Abstract
A major controversy in the area of DNA biochemistry concerns the actual in vivo levels of oxidative damage in DNA. We show here that 8-oxo-2-deoxyguanosine (oxo8dG) generation during DNA isolation is eliminated using the sodium iodide (NaI) isolation method and that the level of oxo8dG in nuclear DNA (nDNA) is almost one-hundredth of the level obtained using the classical phenol method. We found using NaI that the ratio of oxo8dG/105 deoxyguanosine (dG) in nDNA isolated from mouse tissues ranged from 0.032 ± 0.002 for liver to 0.015 ± 0.003 for brain. We observed a significant increase (10-fold) in oxo8dG in nDNA isolated from liver tissue after 2 Gy of γ-irradiation when NaI was used to isolate DNA. The turnover of oxo8dG in nDNA was rapid, e.g. disappearance of oxo8dG in the mouse liver in vivo after γ-irradiation had a half-life of 11 min. The levels of oxo8dG in mitochondrial DNA isolated from liver, heart and brain were 6-, 16- and 23-fold higher than nDNA from these tissues. Thus, our results showed that the steady-state levels of oxo8dG in mouse tissues range from 180 to 360 lesions in the nuclear genome and from one to two lesions in 100 mitochondrial genomes.
INTRODUCTION
The observation that oxidative damage to DNA occurs in vivo was a major discovery because it demonstrated that DNA damage could occur endogenously as a consequence of normal metabolism (1–3). Currently, over 100 oxidative DNA lesions have been characterized (4,5). The 8-oxo-2-deoxyguanosine (oxo8dG) lesion has been studied most extensively because it is the major oxidative lesion (oxo8dG accounts for ∼5% of the total oxidized bases known to occur in DNA) (6) and because it can be detected using a variety of assays (7).
Oxo8dG was initially identified by Kasai et al. (8) following treatment of DNA with fenton-type reagents and γ-irradiation in vitro (9,10). The chemical agents 2-nitropropane, potassium bromate and ciprofibrate, and γ-irradiation, which generate reactive oxygen species (ROS), have also been shown to induce the formation of oxo8dG in vivo (10–12). Oxo8dG is believed to arise through metal-catalyzed oxidation reactions involving metal ions such as iron or copper, both of which are known to bind DNA because of its overall negative charge. For example, the binding affinities of iron (III) and copper (II) for DNA are reported to be 2.1 × 1014 M and 2 × 104 M, respectively (14). Because oxo8dG pairs preferentially with A rather than C, its presence can generate a G→T transversion (15). G→T transversions are frequently found in tumor relevant genes and are among the most common mutations in the p53 gene (16).
Because levels of oxidative DNA damage are small (in the nmole to fmole range per 30–50 µg DNA), it is difficult to detect and quantify these lesions. Over the last 10 years much of the progress in identifying different types of oxidative DNA damage can be attributed to advances in the development of sensitive analytical methods that detect modified nucleotides (17), such as high performance liquid chromatography with electrochemical detection (HPLC-EC) and gas chromatography-mass spectroscopy (GC-MS). Although GC-MS can detect a large number of DNA lesions, the derivatization procedure used to make the nucleosides volatile and stable at high temperatures results in DNA oxidation (18). The HPLC-EC system developed by Floyd (19) in 1986 allows the separation of nucleosides under mild conditions and is a sensitive assay that can detect compounds in the fmole range.
Although there has been much progress in the field of DNA oxidation since the initial observation of the presence of oxidative damage in the genome, significant controversies continue in this area. One of the most troubling problems is the contradictory values reported in the levels of various oxidative lesions present in the nuclear DNA (nDNA) and mitochondrial DNA (mtDNA). For example, oxo8dG levels measured in nDNA from various cells and tissues have shown an almost 5000-fold difference and oxo8dG levels in mtDNA have shown a staggering 60 000-fold variation (1). Such variation has raised important questions concerning the accuracy of various assays used to measure in vivo DNA oxidation levels, e.g. by measuring oxo8dG. During the past 5 years, it has become apparent that a significant amount of DNA oxidation can occur during DNA isolation or preparation of the sample for the assay. For example, oxo8dG levels obtained by GC-MS are considerably higher than levels of oxo8dG obtained using HPLC-EC (18). A report by Claycamp (20) also showed that a significant amount of oxidative damage can occur during the isolation of the DNA, especially in the presence of phenol. Phenol, which is a known reducing agent, is believed to reduce metal ions (e.g. iron) present in biological extracts. After reduction, these ions can enter the Haber–Wiess/Fenton reaction and generate hydroxyl free radicals, which can oxidize the DNA (21). Because various tissues as well as DNA contain relatively high amounts of iron (2–5 nmol/mg protein), this reaction could be very important in generating oxidative damage during DNA isolation (22). In 1995, Nakae et al. (23) reported very low oxo8dG levels in nDNA isolated from liver by a method using sodium iodide (NaI). In this method, NaI, a chaotropic salt, was used to precipitate protein from DNA rather than using an organic solvent, such as phenol (24). In a recent study, Helbock et al. (25) compared levels of DNA oxidation generated by several different DNA isolation methods. When using the NaI method, these investigators found that oxo8dG levels were one-tenth the levels obtained by the other methods studied, including the classic phenol method.
In the present study, we conducted a comprehensive investigation comparing levels of oxo8dG in DNA isolated using the classic phenol method with the newly developed NaI method. We found that the NaI method minimized, if not eliminated, the oxidative damage to DNA that occurs during isolation and that the NaI method was more sensitive than the phenol method. Using the NaI method to isolate mtDNA and nDNA from mouse tissues, we found that the levels of oxo8dG were 6–23-fold higher in mtDNA compared to nDNA.
MATERIALS AND METHODS
Animals
In most of the experiments, tissues from 4–8-month-old male C57BL/6 mice (Jackson Laboratory, Bar Harbor, ME) fed an ad libitum a NIH-31 diet, were used. The mice were euthanized by cervical dislocation and the liver, brain and heart were immediately removed and frozen in liquid nitrogen. The tissues were stored at –80°C and analyzed within 30 days of collection. No difference was observed in the level of oxo8dG after the tissues were stored at –80°C as compared to freshly isolated tissues. In one experiment, we also used tissues collected from 4–6-month-old male F344 rats and C57BL/6 X DBA2 F1 hybrid (B6D2F1) mice, 2–5-year-old parakeets (budgerigar: Melopsittacus undulatus) and benign prostate samples from human subjects (60–78 years of age) undergoing radical prostatectomy. Histological confirmation of the presence or absence of tumors in a particular portion of tissue was established by examination of hematoxylin and eosin stained cryosections of each portion of tissue. The procedures for handling the rodents and parakeets were approved by the Institutional Animal Care and Use Committees of the University of Texas Health Science Center at San Antonio and the University of Idaho. The prostate tissues were obtained from an Institutional Review Board-approved tissue bank housed in the Department of Pathology at The University of Texas Health Science Center at San Antonio that houses both tumor and normal tissues retrieved from radical prostatectomies performed at either the University Hospital or the Audie Murphy VA Hospital.
γ-Irradiation of mice
Mice were exposed to whole-body γ-irradiation (0.5–50 Gy) using a 137Cs GammaCell-40 Irradiator (Atomic Energy of Canada, Mississauga, Ontario, Canada) that produced 136 cGy/min. Mice were either euthanized immediately after the irradiation or at various times (0–90 min) after the irradiation to follow the removal of oxo8dG from the DNA. The tissues were collected and immediately frozen in liquid nitrogen. The procedures for irradiating the mice were approved by the Institutional Animal Care and Use Committees of the University of Texas Health Science Center at San Antonio.
DNA isolation and hydrolysis
nDNA was isolated from liver using two different methods: the phenol method and the NaI method. The classic phenol method used was similar to that originally described by Gupta (26) with the following modifications. Liver tissue (50–100 mg) was homogenized in MSHE buffer (0.21 M mannitol, 0.07 M sucrose, 5 mM HEPES pH 7.4, 10 mM EDTA, 0.1% butylated hydroxytoluene) using a Dounce homogenizer. Nuclei were collected by centrifuging the homogenate at 1000 g for 15 min. The nuclear pellet was washed three times with ice-cold MSHE buffer and once with 0.15 M Tris/1 mM EDTA. Nuclei were resuspended in lysis buffer (10 mM Tris–HCl pH 8.0, 1 mM EDTA, 0.5% SDS, 20 µg/ml RNase) and incubated at 37°C for 45 min. Samples were then treated with 200 µg/ml proteinase K and incubated at 50°C for 1 h. After three extractions with phenol/chloroform and two extractions with chloroform–isoamyl alcohol, DNA was precipitated with ice-cold isopropyl alcohol and suspended in TE buffer (10 mM Tris pH 7.5, 1 mM EDTA). DNA concentrations were determined by measuring the absorbance at 260 nm (27).
nDNA was also isolated by the NaI extraction technique using the DNA Extractor WB Kit (Wako Chemicals USA, Inc., Richmond, VA). The liver and brain were homogenized in a Dounce homogenizer in ice-cold lysis solution and the heart was homogenized in a ground glass homogenizer in ice-cold lysis solution. The human prostate tissue was first pulverized in liquid nitrogen and then homogenized in a Dounce homogenizer in ice-cold lysis solution. The homogenates were then centrifuged at 10 000 g for 20 s and the nuclear pellets resuspended in enzyme reaction solution and 10 µg/ml proteinase K. The resulting solution was incubated at 50°C for 20 min. RNase cocktail (Ambion, Austin, TX) was then added to a final concentration of 20 µg/ml and the resulting solution incubated at 50°C for a further 10 min. Samples were then centrifuged at 10 000 g for 5 min at room temperature. The supernatant was collected and mixed with NaI solution. DNA was precipitated by ice-cold isopropyl alcohol and resuspended in TE buffer. The DNA concentrations were determined by measuring the absorbance at 260 nm (27).
mtDNA was isolated by NaI extraction using the mtDNA Extractor CT Kit (Wako Chemicals USA, Inc.). All steps, except the incubation periods with enzymes, were performed at 4°C. Briefly, tissue was obtained from livers, brains or hearts pooled from 5, 8 or 11 mice, respectively. The liver and brain were homogenized in a Dounce homogenizer, while the heart was homogenized in a ground glass homogenizer in ice-cold homogenizing buffer. The homogenate was centrifuged at 1000 g for 1 min. The supernatant was collected and centrifuged at 10 000 g for 10 min. The pellet, which contained mitochondria, was treated with the appropriate extractor kit solutions and mtDNA was precipitated with ice-cold isopropyl alcohol. The DNA was resuspended in TE buffer and the DNA concentrations were determined by measuring the absorbance at 260 nm (27).
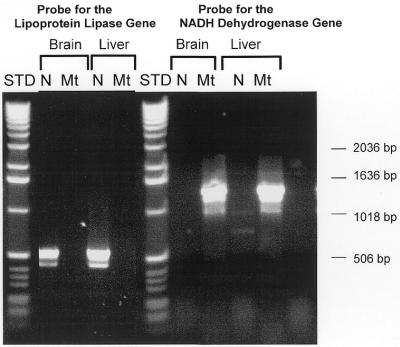
The purity of the nDNA and mtDNA isolated using the DNA Extractor WB Kit or mtDNA Extractor CT Kit was determined by measuring the presence of the lipoprotein lipase gene, which is coded by nDNA, and the NADH dehydrogenase gene, which is coded by mtDNA, in the DNA preparations. As shown in Figure 1, the lipoprotein lipase gene, which was identified as a 550 bp product, was detected only in nDNA from brain and liver. In addition, the data in Figure 1 show that the NADH dehydrogenase gene, which was detected as a 1400 bp product, was present in mtDNA isolated from brain and liver tissues. The NADH dehydrogenase gene was not present in nDNA from the brain and liver. The DNA was identified using 0.5 mg/ml ethidium bromide, which has a lower limit of detection of 1–10 ng of DNA (28). Thus, the data in Figure 1 show no evidence of either nDNA contamination in the mtDNA preparations or mtDNA contamination in the nDNA preparations isolated using the DNA Extractor Kits.
Figure 1.
Characterization of nDNA and mtDNA isolated from liver and brain. nDNA and mtDNA isolated from liver and brain by the NaI method were used as PCR templates for primers to the NADH dehydrogenase gene (found in the mtDNA) and the lipoprotein lipase gene (found in the nDNA). A 1400 bp product was generated with mtDNA using primers for the NADH dehydrogenase gene, and a 500 bp product was generated with nDNA using primers for the lipoprotein lipase gene. A 1 kb DNA Ladder (Gibco/BRL, Santa Fe, NM) is shown as a standard.
nDNA and mtDNA were hydrolyzed as described by Kasai et al. (29). DNA (30–75 µg) was digested with nuclease P1 at 65°C for 11 min. Following digestion, DNA was treated with calf alkaline phosphatase in a Tris buffer (40 mM Tris–HCl pH 8.5, 10 mM MgCl2) for 60 min at 37°C. Samples were filtered using a Microcon“ microconcentrator-10 filtration system (Amicon, Beverly, MA) for 45 min at 4°C. DNA (30–75 µg) was analyzed by HPLC-EC.
Re-extraction of DNA with phenol or NaI
nDNA isolated from mouse liver by the NaI method was re-extracted using either the NaI or phenol method in the presence or absence of either iron (FeCl2), the post-nuclear supernatant (PNS) or mitochondria extracts. The PNS was obtained by centrifugation of the liver homogenate at 1 000 g for 20 s and the mitochondrial extracts were obtained from liver mitochondria isolated as described above. The mitochondria were then treated with DNase (5 µl/ml) to digest the mtDNA and then incubated at 75°C for 5 min to inactivate the DNase. The original DNA (40 µg) was mixed with the following: 1 ml of TE buffer with or without 1 mM FeCl2, 1 ml PNS (35 mg protein/ml) containing no metal chelator or with 1 M EDTA or 1 mM deferoximine, or 1 ml of mitochondrial extract (19 mg protein/ml). The DNA was then re-extracted using either the NaI or phenol methods under air or nitrogen (N2).
Measurement of oxo8dG
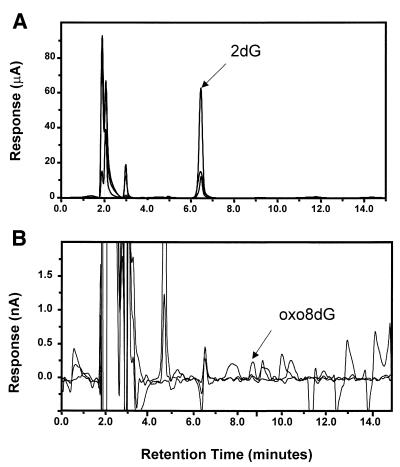
Oxo8dG and 2-deoxyguanosine (2dG) were resolved by HPLC and quantified by electrochemical detection as described by Floyd et al. (30). A CoulArray“ electrochemical detection system (ESA Model 5500/5600; ESA, Inc., Chelmsford, MA) was employed using a 3 µM, 150 × 4.6 C-18 column (YMC, Wilmington, NC). The nucleosides were eluted from the column with an isocratic mobile phase consisting of 50 mM sodium acetate pH 5.2, 5.0% methanol. The mobile phase was filtered and degassed prior to application using a Sep-pak C-18 cartridge (Waters, Milford, MA) followed by filtration using a 0.02 µM nitrocellulose filter (Millipore, Bedford, MA) and sonication for 30 min. Two coulometric cells (Model 6210-Four Channel; ESA) were placed in series and were set to the following potentials: 120, 175, 320, 400, 500, 575, 830 and 890. The HPLC-EC system was calibrated with 500 fmol–1 nmol of oxo8dG and 5 nmol–10 µmol of 2dG using authentic oxo8dG and 2dG standards (Sigma Chemical Co., St Louis, MO). Figure 2 shows the typical chromatograms obtained for DNA hydrolysates using a reverse-phase HPLC-EC system where 2dG had a retention time of 6.5 min and oxo8dG had a retention time of 8.7 min. The identity of 2dG and oxo8dG on the chromatograms were determined by co-injection of standards. Standards were run after every sixth sample for verification and the data were expressed as the ratio of nmoles of oxo8dG to 105 nmoles of 2dG.
Figure 2.
Analysis of DNA hydrolysates by HPLC-EC. nDNA (50 µg) isolated from liver by the NaI method was analyzed. (A) The retention time of 2dG was ∼6.5 min with potential settings across three channels of 550, 650 and 750 mV. (B) The retention time for oxo8dG was ∼8.7 min with potential settings across three channels of 250, 350 and 450 mV. The peaks for 2dG and oxo8dG were identified based on the co-injection of authentic standards as described in the Materials and Methods.
RESULTS AND DISCUSSION
Comparison of phenol and NaI methods
The data in Table 1 compare nDNA isolated from mouse liver by the phenol and NaI methods. The recovery and purity of DNA isolated from liver were similar for the two methods; however, the level of oxo8dG in nDNA isolated by the phenol method was 90-fold greater than that obtained by the NaI method. We also measured the percent co-efficience of variance for the ratio of oxo8dG/dG obtained by the two methods as a gauge of the reproducibility of the methods as described by Wood et al. (31). The percent co-efficience of variance (10 and 12 for the NaI and phenol methods, respectively) was not statistically significant for the two methods. The values we obtained for the oxo8dG/105dG ratio for nDNA (0.035 ± 0.007) isolated by the NaI method are similar to or lower than the values reported in the three previous studies that used NaI to isolate DNA. For example, Nakae et al. (23), Helbock et al. (25) and Beckman et al. (32) reported oxo8dG/105dG ratios of 0.3, 0.04 and 0.04, respectively, for nDNA isolated from rat liver using the NaI method.
Table 1. Comparison of nDNA isolated from mouse liver by the phenol and NaI methodsa.
| µg DNA/g tissue | 260/280 ratio | oxo8dG/105dG | |||
| NaI |
Phenol |
NaI |
Phenol |
NaI |
Phenol |
| 488 |
350 |
1.89 |
1.85 |
0.033 |
3.623 |
| 316 |
290 |
1.92 |
1.83 |
0.039 |
2.822 |
| 387 |
434 |
1.93 |
1.86 |
0.031 |
3.475 |
| 465 |
368 |
1.89 |
1.88 |
0.038 |
3.019 |
| 269 |
482 |
1.90 |
1.91 |
0.030 |
3.248 |
| 366 |
302 |
1.85 |
1.81 |
0.040 |
2.681 |
| 378 |
322 |
1.86 |
1.87 |
0.033 |
3.644 |
| 434 |
500 |
1.86 |
1.83 |
0.029 |
3.308 |
| 510 |
494 |
1.88 |
1.78 |
0.035 |
2.779 |
| 297 |
280 |
1.86 |
1.80 |
0.038 |
3.700 |
| 391 ± 82 | 382 ± 88 | 1.88 ± 0.02 | 1.84 ± 0.02 | 0.035 ± 0.007 | 3.230 ± 0.069b |
aLiver tissue from 10 different mice was divided in half and nDNA was isolated from one half by the NaI method and from the other half by the phenol method. The nDNA was then hydrolyzed and analyzed using HPLC-EC as described in the Materials and Methods. The data for each liver sample is given as well as the mean ± SEM for the 10 animals.
bThis value is significantly (P < 0.001) greater than the value obtained by the NaI method as determined by the Student’s t-test.
The data in Table 1 are consistent with the view that less DNA oxidation occurs during isolation by the NaI method compared to the commonly used phenol method. To study more directly the amount of oxidative damage that occurs to DNA during isolation, we measured the levels of oxo8dG generated in DNA when an original DNA sample, which contained a known amount of oxo8dG, was re-extracted using either NaI or phenol. The data in Table 2 show that when the DNA was re-extracted from the TE buffer, there was no statistically significant increase in the level of oxo8dG in the re-extracted DNA compared to the original sample using either the NaI or phenol method. However, if we added ferrous ions to the buffer, the level of oxo8dG in the DNA isolated by the phenol method was 20-fold higher than the original DNA. When the DNA was isolated under nitrogen, the level of DNA oxidation was reduced by ∼30%. In contrast, no significant increase in oxidative damage to the DNA was observed when the DNA was isolated by the NaI method from the ferrous ion-containing solutions either in the presence or absence of nitrogen.
Table 2. Induction of DNA oxidation by re-extraction with phenol or NaI oxo8dG/105dG ratio.
| Oxo8dG/105dG ratio |
| |
|
NaI |
Phenol |
| Original DNAa |
|
0.037 ± 0.004 |
not done |
| DNA Re-extraction(s): |
|
|
|
| TE buffer |
Control |
0.030 ± 0.008 |
0.042 ± 0.007 |
| |
+Fe (II) |
0.036 ± 0.007b |
0.668 ± 0.060c |
| |
+Fe (II) + N2 |
0.034 ± 0.006b |
0.384 ± 0.047c,d |
| PNS |
–EDTA |
0.041 ± 0.009b |
0.496 ± 0.046c |
| |
+EDTA |
0.040 ± 0.005b |
0.371 ± 0.021c |
| |
+Deferoximine |
0.032 ± 0.007b |
0.310 ± 0.055c,e |
| Mitochondrial extract | +EDTA | 0.045 ± 0.007b | 3.94 ± 0.289c |
The DNA was hydrolyzed and analyzed as described in the Materials and Methods
aThe values for the original DNA represent the mean ± SEM for four separate HPLC analyses. For the re-extraction data, the values represent the mean ± SEM of four separate re-extractions. The data were statistically analyzed using a three-factor ANOVA.
bThe values for the NaI re-extractions were significantly lower at the P < 0.001 level than the phenol re-extractions values
cThe values for the re-extractions were significantly higher at the P < 0.05 level than the original DNA values.
dThe value obtained in the presence of N2 was significantly higher at the P < 0.05 level than the value obtained in air.
eThe value obtained in the presence of deferoximine was significantly lower at the P < 0.05 level than the values obtained in the presence or absence of EDTA.
To carry out the re-extractions under conditions comparable to those used with biological samples, we added the original DNA to solutions containing either the PNS or mitochondrial extracts isolated from mouse liver. When phenol was used to re-extract the DNA from solutions containing PNS, we observed more than a 10-fold increase in the level of oxo8dG in the re-extracted DNA compared to the original DNA. These data demonstrate that substantial DNA oxidation occurs during isolation when using phenol in the presence of biological extracts. In contrast to the phenol method, the NaI method did not show a significant increase in the levels of oxo8dG in the DNA re-extracted from solutions containing PNS. In other words, no detectable DNA oxidation was observed when DNA was isolated by the NaI method. Because DNA oxidation is believed to occur primarily through metal catalyzed oxidation–reduction reactions, which involve metal ions such as iron and/or copper, we also measured the levels of oxo8dG in re-extracted DNA when metal chelators were added to the solutions containing the PNS. Such chelators include EDTA, which is normally added to solutions used to isolate DNA and deferoximine, which has a high affinity (Ka = 1 × 1029) for iron (14). The data in Table 2 show that the addition of either EDTA or deferoximine to the PNS-containing solutions significantly reduced the levels of oxo8dG when phenol was used to re-extract the DNA; and the reduction was greatest (>35%) for deferoximine. However, when DNA was re-extracted by the NaI method, the level of oxo8dG in the re-extracted DNA was not significantly altered by the addition of either EDTA or deferoximine.
We also measured the level of DNA oxidation that occurred during DNA isolation in the presence of mitochondria extracts from liver. As shown in Table 2, a dramatic increase occurred in the level of oxo8dG when DNA was isolated in the presence of the mitochondrial extracts by the phenol method. The level of oxo8dG in the isolated DNA was 100-fold higher than that found in the original DNA and was 10-fold greater than that found in DNA when isolated in the presence of the PNS. Therefore, there are some components in the mitochondria extracts that generate a great deal of oxidative damage to DNA during the isolation procedure in the presence of phenol. The most likely components responsible for the increased oxidative damage are the iron-containing heme proteins and iron–sulfur clusters, which are highly concentrated in the mitochondria. Thus, these mitochondrial components might be at least partially responsible for the high levels of oxidative damage reported for mtDNA isolated by the phenol method (33). In contrast to the phenol method, DNA isolated in the presence of mitochondria extracts by the NaI method showed no significant increase in oxo8dG levels.
The data in Table 2 demonstrate that no detectable DNA oxidation (measured as oxo8dG) was observed when DNA was isolated using the NaI method. In contrast, we found a considerable amount of DNA oxidation can occur during the isolation procedure using phenol. In fact, when DNA was isolated from solutions containing the PNS or mitochondria extracts by the phenol method, 90–99% of the oxo8dG content in the DNA arose during isolation. In addition, the data in Table 2 clearly demonstrate that changes in the type of biological extract, e.g. PNS versus mitochondria, can significantly affect the amount of oxidative damage that occurs during the isolation process using the phenol method. Thus, the NaI method was shown to not only minimize, if not eliminate, oxidative damage to DNA during the isolation process, but also to eliminate the variation in DNA oxidation during isolation that could arise when different cellular extracts are used.
It is possible that the low levels of oxo8dG obtained by the NaI method arise from the loss of oxo8dG because the N-glycosidic bond is sensitive to hydrolysis in the presence of NaI, leading to an under-estimation of the oxo8dG levels in the DNA. Previous studies have found no evidence that an under-estimation of oxo8dG occurs when DNA is isolated by the NaI method. For example, Nakae et al. (23) and Helbock et al. (25) showed that substituting sodium acetate for NaI gave the same values for oxo8dG levels. The data in Table 2 also support the premise that NaI does not result in an under-estimation of oxo8dG levels in DNA. For example, we observed no significant decrease in oxo8dG levels in DNA after re-extraction using NaI. In addition, we have re-extracted DNA by the NaI method that had initially been isolated by the phenol method, i.e. this DNA contained high levels of oxo8dG. The levels of oxo8dG in the re-extracted DNA were the same as that observed in the original DNA (e.g. 3.03 ± 0.07 oxo8dG/105dG versus 2.98 ± 0.06 oxo8dG/105dG). Thus, there is no evidence that the NaI method of DNA isolation results in an under-estimation of oxo8dG in DNA samples.
Induction of DNA oxidation by γ-irradiation
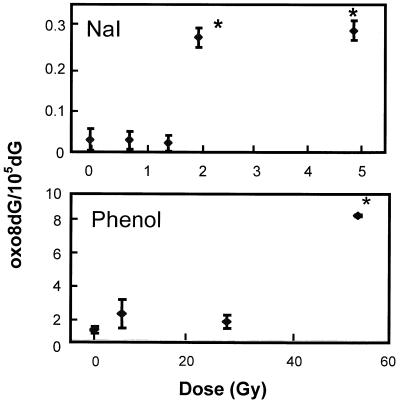
We compared the sensitivity of the two DNA isolation methods for the detection of oxo8dG in vivo by measuring the induction of oxo8dG in nDNA after exposing mice to various doses of γ-irradiation. The data in Figure 3 show that a significant increase in the level of oxo8dG in nDNA from liver was observed only at a dose of 50 Gy when phenol was used to isolate the DNA. It should be noted that the LD50 for γ-irradiation of mice is reported to be between 8 and 10 Gy (34), which is one-fifth of the dose required to see a significant increase in DNA oxidation using the phenol method to isolate DNA. In contrast, a significant increase (10-fold) in oxo8dG was observed at a dose of 2 Gy when NaI was used to isolate DNA (Fig. 3). We also observed a significant increase in oxo8dG levels after 2 Gy of irradiation for nDNA isolated from brain (12-fold, n = 7) and kidney (15-fold, n = 7) by the NaI method. The data in Figure 3 clearly demonstrate that DNA oxidation can be detected at much lower doses of γ-irradiation when DNA is isolated by the NaI method. This observation is important because low doses of γ-irradiation (3–6 Gy) have been shown to alter the expression of a variety of genes, such as GADD45, in human skin fibroblasts (35). In addition, mutations and nDNA strand breaks are detected at doses of 1–2 Gy (36,37). Thus, using the NaI method, we were able to detect alterations in DNA oxidation at levels of γ-irradiation that are potentially important physiologically.
Figure 3.
Effect of whole-body γ-irradiation on the ratio of oxo8dG/105 2dG in nDNA from mouse liver. Mice were exposed to acute whole-body γ-irradiation at doses of 0–50 Gy. Following irradiation, the mice were immediately killed and the liver divided in half. nDNA was isolated from the two halves of the liver using either the phenol or NaI method. Each value is expressed as a mean ± SEM of data from six mice. The data were analyzed statistically using a one-way ANOVA with a Dunnett’s test and the values shown by the asterisks are significantly higher than the control values (no γ-irradiation).
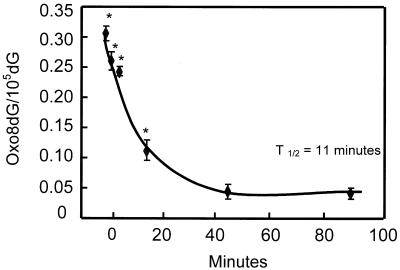
We also measured the ability of the liver to remove oxo8dG from nDNA after exposure to 2 Gy using the NaI isolation method. The data in Figure 4 show that the disappearance of oxo8dG from nDNA in the livers of mice in vivo is very rapid, with a half-life of 11 min. The levels of oxo8dG returned to endogenous levels (0.031 ± 0.004) within 45 min. These data are consistent with the report by Zastawny et al. (38) in which oxo8dG levels in rat liver nDNA were shown to return to endogenous levels 1 h after γ-irradiation. Kasai et al. (29) also reported that the levels of oxo8dG in mouse liver decreased ∼50% 90 min after γ-irradiation; however, the levels of oxo8dG had not yet returned to endogenous levels at this time. Using GC-MS, Jaruga and Dizdaroglu (39) reported that the half-life for the removal of oxo8dG in human lymphoblast cells was 55.2 min. Thus, the oxo8dG lesion represents oxidative damage that is rapidly repaired.
Figure 4.
Time course for the removal of oxo8dG from the nDNA of liver tissue following acute whole-body γ-irradiation. Mice were exposed to 2 Gy of whole-body γ-irradiation and euthanized at 0, 2, 5, 20, 45 and 90 min after irradiation. The nDNA was isolated from the liver using the NaI method. The ratio of oxo8dG/105dG represents the mean ± SEM of data collected from six mice. The data were statistically analyzed using a one-way ANOVA and the values shown by the asterisks are significantly higher (P < 0.05) than the control value (no γ-irradiation).
Comparison of oxo8dG levels in nDNA and mtDNA in tissues of various animals
Using the NaI method, we measured the levels of oxo8dG in nDNA from various tissues of several animal models to determine if the levels of oxo8dG varied significantly both between tissues and between animal models. The data in Table 3 give the levels of oxo8dG in nDNA isolated from liver, brain and heart from mice, rats and parakeets, as well as nDNA isolated from human prostate tissue. The levels of oxo8dG in nDNA from the liver, brain and heart of B6D2F1 mice and F344 rats were identical to those obtained from the same tissues of C57BL/6 mice (Table 4). However, we observed that the level of oxidative damage in nDNA varied significantly from tissue to tissue in rodents. For example, the level of oxo8dG in nDNA from liver was 2–3-fold higher than that found in nDNA from either brain or heart.
Table 3. Comparison of oxo8dG levels in nDNA isolated from tissues of various species.
| Tissue/species |
oxo8dG/105dG ratioa |
| Liver | |
| Mouse (B6D2F1)b | 0.035 ± 0.003d |
| Rat (F344)b | 0.033 ± 0.005d |
| Parakeet (Budgerigar)b | 0.036 ± 0.007d |
| Brain | |
| Mouse (B6D2F1) b | 0.012 ± 0.003e |
| Rat (F344)b | 0.012 ± 0.003e |
| Parakeet (Budgerigar)b | 0.076 ± 0.005f |
| Heart | |
| Mouse (B6D2F1) b | 0.012 ± 0.004e |
| Rat (F344)b | 0.010 ± 0.002e |
| Parakeet (Budgerigar)b | 0.069 ± 0.006f |
| Prostatec | |
| Human | 0.064 ± 0.004c |
aEach value represents the mean ± the SEM.
bThe values for tissues from rodents and birds were obtained from five to seven animals.
cThe value for human prostate tissue was obtained from eight patients.
d,e,fValues with different superscripts are significantly different at the p < 0.05 level.
Table 4. Comparison of oxo8dG levels in nDNA and mtDNA isolated from liver, brain and heart tissues of C57BL/6 mice by the NaI method.
| oxo8dG/105dG ratio | mtDNA/DNA ratiob | ||
| |
NDNAa |
MtDNAa |
|
| Liverc,d |
0.034 |
0.193 |
5.7 |
| |
0.032 |
0.186 |
6.2 |
| |
0.030 |
0.201 |
6.3 |
| |
0.032 ± 0.002 |
0.194 ± 0.050
|
6.1 ± 0.4 |
| Braine,f |
0.019 |
0.366 |
19 |
| |
0.015 |
0.349 |
23 |
| |
0.012 |
0.312 |
26 |
| |
0.015 ± 0.003 |
0.342 ± 0.080 |
23 ± 1 |
| Hearte,f |
0.022 |
0.303 |
14 |
| |
0.020 |
0.336 |
17 |
| |
0.019 |
0.332 |
18 |
| 0.020 ± 0.004 | 0.324 ± 0.095 | 16 ± 1 |
aData for nDNA and mtDNA are expressed as the mean ± SEM for the three groups of animals. The values for mtDNA for all three tissues were statistically different (P < 0.001) from nDNA.
bThe oxo8dG/105dG ratios for the nDNA were obtained by averaging the values obtained from each of the animals in each of the three groups.
cFor the liver, three groups of animals were studied with five animals per group. A small piece from each liver was obtained to isolate the nDNA from each mouse while the remaining liver tissue was pooled to isolate the mtDNA.
dThe values for liver were significantly different (P < 0.05) from either brain or heart as determined by the paired Student’s t-test.
eFor brain and heart, three groups of animals were studied with eight animals per group. A small piece from each brain or heart was obtained to isolate the nDNA while the remaining brain or heart tissue was pooled to isolate the mtDNA.
fThe value for brain was significantly different (P < 0.05) from either liver or heart as determined by the paired Student’s t-test.
We also measured the levels of oxo8dG in liver, brain and heart of parakeets because birds live about three times as long as similar sized mammals, despite having as high, or frequently higher, basal metabolic rates and higher body temperatures (40). Given their greater longevity and assuming that longevity correlates with the ability to resist oxidative stress, one might expect that birds would have reduced steady-state levels of oxidative damage in the face of greater endogenous oxidative stress than similar-sized mammals. Using the phenol/chlorofom extraction method, Herrero and Barja (41) reported that DNA isolated from parakeet hearts contained less oxo8dG than mouse heart and that brain DNA from parakeets and mice exhibited similar levels of oxo8dG. The data in Table 3 show that the level of oxo8dG in nDNA isolated from the livers of parakeets was similar to that observed for rats and mice. However, in contrast to rodents, we observed that the levels of oxo8dG in brain and heart tissue of the parakeets was much higher (∼2-fold) than in liver tissue. In contrast to the report by Herrero and Barja (41), we observed that the level of oxo8dG in nDNA from the brain and heart of the parakeets was ∼6-fold higher than the levels of oxo8dG observed in nDNA from the brain and heart of rodents. Therefore, our data suggest that the brain and heart of parakeets are under higher steady-state levels of oxidative stress than the same organs of rodents.
We also measured the level of oxo8dG in nDNA isolated from prostate tissue from human subjects. The level of oxo8dG in the human prostate tissue was higher than that observed in tissues from the rodents and similar to the levels of oxo8dG observed in the brain and heart of parakeets (Table 3). Because of the limited data available at the present time, we are unable to determine whether the relatively high levels of oxidative damage in nDNA from human prostate tissue is due to species or tissue differences or because the prostate tissue was obtained from relatively old individuals (60–78 years of age) while the rodents and parakeets studied were relatively young (4–6-month-old rodents, which have a mean survival of 24–28 months and 2–5-year-old parakeets, which have a mean survival of 10–15 years). Several reports indicate that the level of oxo8dG in DNA increases with age (24,42–45). However, as the data in Table 3 clearly demonstrate, one must be cautious in extending data obtained on the level of nDNA oxidation obtained from one tissue or species to other tissues or species. Our data suggest that the oxo8dG levels vary significantly from tissue to tissue and from species to species.
We were also interested in comparing the levels of oxo8dG in nDNA and mtDNA using the NaI isolation method because of the current controversy over whether oxidative damage is greater in mtDNA than in nDNA. The initial study by Richter et al. (33) reported that the levels of oxo8dG were 16-fold higher in mtDNA from the livers of rats compared to those found in nDNA. Subsequently, many investigators have reported higher levels of oxo8dG in mtDNA than in nDNA when the DNA was isolated using phenol (46–49). However, Higuchi and Linn (50) reported that the levels of oxo8dG measured in mtDNA isolated using cesium chloride density gradient centrifugation were not only lower than those reported initially by Richter et al. (33) for mtDNA but also were lower than was reported by these investigators for nDNA. This report brought into question the prevailing view that the level of oxidative damage was higher in mtDNA than in nDNA. Recently, Anson et al. (49) also concluded that endogenous oxidative damage to mtDNA had been overestimated when they used an Fpg/EndoIII/Southern blot assay to measure the levels of oxidative damage in mtDNA from rat liver. Because we observed a 100-fold increase in oxo8dG in DNA when it was extracted from solutions containing mitochondria extracts using the phenol method (Table 2), our data would suggest that it is possible that the previous reports showing increased levels of oxo8dG in mtDNA compared to nDNA might be attributed to an artifact from increased oxidative damage to mtDNA during the isolation procedure. Therefore, we measured oxo8dG levels in mtDNA isolated from liver, heart and brain using the NaI isolation method to minimize/eliminate oxidative damage to mtDNA during the isolation procedure. These data are presented in Table 4 and are the first showing oxo8dG levels in mtDNA that were measured using NaI to isolate mtDNA. In these experiments, we pooled mitochondria from several animals to isolate the mtDNA so that the amounts of DNA processed during the isolation procedure and the analysis were identical for both the nDNA and mtDNA preparations. We did this to increase the yield of mtDNA because Beckman and Ames (51) reported a systematic artifact, namely that low quantities of DNA were associated with an apparent elevated level of oxo8dG lesions in mtDNA. The oxo8dG/105dG ratio that we obtained for the mtDNA ranged from 0.19 for liver to 0.34 for brain. Originally, Richter et al. (33) reported an oxo8dG/105dG ratio of 53 for mtDNA isolated from rat liver using the phenol method. More recently, Anson et al. (49) reported a oxo8dG/105dG ratio of 2.3 for mtDNA from crude homogenates of rat liver and a ratio of 8.1 for mtDNA isolated from rat liver mitochondria by the phenol method. Thus, the levels of oxo8dG we observed when mtDNA was isolated by the NaI method were much lower than values previously reported. However, the data in Table 4 show that the levels of oxo8dG in mtDNA isolated by NaI were significantly higher than the levels of oxo8dG in nDNA for all three tissues studied. Thus, our data, in which NaI was used to minimize oxidative damage to DNA during isolation, show that the steady-state levels of oxidative damage in mtDNA are significantly higher than those found in nDNA.
The data in Table 4 also show that the level of oxidative damage in mtDNA varies significantly from tissue to tissue. For example, the level of oxo8dG in mtDNA from liver was 40–50% less than that found in mtDNA isolated from either brain or heart. The tissue differences in the levels of oxidative damage in both mtDNA and nDNA result in dramatic differences in the mtDNA/nDNA ratio of oxo8dG in the three tissues. For example, in liver, the level of oxo8dG in mtDNA was 6-fold higher than in nDNA. This is less than half the value of 16-fold originally reported by Richter et al. (33). However, the levels of oxo8dG in mtDNA from brain and heart were 23- and 16-fold higher, respectively, than nDNA levels. Higher levels of oxidative damage in mtDNA are generally assumed to arise from mtDNA being present in an environment where it is exposed to high levels of ROS, mtDNA having less protein (e.g. histones) associated with it and the reduced ability of mitochondria to repair DNA compared to the nucleus (1,52,53).
Our data show that major differences occur between tissues. The ratio of mtDNA/nDNA oxidative damage was ∼3- to 4-fold higher for heart and brain than for liver. The most likely explanation for the higher levels of oxidative damage in mtDNA from brain and heart is the higher metabolic activity of these tissues compared to liver. For example, Abete et al. (54) showed that brain and heart use more oxygen and produce ATP at a faster rate than liver. In addition, Rolfe et al. (55) reported that proton leak, which is a significant contributor to resting metabolic rates in mammals, was greater in mitochondria from heart and brain than liver. A greater proton leak is believed to be correlated to the enhanced metabolic rate of brain and heart. In addition, Rolfe et al. (55) reported tissue differences in the efficiency of energy production. They observed that the liver was more thermodynamically efficient while the heart and the brain utilize more oxygen and produce ATP at a faster rate than the liver. In other words, liver mitochondria are designed for the economic production of ATP for use in cellular processes while brain and heart mitochondria are designed for the maximal production of ATP.
Based on their data, in which nDNA and mtDNA were isolated from liver by the phenol method, Richter et al. (33) estimated that nDNA contained ∼3.2 oxo8dG lesions for every 105 bases and that mtDNA contained one oxo8dG lesion for every 400 bases. In other words, the 16.5 kb mitochondrial genome would carry a steady-state burden of as many as 80 oxo8dG lesions. These estimates now appear to be a gross overestimation of the steady-state levels of oxo8dG lesions in both nDNA and mtDNA from rodent tissue because of the extensive amount of DNA oxidation that occurs during DNA isolation using the phenol method. Based on the values of oxo8dG we have measured in nDNA and mtDNA from mouse tissues and assuming a GC content of 40% for nDNA and 37% for mtDNA from mice (56), we estimate that the steady-state levels of oxo8dG lesions range from 3 per 108 bases for nDNA from brain to 6 per 108 bases for liver. In other words, the mouse nuclear genome, which contains ∼3 × 109 bp per haploid genome (57), would contain a steady-state level of 180–360 oxo8dG lesions in brain and liver, respectively. For mtDNA, we estimate that the steady-state levels of oxo8dG range from 4 to 7 per 107 bases for liver and brain, respectively. Assuming that the mouse genome contains 33 kb (49), this would mean that steady state levels of 1–2 oxo8dG lesions would be found in every 100 mitochondrial genomes.
We also calculated the de novo formation of oxo8dG from the steady-state level of oxo8dG and the half-life of oxo8dG (11 min) that we measured in nDNA from mouse liver. We estimate that the nuclear genome (diploid) of a liver cell in C57BL/6 mice would be exposed to >47 000 oxo8dG lesions in a 24 h period. Because oxo8dG makes up ∼5% of all oxidative lesions (25), it would appear that a liver cell of a mouse would be exposed to approximately one million oxidative lesions in a day, assuming the generation and removal of all the oxidative lesions occur at similar rates as oxo8dG. Based on a density of 1.2 × 108 cells/g mouse liver (58), we estimate that nDNA in the liver would be exposed to a total of 5–6 × 1012 oxo8dG lesions in a day. Over the life span of the mouse (which is ∼27 months) for C57BL/6 mice (59), this would represent a total of ∼4 × 1015 oxo8dG lesions in nDNA in the liver. This lifetime estimate of the number of oxo8dG lesions in nDNA of the liver is probably an underestimate because it appears that the steady-state levels of oxo8dG increase with age in mouse liver (42,43,45). Thus, while our data show that the steady-state levels of endogenous oxo8dG were grossly overestimated previously, the de novo formation of oxo8dG lesions in the nuclear genome is still considerable, suggesting that oxidative lesions in DNA that arise from normal cellular metabolism are highly relevant.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Dr Christi A.Walter for her input in the writing of this manuscript and Mike Luther for his assistance with the statistical analysis of the data. This work was supported by NIH grants R01-AG13319 and PO1AG14674 and a Merit Review grant from the Department of Veteran Affairs.
References
- 1.Beckman K.B. and Ames,B.N. (1999) Endogenous oxidative damage of mtDNA. Mutat. Res., 424, 51–58. [DOI] [PubMed] [Google Scholar]
- 2.Ames B.N. and Gold,L.S. (1991) Endogenous mutagens and the causes of aging and cancer. Mutat. Res., 250, 3–16. [DOI] [PubMed] [Google Scholar]
- 3.Wagner J.R., Hu,C.C. and Ames,B.N. (1992) Endogenous oxidative damage of deoxycytidine in DNA. Proc. Natl Acad. Sci. USA, 89, 3380–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dizdaroglu M. (1998) Facts about the artifacts in the measurement of oxidative DNA base damage by gas chromatography-mass spectrometry. Free Radic. Res., 29, 551–563. [DOI] [PubMed] [Google Scholar]
- 5.Kaur H. and Halliwell,B. (1996) Measurement of oxidized and methylated DNA bases by HPLC with electrochemical detection. Biochem. J., 318, 21–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dizdaroglu M. (1992) Oxidative damage to DNA in mammalian chromatin. Mutat. Res., 275, 331–342. [DOI] [PubMed] [Google Scholar]
- 7.Helbock H.J., Beckman,K.B. and Ames,B.N. (1999) 8-Hydroxydeoxyguanosine and 8-hydroxyguanine as biomarkers of oxidative DNA damage. Methods Enzymol., 300, 156–165. [DOI] [PubMed] [Google Scholar]
- 8.Kasai H., Hayami,Z., Yamaizumi,Z., Saito,S. and Nishimura,S. (1984) Detection and identification of mutagens and carcinogens as their adducts with guanosine derivatives. Nucleic Acids Res., 12, 2127–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kasai H., Okada,Y., Nishimura,S., Rao,M.S. and Reddy,J.K. (1989) Formation of 8-hydroxydeoxyguanosine in liver DNA of rats following long-term exposure to a peroxisome proliferator. Cancer Res., 49, 2601–2605. [PubMed] [Google Scholar]
- 10.Kasai H. and Nishimura,S. (1984) Hydroxylation of deoxyguanosine at the C-8 position by ascorbic acid and other reducing agents. Nucleic Acids Res., 12, 2137–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kasai H., Tanooka,H. and Nishimura,S. (1984) Formation of 8-hydroxyguanine residues in DNA by X-irradiation. Gann, 75, 1037–1039. [PubMed] [Google Scholar]
- 12.Adachi S., Kawamura,K. and Takemoto,K. (1994) Increased susceptibility to oxidative DNA damage in regenerating liver. Carcinogenesis, 15, 539–543. [DOI] [PubMed] [Google Scholar]
- 13.Fiala E.S., Conaway,C.C. and Mathis,J.E. (1989) Oxidative DNA and RNA damage in the livers of Sprague–Dawley rats treated with the hepatocarcinogen 2-nitropropane. Cancer Res., 49, 5518–5522. [PubMed] [Google Scholar]
- 14.Halliwell B., Gutteridge,J.M. and Cross,C.E. (1992) Free radicals, antioxidants and human disease: where are we now. J. Lab. Clin. Med., 119, 598–620. [PubMed] [Google Scholar]
- 15.Moriya M. and Grollman,A.P. (1993) Mutations in the mutY gene of Escherichia coli enhance the frequency of targeted G:C→T:A transversions induced by a single 8-oxoguanine residue in single-stranded DNA. Mol. Gen. Genet., 239, 72–76. [DOI] [PubMed] [Google Scholar]
- 16.Ho Y.-S., Cheng,H.-T., Wang,Y.-J. and Lin,J.-K. (1995) p53 gene mutational spectra in hepatocellular carcinomas induced by 2-acetylaminofluorene and N-nitroso-2-acetylaminofluorene in rats. Mol. Carcinog., 13, 182–190. [DOI] [PubMed] [Google Scholar]
- 17.Demple B. and Harrison,L. (1994) Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem., 63, 915–948. [DOI] [PubMed] [Google Scholar]
- 18.Ravanat J.-L., Turesky,R.J., Germaud,E., Trudel,L.J. and Stadler,R.H. (1995) Determination of 8-oxoguanine in DNA by gas chromatography-mass spectrometry and HPLC-electrochemical detection: overestimation of the background level of the oxidized base by the gas chromatography-mass spectrometry assay. Chem. Res. Toxicol., 8, 1039–1045. [DOI] [PubMed] [Google Scholar]
- 19.Floyd R.A. (1986) The development of a sensitive analysis for 8-hydroxy-2′-deoxyguanosine. Free Radic. Res. Commun., 8, 139–141. [DOI] [PubMed] [Google Scholar]
- 20.Claycamp H.G. (1992) Phenol sensitization of DNA to subsequent oxidative damage in 8-hydroxyguanine assays. Carcinogenesis, 13, 1289–1292. [DOI] [PubMed] [Google Scholar]
- 21.Aust A.E. and Eveleigh,J.F. (1999) Mechanisms of DNA oxidation. Proc. Soc. Exp. Biol. Med., 222, 246–252. [DOI] [PubMed] [Google Scholar]
- 22.Sohal R.S., Wennberg-Kirch,E., Jaiswal,K., Kwong,L.K. and Forster,M.J. (1999) Effect of age and caloric restriction on bleomycin-chelatable and nonheme iron in different tissues of C57BL/6 mice. Free Radic. Biol. Med., 27, 287–293. [DOI] [PubMed] [Google Scholar]
- 23.Nakae D., Mizumoto,Y., Kobayashi,E., Noguchi,O. and Konishi,Y. (1995) Improved genomic/nuclear DNA extraction for 8-hydroxydeoxyguanosine analysis for small amounts of rat liver tissue. Cancer Lett., 97, 233–239. [DOI] [PubMed] [Google Scholar]
- 24.Sai K., Takagi,A., Umemura,T., Hasegawa,R. and Kurokawa,Y. (1992) Changes of 8-hydroxydeoxyguanosine levels in rat organ DNA during aging process. J. Environ. Pathol. Toxicol. Oncol., 11, 139–143. [PubMed] [Google Scholar]
- 25.Helbock H., Beckman,K., Shigenaga,M., Walter,P.B., Woodall,A., Yeo,H.C. and Ames,B.N. (1998) DNA oxidation matters: The HPLC-electrochemical detection assay of 8-oxo-deoxyguanosine and 8-oxo-guanine. Proc. Natl Acad. Sci. USA, 95, 288–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta R.C. (1984) Nonrandom binding of the carcinogen N-hydroxy-2-acetylaminofluorene to repetitive sequences of rat liver DNA in vivo. Proc. Natl Acad. Sci. USA, 81, 6943–6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ausubel F. (1995) Short Protocols in Molecular Biology. Wiley & Son Inc, New York, NY.
- 28.Sinclair B. (2001) An eye for a dye: safe and sensitive new stains replace ethidium bromide for routine nucleic acid detection. The Scientist, 14, 31. [Google Scholar]
- 29.Kasai H., Crain,P.F., Kuchino,Y., Nishimura,S., Ootsuyana,A. and Tanooka,H. (1986) Formation of 8-hydroxyguanine moiety in cellular DNA by agents producing oxygen radicals and evidence for its repair. Carcinogenesis, 7, 1849–1851. [DOI] [PubMed] [Google Scholar]
- 30.Floyd R.A., West,M.S., Eneff,K.L., Schneider,J.E., Wong,P.K., Tingey,D.T. and Hogsett,W.E. (1990) Conditions influencing yield and analysis of 8-hydroxy-2′-deoxyguanosine in oxidatively damaged DNA. Anal. Biochem., 155–158. [DOI] [PubMed] [Google Scholar]
- 31.Wood S.G., Gedik,C.M. and Collins,A.R. (2000) Controlled oxidation of calf thymus DNA to produce standard samples for 8-oxodeoxyguanosine analysis; effects of freeze-drying, storage and hydrolysis conditions. Free Radic. Res., 32, 327–332. [DOI] [PubMed] [Google Scholar]
- 32.Beckman K.B., Saljoughi,S., Mashiyama,S.T. and Ames,B.N. (2000) A simpler, more robust method for the analysis of 8-oxoguanine in DNA. Free Radic. Biol. Med., 29, 357–367. [DOI] [PubMed] [Google Scholar]
- 33.Richter C., Park,J. and Ames,B.N. (1988) Normal oxidative damge to mitochondrial and nuclear DNA is extensive. Proc. Natl Acad. Sci. USA, 85, 6465–6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schilderman P.A., E.L., Rhijnsburger,E., Zwingmann,I. and Kleinjans,J.C.S. (1995) Induction of oxidative DNA damage and enhancement of cell proliferation in human lymphocytes in vitro by butylated hydroxyanisole. Carcinogenesis, 16, 507–512. [DOI] [PubMed] [Google Scholar]
- 35.de Toledo S.M., Azzam,E.I., Gasmann,M.K. and Mitchel,R.E. (1995) Use of semiquantitative reverse transcription–polymerase chain reaction to study gene expression in normal human skin fibroblasts following low dose-rate irradiation. Int. J. Radiat. Biol., 67, 135–143. [DOI] [PubMed] [Google Scholar]
- 36.Winegar R.A., Lutze,L.H., Hamer,J.D., O’Loughlin,K.G. and Mirsalis,J.C. (1994) Radiation-induced point mutations, deletions and micronuclei in lacI transgenic mice. Mutat. Res., 307, 479–487. [DOI] [PubMed] [Google Scholar]
- 37.Mendiola-Cruz M.T. and Morales-Ramirez,P. (1999) Repair kinetics of γ-ray induced DNA damage determined by the single cell gel electrophoresis assay in murine leukocytes in vivo. Mutat. Res., 433, 45–52. [DOI] [PubMed] [Google Scholar]
- 38.Zastawny T.H., Czerwinska,B., Drzewiecka,B. and Olinski,R. (1996) Radiation induced oxidative DNA base damage and its repair in liver chromatin DNA of rats upon whole body γ-irradiation. Acta Biochim. Pol., 43, 579–582. [PubMed] [Google Scholar]
- 39.Jaruga P. and Dizdaroglu,M. (1996) Repair of products of oxidative DNA base damage in human cells. Nucleic Acids Res., 24, 1389–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holmes D.J. and Austad,S.N. (1995) Birds as animal models for the comparative biology of aging: a prospectus. J. Gerontol. A Biol. Sci. Med. Sci., 50, B59–B66. [DOI] [PubMed] [Google Scholar]
- 41.Herrero A. and Barja,G. (1999) 8-oxo-deoxyguanosine levels in heart and brain mitochondrial and nuclear DNA of two mammals and three birds in relation to their different rates of aging. Aging (Milano), 11, 294–300. [DOI] [PubMed] [Google Scholar]
- 42.Fraga C.G., Shigenaga,M.K., Park,J., Degan,P. and Ames,B.N. (1990) Oxidative damage to DNA during aging: 8-hydroxy-2′-deoxyguanosine in rat organ DNA and urine. Proc. Natl Acad. Sci. USA, 87, 4533–4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaneko T., Tahara,S.M. and Matsuo,M. (1996) Non-linear accumulation of 8-hydroxy-2′-deoxyguanosine, a marker of oxidized DNA damage, during aging. Mutat. Res., 316, 277–285. [DOI] [PubMed] [Google Scholar]
- 44.Sohal R.S., Agarwal,S., Candas,M., Forster,M.J. and Lal,H. (1994) Effect of age and caloric restriction on DNA oxidative damage in different tissues of C57BL/6 mice. Mech. Ageing Dev., 76, 215–224. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y.-J., Ho,Y.-S., Lo,M.-J. and Lin,J. (1995) Oxidative modification of DNA bases in rat liver and lung during chemical carcinogenesis and aging. Chem. Biol. Interact., 94, 135–145. [DOI] [PubMed] [Google Scholar]
- 46.Barja G. and Herrero,A. (2000) Oxidative damage to mitochondrial DNA is inversely related to maximum life span in the heart and brain of mammals. FASEB J., 14, 312–318. [DOI] [PubMed] [Google Scholar]
- 47.Richter C. (1992) Reactive oxygen and DNA damage in mitochondria. Mutat. Res., 275, 249–255. [DOI] [PubMed] [Google Scholar]
- 48.Zastawny T.H., Dabrowska,M., Jaskolski,T., Klimarczyk,M., Kulinski,L., Koszela,A., Szczesniewicz,M., Sliwinska,M., Witkowski,P. and Olinski,R. (1998) Comparision of oxidative base damage in mitochondrial and nuclear DNA. Free Radic. Biol. Med., 24, 722–725. [DOI] [PubMed] [Google Scholar]
- 49.Anson R.M., Hudson,E. and Bohr,V.A. (2000) Mitochondrial endogenous oxidative damage has been overestimated. FASEB J., 14, 355–360. [DOI] [PubMed] [Google Scholar]
- 50.Higuchi Y. and Linn,S. (1995) Purification of all forms of HeLa cell mitochondrial DNA and assessment of damage to it caused by hydrogen peroxide treatment of mitochondria or cells. J. Biol. Chem., 270, 7950–7956. [DOI] [PubMed] [Google Scholar]
- 51.Beckman K.B. and Ames,B.N. (1996) Detection and quantification of oxidative adducts of mitochondrial DNA. Methods Enzymol., 264, 442–453. [DOI] [PubMed] [Google Scholar]
- 52.Bogenhagen D. and Clayton,D.A. (1974) The number of mitochondrial deoxyribonucleic acid genomes in mouse L and human HeLa cells. Quantitative isolation of mitochondrial deoxyribonucleic acid. J. Biol. Chem., 249, 7991–7995. [PubMed] [Google Scholar]
- 53.Yakes F.M. and Van Houten,B. (1997) Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc. Natl Acad. Sci. USA, 94, 514–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abete P., Ferrara,N., Cioppa,A., Ferrara,P., Bianco,S., Calabrese,C., Cacciatore,F., Longobardi,G. and Rengo,F. (1996) Preconditioning does not prevent postischemic dysfunction in aging heart. J. Am. Coll. Cardiol., 27, 1777–1786. [DOI] [PubMed] [Google Scholar]
- 55.Rolfe D.F., Hulbert,A.J. and Brand,M.D. (1994) Characteristics of mitochondrial proton leak and control of oxidative phosphorylation in the major oxygen-consuming tissues of the rat. Biochim. Biophys. Acta, 1188, 405–416. [DOI] [PubMed] [Google Scholar]
- 56.Fasman G. (2000) Handbook of Biochemistry and Molecular Biology. CRC, Cleveland Press, Cleveland, OH.
- 57.Poulsen H.E., Prieme,H. and Loft,S. (1998) Role of oxidative DNA damage in cancer initiation and promotion. Eur. J. Cancer Prev., 7, 9–16. [PubMed] [Google Scholar]
- 58.Atchley W.R., Wei,R. and Crenshaw,P. (2000) Cellular consequences in the brain and liver of age-specific selection for rate of development in mice. Genetics, 155, 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sprott R.L. (1991) Development of animal models of aging at the National Institute on Aging. Neurobiol. Aging, 12, 635–638 [DOI] [PubMed] [Google Scholar]