Abstract
Wolbachia pipientis is an intracellular symbiont of arthropods well known for the reproductive manipulations induced in the host and, more recently, for the ability of Wolbachia to block virus replication in insect vectors. Since Wolbachia cannot yet be genetically manipulated, and due to the constraints imposed when working with an intracellular symbiont, little is known about mechanisms used by Wolbachia for host interaction. Here we employed a bioinformatics pipeline and identified 163 candidate effectors, potentially secreted by Wolbachia into the host cell. A total of 84 of these candidates were then subjected to a screen of growth defects induced in yeast upon heterologous expression which identified 14 top candidates likely secreted by Wolbachia. These predicted secreted effectors may function in concert as we find that their native expression is correlated and is highly upregulated at specific time points during Drosophila development. In addition, the evolutionary histories of some of these predicted effectors are also correlated, suggesting they may function together, or in the same pathway, during host infection. Similarly, most of these predicted effectors are limited to one or two Wolbachia strains—perhaps reflecting shared evolutionary history and strain specific functions in host manipulation. Identification of these Wolbachia candidate effectors is the first step in dissecting the mechanisms of symbiont–host interaction in this important system.
Keywords: symbiosis, evolution, host–microbe interaction, T4SS
Introduction
Wolbachia pipientis is likely the most prevalent intracellular infection on the planet, infecting ∼40% of insect species as well as other invertebrates (Zug and Hammerstein 2012). This recalcitrant, obligate intracellular alpha-proteobacterial symbiont has received much attention recently due to medical relevance. Wolbachia are heavily studied as potential drug targets for filarial nematode infection (Taylor et al. 2000; Hoerauf et al. 2008) and are currently being implemented to prevent transmission of Dengue fever from mosquitoes to humans (Moreira et al. 2009; Turley et al. 2009). Wolbachia may be one answer to controlling some vector borne human diseases—indeed mosquitoes harboring a virus-blocking strain of Wolbachia are presently being released in many parts of the world with this hope in mind. Yet we know little about the molecular determinants required for Wolbachia to infect its hosts.
Previous studies have provided support for Wolbachia interactions with host cytoskeletal elements, the cell cycle, and the host endocytic pathways. Specifically, in flies, Wolbachia require host microtubules and both minus and plus-end motors (dynein and kinesin) for posterior localization in the mature oocyte, positioning themselves for inclusion in the germline of the next generation (Ferree et al. 2005; Serbus and Sullivan 2007). Additionally, Wolbachia use astral microtubules during asymmetric divisions in the developing embryo, leading to the widespread, but uneven, pattern of localization of the bacteria in the adult tissues (Albertson et al. 2009). In both worms and flies, Wolbachia undergo somatic cell to germline transmission, suggesting an ability for the bacterium to alter the host actin cytoskeleton to facilitate transfer between somatic and germ cells (Landmann et al. 2012; Toomey et al. 2013). Indeed, mutations in actin binding proteins that modify F-actin content in Drosophila also alter Wolbachia titer and transmission fidelity (Newton and Sheehan 2015; Newton et al. 2015). From similar studies of host cell biology, it is known that the cellular basis of cytoplasmic incompatibility (CI) may be the result of an asynchrony between the paternal pronucleus and the female cytoplasm upon entry into mitosis (Tram et al. 2003). Wolbachia infected flies produce a larger number of eggs due to increased mitotic activity in infected germ line stem cells (Fast et al. 2011). Finally, Wolbachia rely on host clathrin/dynamin-dependent endocytosis for cell to cell transfer (White et al. 2017). However, to date, only two Wolbachia proteins have been conclusively shown to interact with specific host cell components: TomO (Ote et al. 2016) and WalE1 (Sheehan et al. 2016).
Wolbachia are obligately intracellular and challenging to study. However, obligate intracellularity has not deterred researchers from investigating many important human pathogens, such as Chlamydia (Sisko et al. 2006) and Anaplasma (Adams and Pringle 1984). All intracellular bacteria share a common necessity to control the host cell for survival. Many accomplish this with the help of secretion systems—machinery that inject bacterial proteins into the host cytoplasm (Backert and Meyer 2006). These secreted proteins are termed “effectors” because they effect the host. Although effectors are encoded in the bacterial genome, they act in the eukaryotic context and often contain eukaryotic domains and homologies (de Felipe et al. 2005; Backert and Meyer 2006). From the genomic sequencing of various Wolbachia strains, we know that these bacterial symbionts encode both a Sec (type II) and a Vir (type IVA) secretion system. Both secretion systems are complete (Wu et al. 2004); the type IV system is homologous to that of Agrobacterium (Gillespie et al. 2009), and it is expressed in Wolbachia during host infection (Rances et al. 2008). Wolbachia proteins in sequenced genomes encode a very large number of eukaryotic domains. For example, Wolbachia type A and B strains encode between 23 and 60 ANK domains, a protein–protein interaction domain found in eukaryotes (Siozios et al. 2013), and it is hypothesized that these ANK domain proteins may be secreted by Wolbachia into the host cell (Ishmael et al. 2009). The candidate secreted effectors identified thus far have been implicated in interaction with the host cytoskeleton (Sheehan et al. 2016) and host oogenesis (Ote et al. 2016). The Wolbachia protein TomO interacts directly with nos mRNA in the Drosophila ovary, suggesting this may be the mechanism by which TomO overexpression might rescue Sxl (Ote et al. 2016). The Wolbachia protein (WalE1), directly binds to and bundles actin in vitro, suggesting it functions in the cytoplasm to interact with this host component directly. Finally, a chimeric coupling protein (VirD4), comprised of both Wolbachia and E. coli residues, is able to facilitate the translocation of Wolbachia proteins (including WalE1) via the E. coli conjugation machine (Whitaker et al. 2016). Therefore, we are beginning to identify effectors that may be used by Wolbachia to manipulate the host cell. Further identification and characterization of Wolbachia effectors will allow us to better understand the basic biology of infection, and perhaps also to learn how Wolbachia induces host phenotypes, such as pathogen blocking or reproductive manipulations.
Here, we implement a primary screen in yeast to identify and characterize candidate Wolbachia effectors. A total of 163 initial candidate Wolbachia effectors were selected from strain wMel based on properties indicative of effectors in other model systems, and 84 of these were successfully cloned into a yeast expression vector. We then induced expression, looking for yeast growth defects compared with controls (Lesser and Miller 2001; Kramer et al. 2007; Siggers and Lesser 2008; Slagowski et al. 2008). This technique, used for over 15 years (Lesser and Miller 2001), has been widely implemented to identify and study pathogenic determinants from Legionella (Heidtman et al. 2009), Chlamydia (Sisko et al. 2006), Pseudomonas (Arnoldo et al. 2008), Francisella (Slagowski et al. 2008), and Shigella (Kramer et al. 2007). The growth defect phenotype is more often caused by bacterial effectors and not by core proteins and easily screened in a high-throughput format (Campodonico et al. 2005; Slagowski et al. 2008). About 14 of the 84 proteins causing the most severe growth defects were predicted to be high confidence wMel effectors. Ten of these 14 genes fall into a relatively tight cluster of 105 genes with correlated expression across Drosophila life cycle stages. Similarly, a disproportionate number of the 163 candidate effectors are also present in this cluster, with 29.4% of them in a cluster representing 8.8% of wMel genes. The evolutionary histories of these candidates, many of which encode ankyrin repeat domains, suggest that Wolbachia encode multiple, diverse candidate effectors that are generally restricted to subsets of Wolbachia strains.
Materials and Methods
Bioinformatic Selection of Candidate Effectors
Wolbachia open reading frames from the wMel genome were subjected to a BLAST search against NCBI’s nr database (accessed April 2012) using TBLASTN v2.2.25+ with default options. In addition, we also performed a search of the Pfam-A database (v26.0) using hmmscan v3.0 with default options (http://hmmer.org), identifying Wolbachia proteins with homologies to domains enriched for eukaryote membership. In addition to proteins with eukaryotic homologies, we also included Wolbachia proteins specific to the genus. We then culled the proteins that were predicted to be < 200 amino acids in order to enrich the data set for true open reading frames. This list of 163 candidate genes was then targeted for expression in yeast (see below).
Amplification, Cloning, and Transformation of wMel Genes
Genes from the wMel genome were amplified using modified forward primers to facilitate cloning using the Gateway pENTR-D/TOPO system (as described in the user manual, Invitrogen) and transformed into One Shot Top10 competent cells (Invitrogen) using standard protocols. Transformations were plated on selective plates and entry vector constructs generated by this reaction were sequence verified to confirm that protein products generated were in frame and correctly cloned. Correct entry vectors were used in combination with the pFus yeast destination vector (Huang et al. 2008) in an LR clonase reaction (as described in the user manual, Invitrogen) and these resultant expression vectors were verified by restriction enzyme digests and sequencing. Constructs and strains are available to the community upon request.
Yeast Molecular Biology, Quantitative Growth Assays and Microscopy
Yeast strain S288C (BY4741 MATα) was transformed with sequence-verified expression vectors generated above using the PEG/Lithium acetate method (Gietz and Woods 2002). All manipulations of the yeast colonies and cultures were done via a robotic system (QPExpression, Molecular Devices). Yeast transformants were inoculated into selective synthetic media with 2% (w/v) glucose. These cultures were grown overnight to saturation (at 30 °C) before transfer into media containing 2% raffinose. After cultures reached an OD600 of 0.3–0.4, they were pinned into selective synthetic media containing 2% galactose (to induce expression) or 2% glucose (to repress). These growth assays were performed in triplicate. Optical densities of yeast growing in both conditions were measured using an Epoch plate reader (BioTek instruments, VT) at 24, 36, and 48 h growth at 30 °C.
Some effectors might target conserved cellular processes that are not essential for growth under normal laboratory conditions and thus will not be picked up in the growth assays described above (Sisko et al. 2006). We therefore added four stressors during the expression of the candidate effectors: nocodazole (3 μg/ml), a drug that affects the host microtubule network; sorbitol (0.5 M) and high salt (0.5 M), to increase osmotic stress; and caffeine (6 mM) to alter MAP kinases and calcium channels. Each of these stressors at these concentrations is known to induce sensitivity in yeast expressing candidate effectors (Siggers and Lesser 2008).
To convert absorbance readings to z-score, for each experiment, stressor (or none) and well combination, absorbance readings were averaged across all replicate plates to give abscw, where (c) corresponds to the experiment and stressor combination and (w) corresponds to the well. The mean and standard deviation of all wells in the corresponding replicate plates were calculated to give meanc and sdc. For each condition and well combination for a particular gene z-scorecw = (abscw−meanc)/sdc.
To visualize the localization of the Wolbachia proteins when expressed in Saccharomyces, yeast harboring the expression vectors containing Wolbachia GFP proteins were grown overnight in selective synthetic media containing 2% raffinose. Optical density measurements were taken and the yeast were diluted to an OD600 of 0.1 in synthetic media containing 2% galactose to induce expression. Localization of Wolbachia proteins was monitored in live yeast at 2- to 4-h postinduction and visualized by observation of fixed yeast on a DeltaVision fluorescent microscope (Applied Precision) with 100× oil objective and processed using Softworx (Applied Precision). Yeast were fixed in 4% paraformaldehyde for 20 min at room temperature after a 2-h induction and imaged using a 100× objective.
RNAseq Expression Clustering
For expression correlations between genes, the raw RNAseq counts were divided by (gene length + 99), where 99 corresponds to read length (100)−1. Within a growth stage these values were multiplied by 1e6/(sum of values in stage) (Li and Dewey 2011). A pairwise distance between all genes was defined as (1−R), where the R is the Pearson correlation coefficient between the normalized expression values of two genes. Possible negative correlations would be “penalized” here. Distances were clustered using the Kitsch program of PHYLIP (Felsenstein 1989). This distance tree is shown in figure 3 for the Wolbachia candidate effectors used in the growth experiment and in supplementary figure 2, Supplementary Material online, for all genes. For t-tests on two subsets of genes within a life cycle stage, the above normalized expression values were converted to proportions over all stages within a given gene and the mean proportions compared.
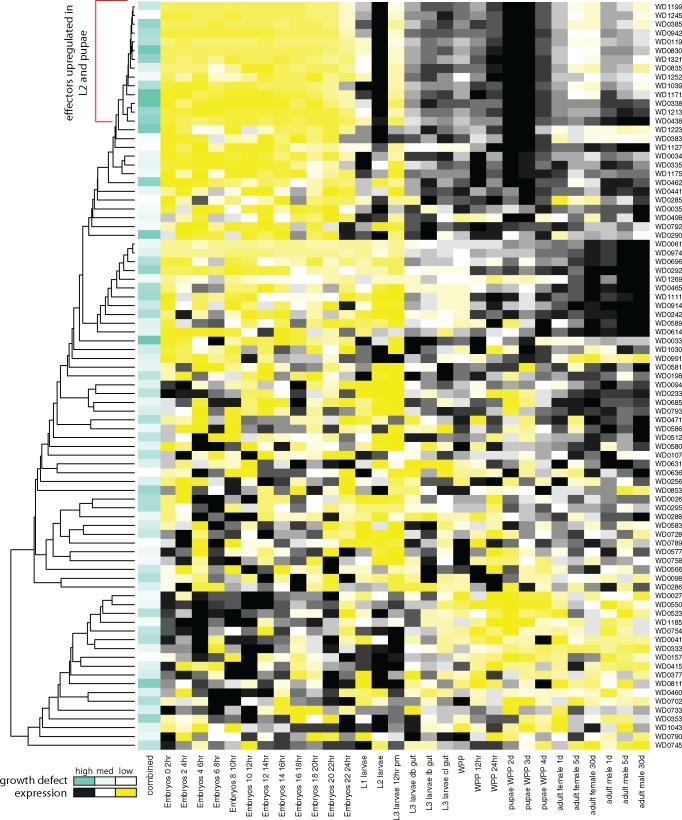
Fig. 3.
—Growth defects induced by expression of Wolbachia proteins are correlated with Wolbachia gene expression during Drosophila development. For each candidate effector, listed to the far right, Wolbachia gene expression across 27 life cycle stages from embryos to adult in Drosophila melanogaster (Gutzwiller et al. 2015) illustrated as a heat map (high (black) to low (yellow) relative expression across stages). Clustering of candidate effectors based on expression correlations between genes depicted by the tree. Extent of growth inhibition in yeast (median z-score over all conditions) shown as extent of teal coloring in first column. Red bracket indicates tight cluster in dendrogram with significant enrichment for growth defect inducing loci.
Wolbachia Ortholog Definitions
Wolbachia orthologs were defined based on reciprocal best BLASTp hits of amino acid sequences between Wolbachia strains. An orthologous group of genes was defined by complete linkage such that all members of the group had to be the reciprocal best hit of all other members of the group, thus a particular strain could have at most one gene per ortholog group.
Global Phylogenetic Tree
Ortholog alignments for all strains were concatenated into a single alignment and columns with less than four taxa represented were removed. A RAxML (Stamatakis 2014) maximum likelihood tree was calculated with the rapid bootstrap analysis and search algorithm, the GTRGAMMA model, and 100 bootstrap replicates. This tree is represented in figure 4.
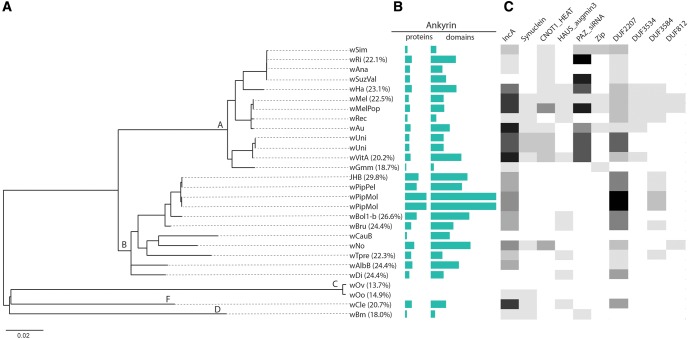
Fig. 4.
—Phylogenetic correlation of domains found in predicted Wolbachia type IV secretion substrates. (A) Phylogeny generated from concatenated gene set of 25 Wolbachia genomes (see Materials and Methods). We recovered 100% support for five Wolbachia supergroups (A, B, C, D, and F). (B) The prevalence of ankyrin repeat domain containing proteins and total domains in each proteome shown as a turquoise bar graph. (C) The abundance of domains found in predicted effectors shown as a heat map where darker shading is equivalent to a higher number of domains in the proteome.
Coevolution Analysis
As a measure of coevolution between orthologs, we used the p-mirrortree approach (Ochoa et al. 2015). Mirrortree uses the correlation of pairwise distances between two trees and p-mirrortree normalizes the significance using genome-wide background distributions of correlations to produce a P value. Pairwise maximum likelihood distances were calculated for each ortholog alignment with RAxML. Protein alignments were generated with MAFFT v7.058 b (Katoh and Standley 2013), converted to the corresponding nucleotide alignments, then hand edited. A P value of 0.05 or less was used as the significance cutoff.
Wolbachia Domain Analysis
Pfam-A version 28.0 was scanned using hmmscan v3.1b2 (hmmer.org) with each Wolbachia gene. The default per domain thresholds of 0.01 for the sequence and conditional domain e-values were used. Overlapping domains for a given gene were filtered by sorting the domains by their sequence e-value and removing domains where >75% of its length overlapped with domains having higher sequence e-values. Domain counts were tallied at the gene, strain, and supergroup level.
Results
Bioinformatic Selection of Wolbachia Candidate Effectors for Experimental Screen
Based on the observation that bacterial effectors are often genus specific and/or encode similarities to eukaryotic proteins and domains (Beare et al. 2009; Burstein et al. 2009; Voth et al. 2009), we searched both NCBI’s nr database and Pfam-A with wMel proteins to evaluate their similarity to other proteins. Selected loci had one or more of the following characteristics: 1) they were unique to Wolbachia, 2) had a top eukaryotic hit with a better score than non Rickettsiales bacteria, or 3) matched a domain normally specific to eukaryotes. This resulted in a list of 163 candidate secreted substrates for wMel (supplementary table 1, Supplementary Material online). The most commonly occurring domain was the ankyrin repeat domain (24/163 candidates) while 65 contained no Pfam-A domain at the default HMMER threshold. Of note, we identified proteins containing homology to eukaryotic domains involved in the interaction with the cytoskeleton (such as the WH2 domain in WD0332 and WD811 [just under HMMER threshold]), and the synuclein domain in WalE1, domains known to associate with actin (Paunola et al. 2002; Esposito et al. 2007; Sheehan et al. 2016). We also identified several proteins with homology to domains involved in the fusion or scission of membranes in eukaryotic cells; three candidate effectors contained dynamin domain homologies (WD0246, WD0743, WD1098, although importantly they have better overlapping matches to domain families FeoB_N, KH_2, and MMR_HSR1), whereas six candidates have homology to the Chlamydia trachomatis IncA domain (WD0073, WD0224, WD0290, WD0353, WD0630, WD0754), thought to associate with the inclusion membrane and control membrane fusion. Finally, candidates for potential involvement in their interaction with small, regulatory RNAs were identified; WD1240 contains a double-stranded RNA binding motif, whereas WD0033, WD0034, and WD0290 contain PAZ-siRNA-binding domains involved in posttranscriptional gene silencing.
Expression of Wolbachia Candidate Effectors in Saccharomyces Induces Growth Defects
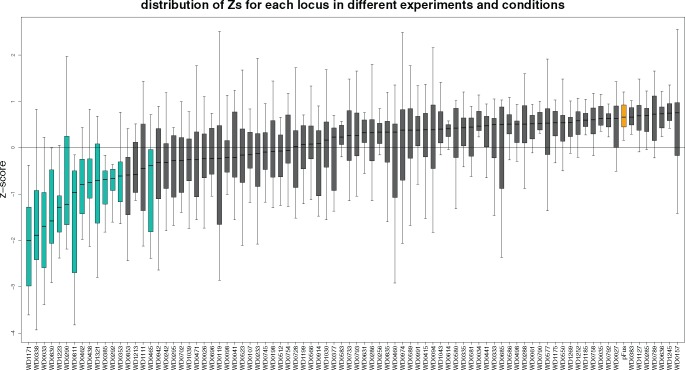
To narrow the candidates identified through bioinformatics, we completed a growth screen in yeast. We successfully cloned 84 candidate Wolbachia effectors into an inducible yeast expression vector. These yeasts were then cultured under inducing conditions and their growth over time (0–72 h) was monitored by optical density readings. Each experiment consisted of three independent technical replicates, including vector only controls and uninduced control plates, resulting in a total of 48 different microtiter plates per experiment. The experiment was repeated a total of three times using independent biological replicates (different yeast colonies). Optical density measurements were compared with vector alone (supplementary fig. 1, Supplementary Material online) or the average and variance within each plate (fig. 1). Without additional stressors, a total of five Wolbachia candidate effectors induced a significant growth defect in yeast (WD1171, WD0338, WD0033, WD1223, WD0290, WD0811; table 1).
Fig. 1.
—Growth defects induced by expression of Wolbachia proteins in Saccharomyces cerevisiae. Yeast cells carrying plasmids that conditionally expressed an N-terminal GFP fusion to 84 different Wolbachia proteins or GFP alone (pFus) were expressed for 72 h under inducing conditions (4% galactose), with or without stressors (caffeine, nocodazole, sodium chloride, and sorbitol). The data for each boxplot are the z-scores for each condition and well combination for a particular gene where z-scorecw = (abscw−meanc)/sdc (see Materials and Methods). Predicted effectors shown in green. Vector alone (pFus) shown in yellow.
Table 1.
Fourteen Predicted Effectors
| Locus | None | noco | caff | salt | sorb | Pfam Domains |
|---|---|---|---|---|---|---|
| WD1171 | −1.32 | −1.54 | −2.11 | −2.64 | −3.00 | DUF3534 |
| WD0338 | −2.13 | −0.71 | −2.05 | −1.67 | −2.31 | – |
| WD0033 | −1.01 | −1.78 | −1.63 | −1.72 | −1.98 | PAZ |
| WalE1 | −0.96 | −1.45 | −1.49 | −1.47 | −1.56 | Synuclein |
| WD1223 | −1.15 | −1.24 | −1.92 | −0.99 | −1.67 | – |
| WD0290 | −0.76 | −0.23 | −1.11 | −0.62 | −1.23 | DUF2207 IncA PAZ |
| WD0811 | −3.14 | −1.07 | −2.46 | −1.66 | −2.67 | – |
| WD0462 | −0.99 | −0.87 | −0.68 | −1.34 | −0.31 | IncA HAUS- augmin3 |
| WD0438 | 0.28 | −0.93 | −1.12 | −0.97 | −1.07 | Ankyrin |
| WD1321 | −0.62 | −0.62 | −1.36 | −0.61 | −0.78 | – |
| WD0385 | −0.26 | −0.83 | −0.47 | −1.60 | −1.13 | Ankyrin |
| WD0292 | 0.56 | −0.49 | −1.08 | −0.77 | −0.76 | Ankyrin |
| WD0353 | −0.87 | −0.54 | −1.11 | −0.57 | −0.08 | IncA Zip |
| WD0465 | 0.11 | −1.25 | −1.31 | −0.62 | −0.56 | DUF812 |
Note.—Column one shows Wobachia pipientis wMel loci that resulted in significant growth defects (z-score less than one standard deviation below the mean under one or more stress conditions) when expressed in yeast. Columns 2–6 are mean z-scores (see Materials and Methods) over three experiments under conditions without (none) or with added stressors nocodazole (noco), caffeine (caff), sodium chloride (salt), and sorbitol (sorb).
In order to identify additional candidate effectors, and potential pathways that these effectors might influence in the eukaryotic cell, we took advantage of a set of stressors, which, when added to growing yeast, perturb cellular physiology. Here we added nocodazole, caffeine, sodium chloride, and sorbitol. An additional nine candidate effectors were identified in this way, as they caused significant growth defects with stressors, but not without (WalE1, WD0290, WD0462, WD0438, WD1321, WD0385, WD0292, WD0353, WD0465; table 1). Seven of these nine were recovered with the use of nocodazole, a microtubule inhibitor, and/or caffeine, which perturbs MAP kinase signaling and calcium channels, whereas two were recovered under sorbitol and/or NaCl (table 1). Interestingly, three of these candidates encode ankyrin repeat domains and have been implicated previously in host interaction based on differential expression in the reproductive tract of male and female insects (WD0292, WD0385, WD0438; Iturbe-Ormaetxe et al. 2005; Yamada et al. 2011; Papafotiou et al. 2011). Two of these (WD0033 and WD0290) contain PAZ-siRNA binding domains. Another candidate, WD0811, has been identified as a secreted substrate using a heterologous secretion assay (Whitaker et al. 2016). We also recovered the known effector WalE1 as part of this large-scale screen (table 1). Importantly, our z-score metric (see Materials and Methods) is likely conservative; because we chose candidates for cloning and expression that have properties common to effectors, the average growth per plate was likely below that of noneffectors. Below, we refer to these 14 Wolbachia proteins, that caused the most severe growth defects in yeast, as predicted effectors, to distinguish them from the initial candidate list.
Localization of Predicted Effectors in Yeast
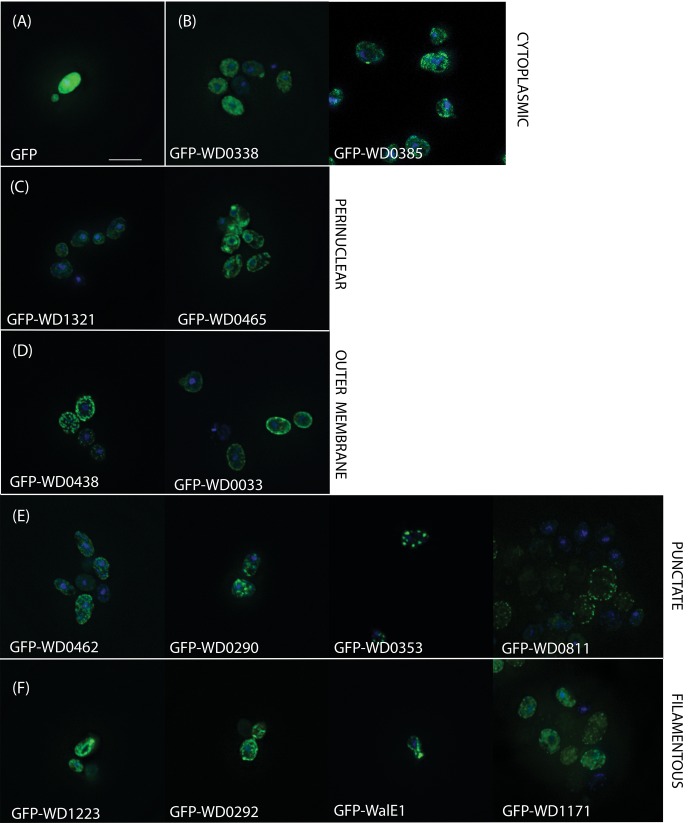
When bacterial effectors are expressed in Saccharomyces, their localization within the yeast cell often mirrors localization in the natural host context; this localization reflects a conservation of eukaryotic cellular processes and structures (Sisko et al. 2006). Indeed, WalE1, an actin bundler, localizes to the yeast actin cytoskeleton (Sheehan et al. 2016). We therefore used an N-terminal GFP tag to track the localization of our predicted Wolbachia effectors in the yeast cell. Using this strategy, we identified five distinct patterns of localization: cytoplasmic, perinuclear, outer membrane, punctate, and filamentous (fig. 2). Although the most common localizations were punctate (four candidates) and filamentous (four candidates), the localization of the GFP tagged Wolbachia predicted effector did not correlate with either domain structure in the protein, degree of growth defect, nor growth inhibition under stressors.
Fig. 2.
—The localization of candidate Wolbachia effectors after yeast overexpression. Wolbachia genes causing significant growth defects (see table 1) were cloned into a GFP-fusion yeast expression vector and, after 3 h of induced expression, yeast were fixed and stained with DAPI to visualize nuclei. Compared with (A) GFP alone, predicted effectors exhibited (B) cytoplasmic (WD0338, WD0385), (C) perinuclear (WD1321, WD0465), (D) outer membrane (WD0438, WD0033), (E) puncate (WD0462, WD0290, WD0353, WD0811), or (F) filamentous (WD1223, WD0292, WalE1, WD1171) localization.
Predicted Effectors Are More Likely to Be Expressed by Wolbachia When Infected Flies Are Larvae (L2) or Pupae
Previously, we showed that a majority of genes encoding the central structure of the type IV secretion system is constitutively expressed in Wolbachia (Gutzwiller et al. 2015). It is possible, however, that secreted effectors are differentially expressed during host development, as is the case with WalE1 (Sheehan et al. 2016). We wanted to know if candidate effectors might be expressed at the same time during host development, suggesting that they might interact with each other or that their function is specific to particular developmental stages. We therefore utilized an existing RNAseq data set for Wolbachia that contained 30 conditions consisting of 24 life cycle stages from embryos to pupae and three-time samplings each for adult male and female in Drosophila melanogaster (Gutzwiller et al. 2015). Clustering of genes based on expression correlations between genes (see Materials and Methods) shows an apparent correspondence between yeast growth inhibiting genes and expression (fig. 3). For example, the top clade in figure 3 contains a disproportionate number of loci with low z-scores (P = 0.006 for post hoc analysis of mean z-scores between the top clade in fig. 3 and the rest of the dendrogram). Interestingly, the predicted effectors that induced the most severe growth defects (WD0385, WD0338, WD0438, WalE1, WD0942, WD1171, WD1223, and WD1321) were more likely to be natively upregulated during both the L2 larval stage and pupation (fig. 3).
Because we identified a trend between growth inhibition in yeast and correlated expression of candidate effectors during host development, we examined all expressed loci in wMel, identifying other genes that follow similar expression patterns. Clustering of these loci based on pairwise expression correlations reveals a large clade containing 105 wMel genes which corresponds to the most tightly clustered clade of its size in the proteome (supplementary fig. 2, Supplementary Material online). A total of 64 of these 105 genes are annotated as “hypothetical proteins” while eight are ankyrin domain containing proteins and another six are predicted to be involved in the secretion of proteins (supplementary fig. 2, Supplementary Material online). In this last category are genes encoding proteins predicted to be involved in the type IV secretion apparatus, either in forming the scaffold for translocation of substrates (virB3 and virB6) or providing the power required for translocation and pilus biogenesis (virB4). In addition to type IV secretion proteins, we identified components of the type I secretion systems as upregulated during host pupation, including genes encoding a type I secretion system permease/ATPase (WD0770) and an HlyD family periplasmic adapter (WD0649). Interestingly, the wsp paralog WD0009 was also found in this cluster of genes. As wsp is known to elicit an immune response in mammalian hosts suffering from filarial disease (Brattig et al. 2004; Turner et al. 2009), and is likely secreted to the bacterial cell surface by Wolbachia.
Because several Wolbachia candidate effectors showed a characteristic expression profile, we were curious to know if any Drosophila loci were similarly expressed. We therefore searched the modENCODE expression data set in Flybase, identifying genes that are of below moderate expression (11–25 RPKM) in L1 and L3 larvae but above moderately high expression (26–50 RPKM) in 2- to 4-day post-WPP. A list of 15 host loci was returned, including Akap200, GstE13, jar, ken, ogre, RanBP3, Sh3β, Ubc6, and Uch-L5. Most interestingly, these host loci are involved in processes known to be important in Wolbachia biology: cytoskeleton binding (jaguar, a myosin), genital formation (the ken and barbie transcription factor), redox signaling (GstE13 and RanBP3), and ubiquitination (Ubc6, Uch-L5).
Candidate Effectors Causing Significant Growth Defects Are Part of Larger Gene Families and Wolbachia Clade Specific
Many secreted substrates are species or genus specific. For example, many Legionella effectors are unique to the Legionella genus (Burstein et al. 2009). Although Wolbachia candidate effectors do contain domain homologies to effectors from other microbes (e.g., IncA domains homologous to Chlamydia inclusion proteins), most of the Wolbachia candidate effectors screened here were specific to Wolbachia (85/163) or the Rickettsiales (108/163). Having identified 14 candidates causing statistically significant growth defects in yeast, we used this reduced data set to determine the prevalence of these effectors across Wolbachia supergroups. To do this, we performed an analysis of homology between the 14 identified wMel effectors and all complete sequenced genomes. Included in this analysis were 11 type A strains (wRi, wAna, wValsugana, wHa, wMel, wMelPop, wAu, wRec, wGmm, wUni, wVitA), 10 type B strains (wPip-JHB, wPip-Pel, wPip-Mol, wBol, wBru, wCauB, wNo, wTpre, wAlbB, wDi), 2 type C strains (wOv, wOo), and one each type D (wBm) and type F (wCle). We included all genomic data available for each strain such that if multiple assemblies existed for each Wolbachia variant (such as in the case of wUni) we included all contigs.
The predicted effectors show similarity across the A and B supergroups, with inconsistent homology found in C, D, and F genomes (table 2, column 3 and fig. 4), whereas strict orthologs are more restricted (table 2, column 2). The supergroup distribution here reflects that of likely effectors present in wMel. Had we begun with a strain in, say, supergroup D, the analysis would have likely revealed other candidates and patterns of evolutionary conservation, reflecting the general strain specificity of effectors. Nine predicted effectors show similarity to other wMel genes (table 2, column 5) and most of these are to other predicted effectors or candidate effectors. As previously observed for the ankyrin repeat domain (Siozios et al. 2013), the most extreme duplications are in the ankyrin domain containing candidates (such as WD0385 with 15 matching genes within wMel and 558 throughout Wolbachia, and WD0292 with 6 and 183, respectively). In addition, three other pairs of predicted effectors show similarity (highlighted in table 2), suggesting propagation of other domains used by effectors.
Table 2.
Homology and Correlated Evolution Between wMel Predicted Effectors and Wolbachia Genes
| Predicted Effector | Ortholog Clade Distributiona | Paralog Clade Distributionb | Totalc | Paralogs in wMeld | Correlated Ortholog Treese | Pfam Domains? |
|---|---|---|---|---|---|---|
| WD0292 | A6 | A90,B88,F5 | 183 | WD0385**, C:4, wMel:1 | WD0290*, WD1171*, C:2, H:4, wMel:21 | Ankyrin |
| WD0353 | A8 | A25,B14,F2 | 41 | wMel:1 | wMel:1 | IncA Zip |
| WD1171 | A10 | A20,B17,D2,F1 | 40 | C:1 | WD0290*, WD0292*, C:1, wMel:6 | DUF3534 |
| WD0811 | A10 | A9,B3,C2,D1,F1 | 16 | – | C:2 | – |
| WD0385 | A7,B6 | A210,B322,D5,F21 | 558 | WD0292**, C:13, wMel:1 | C:1, other:42 | Ankyrin |
| WalE1 | A8,B9 | A13,B9,C2,D1,F1 | 26 | – | C:13, H:7, wMel:61, other:37 | Synuclein |
| WD0338 | A9,B8 | A31,B50,D4,F2 | 87 | C:1, wMel:1 | WD0462*, C:45, H:90, wMel:278, other:29 | – |
| WD0462 | A9,B9 | A37,B19,F1 | 57 | C:1, wMel:1 | WD0338*, C:2, wMel:5, other:27 | IncA HAUS-augmin3 |
| WD1321 | A11,B9 | A10,B9,D1 | 20 | – | C:1, wMel:2, other:30 | – |
| WD0033 | A8,F1 | A18,B17,F1 | 36 | C:1, wMel:1 | C:2, H:5, wMel:19, other:3 | PAZ |
| WD0290 | A6,B4,F1 | A16,B4,F1 | 21 | – | WD0292*, WD1171*, C:3, wMel:8, other:1 | DUF2207 IncA PAZ |
| WD0465 | A7,C2,F1 | A11,B21,C2,F1 | 35 | – | – | DUF812 |
| WD0438 | A10,B8,D1,F1 | A21,B18,D2,F3 | 44 | C:1 | C:1, wMel:6, other:1 | Ankyrin |
| WD1223 | A10,B9,C2,D1,F1 | A11,B9,C2,D1,F1 | 24 | – | C:3, wMel:16, other:12 | – |
Note.—Column 2: The number of strains per supergroup in which this ortholog group is found. Column 3: The number of genes per supergroup that are homologous to this gene. Column 6: Ortholog groups with significantly correlated intraortholog pairwise distances with those of the predicted effector’s distances, based on p-mirrortree. See Materials and Methods.
The numbers after the supergroup letter is the number of strains matching in that supergroup. See figure 4 for supergroup clusters.
The numbers after the supergroup letter is the number of genes matching in that supergroup.
Sum of paralogs in previous column.
wMel genes with BLAST e-value ≤ 1e−6, percent identity ≥ 20, and overlap ≥ 30. *: predicted effector locus, C: candidate effector count, wMel: similarity to other wMel genes.
Orthologs with correlation significance P ≤ 0.05. *: predicted effector locus in correlated tree, C: other candidate effector ortholog count, H: wMel housekeeping gene, wMel: other wMel ortholog count, other: count of non-wMel containing orthologs.
When observing the pattern in domain conservation and expansion across the Wolbachia phylogeny (fig. 4), we hypothesized that the enrichment of some domains might be correlated. Indeed, when subjected to a Pearson analysis of correlation, the abundance of different domains is correlated. Specifically, the occurrence of the PAZ_siRNA, IncA, DUF3534, ZIP, CNOT1_HEAT, and DUF812 domains is correlated (for all pairwise comparisons, Pearson correlation > 0.990 and significance < 0.03). Similarly, the prevalence of the Ank2 domain is correlated with the occurrence of the DUF2207, HAUS_augmin, and DUF3584 domains (Pearson correlation > 0.990 and significance < 0.001 for all comparisons). These correlations are certainly influenced by the sampling on the phylogeny, which is overwhelmingly represented by the A and B Wolbachia clades, and therefore the interpretation of these results should be tempered.
Candidate Wolbachia Effectors Show Evidence of Correlated Evolution
Proteins which function together in the cellular environment may exhibit signatures of correlated evolution. In order to identify if any of our predicted effectors might functionally interact, we calculated pairwise distances between genes within each ortholog group for all Wolbachia orthologs. Using these distances to test for topological correlations with a mirror tree approach (see Materials and Methods), we identified 13 predicted effectors whose evolutionary history correlated with other wMel loci (table 2, column 6). The phylogeny generated from WD0338, for example, significantly correlated with phylogenies generated for 278 other wMel genes, including 45 candidate effector proteins (table 2). About 12 of these 13 predicted effectors had correlations with phylogenies of genes in the original set of candidate effectors. For example, analysis of WD0462 identified correlated evolution with WD0338. Additionally, three predicted effectors, WD0292, WD0290, and WD1171, are correlated in their evolutionary history and interestingly, both WD0290 and WD1171 localize to filaments in yeast (fig. 2).
Discussion
Wolbachia pipientis is the most prevalent infection on Earth and increasingly promoted for its use in disease vector control (LePage and Bordenstein 2013). Due to both the direct effects that Wolbachia may have on the transmission of human pathogens (Moreira et al. 2009) and the myriad effects Wolbachia has on insect populations (Werren et al. 2008), it is important that we identify the mechanisms for symbiosis between Wolbachia and its hosts. Although the type IV secretion system has long been hypothesized to be involved in host interaction (Pichon et al. 2009), here we present the first large-scale screen for proteins likely used by Wolbachia to manipulate host cell biology. A total of 14 candidate effectors from wMel induce significant growth defects when expressed in yeast (fig. 1), suggesting that these proteins are Wolbachia secreted effectors. Further evidence in support of these proteins being true secreted substrates is strong and includes correlated expression during host development for 10/14 (fig. 3) and independent prediction of secretion based on published algorithms (such as T4Pred, supplementary fig. 2, Supplementary Material online).
One intriguing result of this work is that 10/14 predicted effectors are upregulated during host larval (L2) and pupal stages (fig. 3). Expression of a previously characterized candidate effector (WalE1) is also upregulated during host pupation, suggesting that, at this developmental stage, Wolbachia may deploy many secreted substrates (Sheehan et al. 2016). The pupal stage in the fly is coincident with the formation of the reproductive tissues (Dansereau and Lasko 2008; Eliazer and Buszczak 2011); therefore, some of these effectors may be involved in reproductive manipulations. However, it is generally posited that Wolbachia loci involved in reproductive manipulations would be differentially regulated in adult males and females or at least highly expressed in reproductive tissue. The predicted effectors analyzed here (N = 14) do not show this pattern, with average expression in males and females being roughly equal across the data set (avg TPM values for males = 1,384 and females = 1,165). We favor the hypothesis that Wolbachia must manipulate the basic biology of the host in order to colonize all tissues and to persist within the reproductive tract during this dramatic morphological transformation. We therefore hypothesize that many of these identified effectors, upregulated during pupation, will be involved in the basic biology of Wolbachia infection, although it is possible that they influence reproductive manipulations or pathogen blocking. For example, one of the recently identified CI factor loci cifA/cidA (WD0631) was included in this screen (Beckmann and Fallon 2013; Beckmann et al. 2017; LePage et al. 2017). However, it did not induce significant growth defects upon expression in yeast (fig. 1, also observed by Beckmann et al. 2017) and was not upregulated during L2 and pupal stages (fig. 4), suggesting that the loci behind reproductive manipulations in Wolbachia would not be identified by our approach.
Orthologs of these predicted effectors are largely limited to supergroups A and B within Wolbachia with variable presence in the other groups (fig. 4). Further homology is primarily limited to Wolbachia or Rickettsiales, allowing for the possibility of novel domains and functions. Four genes have orthologs only in A, and five only in A and B. This most likely points to the recent acquisition of host specific functions within the Wolbachia genus, analogous to the array of lineage specific effectors predicted in Legionella (Burstein et al. 2016). In contrast, and as has been previously observed (Siozios et al. 2013), the ankyrin repeat domain is found in all supergroups except C, is present in most strains within each group, and is found in multiple copies in many genes per strain. Three of our predicted effectors contain ankyrin repeats and many of these other ankyrin containing proteins, combined in various domain architectures, are likely involved in protein–protein interactions with their arthropod hosts. The only other domains found across all of the other clades is IncA, whereas DUF2207 is conspicuously absent from the nematode-infecting Wolbachia clades.
Interestingly, the predicted effectors are significantly correlated in their evolution. This evidence is based on the comparisons between phylogenetic trees produced by the orthologous gene sets across the Wolbachia genomes analyzed herein. Why would Wolbachia candidate effectors correlate across evolutionary time? Two possibilities are that the coevolution identified here could be the result of either direct (physical) or indirect (functional) interaction between the candidate effectors. Weak evidence in support of this is the colocalization of both WD1171 and WD0292 to filaments—these candidate effectors correlated in their evolutionary history. More evidence comes in the way of correlated gene expression, where these predicted effectors are likely targeting the same host process or working in concert in the host cytosol. Indeed, in the Brugia symbiosis, a similar RNAseq approach revealed a high incidence of Wolbachia differentially expressed genes during female worm development, with many Wolbachia loci involved in chaperone function, energy production, and translation, upregulated (Grote et al. 2017). Future biochemical analyses (such as coimmunoprecipitations and colocalization studies) will determine if these Wolbachia effectors truly interact directly with each other or with host cellular components.
Here we present the first high-throughput screen of Wolbachia candidate secreted effectors. As part of this screen, we identified 163 wMel proteins as candidate secreted substrates of the T4SS. How many of these Wolbachia proteins are likely secreted? Our data, including predictions based on expression correlations and secretion prediction algorithms, suggest that between 50 and 105 Wolbachia proteins may be secreted (4–10% of the proteome). Therefore, why did not more of these proteins cause growth defects in the yeast screen? There are several possible explanations for this result. First, as stated earlier, the Z score metric we used is conservative; pairwise comparisons to vector alone would identify a greater number of candidates (supplementary fig. 1, Supplementary Material online). That said, because we are in the initial stages of identification and characterization of these proteins, we believe it is important to be conservative. Another explanation is that Wolbachia may target Drosophila-specific processes, without homologs in yeast. Finally, it is also possible that these Wolbachia proteins may not be as stable in this heterologous system, reducing their toxicity. Regardless, despite the possibility of some false negatives, because very little is currently known about the biochemistry of Wolbachia effectors, this screen has yielded the largest amount of data on Wolbachia proteins and potential host interactions to date. Our results underscore the importance of the Ankyrin repeat domains in Wolbachia–host interaction and point to correlated gene expression, during critical developmental timepoints, as a signal for candidate secreted effectors.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Dr Carol Anderson for the generous use of her microscope. This work was funded by NSF IOS 1456545 to ILGN.
Literature Cited
- Adams AE, Pringle JR.. 1984. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J Cell Biol. 98(3):934–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson R, Casper-Lindley C, Cao J, Tram U, Sullivan W.. 2009. Symmetric and asymmetric mitotic segregation patterns influence Wolbachia distribution in host somatic tissue. J Cell Sci. 122(Pt 24):4570–4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoldo A, et al. 2008. Identification of small molecule inhibitors of Pseudomonas aeruginosa exoenzyme S using a yeast phenotypic screen. PLoS Genet. 4(2):e1000005.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backert S, Meyer TF.. 2006. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr Opin Microbiol. 9(2):207–217. [DOI] [PubMed] [Google Scholar]
- Beare PA, et al. 2009. Comparative genomics reveal extensive transposon-mediated genomic plasticity and diversity among potential effector proteins within the genus Coxiella. Infect Immun. 77(2):642–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann JF, Fallon AM.. 2013. Detection of the Wolbachia protein WPIP0282 in mosquito spermathecae: implications for cytoplasmic incompatibility. Insect Biochem Mol Biol. 43(9):867–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann JF, Ronau JA, Hochstrasser M.. 2017. A Wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat Microbiol. 2:17007.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brattig NW, et al. 2004. The major surface protein of Wolbachia endosymbionts in filarial nematodes elicits immune responses through TLR2 and TLR4. J Immunol. 173(1):437–445. [DOI] [PubMed] [Google Scholar]
- Burstein D, et al. 2016. Genomic analysis of 38 Legionella species identifies large and diverse effector repertoires. Nat Genet. 48(2):167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein D, et al. 2009. Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog. 5(7):e1000508.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campodonico EM, Chesnel L, Roy CR.. 2005. A yeast genetic system for the identification and characterization of substrate proteins transferred into host cells by the Legionella pneumophila Dot/Icm system. Mol Microbiol. 56(4):918–933. [DOI] [PubMed] [Google Scholar]
- Dansereau DA, Lasko P.. 2008. The development of germline stem cells in Drosophila. Methods Mol Biol. 450:3–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Felipe KS, et al. 2005. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J Bacteriol. 187(22):7716–7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliazer S, Buszczak M.. 2011. Finding a niche: studies from the Drosophila ovary. Stem Cell Res Ther. 2(6):45.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito A, et al. 2007. alpha-Synuclein and its disease-related mutants interact differentially with the microtubule protein tau and associate with the actin cytoskeleton. Neurobiol Dis. 26(3):521–531. [DOI] [PubMed] [Google Scholar]
- Fast EM, et al. 2011. Wolbachia enhance Drosophila stem cell proliferation and target the germline stem cell niche. Science 334(6058):990–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 1989. PHYLIP – Phylogeny inference package (Version 3.2). Cladistics 5: 164–166. [Google Scholar]
- Ferree PM, et al. 2005. Wolbachia utilizes host microtubules and dynein for anterior localization in the Drosophila oocyte. PLoS Pathog. 1(2):e14.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Woods RA.. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods in Enzymol. 350:87–96. [DOI] [PubMed] [Google Scholar]
- Gillespie JJ, et al. 2009. An anomalous type IV secretion system in Rickettsia is evolutionarily conserved. PLoS One 4(3):e4833.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote A, et al. 2017. Defining Brugia malayi and Wolbachia symbiosis by stage-specific dual RNA-seq. PLoS Negl Trop Dis. 11(3):e0005357.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutzwiller F, et al. 2015. Dynamics of Wolbachia pipientis gene expression across the Drosophila melanogaster life cycle. G3 (Bethesda) 5(12):2843–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidtman M, Chen EJ, Moy MY, Isberg RR.. 2009. Large-scale identification of Legionella pneumophila Dot/Icm substrates that modulate host cell vesicle trafficking pathways. Cell Microbiol. 11(2):230–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerauf A, et al. 2008. Wolbachia endobacteria depletion by doxycycline as antifilarial therapy has macrofilaricidal activity in onchocerciasis: a randomized placebo-controlled study. Med Microbiol Immunol. 197(3):295–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Lesser CF, Lory S.. 2008. The essential role of the CopN protein in Chlamydia pneumoniae intracellular growth. Nature 456(7218):112–U110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishmael N, et al. 2009. Extensive genomic diversity of closely related Wolbachia strains. Microbiology 155(Pt 7):2211–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturbe-Ormaetxe I, et al. 2005. Distribution, expression, and motif variability of ankyrin domain genes in Wolbachia pipientis. Bacteriol 187(15):5136–5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer RW, et al. 2007. Yeast functional genomic screens lead to identification of a role for a bacterial effector in innate immunity regulation. PLoS Pathog. 3(2):e21.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landmann F, et al. 2012. Both asymmetric mitotic segregation and cell-to-cell invasion are required for stable germline transmission of Wolbachia in filarial nematodes. Biol Open 1(6):536–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LePage D, Bordenstein SR.. 2013. Wolbachia: can we save lives with a great pandemic? Trends Parasitol. 29(8):385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LePage DP, et al. 2017. Prophage WO genes recapitulate and enhance Wolbachia-induced cytoplasmic incompatibility. Nature 543(7644):243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser CF, Miller SI.. 2001. Expression of microbial virulence proteins in Saccharomyces cerevisiae models mammalian infection. EMBO J. 20(8):1840–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN.. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira LA, et al. 2009. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, chikungunya, and plasmodium. Cell 139(7):1268–1278. [DOI] [PubMed] [Google Scholar]
- Newton IL, Sheehan KB.. 2015. Passage of Wolbachia pipientis through mutant drosophila melanogaster induces phenotypic and genomic changes. Appl Environ Microbiol. 81(3):1032–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton ILG, Savytskyy O, Sheehan KB.. 2015. Wolbachia utilize host actin for efficient maternal transmission in Drosophila melanogaster. PLoS Pathog. 11(4):e1004798.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa D, Juan D, Valencia A, Pazos F.. 2015. Detection of significant protein coevolution. Bioinformatics 31(13):2166–2173. [DOI] [PubMed] [Google Scholar]
- Ote M, Ueyama M, Yamamoto D.. 2016. Wolbachia protein TomO targets nanos mRNA and restores germ stem cells in Drosophila sex-lethal mutants. Curr Biol. 26(17):2223–2232. [DOI] [PubMed] [Google Scholar]
- Papafotiou G, et al. 2011. Regulation of Wolbachia ankyrin domain encoding genes in Drosophila gonads. Res. Microbiol. 162(8):764–772. [DOI] [PubMed] [Google Scholar]
- Paunola E, Mattila PK, Lappalainen P.. 2002. WH2 domain: a small, versatile adapter for actin monomers. FEBS Lett. 513(1):92–97. [DOI] [PubMed] [Google Scholar]
- Pichon S, et al. 2009. Conservation of the type IV secretion system throughout Wolbachia evolution. Biochem Biophys Res Commun. 385(4):557–562. [DOI] [PubMed] [Google Scholar]
- Rances E, Voronin D, Tran-Van V, Mavingui P.. 2008. Genetic and functional characterization of the type IV secretion system in Wolbachia. J Bacteriol. 190(14):5020–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbus LR, Sullivan W.. 2007. A cellular basis for Wolbachia recruitment to the host germline. PLoS Pathog. 3(12):e190.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan KB, Martin M, Lesser CF, Isberg RR, Newton IL.. 2016. Identification and characterization of a candidate Wolbachia pipientis type IV effector that interacts with the actin cytoskeleton. MBio 7(4):e00622–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggers KA, Lesser CF.. 2008. The Yeast Saccharomyces cerevisiae: a versatile model system for the identification and characterization of bacterial virulence proteins. Cell Host Microbe 4(1):8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siozios S, et al. 2013. The diversity and evolution of Wolbachia ankyrin repeat domain genes. PLoS One 8(2):e55390.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisko JL, Spaeth K, Kumar Y, Valdivia RH.. 2006. Multifunctional analysis of Chlamydia-specific genes in a yeast expression system. Mol Microbiol. 60(1):51–66. [DOI] [PubMed] [Google Scholar]
- Slagowski NL, Kramer RW, Morrison MF, LaBaer J, Lesser CF.. 2008. A functional genomic yeast screen to identify pathogenic bacterial proteins. PLoS Pathog. 4(1):e9.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MJ, Bandi C, Hoerauf AM, Lazdins J.. 2000. Wolbachia bacteria of filarial nematodes: a target for control? Parasitol Today 16(5):179–180. [DOI] [PubMed] [Google Scholar]
- Toomey ME, Panaram K, Fast EM, Beatty C, Frydman HM.. 2013. Evolutionarily conserved Wolbachia-encoded factors control pattern of stem-cell niche tropism in Drosophila ovaries and favor infection. Proc Natl Acad Sci U S A. 110(26):10788–10793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tram U, Ferree PM, Sullivan W.. 2003. Identification of Wolbachia–host interacting factors through cytological analysis. Microbes Infect. 5(11):999–1011. [DOI] [PubMed] [Google Scholar]
- Turley AP, Moreira LA, O’Neill SL, McGraw EA.. 2009. Wolbachia infection reduces blood-feeding success in the dengue fever mosquito, Aedes aegypti. Plos Neglected Trop Dis. 3(9):e516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JD, et al. 2009. Wolbachia lipoprotein stimulates innate and adaptive immunity through Toll-like receptors 2 and 6 to induce disease manifestations of filariasis. J Biol Chem. 284(33):22364–22378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voth DE, et al. 2009. The Coxiella burnetii ankyrin repeat domain-containing protein family is heterogeneous, with C-terminal truncations that influence Dot/Icm-mediated secretion. J Bacteriol. 191(13):4232–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, Baldo L, Clark ME.. 2008. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 6(10):741–751. [DOI] [PubMed] [Google Scholar]
- Whitaker N, et al. 2016. Chimeric coupling proteins mediate transfer of heterologous type IV effectors through the Escherichia coli pKM101-encoded conjugation machine. J Bacteriol. 198(19):2701–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PM, et al. 2017. Mechanisms of horizontal cell-to-cell transfer of Wolbachia spp. in Drosophila melanogaster. Appl Environ Microbiol. 83(7):e03425–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, et al. 2004. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2(3):E69.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada R, et al. 2011. Functional test of the influence of Wolbachia genes on cytoplasmic incompatibility expression in Drosophila melanogaster. Insect Mol. Biol. 20(1):75–85. [DOI] [PubMed] [Google Scholar]
- Zug R, Hammerstein P.. 2012. Still a host of hosts for wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS One 7(6):e38544.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.