Abstract
The MalG protein is needed for the transport of maltose in Escherichia coli K12. We present the sequence of gene malG. The deduced amino acid sequence corresponds to a protein of 296 amino acid residues (mol. wt. = 32 188 daltons). This protein is largely hydrophobic (hydrophobic index = 0.83) and is thus presumably an integral inner membrane protein which could span the membrane through six hydrophobic segments. We provide direct evidence from fusion proteins for the translation frame and we also identified the in vitro made MalG protein. We have found a sequence which is highly conserved between MalG and MalF, the other integral inner membrane protein of the maltose transport system. This conserved sequence is also present in all known integral membrane proteins of binding protein-dependent transport systems, always at the same distance (approximately 90 residues) from their COOH terminus. We discuss briefly this finding.
Full text
PDF
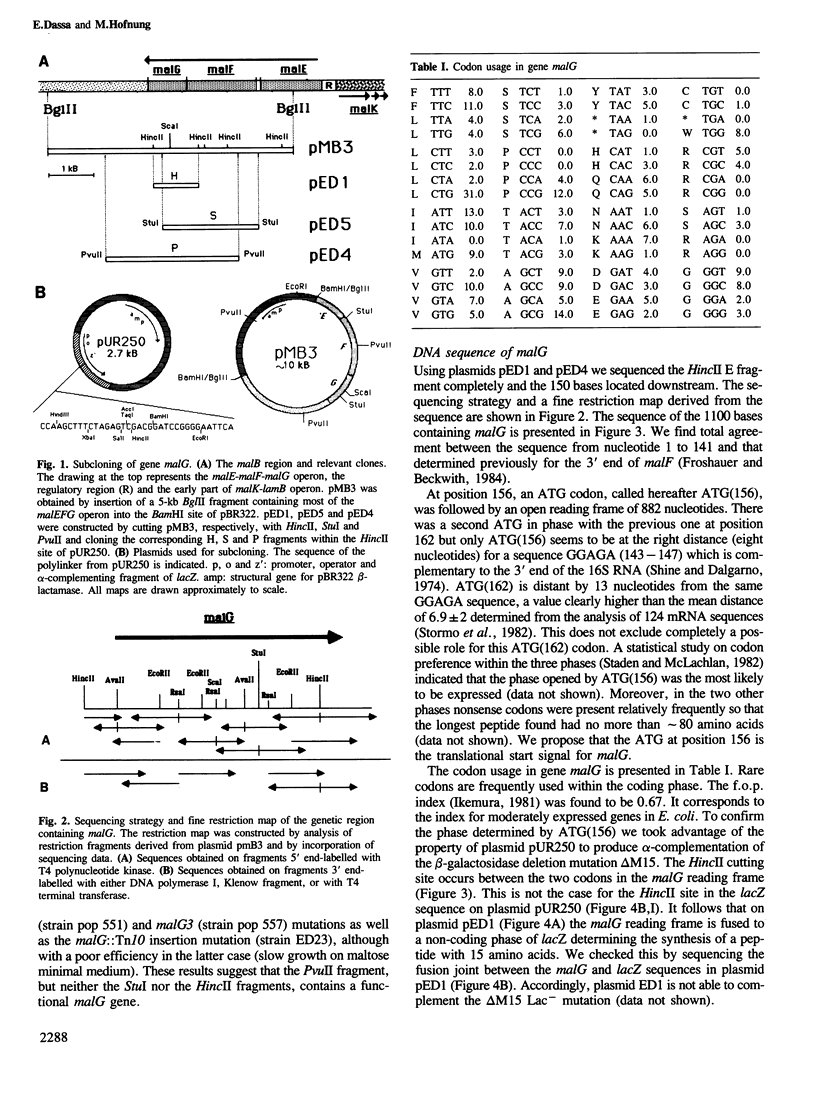

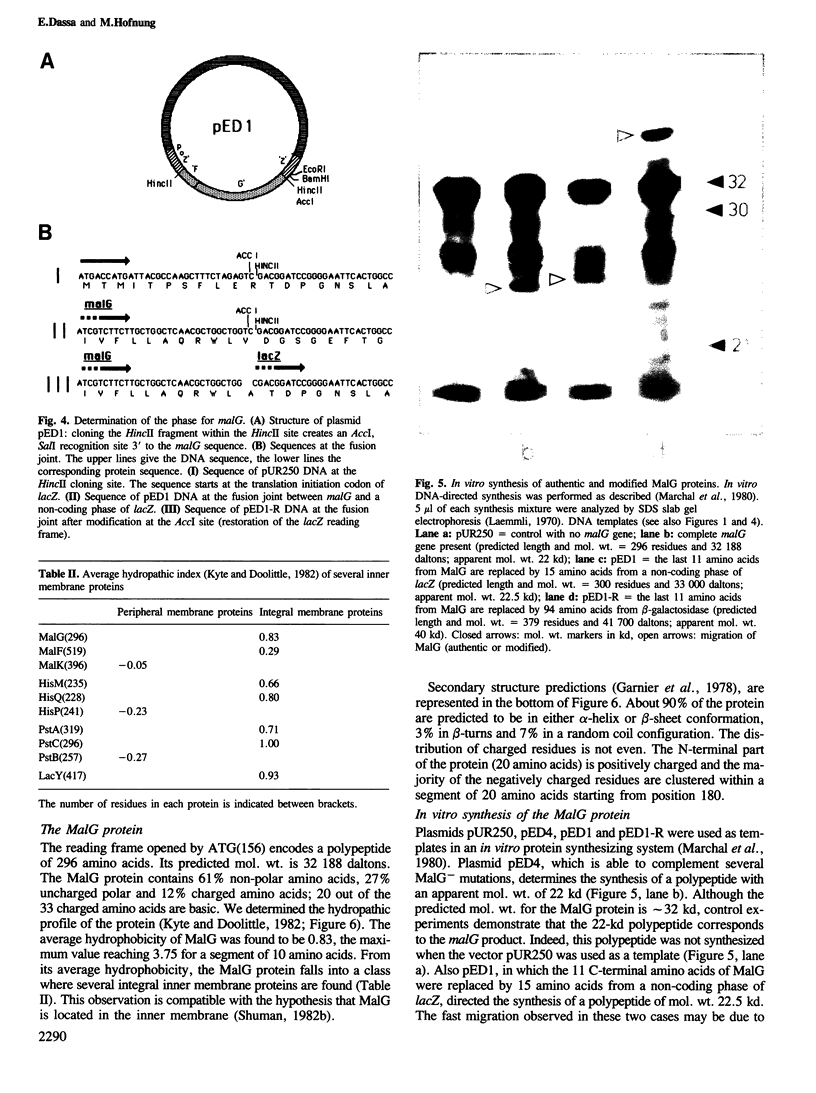
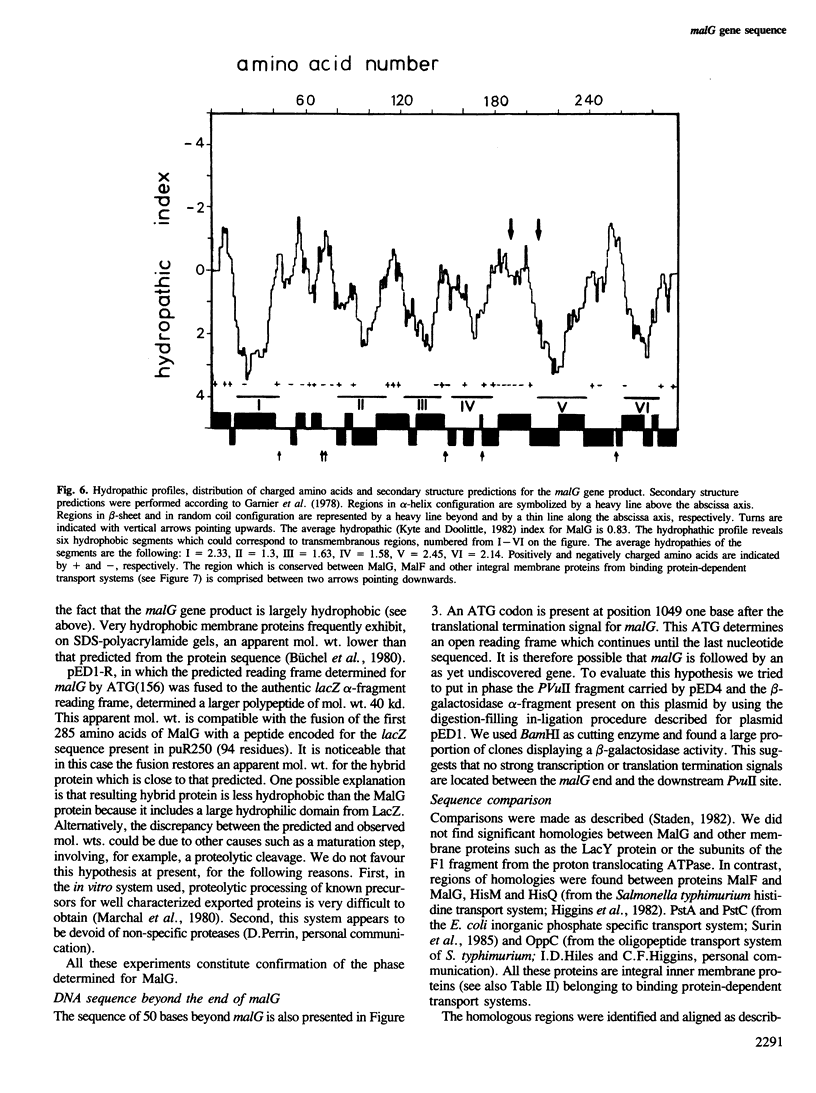


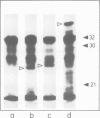
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Backman K., Boyer H. W. Tetracycline resistance determined by pBR322 is mediated by one polypeptide. Gene. 1983 Dec;26(2-3):197–203. doi: 10.1016/0378-1119(83)90190-7. [DOI] [PubMed] [Google Scholar]
- Bavoil P., Hofnung M., Nikaido H. Identification of a cytoplasmic membrane-associated component of the maltose transport system of Escherichia coli. J Biol Chem. 1980 Sep 25;255(18):8366–8369. [PubMed] [Google Scholar]
- Bavoil P., Nikaido H. Physical interaction between the phage lambda receptor protein and the carrier-immobilized maltose-binding protein of Escherichia coli. J Biol Chem. 1981 Nov 25;256(22):11385–11388. [PubMed] [Google Scholar]
- Büchel D. E., Gronenborn B., Müller-Hill B. Sequence of the lactose permease gene. Nature. 1980 Feb 7;283(5747):541–545. doi: 10.1038/283541a0. [DOI] [PubMed] [Google Scholar]
- Claverie J. M. A common philosophy and FORTRAN 77 software package for implementing and searching sequence databases. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):397–407. doi: 10.1093/nar/12.1part1.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clément J. M., Hofnung M. Gene sequence of the lambda receptor, an outer membrane protein of E. coli K12. Cell. 1981 Dec;27(3 Pt 2):507–514. doi: 10.1016/0092-8674(81)90392-5. [DOI] [PubMed] [Google Scholar]
- Dassa E., Hofnung M. Homologies entre les protéins intégrales de membrane interne de systèmes de transport à protéine affine chez les entérobactéries. Ann Inst Pasteur Microbiol. 1985 May-Jun;136A(3):281–288. doi: 10.1016/s0769-2609(85)80090-9. [DOI] [PubMed] [Google Scholar]
- Duplay P., Bedouelle H., Fowler A., Zabin I., Saurin W., Hofnung M. Sequences of the malE gene and of its product, the maltose-binding protein of Escherichia coli K12. J Biol Chem. 1984 Aug 25;259(16):10606–10613. [PubMed] [Google Scholar]
- Froshauer S., Beckwith J. The nucleotide sequence of the gene for malF protein, an inner membrane component of the maltose transport system of Escherichia coli. Repeated DNA sequences are found in the malE-malF intercistronic region. J Biol Chem. 1984 Sep 10;259(17):10896–10903. [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Gilson E., Higgins C. F., Hofnung M., Ferro-Luzzi Ames G., Nikaido H. Extensive homology between membrane-associated components of histidine and maltose transport systems of Salmonella typhimurium and Escherichia coli. J Biol Chem. 1982 Sep 10;257(17):9915–9918. [PubMed] [Google Scholar]
- Gilson E., Nikaido H., Hofnung M. Sequence of the malK gene in E.coli K12. Nucleic Acids Res. 1982 Nov 25;10(22):7449–7458. doi: 10.1093/nar/10.22.7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgpeth J., Clement J. M., Marchal C., Perrin D., Hofnung M. DNA sequence encoding the NH2-terminal peptide involved in transport of lambda receptor, an Escherichia coli secretory protein. Proc Natl Acad Sci U S A. 1980 May;77(5):2621–2625. doi: 10.1073/pnas.77.5.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins C. F., Haag P. D., Nikaido K., Ardeshir F., Garcia G., Ames G. F. Complete nucleotide sequence and identification of membrane components of the histidine transport operon of S. typhimurium. Nature. 1982 Aug 19;298(5876):723–727. doi: 10.1038/298723a0. [DOI] [PubMed] [Google Scholar]
- Higgins C. F., Hiles I. D., Whalley K., Jamieson D. J. Nucleotide binding by membrane components of bacterial periplasmic binding protein-dependent transport systems. EMBO J. 1985 Apr;4(4):1033–1039. doi: 10.1002/j.1460-2075.1985.tb03735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol. 1981 Sep 25;151(3):389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Marchal C., Greenblatt J., Hofnung M. malB region in Escherichia coli K-12: specialized transducing bacteriophages and first restriction map. J Bacteriol. 1978 Dec;136(3):1109–1119. doi: 10.1128/jb.136.3.1109-1119.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Raibaud O., Clément J. M., Hofnung M. Structure of the malB region in Escherichia coli K12. III. Correlation of the genetic map with the restriction map. Mol Gen Genet. 1979 Jul 24;174(3):261–267. doi: 10.1007/BF00267798. [DOI] [PubMed] [Google Scholar]
- Rüther U. pUR 250 allows rapid chemical sequencing of both DNA strands of its inserts. Nucleic Acids Res. 1982 Oct 11;10(19):5765–5772. doi: 10.1093/nar/10.19.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K., Weisberg R. A., Gottesman M. E. Prophage lambda at unusual chromosomal locations. I. Location of the secondary attachment sites and the properties of the lysogens. J Mol Biol. 1972 Feb 14;63(3):483–503. doi: 10.1016/0022-2836(72)90443-3. [DOI] [PubMed] [Google Scholar]
- Shuman H. A. Active transport of maltose in Escherichia coli K12. Role of the periplasmic maltose-binding protein and evidence for a substrate recognition site in the cytoplasmic membrane. J Biol Chem. 1982 May 25;257(10):5455–5461. [PubMed] [Google Scholar]
- Shuman H. A., Silhavy T. J., Beckwith J. R. Labeling of proteins with beta-galactosidase by gene fusion. Identification of a cytoplasmic membrane component of the Escherichia coli maltose transport system. J Biol Chem. 1980 Jan 10;255(1):168–174. [PubMed] [Google Scholar]
- Shuman H. A., Silhavy T. J. Identification of the malK gene product. A peripheral membrane component of the Escherichia coli maltose transport system. J Biol Chem. 1981 Jan 25;256(2):560–562. [PubMed] [Google Scholar]
- Shuman H. A. The maltose-maltodextrin transport system of Escherichia coli. Ann Microbiol (Paris) 1982 Jan;133A(1):153–159. [PubMed] [Google Scholar]
- Silhavy T. J., Brickman E., Bassford P. J., Jr, Casadaban M. J., Shuman H. A., Schwartz V., Guarente L., Schwartz M., Beckwith J. R. Structure of the malB region in Escherichia coli K12. II. Genetic map of the malE,F,G operon. Mol Gen Genet. 1979 Jul 24;174(3):249–259. doi: 10.1007/BF00267797. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 1982 May 11;10(9):2951–2961. doi: 10.1093/nar/10.9.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R., McLachlan A. D. Codon preference and its use in identifying protein coding regions in long DNA sequences. Nucleic Acids Res. 1982 Jan 11;10(1):141–156. doi: 10.1093/nar/10.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L. M. Characterization of translational initiation sites in E. coli. Nucleic Acids Res. 1982 May 11;10(9):2971–2996. doi: 10.1093/nar/10.9.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surin B. P., Rosenberg H., Cox G. B. Phosphate-specific transport system of Escherichia coli: nucleotide sequence and gene-polypeptide relationships. J Bacteriol. 1985 Jan;161(1):189–198. doi: 10.1128/jb.161.1.189-198.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szmelcman S., Hofnung M. Maltose transport in Escherichia coli K-12: involvement of the bacteriophage lambda receptor. J Bacteriol. 1975 Oct;124(1):112–118. doi: 10.1128/jb.124.1.112-118.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szmelcman S., Schwartz M., Silhavy T. J., Boos W. Maltose transport in Escherichia coli K12. A comparison of transport kinetics in wild-type and lambda-resistant mutants as measured by fluorescence quenching. Eur J Biochem. 1976 May 17;65(1):13–19. doi: 10.1111/j.1432-1033.1976.tb10383.x. [DOI] [PubMed] [Google Scholar]
- Wandersman C., Schwartz M., Ferenci T. Escherichia coli mutants impaired in maltodextrin transport. J Bacteriol. 1979 Oct;140(1):1–13. doi: 10.1128/jb.140.1.1-13.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]