Abstract
Agenesis of the corpus callosum (AgCC) is a congenital brain malformation that occurs in approximately 1:1,000–1:6,000 births. Several syndromes associated with AgCC have been traced to single gene mutations; however, the majority of AgCC causes remain unidentified. We investigated a mother and two children who all shared complete AgCC and a chromosomal deletion at 1q42. We fine mapped this deletion and show that it includes Disrupted-in-Schizophrenia 1 (DISC1), a gene implicated in schizophrenia and other psychiatric disorders. Furthermore, we report a de novo chromosomal deletion at 1q42.13 to q44, which includes DISC1, in another individual with AgCC. We resequenced DISC1 in a cohort of 144 well-characterized AgCC individuals and identified 20 sequence changes, of which 4 are rare potentially pathogenic variants. Two of these variants were undetected in 768 control chromosomes. One of these is a splice site mutation at the 5’ boundary of exon 11 that dramatically reduces full-length mRNA expression of DISC1, but not of shorter forms. We investigated the developmental expression of mouse DISC1 and find that it is highly expressed in the embryonic corpus callosum at a critical time for callosal formation. Taken together our results suggest a significant role for DISC1 in corpus callosum development.
Keywords: agenesis of the corpus callosum, schizophrenia, genetics, DISC1
INTRODUCTION
Agenesis of the corpus callosum (AgCC), a congenital malformation resulting from a failure to develop the brain’s largest cerebral commissure, occurs in approximately 1:1,000–1:6,000 individuals [Guillem et al., 2003; Wang et al., 2004; Glass et al., 2008]. While identifiable causes of AgCC are only known for an estimated 25% of cases [Paul et al., 2007], the etiology of AgCC is likely to contain a large as yet unidentified genetic component. Even the known genetic causes are variable, reflecting the complex and multigenic nature of callosal development. Patients with AgCC present with a wide variety of cognitive deficits that often fall within the autism spectrum [Badaruddin et al., 2007; Paul et al., 2007]. Volume based morphometry (VBM) analyses of the brain in autism consistently show a smaller corpus callosum [Boger-Megiddo et al., 2006; Just et al., 2007; Frazier and Hardan, 2009; Keary et al., 2009; Hardan et al., 2009], and recent studies of the most common copy number variant in autism, 16p11.2, show AgCC in a subset of these patients [Weiss et al., 2008; Bedoyan et al., 2010; Rosenfeld et al., 2010]. There may also be behavioral symptom overlap between AgCC and other neurodevelopmental disorders. Individuals with AgCC, similar to those with schizophrenia or autism, tend to exhibit concrete thinking, impaired social skills, lack of introspection, and poor social judgment [Paul et al., 2007]. Abnormal interhemispheric transfer has been proposed as an explanation for some symptoms within schizophrenia [David, 1994], and several manuscripts report identification of patients with AgCC in cohorts of schizophrenia screened for anatomic brain changes [David et al., 1993; Motomura et al., 2002; Chinnasamy et al., 2006; Hallak et al., 2007]. Moreover, we have shown that AgCC patients have a reduction in the size and in the fractional anisotropy (FA) of the cingulum bundle, as compared to matched controls [Nakata et al., 2009a]. Impairment of the cingulum bundle has also been implicated in schizophrenia (with a similarly reduced size and FA), suggesting a possible mechanistic overlap between a disorder grouped by its most visible anatomic change(AgCC) and one defined by its constellation of clinical symptoms (schizophrenia) [Kubicki et al., 2005; Nestor et al., 2008].
Millar et al.[2000] originally reported a large Scottish family with a balanced translocation with one chromosomal breakpoint at 1q42.1, which disrupts a gene eventually termed Disrupted-in-Schizophrenia 1 (DISC1). The majority of the family members with this translocation, and none without the translocation, presented with schizophrenia or related psychiatric disorders including schizoaffective disorder and bipolar disorder, although brain imaging for this family has not been reported. Since this report, additional studies have strengthened the argument that DISC1 plays an important role in schizophrenia, and recent work has suggested that DISC1 mutations disrupt neurite outgrowth and normal cerebral cortex development [Ozeki et al., 2003; Kamiya et al., 2005]. Furthermore, DISC1 has been implicated as a candidate gene for autism [Kilpinen et al., 2008; Williams et al., 2009; Crepel et al., 2010]. The phenotypic similarities between patients with AgCC and those with autism reinforce the need to investigate DISC1 as a potential candidate gene for callosal development as well.
Puthuran et al. [2005] described a mother and two children who all carried an interstitial 1q42 deletion and had AgCC. In this present study, we have performed further molecular analysis on the affected mother of the family using a genome-wide microarray and FISH to define the boundaries and architecture of the deletion in more detail. In addition to this family, we report on another individual with AgCC and a de novo interstitial deletion that includes DISC1. We also sequenced all 13 exons of the full-length DISC1 gene in a cohort of 144 individuals with MRI characterized AgCC and identified four potentially deleterious mutations, two of which were not found in a large control population (n = 768 chromosomes). Finally, we are the first to show that DISC1 is highly expressed in the developing corpus callosum in embryonic mice. Taken together, these data suggest an important functional role for DISC1 in callosal development.
MATERIALS AND METHODS
FISH and Microarray Analysis
Protocols from the BlueGnome Cytochip reference manual 0.9 were used throughout to label and purify patient DNA and reference DNA (female pooled, Promega, Madison, WI) (Invitrogen BioPrime labelling kit followed by GE Healthcare Autoseg G50 columns) and hybridized to version v1.1 CytoChip. Perkin Elmer Proscanarray captured images were processed using Blue Fuse v3.4. Clinical microarray analysis in the individual identified in Figure 1D was performed at Emory Genetics Laboratory on the EmArray 60K platform.
FIG. 1.
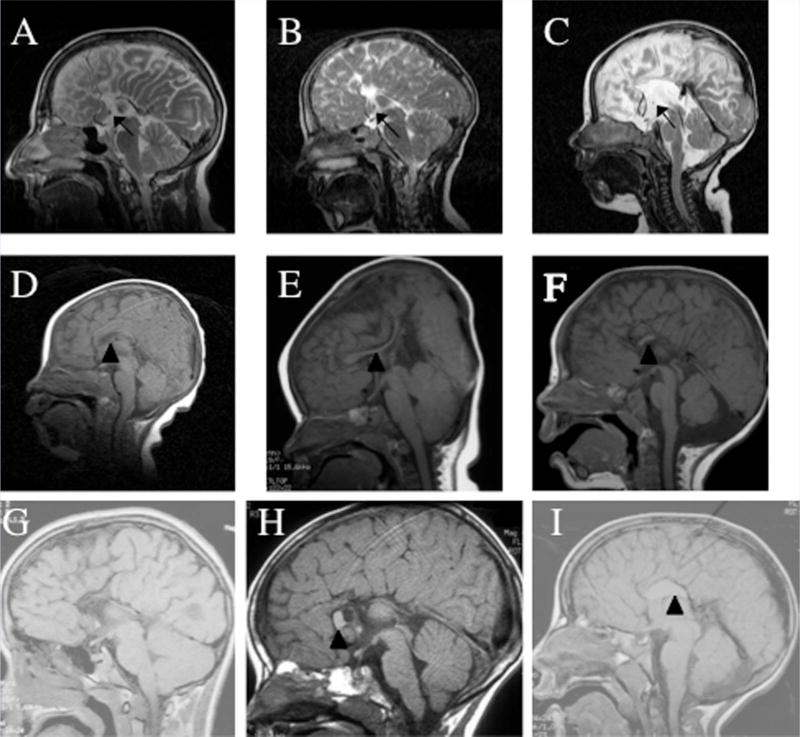
Patients with corpus callosum abnormalities and genetic aberrations of DISC1. Midline sagittal MRI of mother (A) and two sons (B,C) with 1q42 deletion demonstrate complete absence of the corpus callosum. The anterior commissure (arrows) is present in these three family members. D: A residual corpus callosum (arrowhead) is present in proposita 1650-0 with 1q42.13 to q44 deletion. E–I: Corpus callosum abnormalities are also present in individuals with single nucleotide changes affecting DISC1. E: Propositus 1148-0 with a thin and morphologically distorted corpus callosum (arrowhead). F:Propositus 1132-0 with only the superior genu and anterior body of the corpus callosum present (arrowhead). G: Propositus 1294-0 with isolated AgCC. H: Propositus 1145-0 with an absent corpus callosum except for the anterior aspect of the genu (arrowhead). I:Proposita 1058-0 with only a residual component of the posterior body of the corpus callosum present (arrowhead).
DISC1 Sequencing and Variant Genotyping
DNA was prepared from whole blood samples using the Puregene DNA extraction method according to standard protocols. Primers for DISC1 exon sequencing were chosen using Primer3 software (sequences available upon request). Rare variant alleles were screened in controls using the Custom Taqman SNP Genotyping Assay (Applied Biosystems, Carlsbad, CA). The genotyping PCRs were run on a Dual GeneAmp PCR System 9700 and allelic discrimination was called with SDS 2.0 software on an ABI 7900 Analyzer. SNPs rs3738401 (R264Q), rs6675281 (L607F), and rs821616 (S704C) were genotyped in controls using the Sequenom iPLEX genotyping assay on the MassARRAY system. P-values for allele frequencies were determined using a two-tailed Fisher’s exact test. Amino acid sequence alignments were created using the HomoloGene program (http://www.ncbi.nlm.nih.gov/homologene). DNA sequence alignments were created using the ClustalW2 program (http://www.ebi.ac.uk/Tools/clustalw2/index.html).
Real-Time PCR
Total RNA was isolated using the PAXgene Blood RNA Kit (Qiagen, Valencia, CA), and 1 µg total RNA was used for first-strand cDNA synthesis using the SuperScript First-Strand Synthesis System (Invitrogen, Carlsbad, CA). Real-time reverse transcriptase PCR was performed by iCycler (Bio-Rad, Hercules, CA) in a SYBR green I mix. PCR conditions were 95°C for 15 sec, 57°C for 30 sec and 72° C for 30 sec. The forward and reverse primer sequences used for DISC1 exon 10 were AGCACCAGGAGACTGCCTATGAAA and AGT CAGCTT CCC ACA CTT TCC CAA; for exon 11 of DISC1 Lv isoform the primers were TTG GGA AAGTGTGGG AAG CTG ACT and AGC AGC TCC CTC TAA GTC ATC CAT; for exon 11 of DISC1 L isoform the primers were TTG GGA AAG TGT GGG AAG CTG ACT and CTC TTT CTG TTC ACC TCC AGC ACA G; for GAPDH the primers were TGC ACC ACC AAC TGC TTA GC and GGC ATG GAC TGT GGT CAT GAG. PCR specificity was examined on a 2% agarose gel using 10 ml from each reaction and each sample was run in triplicate. Statistical analysis was performed using the 2−ΔΔCT method, as described by a previous protocol [Livak and Schmittgen, 2001].
Immunohistochemistry
Whole brains were fixed in 4% formalin solution and embedded in paraffin, and sections were cut at 5 mm. After dewaxing and hydration, endogenous peroxidase was quenched with 0.3% hydrogen peroxide in methanol for 20 min at room temperature. Rabbit polyclonal antibodies specific for DISC1 (c-terminus) (Zymed, Carlsbad, CA) and for the neuronal marker PGP9.5 (Abcam, Cambridge, MA) were used at 1:200 dilution and expression was detected with the ABC kit (Vector Laboratories, Burlingame, CA) and 3,3’-diaminobenzidine (DAB) (Vector Laboratories). All slides were counterstained with cresyl violet.
RESULTS
Clinical Reports
Proposita 1650-0
This patient is now a 3-year-old female who was born at term but was small for her gestational age throughout the pregnancy. She is noted to have multiple cardiac abnormalities, including an atrial septal defect, two ventricular septal defects, and Wolff–Parkinson–White syndrome. Additionally, the patient was born with microcephaly and bilateral dislocated hips, and has severe gastro-esophageal reflux as well as grade IV vesico-ureteral reflux. EEG at 4 months of age appeared normal. MRI showed a thin and short corpus callosum with absent rostrum, minimal genu, and absent splenium.
Propositus 1148-0
This patient is now a 9-year-old male, born at term via cesarean indicated by hydrocephalus. A ventricular peritoneal shunt was placed shortly after birth. MRI revealed partial AgCC with possible polymicrogyria in the perisylvian regions. The patient also was noted to have right eye esotropia with nystagmus, and global hypotonia.
Propositus 1132-0
This patient is now an 8-year-old male with craniofacial dysmorphisms (including hypertelorism, frontal bossing, and simplified low set helices), global developmental delay, and is non-verbal. He was noted to have infantile spasms and more recently has been evaluated for possible absence seizures. He has severe chronic constipation requiring frequent enemas, kyphoscoliosis, and short stature. Imaging studies show partial AgCC with only the superior genu and the anterior body of the corpus callosum present, inferior placement of the posterior pituitary, bilateral perisylvian polymicrogyria, and a sphenoid encephalocele.
Propositus 1294-0
This patient is now a 17-year-old male born to a 28-year-old mother. He was noted to have testicular hydrocele at birth. He has ataxia and developed partial-complex and absence seizures at age 9. His full-scale IQ is 73 on the WISC-III. MRI studies revealed isolated AgCC with no other remarkable findings.
Propositus 1145-0
This patient is now a 9-year-old male with no prenatal complications, although he had respiratory distress requiring resuscitation after delivery. He demonstrated left arm paralysis and moderate developmental delays with walking at 20 months, crawling at 10–11 months, and speaking after 1 year. Chromosome analysis including subtelomeric FISH revealed no deletions, duplications, or translocations. MRI showed isolated partial AgCC.
Proposita 1058-0
This patient is now an 18-year-old female with severe language delays, a non-verbal IQ of 44 and behavior on the autism spectrum. She was found to have a bicuspid aortic valve and secondary mitral valve regurgitation. MRI reveals partial AgCC, with only a residual component of the posterior body. She also does not have probst bundles.
Refinement of 1q42 Deletion in Family With Callosal Agenesis and Identification of 1q42.13 to q44 Deletion in Individual With AgCC
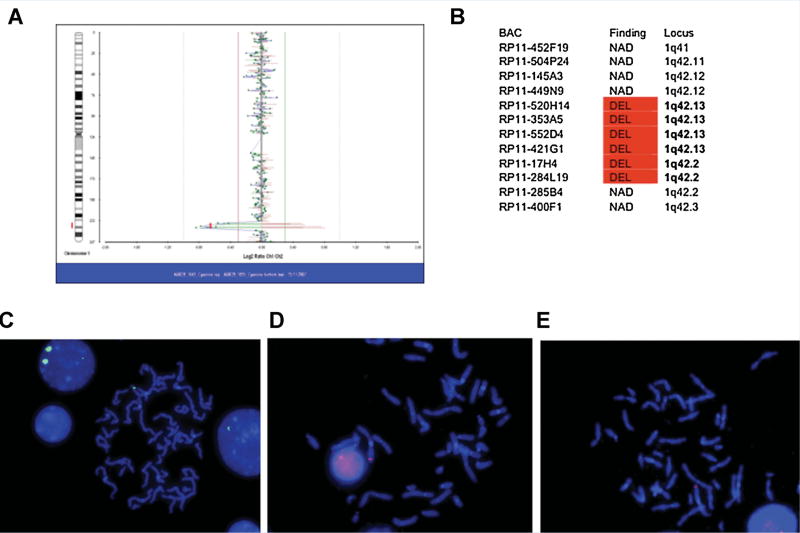
The clinical details of three members of the same family (mother and two children) with AgCC (Fig. 1A – C) and chromosome 1q42 deletion have been described in a previous report [Puthuran et al. [2005]]. Using standard cytogenetic analysis with FISH the maternal karyotype was as follows: 46,XX,del(1)(q42.1-q42.3).ish del(1)(q42.13-q42.2)(RP11–449N9+, RP11–520H14–, RP11–353A5–, RP11–284L19–,RP11–285B4+). Follow-up BAC microarray analysis of her DNA confirmed the deletion and refined its size and boundaries to a 5.8 Mb region spanning BAC clones RP11-375H24 to RP11-87P4 (Figs. 2A and 3). Metaphase spreads counterstained with DAPI illustrate the hybridization of probes RP11-520H14, RP11-353A5, and RP11-284L19 (Fig. 2C–E). Detailed information about the clones used in this analysis (Fig. 2B) can be obtained from the Ensembl database. In addition to this family, clinical microarray and confirmatory G-banding analyses in a separate individual with partial AgCC (proposita 1650-0) (Figs. 1D and 3) identified a 13.7 Mb deletion at 1q42.13 to q44 (between 226.5 and 240.2 Mb). G-banding analysis of both parents indicated that the deletion occurred de novo.
FIG. 2.
Refinement of 1q42 deletion in family with agenesis of the corpus callosum. Follow-up BAC clone microarray analysis of the mother of the family with AgCC and 1q42 deletion showed a deletion spanning six clones over approximately 5.8 Mb—arr cgh 1q42.13 → 1q42.2 (RP11-375H24 → RP11-87P4)x1 which refines the earlier karyotype results (A). The table in (B) gives the BAC clones used in microarray analysis. Detailed information about the clones can be obtained from the Ensembl database. Metaphase spreads counterstained with DAPI illustrate the hybridization of probes RP11-520H14 (C), RP11-353A5 (D), and RP11-284L19 (E).
FIG. 3.
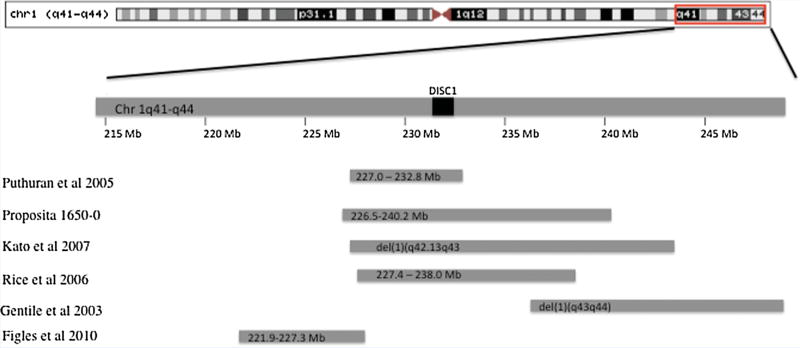
Genomic map of deletions in chromosomal region 1q42. Deletions in the chromosome region 1q42 including and adjacent to DISC1 are found in patients who demonstrate callosal abnormalities. The deletion in the family with AgCC reported by Puthuran et al. spans a 5.8 Mb region between 227.0 and 232.8 Mb on chromosome 1q. Proposita 1650-0 with partial AgCC has a 13.7 Mb deletion between 226.5 and 240.2 Mb. Cytogenetic analysis by Kato et al. and Gentile et al. locate deletions in patients with partial AgCC at 1q42.13-q43 and 1q43-q44, respectively. The deletion described by Rice et al. is found in a patient with a normal corpus callosum and is located between 227.4 and 238.0 Mb. The deletion in a patient with AgCC reported by Filges et al. is located between 221.9 and 227.3 Mb.
Detection of DISC1 Nucleotide Changes in AgCC Population
The two-generation family with AgCC and 1q42 deletion, the prevalence of schizophrenia among AgCC individuals, and in vitro evidence linking DISC1 to neurite outgrowth highlighted DISC1 as an important AgCC candidate gene. To address this hypothesis, we sequenced all 13 exons and intron–exon boundaries of the long form of the DISC1 gene in a cohort of 144 MRI-verified AgCC individuals [Hetts et al., 2006]. A total of 20 nucleotide changes were observed in the sequencing, 10 of which were previously unreported in dbSNP (Table I). Three of these nucleotide changes result in non-conservative amino acid substitutions (P287L in propositus 1148-0, T453M [rs28930675] in propositus 1132-0 and 1294-0, and P540Q in propositus 1145-0), and are predicted to impair DISC1 protein function using the PolyPhen and SIFT protein folding prediction programs. The wild type amino acid at these three positions appears to be highly conserved in mammals (Table II). Additionally, we detected a novel change at the 5’ intron–exon boundary of exon 11 (IVS10 –2; A–G nucleotide change) in proposita 1058-0 that is predicted to disrupt mRNA splicing, as it occurs at the invariant second position in the splice acceptor site (see below). In mammals that express exon 11 of DISC1, this IVS10 –2 A nucleotide at the splice site is highly conserved (Table III). In all cases, we confirmed the detection of these nucleotide changes by resequencing fresh DNA samples. Parents were also genotyped to determine whether the mutations were inherited or occurred de novo; all four changes were inherited, with no obvious gender predilection. To determine the population frequency of these genetic changes, we then screened a Caucasian control population. P287L and rs28930675 were detected at low frequencies (1/764 and 3/694 chromosomes, respectively). However, P540Q and IVS10 –2 were absent in all genotyped controls (0/756 and 0/706, respectively).
Table I.
Summary of DISC1 Nucleotide Changes in AgCC Patients and Controls
| AA change | Rs# | Location | PolyPhen prediction |
SIFT prediction |
Patient genotype frequency |
Control genotype frequency |
Minor allele frequency |
Pvalue |
|---|---|---|---|---|---|---|---|---|
| V71L | Exon 2 | GG = 139 | ||||||
| GT = 1 | ||||||||
| TT = 0 | ||||||||
| A83V | Exon 2 | CC = 139 | ||||||
| CT = 1 | ||||||||
| TT = 0 | ||||||||
| R264Q | rs3738401 | Exon 2 | GG = 55 | GG = 161 | Patients = 0.384 | 0.23 | ||
| AG = 60 | AG = 126 | Controls = 0.337 | ||||||
| AA = 23 | AA = 51 | |||||||
| P287L | Exon 2 | Probably damaging | Tolerated | CC = 137 | CC = 381 | |||
| CT = 1 | CT = 1 | |||||||
| TT = 0 | TT = 0 | |||||||
| IVS4–36 | Intron 4 | TT = 139 | ||||||
| CT = 2 | ||||||||
| CC = 0 | ||||||||
| T453M | rs28930675 | Exon 5 | Possibly damaging | Damaging [low confidence] | CC = 139 | CC = 344 | ||
| CT = 2 | CT = 3 | |||||||
| TT = 0 | TT = 0 | |||||||
| L465L | rs3738402 | Exon 5 | CC = 132 | |||||
| CT = 9 | ||||||||
| TT = 0 | ||||||||
| I4691 | rs2492367 | Exon 6 | CC = 115 | |||||
| CT = 20 | ||||||||
| TT = 5 | ||||||||
| P540Q | Exon 6 | Probably damaging | Damaging (low confidence) | CC = 138 | CC = 378 | |||
| AC = 1 | AC = 0 | |||||||
| AA = 0 | AA = 0 | |||||||
| IVS6 + 31 | rs16854940 | Intron 6 | CC = 128 | |||||
| CT = 8 | ||||||||
| TT = 0 | ||||||||
| IVS6 - 89 | rs41271515 | Intron 6 | GG = 138 | |||||
| AG = 1 | ||||||||
| AA = 0 | ||||||||
| IVS7 + 6 | Intron 7 | TT = 104 | ||||||
| CT = 32 | ||||||||
| CC = 4 | ||||||||
| IVS8 + 60 | Intron 8 | GG = 128 | ||||||
| AG = 2 | ||||||||
| AA = 0 | ||||||||
| L607F | rs6675281 | Exon 9 | CC = 109 | CC = 250 | Patients = 0.124 | 0.36 | ||
| CT = 29 | CT = 88 | Controls = 0.148 | ||||||
| TT = 3 | TT = 7 | |||||||
| L621L | rs12133766 | Exon 9 | GG = 126 | |||||
| AG = 15 | ||||||||
| AA = 0 | ||||||||
| IVS10 + 95 | Intron 10 | GG = 72 | ||||||
| AG = 53 | ||||||||
| AA = 13 | ||||||||
| IVS10–2 | Intron 10 | AA = 140 | AA = 353 | |||||
| AG = 1 | AG = 0 | |||||||
| GG = 0 | GG = 0 | |||||||
| S704C | rs821616 | Exon 11 | AA = 85 | AA = 185 | Patients = 0.225 | 0.15 | ||
| AT = 50 | AT = 133 | Controls = 0.271 | ||||||
| TT = 7 | TT = 27 | |||||||
| L792L | Exon 12 | GG = 140 | ||||||
| AG = 1 | ||||||||
| AA = 0 | ||||||||
| IVS12 – 64 | rs17773715 | Intron 12 | GG = 92 | |||||
| AG = 43 | ||||||||
| AA = 4 |
Resequencing of all 13 exons in 144 individuals with AgCC detected several SNPs and previously unreported single nucleotide variants. The rare variants indicated in bold result in an amino acid substitution with significantly different biochemical properties from the wild type amino acid at that position. The IVS10 – 2 mutation was detected in a splice site two bases upstream of exon 11. These rare variants as well as other selected SNPs were then genotype-screened in a control population. While P287L and rs28930675 [T453M] were detected at low frequencies, P540Q and IVS10 – 2 were found only in the AgCC cohort. The variation in minor allele frequencies of rs3738401 (R264Q), rs6675281 (L607F), and rs821616 (S704C) between patients and controls did not reach statistical significance.
Table II.
Alignment of DISCI Amino Acid Sequences in Mammals
| Species | Amino Acid Sequence |
|---|---|
| Homo sapiens | 270 ATRVSADLAQAAR–NSSRPER 289 |
| Pan troglodytes | 270 ATRVSADLAQAAR–NSSRPER 289 |
| Canis lupus familiaris | 304 TIPSLADSAQTTG–GSHRPEC 323 |
| Bos taurus | --------------------- |
| Mus musculus | 271 AAPGLADLAQVTRSSSRQSEC 290 |
| Rattus norvegicus | 266 AAPGLVDLAQGTR-SNRQPEC 285 |
| Homo sapiens | 440 LEPTAQDSLHV–SITRRDWLL 459 |
| Pan troglodytes | 440 LEPTAQDSLHV–SITRRDWLL 459 |
| Canis lupus familiaris | 469 WEPTAQDTLRV–SITRRDWLL 488 |
| Bos taurus | 65 LEAAAQDSLRV–SITRRDWLL 84 |
| Mus musculus | 440 ---TAQDSLPA–SITRRDWLI 456 |
| Rattus norvegicus | 432 ---TAQDSLPGLAVTRRDWLM 449 |
| Homo sapiens | 530 AGQIPFHAEPPETIRSLQER 549 |
| Pan troglodytes | 530 AGQIPFHAEPPETIRSLQER 549 |
| Canis lupus familiaris | 559 ANQIPICAEPPETIRSLQER 578 |
| Bos taurus | 153 AEQIPLHAEPPETIRSLQER 172 |
| Mus musculus | 527 ANQAPFQVEPPETLRSLRER 546 |
| Rattus norvegicus | 520 ARWAPFRVEPPETLRSLRER 539 |
Protein sequence alignment using HomoloGene reveals that amino acids at positions 287, 453, and 540 (bold) in humans are highly conserved in several other mammal species.
Table III.
DNA Sequence Alignment of the 5° Splice Site of Exon 11 of DISC1 in Mammals
| Species | DNA sequence |
|---|---|
| Homo sapiens | ctcagCTGCAAGTGTCCACTGCTTG |
| Pan troglodytes | ctgacCTGCAAGTGTCCACTGCTCG |
| Pongo pygmaeus | ctcagCTGCAAGTGTCCACTGCTTG |
| Macaca mulatta | ctcagCTGCAAGTGTCCACTGCTTG |
| Equusferus caballus | cttagCTGCAAGTGTCCGCTGCTTG |
| Canis lupus familiaris | ---agCTGCAAGTGTCCACTGCTTG |
| Cavia porcellus | cttagCTGCAAGTGTCCGCTGCTTG |
The “A nucleotide” (bold) at the 5′ splice site of exon 11 if DISC1 is highly conserved in primates and other mammals.
DISC1 variants R264Q, L607F, and S704C have been associated with a higher risk for developing schizophrenia [Zhang et al., 2006; Qu et al., 2007; Song et al., 2008; Nakata et al., 2009b]. We compared the allele frequencies at these genetic loci between AgCC patients and controls to inquire whether these variants also segregate with the AgCC phenotype, While the allele frequencies differed somewhat between the two groups, the associations did not reach a threshold of statistical significance.
Mutation at DISC1 Exon 11 Splice Site and Reduction of mRNA Levels
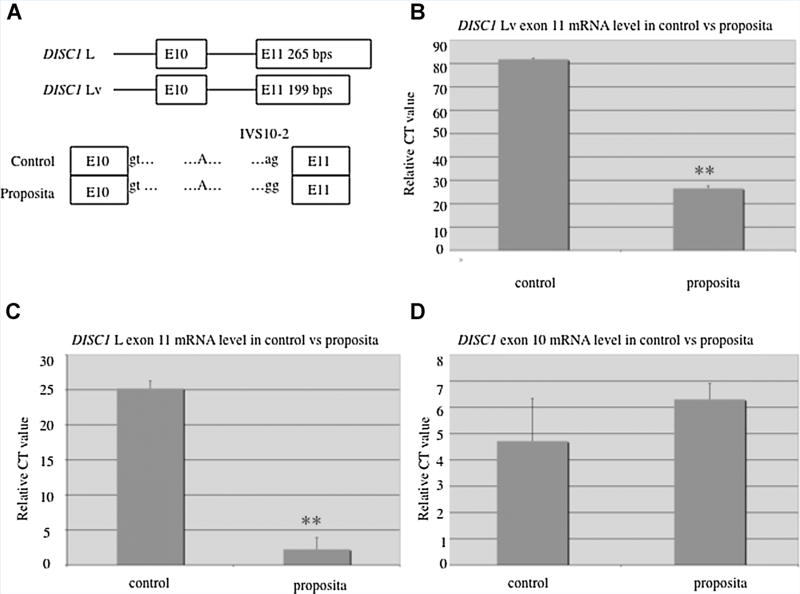
As IVS10 – 2 is located two bases upstream of the start of exon 11, we hypothesized that the mutant G allele at this locus may disrupt expression of the long forms of DISC1 mRNA (L and Lv, which differ by a length of 66 base pairs in exon 11). To address the functional significance of this change in vitro, we conducted realtime PCR to measure the steady state levels of the three main DISCI mRNA splice products in total RNA freshly isolated from whole blood of a matched healthy control and of proposita 1058-0 with the IVS10 – 2 mutation. This analysis demonstrated a dramatic reduction of exon 11 mRNA levels (Fig. 4B,C) in proposita 1058-0 compared to the control, with no significant difference in the overall mRNA levels of exon 10 (Fig. 4D). We propose that the reduction in exon 11 mRNA expression in the proposita corresponds to a reduction in both DISCI L and DISCI Lv mRNA expression, whereas the unaffected expression of exon 10 signifies normal levels of short DISCI isoforms. This differential expression between total mRNA abundance and the abundance of specific isoforms serves as an internal control for this measurement.
FIG. 4.
Splice site mutation resulting in reduced expression of DISC1 exon 11 mRNA. A: Top, The two DISC1 long isoforms only differ for the length of exon 11: 265 bp for DISC1 L and 199 bp for DISC1 Lv. Bottom, Sequence analysis revealed a splice site mutation found in proposita 1058-0 where one nucleotide at the acceptor site changed from A to G in intron 10 (SNP IVS10 2). B–D: Quantitative measurement of DISC1 mRNA levels from whole blood samples in control and proposita. Compared with the control, both DISC1 Lv and DISC1 L exon 11 mRNA are significantly decreased in the proposita [mean + SD, P < 0.01] (B,C). However, exon 10 expression was similar in the proposita and control (D).
DISC1 Expression in Mouse Developing Corpus Callosum
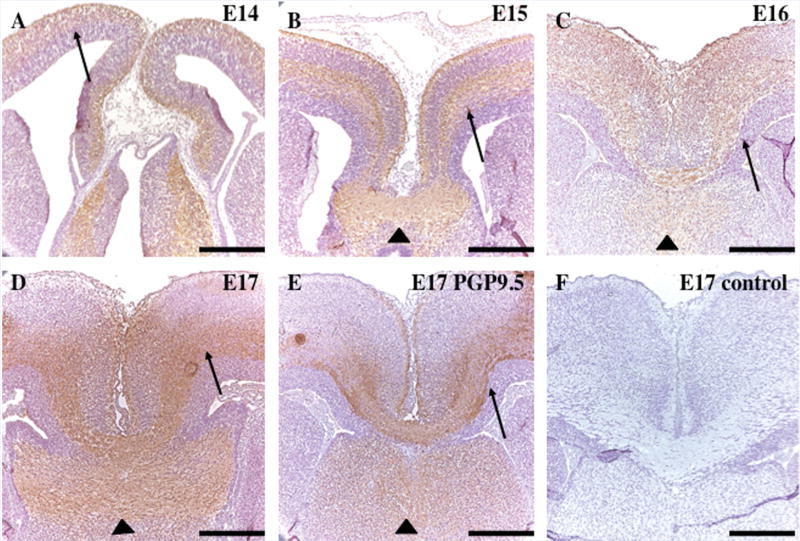
To address whether and when DISC1 is expressed in the developing corpus callosum, we performed immunohistochemical staining with a DISC1 polyclonal antibody in E14–E17 mice. A high level of DISC1 expression was observed throughout the corpus callosum during E14–E17 (Fig. 5A – D). In addition to being expressed in commissural axons, DISC1 is also found in the cerebral cortex and in the dorsal hippocampal commissure and the fornices, regions that have been previously reported to show DISC1 expression [Schurov et al., 2004]. While the hippocampus and cerebral cortex have been previously shown to express DISC1, our findings are the first to show that DISC1 is highly expressed in the developing corpus callosum.
FIG. 5.
Developmental expression of DISC1 in mouse corpus callosum. Immunohistochemistry demonstrates expression of DISC1 in the cerebral cortex as early as E14 (A, arrow). During corpus callosum development from E16–17 (B–D), strong expression of DISC1 is found in commissural neurons (arrows), the dorsal hippocampus commissure, and the fornices (arrowheads). Similar expression is also found for neuronal marker PGP9.5 (E, arrow and arrowhead). Panel F shows the negative control with secondary anti-rabbit antibody only. Scale bar, 50 µm.
DISCUSSION
In this present study, we provide multiple lines of evidence to support a role for DISC1 in callosal development. This includes refinement of the 1q42 deletion that contains DISC1 in a family with AgCC [Puthuran et al. [2005]] as well as identification of a different interstitial deletion including DISC1 in an unrelated individual with AgCC. Our findings also encompass identification of multiple rare, likely pathogenic DISC1 variants in a cohort of 144 well-characterized ACC patients, including a splice site mutation that we demonstrated diminishes the long forms of DISC1 mRNA. We also provide evidence that DISC1 is highly expressed in the developing corpus callosum, a previously unreported finding.
Others have also shown deletions in or near the chromosomal region of 1q42 in patients with readily apparent callosal abnormalities (Fig. 3) [Gentile et al., 2003; Kato et al., 2007; Filges et al., 2010]. However, Rice et al. [2006] report a patient with an interstitial deletion in this region that includes DISC1 but with normal callosal development. Furthermore, patients with callosal abnormalities and deletions just proximal [Filges et al., 2010] or distal [Gentile et al., 2003] to DISC1 have been reported in the literature as well. It is also possible that genes deleted in this region other than DISC1 are responsible for the callosal malformations in these patients. For example, CDC42 binding protein kinase A (CDC42BPA), a gene included in both deleted regions in the family with AgCC originally reported by Puthuran et al. and proposita 1650-0 in this study, is expressed in the developing CNS. CDC42 has been implicated in establishing neuronal polarity, and CDC42-null mice show defects in developing axonal tracts [Garvalov et al., 2007]; however, little information exists for CDC42BPA itself. Exocyst complex component 8 (EXOC8) is also included within these deleted intervals, and the exocyst complex has been shown to promote neurite outgrowth [Vega and Hsu, 2001]. Nevertheless, we hypothesize that AgCC is likely a polygenic phenotype, and thus we would anticipate that deletion of DISC1 could quite likely have variable penetrance for the acallosal phenotype. Additionally, a deletion adjacent to DISC1 (as in the Filges et al. and Gentile et al. manuscripts) may alter DISC1 transcription through a secondary effect on the chromatin structure of the DISC1 gene.
Recently, Shen et al. [2008] created a transgenic mouse, that in addition to expressing one allele of the endogenous DISC1, expressed a truncated DISC1 gene encoding only the first eight exons to model the DISC1 translocation originally identified in the large Scottish pedigree. These mice exhibited reduced neuronal proliferation as well as reduced neurite outgrowth, consistent with earlier data [Ozeki et al., 2003]. Morphological analysis also showed partial AgCC in the transgenic mice, with thinning at the rostral portion of the corpus callosum and failure to cross the midline caudally. These findings provide potential corroboration for the findings in our AgCC patient cohort. The IVS10 – 2 mutation found in one AgCC individual in our study also nearly mimics the DISC1 translocation in the original Scottish family as it selectively diminishes expression of the long forms of DISC1. There are currently no reports of brain imaging in this family, but we hypothesize that callosal anomalies are likely to be identified in this and similar individuals. Another genetic locus that has been associated with schizophrenia, duplication of 16p11.2, has also been reported to have callosal anomalies [McCarthy et al., 2009; Rosenfeld et al., 2010].
This connection between schizophrenia and callosal anomalies may be more widespread. Diffusion tensor imaging has shown reduced interhemispheric connectivity (IHC) involving callosal fibers in schizophrenia [Kubicki et al., 2008]. Moreover, a recent meta-analysis of 28 studies that used magnetic resonance imaging to analyze brain morphology of schizophrenic individuals showed that over all, corpus callosum area was reduced when compared to healthy controls [Arnone et al., 2008]. Moreover, callosal agenesis has been reported in many case studies [David et al., 1993; Motomura et al., 2002; Chinnasamy et al., 2006; Hallak et al., 2007] and in one study callosal agenesis was found in two patients of 140 schizophrenics systematically imaged for observed structural changes. The callosal phenotype was the only notable brain finding [Swayze et al., 1990]. Another series of 52 cases of schizophrenia found AgCC in one individual [Scott et al, 1993]. Analogously, a population study of callosal agenesis in the United Kingdom found that, of the 56 adults with AgCC identified, 35% had psychiatric histories of which 8% had schizophrenia or bipolar psychosis [Taylor and David, 1998]. These observations suggest that callosal malformations may be an important underlying biological mechanism for schizophrenia.
Resequencing candidate genes in a large cohort of patients and controls provides an ideal method for identifying variants contributing to a disease phenotype, and recent analysis has suggested that the overwhelming majority of rare missense or splice site mutations are in fact detrimental and likely contribute to the phenotype of such diseases [Kryukov et al., 2007]. Indeed, rare variants have been shown to be pathogenic in more common diseases such as cardiovascular disease and autism [Barnby et al., 2005; Durand et al., 2007; Romeo et al., 2007]. Rare DISC1 variants have also been shown to correlate with disease. Certain ENU-induced DISC1 missense mutations are associated with phenotypes that attempt to model psychiatric symptoms in mice. Animals with Q31L DISC1 mutation perform poorly on the forced swim test, a paradigm that has been used to demonstrate depressive behavior in rodents. These mice improved their performance after the administration of the antidepressant bupropion. Mice with L100P mutations have deficits in prepulse inhibition as well as latent inhibition, models used to demonstrate information-processing deficits in patients with schizophrenia. The deficits in these mice partially improved after antipsychotic treatment with haloperidol or clozapine [Clapcote et al., 2007].
A recent study reported resequencing DISC1 in a cohort of 288 schizophrenia patients and found seven non-synonymous variants that were not found in 288 controls. Direct sequencing of the controls revealed only one novel variant, suggesting that rare DISC1 variants occur at a lower frequency in phenotypically normal controls than in schizophrenic patients. Five of the variants (G14A, R37W, S90L, R418H, and T603I) were absent in 10,000 screened control alleles, and authors estimated a 2% schizophrenia attributable-risk for these rare variants [Song et al., 2008]. This frequency of rare DISC1 alleles is comparable to the rate found in our AgCC patients, in which 2 variants (P540Q and IVS10 –2) were present in the 144 AgCC individuals but were absent in all 764 genotyped control chromosomes.
Neuronal migration during development is regulated in part by microtubule dynamics [Smith et al, 2000; Hatten, 2002; Tischfield et al., 2010]. In support of this, patients with TUBB3 mutations and mice null for the microtubule-associated protein 1B (MAP1B) display AgCC [Meixner et al, 2000; Tischfield et al., 2010]. Since DISC1 is a component of the dynein protein motor complex that participates in microtubule organization and dynamics [Kamiya et al., 2005] and has been shown to bind another microtubule associated protein, MAP1A [Morris et al., 2003], it is reasonable to hypothesize that a mutant dynein motor complex could affect microtubule dynamics in the axon and prevent axons from traversing the midline, causing callosal agenesis.
In conclusion, researchers have long proposed a link between corpus callosum morphology and function and schizophrenia. Our data implicating DISC1 mutations in callosal development also supports the hypothesis that abnormal callosal formation correlates with schizophrenia, and may provide insight into the correlation between aberrant cerebral connectivity and neurodevelopmental disorders more generally.
Acknowledgments
This work was supported by NIH/NCRR UCSF-CTSI Grant Number UL1 RR024131 and by the NIHR through the Manchester Biomedical Research Centres. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. E.H.S. is supported by grants NS052192 and NS058721 from the NINDS.M.C.O’D. is funded by the Manchester NIHR Biomedical Research Centre and the Wellcome Trust.
References
- Arnone D, McIntosh AM, Tan GM, Ebmeier KP. Meta-analysis of magnetic resonance imaging studies of the corpus callosum in schizophrenia. Schizophr Res. 2008;101:124–132. doi: 10.1016/j.schres.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Badaruddin DH, Andrews GL, Bolte S, Schilmoeller KJ, Schilmoeller G, Paul LK, Brown WS. Social and behavioral problems of children with agenesis of the corpus callosum. Child Psychiatry Hum Dev. 2007;38:287–302. doi: 10.1007/s10578-007-0065-6. [DOI] [PubMed] [Google Scholar]
- Barnby G, Abbott A, Sykes N, Morris A, Weeks DE, Mott R, Lamb J, Bailey AJ, Monaco AP. International Molecular Genetics Study of Autism Consortium Candidate-gene screening and association analysis at the autism-susceptibility locus on chromosome 16p: Evidence of association at GRIN2A and ABAT. Am J Hum Genet. 2005;76:950–966. doi: 10.1086/430454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedoyan JK, Kumar RA, Sudi J, Silverstein F, Ackley T, Iyer RK, Christian SL, Martin DM. Duplication 16p11.2 in a child with infantile seizure disorder. Am J Med Genet Part A. 2010;152A:1567–1574. doi: 10.1002/ajmg.a.33415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boger-Megiddo I, Shaw DW, Friedman SD, Sparks BF, Artru AA, Giedd JN, Dawson G, Dager SR. Corpus callosum morphometrics in young children with autism spectrum disorder. J Autism Dev Disord. 2006;36:733–739. doi: 10.1007/s10803-006-0121-2. [DOI] [PubMed] [Google Scholar]
- Chinnasamy D, Rudd R, Velakoulis D. A case of schizophrenia with complete agenesis of the corpus callosum. Australas Psychiatry. 2006;14:327–330. doi: 10.1080/j.1440-1665.2006.02299.x. [DOI] [PubMed] [Google Scholar]
- Clapcote SJ, Lipina TV, Millar JK, Mackie S, Christie S, Ogawa F, Lerch JP, Trimble K, Uchiyama M, Sakuraba Y, Kaneda H, Shiroishi T, Houslay MD, Henkelman RM, Sled JG, Gondo Y, Porteous DJ, Roder JC. Behavioral phenotypes of DISC1 missense mutations in mice. Neuron. 2007;54:387–402. doi: 10.1016/j.neuron.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Crepel A, Breckpot J, Fryns JP, De la Marche W, Steyaert J, Devriendt K, Peeters H. DISC1 duplication in two brothers with autism and mild mental retardation. Clin Genet. 2010;77:389–394. doi: 10.1111/j.1399-0004.2009.01318.x. [DOI] [PubMed] [Google Scholar]
- David AS. Schizophrenia and the corpus callosum: Developmental, structural and functional relationships. Behav Brain Res. 1994;64:203–211. doi: 10.1016/0166-4328(94)90132-5. [DOI] [PubMed] [Google Scholar]
- David AS, Wacharasindhu A, Lishman WA. Severe psychiatric disturbance and abnormalities of the corpus callosum: Review and case series. J Neurol Neurosurg Psychiatry. 1993;56:85–93. doi: 10.1136/jnnp.56.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsäter H, Sponheim E, Goubran-Botros H, Delorme R, Chabane N, Mouren-Simeoni MC, de Mas P, Bieth E, Rogé B, Héron D, Burglen L, Gillberg C, Leboyer M, Bourgeron T. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filges I, Röthlisberger B, Boesch N, Weber P, Wenzel F, Huber AR, Heinimann K, Miny P. Interstitial deletion 1q42 in a patient with agenesis of corpus callosum: Phenotype-genotype comparison to the 1q41q42 microdeletion suggests a contiguous 1q4 syndrome. Am J Med Genet Part A. 2010;152A:987–993. doi: 10.1002/ajmg.a.33330. [DOI] [PubMed] [Google Scholar]
- Frazier TW, Hardan AY. A meta-analysis of the corpus callosum in autism. Biol Psychiatry. 2009;66:935–941. doi: 10.1016/j.biopsych.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvalov BK, Flynn KC, Neukirchen D, Meyn L, Teusch N, Wu X, Brakebusch C, Bamburg JR, Bradke F. Cdc42 regulates coflin during the establishment of neuronal polarity. J Neurosci. 2007;27:13117–13129. doi: 10.1523/JNEUROSCI.3322-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile M, Di Carlo A, Volpe P, Pansini A, Nanna P, Valenzano MC, Buonadonna AL. FISH and cytogenetic characterization of a terminal chromosome 1q deletion: Clinical case report and phenotypic implications. Am J Med Genet Part A. 2003;117A:251–254. doi: 10.1002/ajmg.a.10018. [DOI] [PubMed] [Google Scholar]
- Glass HC, Shaw GM, Ma C, Sherr EH. Agenesis of the corpus callosum in California 1983–2003: A population-based study. Am J Med Genet Part A. 2008;146A:2495–2500. doi: 10.1002/ajmg.a.32418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillem P, Fabre B, Cans C, Robert-Gnansia E, Jouk PS. Trends in elective terminations of pregnancy between 1989 and 2000 in a French county (the Isére) Prenat Diagn. 2003;23:877–883. doi: 10.1002/pd.711. [DOI] [PubMed] [Google Scholar]
- Hallak JE, Crippa JA, Pinto JP, Machado de Sousa JP, Trzesniak C, Dursun SM, McGuire P, Deakin JF, Zuardi AW. Total agenesis of the corpus callosum in a patient with childhood-onset schizophrenia. Arq Neuropsiquiatr. 2007;65:1216–1219. doi: 10.1590/s0004-282x2007000700024. [DOI] [PubMed] [Google Scholar]
- Hardan AY, Pabalan M, Gupta N, Bansal R, Melhem NM, Fedorov S, Keshavan MS, Minshew NJ. Corpus callosum volume in children with autism. Psychiatry Res. 2009;174:57–61. doi: 10.1016/j.pscychresns.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatten ME. New directions in neuronal migration. Science. 2002;297:1660–1663. doi: 10.1126/science.1074572. [DOI] [PubMed] [Google Scholar]
- Hetts SW, Sherr EH, Chao S, Gobuty S, Barkovich AJ. Anomalies of the corpus callosum: An MR analysis of the phenotypic spectrum of associated malformations. AJR Am J Roentgenol. 2006;187:1343–1348. doi: 10.2214/AJR.05.0146. [DOI] [PubMed] [Google Scholar]
- Just MA, Cherkassky VL, Keller TA, Kana RK, Minshew NJ. Functional and anatomical cortical underconnectivity in autism: Evidence from an FMRI study of an executive function task and corpus callosum morphometry. Cereb Cortex. 2007;17:951–961. doi: 10.1093/cercor/bhl006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya A, Kubo K, Tomoda T, Takaki M, Youn R, Ozeki Y, Sawamura N, Park U, Kudo C, Okawa M, Ross CA, Hatten ME, Nakajima K, Sawa A. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat Cell Biol. 2005;7:1167–1178. doi: 10.1038/ncb1328. [DOI] [PubMed] [Google Scholar]
- Kato Z, Yamagishi A, Kondo N. Interstitial deletion of 1q42.13-q43 with Duane retraction syndrome. J AAPOS. 2007;11:62–64. doi: 10.1016/j.jaapos.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Keary CJ, Minshew NJ, Bansal R, Goradia D, Fedorov S, Keshavan MS, Hardan AY. Corpus callosum volume and neurocognition in autism. J Autism Dev Disord. 2009;39:834–841. doi: 10.1007/s10803-009-0689-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpinen H, Ylisaukko-Oja T, Hennah W, Palo OM, Varilo T, Vanhala R, Nieminen-von Wendt T, von Wendt L, Paunio T, Peltonen L. Association of DISC1 with autism and Asperger syndrome. Mol Psychiatry. 2008;13:187–196. doi: 10.1038/sj.mp.4002031. [DOI] [PubMed] [Google Scholar]
- Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: Implications for complex disease and association studies. Am J Hum Genet. 2007;80:727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicki M, Park H, Westin CF, Nestor PG, Mulkern RV, Maier SE, Niznikiewicz M, Connor EE, Levitt JJ, Frumin M, Kikinis R, Jolesz FA, McCarley RW, Shenton ME. DTI and MTR abnormalities in schizophrenia: Analysis of white matter integrity. Neuroimage. 2005;26:1109–1118. doi: 10.1016/j.neuroimage.2005.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicki M, Styner M, Bouix S, Gerig G, Markant D, Smith K, Kikinis R, McCarley RW, Shenton ME. Reduced interhemispheric connectivity in schizophrenia-tractography based segmentation of the corpus callosum. Schizophr Res. 2008;106:125–131. doi: 10.1016/j.schres.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- McCarthy SE, Makarov V, Kirov G, Addington AM, McClellan J, Yoon S, Perkins DO, Dickel DE, Kusenda M, Krastoshevsky O, Krause V, Kumar RA, Grozeva D, Malhotra D, Walsh T, Zackai EH, Kaplan P, Ganesh J, Krantz ID, Spinner NB, Roccanova P, Bhandari A, Pavon K, Lakshmi B, Leotta A, Kendall J, Lee YH, Vacic V, Gary S, Iakoucheva LM, Crow TJ, Christian SL, Lieberman JA, Stroup TS, Lehtimäki T, Puura K, Haldeman-Englert C, Pearl J, Goodell M, Willour VL, Derosse P, Steele J, Kassem L, Wolff J, Chitkara N, McMahon FJ, Malhotra AK, Potash JB, Schulze TG, Nöthen MM, Cichon S, Rietschel M, Leibenluft E, Kustanovich V, Lajonchere CM, Sutcliffe JS, Skuse D, Gill M, Gallagher L, Mendell NR, Wellcome Trust Case Control Consortium. Craddock N, Owen MJ, O’Donovan MC, Shaikh TH, Susser E, Delisi LE, Sullivan PF, Deutsch CK, Rapoport J, Levy DL, King MC, Sebat J. Micro-duplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meixner A, Haverkamp S, Wässle H, Führer S, Thalhammer J, Kropf N, Bittner RE, Lassmann H, Wiche G, Propst F. MAP1B is required for axon guidance and is involved in the development of the central and peripheral nervous system. J Cell Biol. 2000;151:1169–1178. doi: 10.1083/jcb.151.6.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, Devon RS, Clair DM, Muir WJ, Blackwood DH, Porteous DJ. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000;9:1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- Morris JA, Kandpal G, Ma L, Austin CP. DISC1 (Disrupted-In-Schizophrenia 1) is a centrosome-associated protein that interacts with MAP1A, MIPT3, ATF4/5 and NUDEL: Regulation and loss of interaction with mutation. Hum Mol Genet. 2003;12:1591–1608. doi: 10.1093/hmg/ddg162. [DOI] [PubMed] [Google Scholar]
- Motomura N, Satani S, Inaba M. Monozygotic twin cases of the agenesis of the corpus callosum with schizophrenic disorder. Psychiatry Clin Neurosci. 2002;56:199–202. doi: 10.1046/j.1440-1819.2002.00944.x. [DOI] [PubMed] [Google Scholar]
- Nakata Y, Barkovich A, Wahl M, Strominger Z, Jeremy RJ, Wakahiro M, Mukherjee P, Sherr EH. Diffusion abnormalities and reduced volume of the ventral cingulum bundle in agenesis of the corpus callosum: A 3 tesla imaging study. AJNR Am J Neuroradiol. 2009a;30:1142–1148. doi: 10.3174/ajnr.A1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata K, Lipska BK, Hyde TM, Ye T, Newburn EN, Morita Y, Vakkalanka R, Barenboim M, Sei Y, Weinberger DR, Kleinman JE. DISC1 splice variants are upregulated in schizophrenia and associated with risk polymorphisms. Proc Natl Acad Sci USA. 2009b;106:15873–15878. doi: 10.1073/pnas.0903413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestor PG, Kubicki M, Niznikiewicz M, Gurrera RJ, McCarley RW, Shenton ME. Neuropsychological disturbance in schizophrenia: A diffusion tensor imaging study. Neuropsychology. 2008;22:246–254. doi: 10.1037/0894-4105.22.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozeki Y, Tomoda T, Kleiderlein J, Kamiya A, Bord L, Fujii K, Okawa M, Yamada N, Hatten ME, Snyder SH, Ross CA, Sawa A. Disrupted-in-Schizophrenia-1 (DISC-1): Mutant truncation prevents binding to NudE-like (NUDEL) and inhibits neurite outgrowth. Proc Natl Acad Sci USA. 2003;100:289–294. doi: 10.1073/pnas.0136913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul LK, Brown WS, Adolphs R, Tyszka JM, Richards LJ, Mukherjee P, Sherr EH. Agenesis of the corpus callosum: Genetic, developmental and functional aspects of connectivity. Nat Rev Neurosci. 2007;8:287–299. doi: 10.1038/nrn2107. [DOI] [PubMed] [Google Scholar]
- Puthuran MJ, Rowland-Hill CA, Simpson J, Pairaudeau PW, Mabbott JL, Morris SM, Crow YJ. Chromosome 1q42 deletion and agenesis of the corpus callosum. Am J Med Genet Part A. 2005;138A:68–69. doi: 10.1002/ajmg.a.30888. [DOI] [PubMed] [Google Scholar]
- Qu M, Tang F, Yue W, Ruan Y, Lu T, Liu Z, Zhang H, Han Y, Zhang D, Wang F, Zhang D. Positive association of the Disrupted-in-Schizophrenia-1 gene (DISC1) with schizophrenia in the Chinese Han population. Am J Med Genet Part B. 2007;144B:266–270. doi: 10.1002/ajmg.b.30322. [DOI] [PubMed] [Google Scholar]
- Rice GM, Qi Z, Selzer R, Richmond T, Thompson K, Pauli RM, Yu J. Microdissection-based high-resolution genomic array analysis of two patients with cytogenetically identical interstitial deletions of chromosome 1q but distinct clinical phenotypes. Am J Med Genet Part A. 2006;140A:1637–1643. doi: 10.1002/ajmg.a.31349. [DOI] [PubMed] [Google Scholar]
- Romeo S, Pennacchio LA, Fu Y, Boerwinkle E, Tybjaerg-Hansen A, Hobbs HH, Cohen JC. Population-based resequencing of ANGPTL4 uncovers variations that reduce triglycerides and increase HDL. Nat Genet. 2007;39:513–516. doi: 10.1038/ng1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld JA, Coppinger J, Bejjani BA, Girirajan S, Eichler EE, Shaffer LG, Ballif BC. Speech delays and behavioral problems are the predominant features in individuals with developmental delays and 16p11.2 microdeletions and microduplications. J Neurodevelop Disord. 2010;2:26–38. doi: 10.1007/s11689-009-9037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurov IL, Handford EJ, Brandon NJ, Whiting PJ. Expression of disrupted in schizophrenia 1 (DISC1) protein in the adult and developing mouse brain indicates its role in neurodevelopment. Mol Psychiatry. 2004;9:1100–1110. doi: 10.1038/sj.mp.4001574. [DOI] [PubMed] [Google Scholar]
- Scott TF, Price TR, George MS, Brillman J, Rothfus W. Midline cerebral malformations and schizophrenia. J Neuropsychiatry Clin Neurosci. 1993;5:287–293. doi: 10.1176/jnp.5.3.287. [DOI] [PubMed] [Google Scholar]
- Shen S, Lang B, Nakamoto C, Zhang F, Pu J, Kuan SL, Chatzi C, He S, Mackie I, Brandon NJ, Marquis KL, Day M, Hurko O, McCaig CD, Riedel G, St Clair D. Schizophrenia-related neural and behavioral phenotypes in transgenic mice expressing truncated DISC1. J Neurosci. 2008;28:10893–10904. doi: 10.1523/JNEUROSCI.3299-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DS, Niethammer M, Ayala R, Zhou Y, Gambello MJ, Wynshaw-Boris A, Tsai LH. Regulation of cytoplasmic dynein behaviour and microtubule organization by mammalian Lis1. Nat Cell Biol. 2000;2:767–775. doi: 10.1038/35041000. [DOI] [PubMed] [Google Scholar]
- Song W, Li W, Feng J, Heston LL, Scaringe WA, Sommer SS. Identification of high risk DISC1 structural variants with a 2% attributable risk for schizophrenia. Biochem Biophys Res Commun. 2008;367:700–706. doi: 10.1016/j.bbrc.2007.12.117. [DOI] [PubMed] [Google Scholar]
- Swayze VW, II, Andreasen NC, Ehrhardt JC, Yuh WT, Alliger RJ, Cohen GA. Developmental abnormalities of the corpus callosum in schizophrenia. Arch Neurol. 1990;47:805–808. doi: 10.1001/archneur.1990.00530070103018. [DOI] [PubMed] [Google Scholar]
- Taylor M, David AS. Agenesis of the corpus callosum: A United Kingdom series of 56 cases. J Neurol Neurosurg Psychiatry. 1998;64:131–134. doi: 10.1136/jnnp.64.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischfield MA, Baris HN, Wu C, Rudolph G, Van Maldergem L, He W, Chan WM, Andrews C, Demer JL, Robertson RL, Mackey DA, Ruddle JB, Bird TD, Gottlob I, Pieh C, Traboulsi EI, Pomeroy SL, Hunter DG, Soul JS, Newlin A, Sabol LJ, Doherty EJ, de Uzcátegui CE, de Uzcátegui N, Collins ML, Sener EC, Wabbels B, Hellebrand H, Meitinger T, de Berardinis T, Magli A, Schiavi C, Pastore-Trossello M, Koc F, Wong AM, Levin AV, Geraghty MT, Descartes M, Flaherty M, Jamieson RV, Møller HU, Meuthen I, Callen DF, Kerwin J, Lindsay S, Meindl A, Gupta ML, Jr, Pellman D, Engle EC. Human TUBB3 mutations perturb microtubule dynamics, kinesin interactions, and axon guidance. Cell. 2010;140:74–87. doi: 10.1016/j.cell.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega IE, Hsu SC. The exocyst complex associates with microtubules to mediate vesicle targeting and neurite outgrowth. J Neurosci. 2001;21:3839–3848. doi: 10.1523/JNEUROSCI.21-11-03839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LW, Huang CC, Yeh TF. Major brain lesions detected on sonographic screening of apparently normal term neonates. Neuroradiology. 2004;46:368–373. doi: 10.1007/s00234-003-1160-4. [DOI] [PubMed] [Google Scholar]
- Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, Platt OS, Ruderfer DM, Walsh CA, Altshuler D, Chakravarti A, Tanzi RE, Stefansson K, Santangelo SL, Gusella JF, Sklar P, Wu BL, Daly MJ. Autism Consortium. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- Williams JM, Beck TF, Pearson DM, Proud MB, Cheung SW, Scott DA. A 1q42 deletion involving DISC1, DISC2, and TSNAX in an autism spectrum disorder. Am J Med Genet Part A. 2009;149A:1758–1762. doi: 10.1002/ajmg.a.32941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Sarginson J, Crombie C, Walker N, St Clair D, Shaw D. Genetic association between schizophrenia and the DISC1 gene in the Scottish population. Am J Med Genet Part B. 2006;141B:155–159. doi: 10.1002/ajmg.b.30274. [DOI] [PubMed] [Google Scholar]