Abstract
Background and Aims
Angiogenesis is associated with neoplastic progression of Barrett’s esophagus (BE). Volumetric optical coherence tomography angiography (OCTA) visualizes subsurface microvasculature without exogenous contrast agents. We investigated the association of OCTA microvascular features with low-grade dysplasia (LGD) and high-grade dysplasia (HGD).
Methods
Fifty-two patients undergoing BE surveillance or endoscopic eradication therapies for dysplasia were imaged using volumetric OCTA and corresponding histological diagnoses obtained, to yield 97 data sets (non-dysplastic BE (NDBE): N=74; LGD: N=10; HGD: N=13). After evaluating OCTA image quality, 54 datasets (NDBE: N=35; LGD: N=8; HGD: N=11) from 32 patients were used to develop a training and reading protocol. The association of abnormal vessel branching and heterogeneous vessel size with LGD/HGD, and a regular honeycomb vessel pattern with NDBE was investigated.
Results
Blinded OCTA reading of 41 OCTA datasets (NDBE: N=27; LGD: N=7; HGD: N=7) was performed by readers with various levels of OCT/OCTA experience including 3 OCT trainees, one gastroenterologist, and 2 gastroenterology fellows. Among the six readers, OCTA features of abnormal vessel branching and heterogeneous vessel size had an overall 94% sensitivity (95% CI, 89–99) and 69% specificity (95% CI, 62–76) for differentiating LGD/HGD vs. NDBE with a mean reading time of 45 seconds per data set and moderate (kappa: 0.58) interobserver agreement.
Conclusions
Volumetric en face OCTA imaging enables rapid examination of depth resolved microvascular features with near-microscopic resolution. OCTA can visualize microvascular features associated with LGD/HGD with high accuracy, which motivates new technological advances as well as future studies investigating the diagnostic performance of OCTA.
Keywords: Barrett’s esophagus, dysplasia, optical coherence tomography, angiography, angiogenesis
INTRODUCTION
Barrett’s esophagus (BE) is a precursor in the progression of esophageal adenocarcinoma (EAC), which is among the most lethal diseases with a 5-year survival rate <20%1. EAC accounts for the majority of esophageal cancers and its incidence has increased ~300% to 500% in the last 40 years2, 3. Neoplastic progression from BE to EAC involves a multi-step process from non-dysplastic BE (NDBE) to low-grade dysplasia (LGD), high-grade dysplasia (HGD), and finally EAC4. The incidence of EAC is significantly increased in patients with any grade of dysplasia5, and HGD is associated with 10% to 60% increased risk of developing into EAC within 3 to 5 years4, 6, 7. Although various advanced endoscopic imaging modalities have been widely investigated, detecting dysplasia in BE with high diagnostic accuracy remains a challenge8.
Angiogenesis is associated with tumor progression, and vascular alterations often precede neoplastic transformation, suggesting that changes in vascular patterns may be indicators for early stage neoplasms9–11. Increased microvessel density in the progression from NDBE to LGD and HGD was recently reported based on ex vivo pathology specimens12. Narrow band imaging (NBI)13, 14 and confocal laser endomicroscopy (CLE)15, 16 were used to investigate microvascular changes as a potential marker for dysplasia. However, NBI has limited resolution and only visualizes surface vascular patterns. CLE has a limited field of view (<0.2 mm2), is less suited for surveying wide areas of the esophagus and requires contrast agents such as fluorescein17.
Endoscopic optical coherence tomography (OCT) provides near-microscopic, real-time, volumetric imaging of esophageal mucosa with an imaging depth of 1 to 2 mm18, 19. Previous studies investigated structural features for detecting dysplasia using cross-sectional OCT images, but diagnostic accuracy and interobserver agreement were limited20–22. OCT angiography (OCTA) is an extension of OCT, which can visualize subsurface 3-dimensional (3D) microvasculature without requiring contrast agents19, 23. However, the limited imaging speed and optical scanning instability in previous endoscopic OCT systems have made clinical OCTA difficult. Recently, our group demonstrated endoscopic OCTA imaging of subsurface microvasculature in the human esophagus using an ultrahigh speed endoscopic OCT system and micromotor imaging catheters that address these limitations24. Compared with other endoscopic imaging modalities providing vascular contrast, OCTA can image a larger area than CLE and does not require contrast agent administration. Furthermore, OCTA allows subsurface imaging of the esophagus with higher imaging resolution than NBI. Therefore, OCTA is a promising technique for assessing mucosal microvascular features of dysplasia. In this study, we investigated the feasibility of using endoscopic OCTA to differentiate dysplasia from NDBE. We performed a pilot study investigating microvascular features associated with NDBE and dysplasia in OCTA datasets obtained from patients with BE and developed OCTA criteria to detect LGD/HGD. Preliminary results on the accuracy of the OCTA criteria using blinded reading of the OCTA datasets by multiple readers are reported.
METHODS
Patient enrollment
The imaging procedures were performed at the Veteran Affairs Boston Healthcare System (VABHS, Jamaica Plain Campus) with approvals from the institutional review boards at VABHS, Harvard Medical School, and Massachusetts Institute of Technology. Fifty-two patients undergoing BE surveillance or endoscopic eradication therapy (EET, including radiofrequency ablation, endoscopic mucosal resection (EMR) or cryospray ablation) for dysplasia were recruited from March 2014 to February 2016.
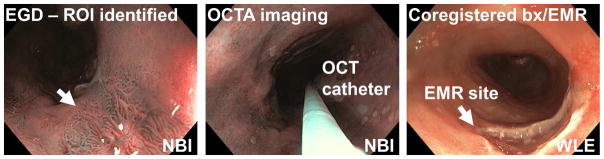
After providing written informed consent, patients underwent standard EGD. Regions of interest identified by white light endoscopy (WLE) or NBI per standard clinical practice were imaged with endoscopic OCTA using a micromotor imaging catheter introduced through one instrument channel of a high-definition, dual-channel endoscope (GIF-2TH180, Olympus). After OCTA imaging, either biopsies using standard biopsy forceps or EMRs (Duette, Cook Medical) were taken from the imaged sites for histopathology analysis based on clinical indication (Figure 1). The spatial location of the specimens (eg, clock and longitudinal location) was documented per standard clinical practice and the histopathological diagnosis was used to classify the associated OCTA dataset. This protocol enabled accurate registration of the OCTA image data with histology, but the OCT catheter had limited area coverage compared with imaging balloons25.
Figure 1.
Flow chart illustrating the OCT/OCTA imaging procedure and collection of the corresponding histology.
OCTA datasets corresponding to NDBE were obtained from patients undergoing BE surveillance who had all histopathological diagnoses negative for dysplasia, or from patients undergoing EET with prior dysplasia diagnosis, who had all histopathological diagnoses negative for dysplasia in the same visit as the OCTA imaging session. All histopathological diagnoses from any neighboring regions were required to be negative for dysplasia in order to assure the integrity of the OCTA datasets corresponding to NDBE. OCTA datasets corresponding to dysplasia were obtained from patients with a history of dysplasia having histopathological diagnoses positive for dysplasia, or from patients undergoing BE surveillance who had incidental histopathological diagnoses positive for dysplasia. Overall, 97 OCTA datasets with corresponding histological diagnoses (NDBE: N=74; LGD: N=10; HGD: N=13) were collected from 52 patients (NDBE: N=41; LGD: N=7: HGD: N=4, based on the baseline pathology) enrolled to the study.
Endoscopic OCT imaging system
The study used a prototype, ultrahigh speed endoscopic OCT instrument and micromotor imaging catheters to perform volumetric imaging of the esophageal microvasculature. The technology has been described in detail previously24, 26. Briefly, the OCT system and micromotor catheter had an imaging speed of 600,000 depth (axial) scans per second and a frame rate of 400 frames per second, >10 times faster than commercially available endoscopic OCT systems. The axial and lateral image resolution was ~8 μm and ~20 μm in tissue, respectively. Each volumetric OCT acquisition imaged an area of 10 mm (circumferential) x 16 mm (longitudinal) in ~8 seconds using a helical (pullback) optical scan pattern.
Endoscopic OCT angiography (OCTA) and data visualization
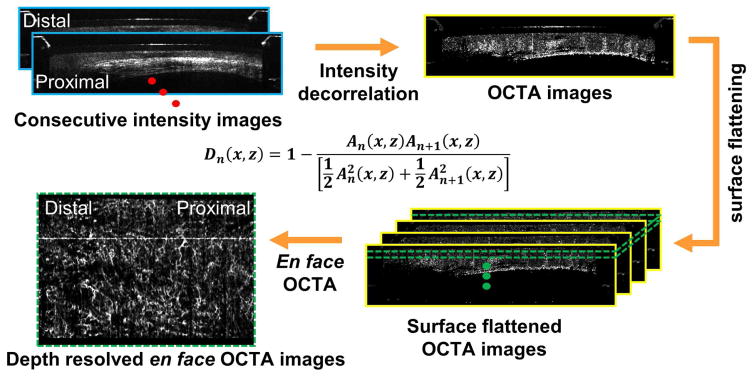
OCTA visualizes microvasculature by using motion contrast, without requiring exogenous contrast agents. Figure 2 shows a flow chart summarizing the image processing steps for generating depth resolved en face OCT angiography (OCTA) images26, 27. Before computing the volumetric OCTA datasets, a motion correction algorithm was applied offline in post-processing to remove the nonuniform rotation distortion (NURD)27. Volumetric OCTA datasets were generated afterward by computing pixel-by-pixel differences/variations of the OCT signal intensities between consecutive OCT frames in NURD-corrected volumetric OCT dataset. Moving erythrocytes in microvasculature cause the OCT signal intensity to vary with time, which can be quantified by calculating a decorrelation (D). Conversely, static tissue has a constant OCT signal. Depth resolved en face OCTA images were generated by using mean projection over a depth range of 100 μm, at various depth levels beneath the tissue surface. Faster vs. slower blood flows are associated with higher vs. lower OCTA decorrelation signals, however it is important to note that OCTA does not measure absolute flow. In addition, OCTA is more sensitive to tissue motion than OCT and respiration or cardiac motion can generate artifacts, compromising data quality26. The tissue motion can potentially result in varying catheter-tissue contact, which decreases the effective OCTA imaging coverage. Furthermore, the excessive pressure exerted by the catheter over the tissue surface can suppress the blood flow and hence results in low OCTA signals.
Figure 2.
Flow chart summarizing the image processing steps for generating depth-resolved en face OCT angiography (OCTA) images from the structural volumetric OCT dataset. An is linear OCT signal amplitude in individual OCT frames. A nonuniform rotational distortion (NURD) correction algorithm was used before calculating decorrelation between consecutive cross-sectional OCT images.
OCTA reading criteria and protocol
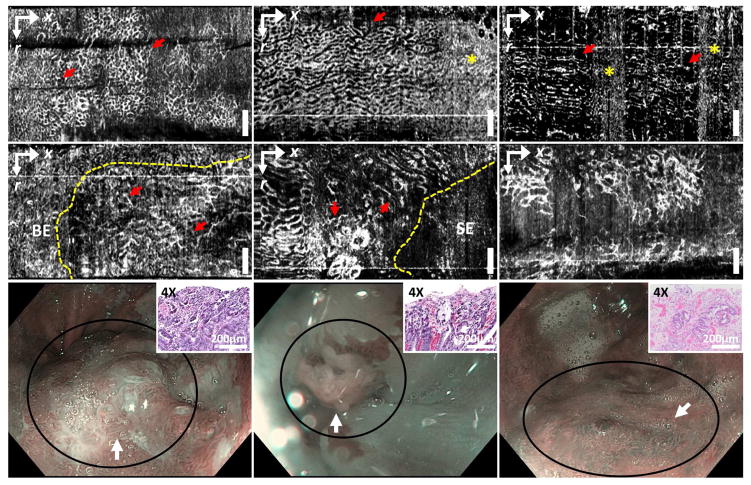
The 97 OCTA datasets from 52 patients were collected and reviewed to assess image quality by a single investigator, not involved in the reading. A total of 54 datasets (NDBE: N = 35; LGD: N = 8; HGD: N = 11) from 32 patients (NDBE: N = 22; LGD: N = 6: HGD: N = 4, based on the baseline pathology) were retained for training and reading, whereas 43 OCTA datasets (NDBE: N = 39; LGD: N = 2; HGD: N = 2) were not used because of inadequate image quality due to artifacts from tissue motion, decreased imaging coverage from varying catheter-tissue contact, or excessive pressures shown in the example OCTA images (Supplemental figure 1). The inadequate image quality resulted in part because OCTA images were generated by post processing and were not available in real time during endoscopy. Most of the dysplasia OCTA datasets exhibited adequate image quality due to the attempts to carefully perform OCTA imaging over the regions exhibiting irregular mucosa or vascular pattern under WLE or NBI. The 54 volumetric OCTA datasets were examined by the same investigator to identify features associated with dysplasia in order to develop the OCTA reading criteria. Details are provided in the supplemental section (Development of the OCTA criteria – initial learning phase). A honeycomb/oval-like microvascular pattern of varying size was observed in NDBE OCTA datasets (Figures 3A–C). Although the size/shape of the honeycombs could vary along the longitudinal (pullback) direction due to motion artifacts, the distribution of the honeycombs was in general relatively regular. In the LGD/HGD datasets, OCTA exhibited microvasculature features of (1) abnormal vessel branching with crowding or corkscrew appearance, and (2) heterogeneous vessel size, i.e. presence of vessels with different calibers (Figures 3D–F), similar to those previously reported with magnification NBI13, 14. However, unlike NBI, OCTA enables volumetric visualization of the subsurface microvasculature.
Figure 3. En face.
OCT angiography (OCTA) images of (A–C) non-dysplastic BE (NDBE) and (D–F) dysplastic BE (LGD: F; HGD: D, E) from ~180 μm beneath the tissue surface. NDBE exhibited regular honeycomb microvascular pattern (arrows, A–C), similar to previously reported with NBI. The shape of the honeycomb features may be compressed or stretched along the longitudinal direction due to motion artifacts. High decorrelation noise from physiological motion can also be observed (stars, B, C). Abnormal vascular features including (1) abnormal vessel branching (arrows, D), (2) heterogeneous vessel size (arrows, E) or both (F) were shown. OCTA allowed delineation of the boundary between abnormal microvasculature and neighboring non-dysplastic regions (dashed line, D, E). (G–I) NBI images near the imaged sites (D–F) respectively (circles). Scale bars: 1 mm. SE: squamous epithelium. Insets (G, H, I): H&E stained histopathology images of the specimens from the imaged sites corresponding to the histological diagnosis of HGD, HGD, and LGD, respectively. r: rotary direction; x: longitudinal (pullback) direction.
Training/validation session
The two-feature OCTA reading criteria was developed based on these observations and independently validated by blinded readers with various levels of OCT/OCTA experience, including: 3 OCT trainees, one gastroenterologist, and 2 gastroenterology (GI) fellows. Before the validation reading, each reader received a training session including a ~40 minute interactive presentation consisting of six volumetric OCTA datasets. During the training session, examples of OCTA datasets corresponding to NDBE and LGD/HGD that exhibited characteristic microvascular features, as well as datasets having minor artifacts from respiration or cardiac motion, or a slightly decreased imaging coverage from varying catheter-tissue contact were presented (Figure 3).
The training session was followed by a pretest with seven volumetric OCTA datasets before the validation reading. During the pretest, each reader assessed the presence of abnormal microvascular features in the volumetric OCTA datasets using the software viewer ImageJ (National Institutes of Health) following the workflow described in the supplemental section (Workflow of the OCTA reading protocol). The pretest results of individual readers were immediately reviewed and discussed with an investigator to ensure that each reader understood the reading criteria before performing the validation reading. For the validation, the readers followed the same protocol as the pretest, but without discussion of results. In addition, after reading all volumetric OCTA datasets sequentially, each reader was asked to review the datasets and allowed to adjust their assessments made during the initial reading. This consolidated readers’ understanding on the OCTA features from the initial reading and therefore reduced possible inconsistency in the assessment of features. Furthermore, during the review process (final reading), each reader was asked to rate the confidence level of his/her feature assessment in each dataset as “high” or “low.” During the pretest and validation, readers were blinded to the endoscopic and histopathological findings. The reading time and individual readers’ confidence level for each OCTA dataset were recorded.
Statistical analysis
Measures of the OCTA criteria accuracy including the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated for each reader separately as well as all six readers combined, along with the binomial 95% confidence interval. The interobserver agreement on the assessment of abnormal microvascular features as well as the accuracy among six readers was calculated using unweighted kappa statistics28. The level of agreement was interpreted as a kappa value, where 0.41 to 0.60 was defined as moderate agreement, and 0.61 to 0.80 as substantial agreement29.
RESULTS
Baseline characteristics
Table 1 presents the demographics and baseline characteristics of the patient enrollment. After excluding the datasets with inadequate image quality, 54 volumetric OCTA datasets remained from 32 patients (all male, age (y) 67±8). A median of 1 (range: 1–7) OCTA dataset was obtained per patient. 14 out of 35 NDBE OCTA datasets (40%) were from patients without a history of dysplasia or EET treatment. The number of the OCTA datasets corresponding to NDBE and LGD/HGD pathologies for the training/pretest/validation sessions were N=4, 4, 27, and N=2, 3, 14, respectively (Supplemental table 1).
Table 1.
Patient demographics and baseline characteristics.
| Patient characteristics (N = 32) | |
| Age, mean (± SD) | 67 (8) |
| Sex, male, no. (%) | 32 (100) |
| BMI, mean (± SD) | 29 (7.1) |
| Race, white, no. (%) | 30 (94) |
| Baseline pathology | |
| NDBE, subjects, no. (%) | 22 (68.8) |
| LGD/HGD, subjects, no. (%) | 6/4 (31.2) |
| Length of BE (cm) | |
| circumferential extent (C), mean (± SD) | 3.3 (4.0) |
| maximum extent (M), mean (± SD) | 4.7 (4.0) |
| Presence of Hiatal Hernia (HH) (N=32) | |
| Subjects, no. (%) | 28 (87.5) |
| HH length (cm), mean (± SD) | 2.1 (2.2) |
| Number of biopsy/EMR-correlated OCTA dataset per subject | |
| median (range) | 1 (1–7) |
| Biopsy/EMR specimen characteristics (N=54) | |
| NDBE, no. (%) | 35 (64.8) |
| from patients with prior EET treatment history, no. (%) | 21 (60) |
| from patients without prior EET treatment history, no. (%) | 14 (40) |
| LGD/HGD, no. (%) | 8/11 (35.2) |
SD, standard deviation; EMR, endoscopic mucosal resection; EET: endoscopic eradication therapy (including EMR, radiofrequency ablation (RFA), and cryospray ablation (CSA)).
Diagnostic performance of the OCTA criteria
The overall diagnostic performance of individual microvascular features associated with dysplasia is summarized in Table 2. A scoring index was used to assess the accuracy of the OCTA criteria by applying different thresholds. A score of 1 or 2 was assigned if one or two of the features were present, respectively. A score threshold of ≥1 resulted in an overall 94% (95% CI, 89–99) sensitivity, 69% (95% CI, 62–76) specificity and 96% (95% CI, 92–99) NPV. The accuracies of the OCTA criteria for individual readers are summarized in Table 3. Table 4 lists the number of datasets corresponding to the histopathological diagnosis of NDBE/LGD/HGD assessed by readers with high confidence. The majority of the LGD/HGD pathologies were assessed with high confidence and low confidence readings were mostly associated with false positive assessment of NDBE as LGD/HGD. For the datasets that were read with high confidence, an overall 93% (95% CI, 87–99) sensitivity, 81% (95% CI, 75–88) specificity and 95% (95% CI, 92–99) NPV was obtained (Table 2).
Table 2.
The performance of the OCTA features for detecting dysplasia in Barrett’s esophagus.
| Microvascular features | Sensitivity, % (95% CI) | Specificity, % (95% CI) | PPV, % (95% CI) | NPV, % (95% CI) |
|---|---|---|---|---|
| Feature 1 (branching) | 86.9 (79.7–94.1) | 77.2 (70.7–83.6) | 66.4 (57.5–75.2) | 91.9 (87.3–96.5) |
| Feature 2 (vessel size) | 73.8 (64.4–83.2) | 87.7 (82.6–92.7) | 75.6 (66.3–84.9) | 86.6 (81.3–91.8) |
| Overall | ||||
| score ≥2 | 66.7 (56.6–76.7) | 95.7 (92.5–98.8) | 88.9 (81.1–96.6) | 84.7 (79.5–89.9) |
| score ≥1 | 94.0 (89.0–99.1) | 69.1 (62.0–76.2) | 61.2 (52.8–69.6) | 95.7 (92.1–99.4) |
| High confidence | ||||
| score ≥2 | 69.9 (59.3–80.4) | 96.9 (93.9–99.9) | 92.7 (85.9–99.6) | 84.9 (79.1–90.7) |
| score ≥1 | 93.2 (87.4–98.9) | 81.3 (74.5–88.0) | 73.9 (64.9–82.9) | 95.4 (91.5–99.3) |
Overall = score (feature 1) + score (feature 2); PPV: positive predictive value; NPV: negative predictive value.
Table 3.
Accuracy of individual readers using OCTA features to detect dysplasia in Barrett’s esophagus.
| score ≥1 | Reader 1 | Reader 2 | Reader 3 | Reader 4 | Reader 5 | Reader 6 |
|---|---|---|---|---|---|---|
| Overall | ||||||
| Sensitivity, % | 92.9 | 92.9 | 92.9 | 100 | 92.9 | 92.9 |
| Specificity, % | 77.8 | 63 | 70.4 | 70.4 | 63 | 70.4 |
| PPV, % | 68.4 | 56.5 | 61.9 | 63.6 | 56.5 | 61.9 |
| NPV, % | 95.5 | 94.4 | 95 | 100 | 94.4 | 95 |
| High-confidence | ||||||
| Sensitivity, % | 90.9 | 92.3 | 92.9 | 100 | 91.7 | 91.7 |
| Specificity, % | 80.8 | 85 | 89.5 | 73.1 | 81 | 81.3 |
| PPV, % | 66.7 | 80 | 86.7 | 61.1 | 73.3 | 78.6 |
| NPV, % | 95.5 | 94.4 | 94.4 | 100 | 94.4 | 92.9 |
PPV: positive predictive value; NPV: negative predictive value.
Table 4.
Number of OCTA dataset with corresponding histopathological diagnosis that readers assess with high confidence
| High confidence | Reader 1 | Reader 2 | Reader 3 | Reader 4 | Reader 5 | Reader 6 | Overall |
|---|---|---|---|---|---|---|---|
| Histopathology | |||||||
| 27 NDBE, no. (%) | 26 (96.3) | 20 (74.1) | 19 (70.4) | 26 (96.3) | 21 (77.8) | 16 (59.3) | 128(79.0) |
| 7 LGD, no. (%) | 6 (85.7) | 7 (100) | 7 (100) | 6 (85.7) | 6 (85.7) | 6 (85.7) | 38(90.5) |
| 7 HGD, no. (%) | 5 (71.4) | 6 (85.7) | 7 (100) | 5 (71.4) | 6 (85.7) | 6 (85.7) | 35(83.3) |
| 41 Combined, no. (%) | 37 (90.2) | 33 (80.5) | 33 (80.5) | 37 (90.2) | 33 (80.5) | 28 (68.3) | 201 (81.7) |
Among the 6 readers, the interobserver agreement was moderate (kappa 0.58) using a threshold of ≥1. The interobserver agreement was moderate for abnormal vessel branching (kappa 0.53) and heterogeneous vessel size (kappa 0.43). A subset analysis using only NDBE datasets from patients without a history of dysplasia showed similar diagnostic performance to using only NDBE datasets from patients with a history of dysplasia/EET (p=0.71, Supplemental table 2). The mean reading time per volumetric OCTA dataset was 45 ± 25 seconds and breakdown of the overall time into initial and final reading times by individual readers is shown in Table 5.
Table 5.
Reading time per reader in the validation session.
| score ≥1 | Reader 1 | Reader 2 | Reader 3 | Reader 4 | Reader 5 | Reader 6 | Overall |
|---|---|---|---|---|---|---|---|
| Per OCTA dataset* | |||||||
| mean (SD), secs | 29 (19) | 59 (26) | 43 (26) | 33 (14) | 57 (26) | 46 (23) | 45 (25) |
| Overall reading time | |||||||
| initial reading, mins | 20 | 40 | 29 | 23 | 39 | 32 | 30.5 (7.5) |
| final reading, mins | 19 | 22 | 21 | 15 | 31 | 30 | 23.0 (5.7) |
analysis based on the initial readings; SD: standard deviation; secs: seconds; mins: minutes.
A similar distribution of OCTA microvascular features was observed between LGD and HGD using either abnormal vessel branching (p=0.33) or heterogeneous vessel size (p=0.62) (Supplemental table 3). Analysis of OCTA features associated with LGD and HGD subgroups shows 88.1% sensitivity and 69.1% specificity for differentiating LGD from NDBE and 100% sensitivity and 69.1% specificity for differentiating HGD from NDBE (Supplemental table 4). Finally, the accuracy for differentiating dysplasia from NDBE after the initial reading (before reviewing the datasets (final reading)) by individual readers was 88.1% sensitivity and 67.3% specificity using a score threshold of ≥1 (Supplement table 5). A moderate interobserver agreement (kappa: 0.46) was found on the initial readings among the 6 readers using a threshold of ≥1.
DISCUSSION
The detection of early dysplastic progression toward adenocarcinoma is a major unmet need in the assessment of Barrett’s esophagus. Among various advanced endoscopic imaging modalities, endoscopic OCTA has the unique advantage of visualizing subsurface microvasculature in three dimensions without contrast agents. Although previous endoscopic studies have demonstrated OCT vascular contrast using the Doppler effect, results were limited to measuring blood flow in large vessels either within or below the muscular layer19, 23 and did not visualize microvasculature in the superficial BE mucosa24. It was challenging to perform OCTA using earlier-generation endoscopic OCT systems because of insufficient imaging speeds. Thus, most OCT studies focused on investigating tissue architectural features. To the best of our knowledge, this is the first study investigating in vivo microvasculature identified by OCTA as a potential marker for pathology. In particular, this study focused on identifying OCTA features associated with dysplasia vs. NDBE.
A recent study using commercial endoscopic OCT, volumetric laser endomicroscopy (VLE), investigated dysplasia detection performance in 27 patients by analyzing the architectural features in multiple cross-sectional images from volumetric OCT datasets30. This study achieved 86% sensitivity and 88% specificity for detecting dysplasia on ex vivo EMR specimens using a new algorithm based on OCT structural features. However, further validation on in vivo volumetric OCT datasets is still required. In our study, volumetric structural OCT datasets with higher sampling density than commercial endoscopic OCT technology, co-registered to the volumetric OCTA datasets, were also available. However, the optimal method for reading volumetric structural OCT data is complex and still under investigation30, 31. Therefore, our study focused on OCTA and investigated the accuracy for differentiating dysplasia from NDBE using microvascular features.
Given the difficulties in diagnosing LGD by imaging as well as histopathology, in the majority of previously reported endoscopic imaging studies LGD was either categorized together with NDBE or was excluded from the study. During the development of the OCTA criteria, we observed that OCTA can differentiate LGD from NDBE in the cases collected for this study. We also observed that OCTA features associated with LGD were similar to those associated with HGD (Supplemental table 3). Furthermore, the pathological diagnosis of all LGD cases was made by a specialized pathologist with >15 years’ experience in GI pathology. A third party confirmation from expert referral centers was obtained when necessary. Therefore, in this study we have grouped LGD cases together with HGD, given the malignant potential of confirmed LGD that necessitates RFA treatment32. Nevertheless, we also investigated the accuracy of using OCTA to differentiate LGD from NDBE independently and showed comparable performance to HGD vs. NDBE (Supplemental table 4). However, the sample size is small and further larger scale studies are warranted.
Due to the limited sample size and readers’ varying OCT/OCTA experience, individual readers were also asked to review the datasets (final reading) after their initial reading and rate the confidence level in their assessment of features during the final reading, similar to previously reported NBI33, 34 and CLE35 imaging studies. Some of the low confidence readings might be associated with cases where features were less clear and thus more difficult to diagnose using the proposed two-feature OCTA criteria alone. We observed an increase in the overall accuracy (sensitivity/specificity: 88.1%/67.3% vs 94%/69.1%, respectively) and interobserver agreement (0.46 vs 0.58) between the initial and final readings (Table 2 & Supplemental table 5). These results suggest a learning curve in the current study whereby the readers consolidated their understanding of the OCTA features from the initial reading. Nevertheless, the accuracy was comparable among the six readers including the two GI fellows who had no prior OCT/OCTA experience, suggesting that the capability to learn and implement the OCTA criteria did not vary with readers’ baseline OCT/OCTA experience levels (Table 3). We hypothesize that both the learning curve and interobserver agreement could be further improved with increased sample size.
In addition, because the effect of prior EET on the microvascular features has not been investigated completely, a subgroup analysis was performed to compare the assessment of NDBE OCTA datasets obtained from patients without a history of dysplasia versus NDBE datasets obtained from patients with a history of dysplasia/EET (Supplemental table 2). A comparable detection accuracy was observed (p=0.71).
Approximately 44% of the datasets (N=43) were not used for analysis due to inadequate image quality: (1) artifacts from respiration or cardiact motion, (2) decreased imaging coverage from varying tissue contact, and (3) excessive pressure exerted by the catheter over the tissue surface suppressing blood flow (Supplemental figure 1). The low yield of OCTA data occurred in part because our prototype instrument did not generate OCTA images in real time. Although, structural OCT was displayed during endoscopy immediately as the images were acquired, OCTA required post processing. Real time OCTA display can potentially be implemented, but requires significant software development efforts.
The imaging coverage of OCTA is currently limited by the small catheter size. In this study we used an imaging catheter introduced in one instrument channel of a dual-channel endoscope so the second channel could be used for biopsy. Commercial endoscopic OCT instruments use a balloon catheter which images a 6 cm circumference x 6 cm length of the esophagus in ~90 seconds. Our current prototype instrument could generate a comparable structural image ~10x faster; however, because OCTA uses motion contrast to visualize blood flow, the same region must be scanned very densely and there are trade-offs between imaging speed and area coverage. Our prototype instrument and catheter can perform OCTA of a 1 cm circumference x 1.6 cm length in 8 seconds. A recent study by our group using an ultrahigh speed OCT system and a micromotor balloon catheter demonstrated circumferential OCTA imaging of the swine esophagus over a 5 cm circumference x 2.6 cm length <18 seconds36. This result suggests that wide area OCTA in human subjects is feasible.
In conclusion, volumetric en face OCTA imaging enables rapid examination of depth resolved microvascular features with near-microscopic resolution. This study identifies microvascular features which are associated with dysplasia in Barrett’s esophagus and suggests that OCTA information can serve as an adjunct to volumetric structural OCT. This new imaging modality also provides additional information on subsurface microvasculature, which could help the study of dysplasia pathogenesis. These promising results motivate the need for future technology improvements and larger scale prospective studies investigating the diagnostic accuracy of OCTA.
Supplementary Material
Supplemental table 1. Demographics of the OCTA datasets.
Supplemental table 2. The accuracy of the OCTA criteria including all NDBE datasets as well as datasets from patients without a history of dysplasia and history of dysplasia/EET.
Supplemental table 3. The overall distribution of the abnormal microvascular features on the subgroups of LGD and HGD pathologies.
Supplemental table 4. The overall accuracy of using OCTA on the subgroups of LGD and HGD pathologies.
Supplemental table 5. The accuracy of individual readers using OCTA features to detect dysplasia in the OCTA datasets of BE from the initial reading.
Example OCTA images being removed from the study due to inadequate image quality. (A) OCTA image exhibited high noise because of the motion artifacts. (B) OCTA image showed a limited contact between the tissue and the imaging catheter which decreases coverage over the imaged site. Also, the varying tissue contact along the pullback direction can be observed (dashed line) and might be related to the motion issue as well. (C) Low OCTA signals were observed in the middle of the FOV because of the excess pressures exerted by the imaging catheter, which suppresses blood flow of the imaged site. (D) OCTA image exhibited inadequate image quality due to motion issue (stars, high noise) and potentially the pressure issue (arrows, loss of the OCTA signals). Scale bars: 1 mm. r: rotary direction; x: longitudinal (pullback) direction.
Acknowledgments
Supportive foundations: This work was supported by the National Institutes of Health (NIH) grants R01-CA075289-19 (JGF and HM), R44-CA101067-05 (JGF), R01-CA178636-04 (JGF), R01-EY011289-30 (JGF), F32-CA183400 (MGG), Air Force Office of Scientific Research (AFOSR) FA9550-15-1-0473 and FA9550-12-1-0499 (JGF), and graduate fellowship (KCL) from Agency for Science, Technology and Research (Singapore).
ABBREVIATIONS and ACRONYMS
- 3D
three-dimensional
- BE
Barrett’s esophagus
- CI
confidence interval
- CLE
confocal laser endomicroscopy
- CSA
cryospray ablation
- EAC
esophageal adenocarcinoma
- EET
endoscopic eradication therapy
- EGD
esophagogastroduodenoscopy
- EMR
endoscopic mucosal resection
- GEJ
gastroesophageal junction
- GI
gastrointestinal
- HGD
high grade dysplasia
- LGD
low grade dysplasia
- NBI
narrow band imaging
- NDBE
non-dysplastic Barrett’s esophagus
- NPV
negative predictive value
- OCT
optical coherence tomography
- OCTA
optical coherence tomography angiography
- PPV
positive predictive value
- RFA
radiofrequency ablation
- SD
standard deviation
- VABHS
Veterans Affairs Boston Healthcare System
- VLE
volumetric laser endomicroscopy
Footnotes
Author contributions: JGF, HM, HCL, and OOA designed the study; HCL, OOA, KCL, and BP developed the technical instrumentation; HCL, OOA, KCL, ZW, MGG, and HM acquired data; HCL, OOA, KCL, and QH analyzed and interpreted data; MF provided administrative support; HCL, OOA, KCL, JGF, and HM wrote the manuscript; JGF and HM obtained funding for this study. All authors read the manuscript; JGF and HM are principal investigators and supervised this study.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Hsiang-Chieh Lee, Department of Electrical Engineering and Computer Science and Research Laboratory of Electronics, Massachusetts Institute of Technology, Cambridge, MA, USA.
Osman O. Ahsen, Department of Electrical Engineering and Computer Science and Research Laboratory of Electronics, Massachusetts Institute of Technology, Cambridge, MA, USA.
Kaicheng Liang, Department of Electrical Engineering and Computer Science and Research Laboratory of Electronics, Massachusetts Institute of Technology, Cambridge, MA, USA.
Zhao Wang, Department of Electrical Engineering and Computer Science and Research Laboratory of Electronics, Massachusetts Institute of Technology, Cambridge, MA, USA.
Marisa Figueiredo, VA Boston Healthcare System, Boston, MA, USA.
Michael G. Giacomelli, Department of Electrical Engineering and Computer Science and Research Laboratory of Electronics, Massachusetts Institute of Technology, Cambridge, MA, USA.
Benjamin Potsaid, Department of Electrical Engineering and Computer Science and Research Laboratory of Electronics, Massachusetts Institute of Technology, Cambridge, MA, USA. Advanced Imaging Group, Thorlabs, Inc., Newton, NJ, USA.
Qin Huang, VA Boston Healthcare System, Boston, MA, USA. Harvard Medical School, Boston, MA, USA.
Hiroshi Mashimo, VA Boston Healthcare System, Boston, MA, USA. Harvard Medical School, Boston, MA, USA.
James G. Fujimoto, Department of Electrical Engineering and Computer Science and Research Laboratory of Electronics, Massachusetts Institute of Technology, Cambridge, MA, USA.
References
- 1.Wang KK, Sampliner RE. Updated Guidelines 2008 for the Diagnosis, Surveillance and Therapy of Barrett’s Esophagus. Am J Gastroenterol. 2008;103(3):788–797. doi: 10.1111/j.1572-0241.2008.01835.x. [DOI] [PubMed] [Google Scholar]
- 2.Devesa SS, Blot WJ, Fraumeni JF. Changing patterns in the incidence of esophageal and gastric carcinoma in the United States. Cancer. 1998;83(10):2049–2053. [PubMed] [Google Scholar]
- 3.Shaheen NJ. Advances in Barrett’s esophagus and esophageal adenocarcinoma. Gastroenterology. 2005;128(6):1554–1566. doi: 10.1053/j.gastro.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 4.Spechler SJ. Dysplasia in Barrett’s esophagus: limitations of current management strategies. Am J Gastroenterol. 2005;100(4):927–935. doi: 10.1111/j.1572-0241.2005.41201.x. [DOI] [PubMed] [Google Scholar]
- 5.Hvid-Jensen F, Pedersen L, Drewes AM, et al. Incidence of Adenocarcinoma among Patients with Barrett’s Esophagus. N Engl J Med. 2011;365(15):1375–1383. doi: 10.1056/NEJMoa1103042. [DOI] [PubMed] [Google Scholar]
- 6.Reid BJ, Levine DS, Longton G, et al. Predictors of progression to cancer in Barrett’s esophagus: Baseline histology and flow cytometry identify low- and high-risk patient subsets. Am J Gastroenterol. 2000;95(7):1669–1676. doi: 10.1111/j.1572-0241.2000.02196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buttar NS, Wang KK, Sebo TJ, et al. Extent of high-grade dysplasia in Barrett’s esophagus correlates with risk of adenocarcinoma. Gastroenterology. 2001;120(7):1630–1639. doi: 10.1053/gast.2001.25111. [DOI] [PubMed] [Google Scholar]
- 8.Thosani N, Abu Dayyeh BK, Sharma P, et al. ASGE Technology Committee systematic review and meta-analysis assessing the ASGE Preservation and Incorporation of Valuable Endoscopic Innovations thresholds for adopting real-time imaging-assisted endoscopic targeted biopsy during endoscopic surveillance of Barrett’s esophagus. Gastrointest Endosc. 2016;83(4):684–698. doi: 10.1016/j.gie.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Couvelard A, Paraf F, Gratio V, et al. Angiogenesis in the neoplastic sequence of Barrett’s oesophagus. Correlation with VEGF expression. J Pathol. 2000;192(1):14–18. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH709>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 10.Auvinen MI, Sihvo EIT, Ruohtula T, et al. Incipient angiogenesis in Barrett’s epithelium and lymphangiogenesis in Barrett’s adenocarcinoma. J Clini Oncol. 2002;20(13):2971–2979. doi: 10.1200/JCO.2002.09.011. [DOI] [PubMed] [Google Scholar]
- 11.Mobius C, Stein HJ, Becker I, et al. The ‘angiogenic switch’ in the progression from Barrett’s metaplasia to esophageal adenocarcinoma. Eur J Surg Oncol. 2003;29(10):890–894. doi: 10.1016/j.ejso.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Konda VJA, Hart J, Lin S, et al. Evaluation of microvascular density in Barrett’s associated neoplasia. Mod Pathol. 2013;26(1):125–130. doi: 10.1038/modpathol.2012.146. [DOI] [PubMed] [Google Scholar]
- 13.Kara MA, Ennahachi M, Fockens P, et al. Detection and classification of the mucosal and vascular patterns (mucosal morphology) in Barrett’s esophagus by using narrow band imaging. Gastrointest Endosc. 2006;64(2):155–166. doi: 10.1016/j.gie.2005.11.049. [DOI] [PubMed] [Google Scholar]
- 14.Sharma P, Bansal A, Mathur S, et al. The utility of a novel narrow band imaging endoscopy system in patients with Barrett’s esophagus. Gastrointest Endosc. 2006;64(2):167–175. doi: 10.1016/j.gie.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 15.Kiesslich R, Gossner L, Goetz M, et al. In vivo histology of Barrett’s esophagus and associated neoplasia by confocal laser endomicroscopy. Clin Gastroenterol Hepatol. 2006;4(8):979–987. doi: 10.1016/j.cgh.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 16.Wallace M, Lauwers GY, Chen Y, et al. Miami classification for probe-based confocal laser endomicroscopy. Endoscopy. 2011;43(10):882–891. doi: 10.1055/s-0030-1256632. [DOI] [PubMed] [Google Scholar]
- 17.Shahid MW, Crook JE, Meining A, et al. Exploring the optimal fluorescein dose in probe-based confocal laser endomicroscopy for colonic imaging. J Interv Gastroenterol. 2011;1(4):166–171. doi: 10.4161/jig.19953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yun SH, Tearney GJ, Vakoc BJ, et al. Comprehensive volumetric optical microscopy in vivo. Nat Med. 2006;12(12):1429–1433. doi: 10.1038/nm1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vakoc BJ, Shishko M, Yun SH, et al. Comprehensive esophageal microscopy by using optical frequency-domain imaging (with video) Gastrointest Endos. 2007;65(6):898–905. doi: 10.1016/j.gie.2006.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isenberg G, Sivak MV, Jr, Chak A, et al. Accuracy of endoscopic optical coherence tomography in the detection of dysplasia in Barrett’s esophagus: a prospective, double-blinded study. Gastrointest Endosc. 2005;62(6):825–831. doi: 10.1016/j.gie.2005.07.048. [DOI] [PubMed] [Google Scholar]
- 21.Evans JA, Poneros JM, Bouma BE, et al. Optical coherence tomography to identify intramucosal carcinoma and high-grade dysplasia in Barrett’s esophagus. Clin Gastroenterol Hepatol. 2006;4(1):38–43. doi: 10.1053/S1542-3565(05)00746-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y, Aguirre AD, Hsiung PL, et al. Ultrahigh resolution optical coherence tomography of Barrett’s esophagus: preliminary descriptive clinical study correlating images with histology. Endoscopy. 2007;39(7):599–605. doi: 10.1055/s-2007-966648. [DOI] [PubMed] [Google Scholar]
- 23.Yang VXD, Tang S-J, Gordon ML, et al. Endoscopic Doppler optical coherence tomography in the human GI tract: initial experience. Gastrointest Endosc. 2005;61(7):879–890. doi: 10.1016/s0016-5107(05)00323-8. [DOI] [PubMed] [Google Scholar]
- 24.Tsai T-H, Ahsen OO, Lee H-C, et al. Endoscopic Optical Coherence Angiography Enables Three Dimensional Visualization of Subsurface Microvasculature. Gastroenterology. 2014;147(6):1219–1221. doi: 10.1053/j.gastro.2014.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vakoc BJ, Shishko M, Yun SH, et al. Comprehensive esophageal microscopy by using optical frequency–domain imaging (with video) Gastrointest Endosc. 2007;65(6):898–905. doi: 10.1016/j.gie.2006.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai TH, Lee HC, Ahsen OO, et al. Ultrahigh speed endoscopic optical coherence tomography for gastroenterology. Biomed Opt Exp. 2014;5(12):4387–404. doi: 10.1364/BOE.5.004387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahsen OO, Lee H-C, Giacomelli MG, et al. Correction of rotational distortion for catheter-based en face OCT and OCT angiography. Opt Lett. 2014;39(20):5973–5976. doi: 10.1364/OL.39.005973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fleiss JL, Levin B, Paik MC. Statistical methods for rates and proportions. 3. Hoboken, N.J: J. Wiley; 2003. p. xxvii.p. 760. [Google Scholar]
- 29.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33(1):159–174. [PubMed] [Google Scholar]
- 30.Leggett CL, Gorospe EC, Chan DK, et al. Comparative diagnostic performance of volumetric laser endomicroscopy and confocal laser endomicroscopy in the detection of dysplasia associated with Barrett’s esophagus. Gastrointest Endosc. 2016;83(5):880–888. doi: 10.1016/j.gie.2015.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swager A, Tearney GJ, Leggett CL, et al. Identification of Volumetric Laser Endomicroscopy features predictive for early neoplasia in Barrett’s esophagus using high-quality histological correlation. Gastrointest Endosc. 2016 doi: 10.1016/j.gie.2016.09.012. in press. [DOI] [PubMed] [Google Scholar]
- 32.Phoa KN, van Vilsteren FG, Weusten BL, et al. Radiofrequency ablation vs endoscopic surveillance for patients with Barrett esophagus and low-grade dysplasia: a randomized clinical trial. JAMA. 2014;311(12):1209–1217. doi: 10.1001/jama.2014.2511. [DOI] [PubMed] [Google Scholar]
- 33.Hewett DG, Huffman ME, Rex DK. Leaving distal colorectal hyperplastic polyps in place can be achieved with high accuracy by using narrow-band imaging: an observational study. Gastrointest Endosc. 2012;76(2):374–380. doi: 10.1016/j.gie.2012.04.446. [DOI] [PubMed] [Google Scholar]
- 34.Sharma P, Bergman JJGHM, Goda K, et al. Development and Validation of a Classification System to Identify High-Grade Dysplasia and Esophageal Adenocarcinoma in Barrett’s Esophagus Using Narrow-Band Imaging. Gastroenterology. 2016;150(3):591–598. doi: 10.1053/j.gastro.2015.11.037. [DOI] [PubMed] [Google Scholar]
- 35.Kuiper T, van den Broek FJC, van Eeden S, et al. Feasibility and Accuracy of Confocal Endomicroscopy in Comparison With Narrow-Band Imaging and Chromoendoscopy for the Differentiation of Colorectal Lesions. Am J Gastroenterol. 2012;107(4):543–550. doi: 10.1038/ajg.2012.14. [DOI] [PubMed] [Google Scholar]
- 36.Lee HC, Ahsen OO, Liang KC, et al. Circumferential optical coherence tomography angiography imaging of the swine esophagus using a micromotor balloon catheter. Biomed Opt Exp. 2016;7(8):2927–2942. doi: 10.1364/BOE.7.002927. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental table 1. Demographics of the OCTA datasets.
Supplemental table 2. The accuracy of the OCTA criteria including all NDBE datasets as well as datasets from patients without a history of dysplasia and history of dysplasia/EET.
Supplemental table 3. The overall distribution of the abnormal microvascular features on the subgroups of LGD and HGD pathologies.
Supplemental table 4. The overall accuracy of using OCTA on the subgroups of LGD and HGD pathologies.
Supplemental table 5. The accuracy of individual readers using OCTA features to detect dysplasia in the OCTA datasets of BE from the initial reading.
Example OCTA images being removed from the study due to inadequate image quality. (A) OCTA image exhibited high noise because of the motion artifacts. (B) OCTA image showed a limited contact between the tissue and the imaging catheter which decreases coverage over the imaged site. Also, the varying tissue contact along the pullback direction can be observed (dashed line) and might be related to the motion issue as well. (C) Low OCTA signals were observed in the middle of the FOV because of the excess pressures exerted by the imaging catheter, which suppresses blood flow of the imaged site. (D) OCTA image exhibited inadequate image quality due to motion issue (stars, high noise) and potentially the pressure issue (arrows, loss of the OCTA signals). Scale bars: 1 mm. r: rotary direction; x: longitudinal (pullback) direction.