strong resistance. PLoS One 8: e61964. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.