Abstract
Purpose
To establish prognostic genomic biomarkers for patients with metastatic clear cell renal cell carcinoma (ccRCC).
Materials and Methods
We identified 60 patients who presented with metastatic ccRCC at our institution between 2001 and 2015 and had genomic sequencing on their primary tumor. We pooled these patients with 107 other patients with the same inclusion criteria from three well-known public databases. Five commonly mutated genes were chosen for analysis: VHL, PBRM1, BAP1, SETD2, and KDM5C. Overall survival was estimated using the Kaplan-Meier method and the log-rank test was used for comparisons between groups.
Results
Median overall survival in the cohort was 2.5 years. Higher Fuhrman grade was associated with decreased median overall survival (p<0.001). Mutations in SETD2 (p=0.027) and KDM5C (p=0.019) were associated with reduced risk of death (HR 0.58 [95% CI 0.35-0.94] and HR 0.43 [95% CI 0.22-0.85] respectively). BAP1 mutations (p=0.008) were associated with increased risk of death (HR 1.81 [95% CI 1.16-2.83]. There were significantly more female patients with a BAP1 mutation than females in the overall cohort (p=0.001).
Conclusions
Mutations in BAP1 negatively affected overall survival, while SETD2 and KDM5C mutations were associated with prolonged overall survival in our pooled cohort of 167 metastatic ccRCC patients. Our results expand upon efforts at understanding genomic biomarkers in localized disease. Those efforts set the stage for our novel investigation examining associations of select recurrent somatic mutations in stage IV ccRCC patients.
Keywords: Renal Cell Carcinoma, Neoplasm metastasis, Prognosis, Mortality
Introduction
Clear cell renal cell carcinoma (ccRCC) is the most common and aggressive histologic subtype of renal cell carcinoma.[1] Metastatic disease is present in ∼33% of patients at time of presentation, with a 5-year survival rate of ∼12% for patients with distant metastases.[2]
The foundational genomic aberrations in ccRCC are understood to take place in concert with chromosome 3p loss.[3, 4] While VHL alterations are common in ccRCC, it has been shown that VHL alterations alone are inadequate for the development of ccRCC: other alterations are required for tumorigenesis.[5] Recurrently mutated genes have been found in several large ccRCC cohorts with the following frequencies: VHL—75-80%, Polybromo-1 (PBRM1)—35-45%, Set domain containing 2 (SETD2)—5-15%, and BRCA1 associated protein-1 (BAP1)—5-15%.[4, 6] We and others have reported associations between these genomic alterations and inferior clinical outcomes, mainly in the localized disease setting.[7-13] Several studies have attempted to use the mutation status of these genes to stratify patients and predict their risk of poor clinical outcomes.[14-16]
One such stratification scheme has been based on the mutation status of PBRM1 and BAP1 in patients with ccRCC. Joseph et al. reported the significance of these genes in a large cohort of mostly-localized ccRCC patients using immunohistochemistry. They found that patients with neither mutation had the best clinical outcomes, while patients with mutations in PBRM1, BAP1, or both, had sequentially worse outcomes.[10] Numerous other studies have identified inferior overall survival (OS) in patients carrying BAP1 mutations.[6, 14]
To our knowledge the relevance of these mutations in patients undergoing cytoreductive nephrectomy (CN) remains unknown. In our study, we analyze the effects of recurrent somatic mutations, in particular BAP1 and PBRM1, in patients' primary tumors and the mutations' associations with survival in the setting of ccRCC patients undergoing CN.
Methods
Patient Selection
Upon approval by the institutional review board at Memorial Sloan Kettering Cancer Center (MSKCC), we queried our prospectively collected institutional kidney cancer genomic database for patients with AJCC stage IV ccRCC between January 1st, 2001 and December 31st, 2015 (461 patients). This database includes pathological data including histology, grade, and stage, as well as clinical data including survival data. Of those queried patients, 70 patients had their primary tumors sequenced at time of CN, and were thus eligible for our analysis. We excluded seven patients who received systemic therapy prior to CN (n=63). Three patients underwent biopsy of their primary tumor without CN, and thus were excluded from our cohort (n=60). These patients represent a cohort that has not been previously reported. Most genomic information on these patients was provided via MSK-IMPACT™ (Integrated Mutation Profiling of Actionable Cancer Targets), a next generation sequencing modality using hybridization-based exon capture assays of select introns and commonly altered oncogenes and tumor suppressor genes[17]. Those samples not sequenced using MSK-IMPACT™ were sequenced using Sanger sequencing that targeted our five genes of interest: VHL, PBRM1, BAP1, SETD2, and KDM5C.
To expand our cohort for analysis, we pooled institutional data with three previously published, publicly available ccRCC cohorts.[4, 6, 18] Applying the same search criteria yielded 108 patients, but one was excluded for insufficient pathological information (n=107). Molecular profiling of these public cohorts included whole-genome sequencing, whole-exome sequencing, RNA sequencing, miRNA sequencing, DNA methylation arrays, and reverse phase protein arrays. Further sequencing details for the previously published cohorts are described within their respective methods.
Overall, our composite cohort consisted of 167 patients who presented with AJCC stage IV ccRCC and underwent CN with sequencing of their primary tumors.
Statistical analysis
A panel of five commonly mutated genes (VHL, PBRM1, BAP1, SETD2, KDM5C) was chosen for analysis based on the published literature indicating their high mutational frequencies [4, 6] and clinical significance in localized disease. [7, 9, 10, 19] Patient and disease characteristics were summarized using frequency and percentage for categorical variables, and median and range were used for continuous variables. Comparisons between the groups were conducted using Fisher's exact test for categorical variables and the Wilcoxon rank sum test for continuous variables. Follow-up time was calculated from the date of CN to death or last follow-up. The Kaplan-Meier method estimated OS and the log-rank test was used for survival comparisons between groups. Cox regression was used for univariable and multivariable analysis.
As the majority of patients in our cohort received systemic therapy after surgery, we investigated the possible influence of systemic treatment on OS. We conducted a sensitivity analysis for possible therapeutic influences using 80 patients in our cohort for whom postoperative first line systemic therapy data was available. Therapies were categorized as VEGF inhibitors or ‘other’, the latter including interleukin-2, interferon, mTOR inhibitors, gemcitabine/doxorubicin, and PD-1 inhibitors.
A p-value < 0.05 defined statistical significance for all analyses. All statistical analyses were conducted using R software version 3.2.5 (R Core Development Team, Vienna, Austria) including the ‘survival’ package.
Results
Demographics
Demographic and clinicopathological characteristics of our cohort of 167 patients are described in Table 1, along with univariable associations with OS. Median age was 60 years (range 39-84) and the majority were male (69.5%). Median follow-up time was 3 years (range 0.3-10.6); during follow-up, 105 patients died from any cause. Median OS was 2.5 years (95% confidence interval (CI): 2.0 – 3.4). On univariable analysis, the only characteristic found to be significantly associated with decreased OS was worsening Fuhrman grade of the primary tumor (p<0.001).
Table 1.
Baseline demographic, clinical, and pathologic characteristics and their univariable association with OS.
| Characteristic | Median OS, years (95% CI) | HR (95% CI) | p-value* | |
|---|---|---|---|---|
| Median age at diagnosis (range) | 60 (39–84) | |||
| Median cm maximum tumor dimension (range) | 9 (2–25) | NA | 0.976 | |
| No. sex (%) | 0.145 | |||
| Male | 116 (69.5) | 4.07 (0.38–3.27) | 0.74 (0.49–1.11) | |
| Female | 51 (30.5) | 3.06 (0.43–2.01) | 1.00 | |
| No. race (%) | 0.698 | |||
| White | 139 (83.2) | 2.93 (0.2–2.17) | 1.48 (0.2–10.68) | |
| Other | 3 (1.8) | 3.89 (1.64–1.57) | 1.00 | |
| NA | 25 (15) | |||
| No. primary tumor laterality (%) | 0.272 | |||
| Left | 80 (47.9) | 4.11 (0.47–3.29) | 1.00 | |
| Right | 79 (47.3) | 3.44 (0.37–2.32) | 1.25 (0.84–1.86) | |
| NA | 8 (4.8) | |||
| No. primary tumor grade (%) | <0.001 | |||
| 2 | 30 (18) | 5.25 (0.67–5.24) | 1.00 | |
| 3 | 62 (37.1) | 4.4 (0.55–3.66) | 1.38 (0.75–2.57) | |
| 4 | 75 (44.9) | 2.57 (0.32–1.57) | 2.66 (1.5–4.72) | |
| No. T stage (%) | 0.412 | |||
| T1 | 16 (9.6) | 4.69 (0.92–3.41) | 1.00 | |
| T2 | 20 (12) | 4.88 (0.86–6.1) | 1.04 (0.42–2.59) | |
| T3 | 112 (67.1) | 3.47 (0.33–2.17) | 1.58 (0.76–3.29) | |
| T4 | 15 (9) | 3.47 (0.82–2.79) | 1.46 (0.57–3.72) | |
| NA | 4 (2.4) | |||
| No. N stage (%) | 0.086 | |||
| N0 | 75 (44.9) | 3.92 (0.45–2.54) | 1.00 | |
| N1 | 24 (14.4) | 2.53 (0.58–1.19) | 1.65 (0.95–2.86) | |
| NX | 64 (38.3) | 3.92 (0.39–3.39) | 0.89 (0.58–1.37) | |
| NA | 4 (2.4) | |||
| No. KPS (%) | 0.652 | |||
| 60 | 2 (1.2) | 4.12 (1.4–4.12) | 1.00 | |
| 70 | 4 (2.4) | 2.71 (0.6–3.41) | 1.73 (0.23–13.11) | |
| 80 | 11 (6.6) | 4.18 (0.71–4.45) | 0.65 (0.12–3.6) | |
| 90 | 18 (10.8) | 3.52 (0.63–3.95) | 1.08 (0.21–5.47) | |
| 100 | 11 (6.6) | 4.27 (0.68–NA) | 0.56 (0.1–3.12) | |
| NA | 121 (72.5) | |||
| No. size | 0.245 | |||
| < 7 cm | 44 (26.3) | 4.2 (0.59–3.39) | 1.00 | |
| ≥ 7 cm | 123 (73.7) | 3.53 (0.33–2.08) | 1.31 (0.83–2.07) |
p-value from log-rank test.
Mutational Impact on Clinical Outcomes
Associations between mutation status and OS are shown in Table 2. Mutations in SETD2 (p=0.027) and KDM5C (p=0.019) were associated with a reduced risk of death, with hazard ratios (HR) of 0.58 (95% CI 0.35-0.94) and 0.45 (95% CI 0.23-0.89), respectively. BAP1 (p=0.008) mutations were associated with increased risk of death: HR 1.85 (95% CI 1.17-2.92). There was no significant association between PBRM1 and OS. Corresponding Kaplan-Meier plots for BAP1, PBRM1, SETD2, and KDM5C are shown in Figure 1.
Table 2.
Frequency of gene mutations overall, with univariable associations between mutations and OS.
| Characteristic | No. (%) | HR (95% CI) | p-value |
|---|---|---|---|
| VHL | 0.268 | ||
| WT | 65 (38.9) | 1 | |
| Mut | 102 (61.1) | 0.80 (0.54–1.18) | |
| PBRM1 | 0.494 | ||
| WT | 103 (61.7) | 1 | |
| Mut | 64 (38.3) | 0.87 (0.59–1.29) | |
| BAP1 | 0.008 | ||
| WT | 138 (82.6) | 1 | |
| Mut | 29 (17.4) | 1.85 (1.17–2.92) | |
| SETD2 | 0.027 | ||
| WT | 129 (77.2) | 1 | |
| Mut | 38 (22.8) | 0.58 (0.35–0.94) | |
| KDM5C | 0.019 | ||
| WT | 142 (85) | 1 | |
| Mut | 25 (15) | 0.45 (0.23–0.89) |
Figure 1.
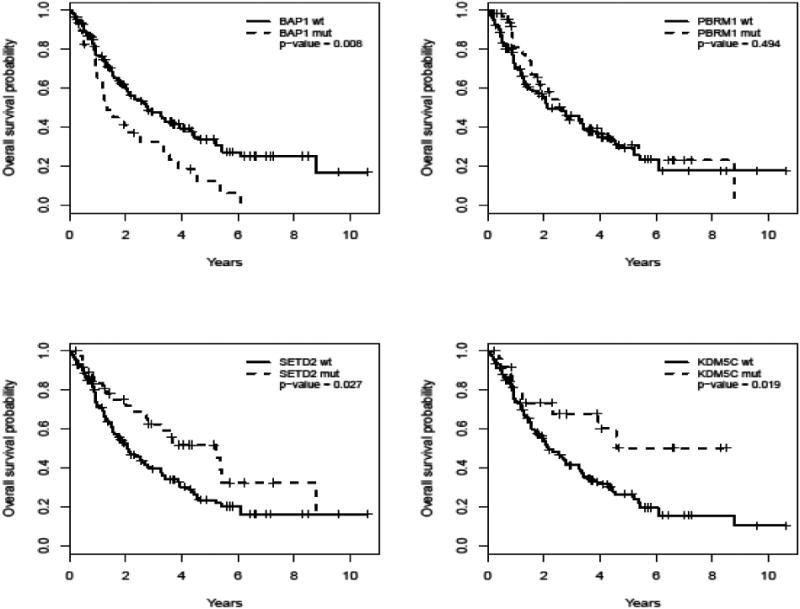
Kaplan-Meier survivorship curves for stage IV ccRCC patients, by mutation status across four recurrent somatic mutations (BAP1, PBRM1, SETD2, and KDM5C).
There was a significant association within our cohort between BAP1 mutations and sex. Females made up 24.6% of all those without a BAP1 mutation, but 58.6% of those with a BAP1 mutation (p=0.001). No significant association was found between BAP1 mutation status and changes in OS when stratified by sex.
BAP1 and PBRM1
To investigate the relationship of BAP1 and PBRM1 mutational status and OS, we stratified the patients in our cohort into four groups based on these mutations. The number of patients within each group, along with their clinicopathologic features, can be found in Table 3. Our analysis found no statistically significant association with OS when patients were stratified by their combined BAP1/PBRM1 mutational status, though the results may be trending towards significance (p=0.054) (Table 4 and Figure 2).
Table 3.
Stratification of patients into four groups on the basis of BAP1 and PBRM1 mutations.
| Characteristic | BAP1 wt & PBRM1 wt (n=80) | BAP1 wt & PBRM1 mut (n=58) | BAP1 mut & PBRM1 wt (n=23) | BAP1 mut & PBRM1 mut (n=6) | p-value* |
|---|---|---|---|---|---|
| Median age at Surgery (range) | 61 (39–84) | 60 (40–81) | 60 (43–76) | 64 (51–73) | 0.946 |
| No. sex (%) | <0.001 | ||||
| Male | 59 (73.8) | 45 (77.6) | 12 (52.2) | 0 | |
| Female | 21 (26.2) | 13 (22.4) | 11 (47.8) | 6 (100) | |
| No. race (%) | 0.626 | ||||
| White | 72 (90) | 40 (69) | 21 (91.3) | 6 (100) | |
| Other | 1 (1.2) | 2 (3.4) | 0 | 0 | |
| NA | 7 (8.8) | 16 (27.6) | 2 (8.7) | 0 | |
| Median cm maximum pathological tumor diameter (range) | 9 (2–25) | 8 (2.4–19) | 10 (2.7–15) | 8.75 (7.5–17.5) | 0.154 |
| No. T Stage (%) | NA | ||||
| T1 | 5 (6.2) | 9 (15.5) | 2 (8.7 | 0 | |
| T2 | 7 (8.8) | 9 (15.5) | 3 (13) | 1 (16.7) | |
| T3 | 53 (66.2) | 37 (63.8 | 17 (73.9) | 5 (83.3) | |
| T4 | 12 (15) | 3 (5.2) | 0 | 0 | |
| NA | 3 (3.8) | 0 | 1 (4.3) | 0 | |
| No. N Stage (%) | NA | ||||
| N0 | 32 (40) | 31 (53.4) | 9 (39.1) | 3 (50) | |
| N1 | 10 (12.5) | 7 (12.1) | 6 (26.1) | 1 (16.7) | |
| NX | 35 (43.8) | 20 (34.5) | 7 (30.4) | 2 (33.3) | |
| NA | 3 (3.8) | 0 | 1 (4.3) | 0 | |
| No. primary tumor grade (%) | 0.011 | ||||
| 2 | 12 (15) | 17 (29.3) | 1 (4.3) | 0 | |
| 3 | 32 (40) | 23 (39.7) | 6 (26.1) | 1 (16.7) | |
| 4 | 36 (45) | 18 (31) | 16 (69.6) | 5 (83.3) |
P-value is from Fisher's exact test when categorical and Wilcoxon rank sum test when continuous.
Table 4. Univariable association between groups formed by the combination of BAP1 and PBRM1 mutations and OS.
| Characteristic | No. (%) | HR (95% CI) | p-value |
|---|---|---|---|
| BAP1 & PBRM1 | 0.054 | ||
| BAP1 wt & PBRM1 wt | 80 (47.9) | 1.00 | |
| BAP1 wt & PBRM1 mut | 58 (34.7) | 0.97 (0.62–1.51) | |
| BAP1 mut & PBRM1 wt | 23 (13.8) | 1.95 (1.14–3.34) | |
| BAP1 mut & PBRM1 mut | 6 (3.6) | 1.46 (0.58–3.67) |
Figure 2.
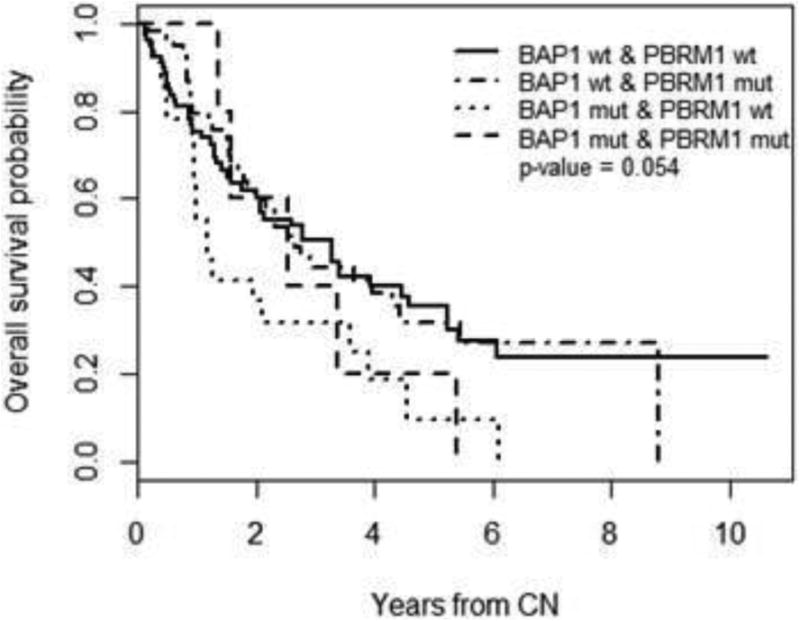
Kaplan-Meier curves for stage IV ccRCC patients stratified across 4 possible combinations of BAP1/PBRM1 mutation statuses.
Effects of First-line Therapy
Subset analysis among 80 patients with available first-line systemic therapy data showed that there appeared to be no statistically significant difference with respect to OS between the two treatment groups (p=0.160). Due to sample size restraints (54 patients on VEGF inhibitors, 26 on ‘other’ therapies), individual analysis by therapeutic class was not possible.
Discussion
To our knowledge this is the first analysis of the role of genomic alterations on clinical outcomes among a cohort of cytoreductive ccRCC patients. In this study we evaluated associations between select recurrent somatic mutations and OS for cytoreductive patients. We found BAP1 mutations were associated with inferior OS, whereas SETD2 and KDM5C were associated with improved OS.
Using a BAP1/PBRM1 stratification scheme for prognosis, we found no statistically significant difference in OS according to the four subgroups based on BAP1 and PBRM1 mutations. This finding is distinct from a recent analysis among patients with mostly localized tumors.[10] This result is likely due to the fact that PBRM1 mutations did not have prognostic value in this cohort of cytoreductive patients. It is also worth noting that patients with PBRM1 loss may possess some element of clinical benefit from their mutational status, as being a PBRM1 mutant often precludes the existence of BAP1 mutations, given these two mutations' tendency towards mutual exclusivity.[8, 19] Our data showed a similar tendency amongst males, further strengthening this possibility. Amongst females, no exclusivity was seen within a limited cohort size, limiting our analysis.
Interestingly, females made up a significantly disproportionate number of patients with BAP1 mutations than they did BAP1 wild type or the cohort overall. A previous study suggested that BAP1 mutations only carry significance for decreased OS among females, as well as increased frequency between female sex and BAP1 mutations.[20] However, our analysis did not see similar results between sex, BAP1, and survival within our patient population. These findings warrant further investigation within both the localized disease and cytoreductive settings.
Mutations in SETD2 and KDM5C conferred improved OS in our cohort. One possible reason for a survival advantage in patients with KDM5C mutations may be an improved response to VEGF inhibitor therapy. We have recently published data showing prolonged progression-free survival among these patients (20.6 months for patients with KDM5C mutations vs 8.3 months for KDM5C wild-type),[21] and other recent research has yielded similar results [22]. In regard to SETD2 mutations, previous studies have indicated these mutations are associated with advanced or recurrent disease.[23] However, in our study, SETD2 mutated patients had a survival advantage. Explanations for this include the natural history of these patients' disease (i.e. patients with low volume, metastatic site, or stable metastatic disease) or an unknown benefit from a specific systemic therapy. Further research is needed to confirm this phenomenon and determine its exact nature.
The frequency of patients with VHL mutations in our study (61.1%) is less than what is commonly found in other large cohorts of ccRCC patients (74%) [18]. This result warrants further investigation, as the sequencing coverage depth of our pooled cohort was improved by including MSK-IMPACT. MSK-IMPACT's depth of coverage ranged from 300×-600× within our cohort, while the public studies included in our pooled cohort posses depths of coverage ranging from approximately 50×-130×, depending on the analysis being done.
Our study is not without limitations. Our institutional results (60 patients) are from a tertiary referral center and as such, may not accurately reflect the more general pathological and clinical course of stage IV ccRCC patients outside our institution. We did not adjust for multiple testing within our analysis because our analysis was exploratory in nature. As such, any results found are intended as hypothesis-generating findings, for future studies. Our study also does not account for other possible driver mutations present in ccRCC at lower frequencies (e.g. TP53, PIK3CA, NF2), which could further stratify our cohort. In testing for significance between systemic therapies and OS, we found no significant association between prolonged OS and either the VEGF inhibitor or ‘other’ category. This result allows us to accept our conclusions about the effects of a particular mutation more confidently, but we still must remain concerned that our data is influenced by the effects of therapy. Furthermore, we were not able to take advantage of the MSKCC risk score [24] or Heng criteria [25] due to a lack of complete laboratory data. Additionally, different genome sequencing platforms were used across our pooled cohort. This runs the risk of overestimation or underestimation of the true frequencies of respective mutations, particularly when compared across different cohorts. Tumor heterogeneity, a well-known concern for evaluating biomarkers, particularly in ccRCC, [26] could have impacted our results. At present, current employment of genome sequencing prevents the practical utility of multi-site sampling, leaving single-site sampling as the most practical and best available choice. We also were unable to stratify our pooled cohort by sarcomatoid subtype, a critically impactful prognosticator of OS in the setting of metastatic ccRCC [27].
Conclusion
In our study of 167 cytoreductive ccRCC patients, we found that mutations in BAP1 negatively affected OS, while SETD2 and KDM5C mutations were associated with prolonged OS. Our results expand upon previously described efforts at understanding genomic biomarkers in localized disease. Those efforts set the stage for our novel investigation examining the specific association of selected recurrent somatic mutations in stage IV ccRCC patients ideally in the clinical trial setting. The favorable prognostic influence of PBRM1 mutations seen in multiple studies of localized ccRCC appears to be lost in the Stage IV setting. While examining the combined influence of PBRM1 and BAP1 mutations, the BAP1 mutations appeared to be the catalyst of worsening OS among this cohort of cytoreductive ccRCC patients.
Acknowledgments
Supported by the Sidney Kimmel Center for Prostate and Urologic Cancers and the NIH/NCI Cancer Center Support Grant P30 CA008748 and Ruth L. Kirschstein National Research Service Award T32CA082088.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fitzmaurice C, et al. The Global Burden of Cancer 2013. JAMA Oncol. 2015;1(4):505–27. doi: 10.1001/jamaoncol.2015.0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.SEER Stat Fact Sheets: Kidney and Renal Pelvis Cancer. 03/17/2016] Available from: http://seer.cancer.gov/statfacts/html/kidrp.html.
- 3.Hakimi AA, Pham CG, Hsieh JJ. A clear picture of renal cell carcinoma. Nat Genet. 2013;45(8):849–50. doi: 10.1038/ng.2708. [DOI] [PubMed] [Google Scholar]
- 4.The Cancer Genome Atlas Research, N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Young AP, et al. VHL loss actuates a HIF-independent senescence programme mediated by Rb and p400. Nat Cell Biol. 2008;10(3):361–9. doi: 10.1038/ncb1699. [DOI] [PubMed] [Google Scholar]
- 6.Sato Y, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013;45(8):860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 7.Cowey CL, Rathmell WK. VHL gene mutations in renal cell carcinoma: role as a biomarker of disease outcome and drug efficacy. Curr Oncol Rep. 2009;11(2):94–101. doi: 10.1007/s11912-009-0015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pena-Llopis S, et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet. 2012;44(7):751–9. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piva F, et al. BAP1, PBRM1 and SETD2 in clear-cell renal cell carcinoma: molecular diagnostics and possible targets for personalized therapies. Expert Rev Mol Diagn. 2015;15(9):1201–10. doi: 10.1586/14737159.2015.1068122. [DOI] [PubMed] [Google Scholar]
- 10.Joseph RW, et al. Clear Cell Renal Cell Carcinoma Subtypes Identified by BAP1 and PBRM1 Expression. J Urol. 2016;195(1):180–7. doi: 10.1016/j.juro.2015.07.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dagher J, et al. Wild-type VHL Clear Cell Renal Cell Carcinomas Are a Distinct Clinical and Histologic Entity: A 10-Year Follow-up. European Urology Focus. 2016;1(3):284–290. doi: 10.1016/j.euf.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 12.Ho TH, et al. High-resolution profiling of histone h3 lysine 36 trimethylation in metastatic renal cell carcinoma. Oncogene. 2016;35(12):1565–74. doi: 10.1038/onc.2015.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gossage L, et al. Clinical and pathological impact of VHL, PBRM1 BAP1, SETD2, KDM6A, and JARID1c in clear cell renal cell carcinoma. Genes Chromosomes Cancer. 2014;53(1):38–51. doi: 10.1002/gcc.22116. [DOI] [PubMed] [Google Scholar]
- 14.Hakimi AA, et al. Adverse outcomes in clear cell renal cell carcinoma with mutations of 3p21 epigenetic regulators BAP1 and SETD2: a report by MSKCC and the KIRC TCGA research network. Clin Cancer Res. 2013;19(12):3259–67. doi: 10.1158/1078-0432.CCR-12-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–9. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hakimi AA, et al. Clinical and pathologic impact of select chromatin-modulating tumor suppressors in clear cell renal cell carcinoma. Eur Urol. 2013;63(5):848–54. doi: 10.1016/j.eururo.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng DT, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn. 2015;17(3):251–64. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scelo G, et al. Variation in genomic landscape of clear cell renal cell carcinoma across Europe. Nat Commun. 2014;5:5135. doi: 10.1038/ncomms6135. [DOI] [PubMed] [Google Scholar]
- 19.Kapur P, et al. Effects on survival of BAP1 and PBRM1 mutations in sporadic clear-cell renal-cell carcinoma: a retrospective analysis with independent validation. Lancet Oncol. 2013;14(2):159–67. doi: 10.1016/S1470-2045(12)70584-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ricketts CJ, Linehan WM. Gender Specific Mutation Incidence and Survival Associations in Clear Cell Renal Cell Carcinoma (CCRCC) PLoS One. 2015;10(10):e0140257. doi: 10.1371/journal.pone.0140257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsieh JJ, et al. Genomic Biomarkers of a Randomized Trial Comparing First-line Everolimus and Sunitinib in Patients with Metastatic Renal Cell Carcinoma. European Urology. 2016 doi: 10.1016/j.eururo.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho TH, et al. Correlation Between Molecular Subclassifications of Clear Cell Renal Cell Carcinoma and Targeted Therapy Response. European Urology Focus. 2016;2(2):204–209. doi: 10.1016/j.euf.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, et al. Prognostic value of SETD2 expression in patients with metastatic renal cell carcinoma treated with tyrosine kinase inhibitors. J Urol. 2016 doi: 10.1016/j.juro.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 24.Motzer RJ, et al. Prognostic factors for survival in previously treated patients with metastatic renal cell carcinoma. J Clin Oncol. 2004;22(3):454–63. doi: 10.1200/JCO.2004.06.132. [DOI] [PubMed] [Google Scholar]
- 25.Heng DY, et al. Prognostic factors for overall survival in patients with metastatic renal cell carcinoma treated with vascular endothelial growth factor-targeted agents: results from a large, multicenter study. J Clin Oncol. 2009;27(34):5794–9. doi: 10.1200/JCO.2008.21.4809. [DOI] [PubMed] [Google Scholar]
- 26.Sankin A, et al. The impact of genetic heterogeneity on biomarker development in kidney cancer assessed by multiregional sampling. Cancer Med. 2014;3(6):1485–92. doi: 10.1002/cam4.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shuch B, et al. Impact of pathological tumor characteristics in patients with sarcomatoid renal cell carcinoma. BJU International. 109:1600–1606. doi: 10.1111/j.1464-410X.2011.10785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


