Abstract
MicroRNA (miRNA) are major regulators of cell responses, particularly in stressed cell states and host immune responses. Some miRNA have a role in pathogen defense, including regulation of immune responses to Plasmodium parasite infection. Using a non-lethal mouse model of blood stage malaria infection we have found that miR-451−/− mice infected with Plasmodium yoelii XNL cleared infection at a faster rate than did wild type (WT) mice. MiR-451−/− mice had an increased leukocyte response to infection, with the protective phenotype primarily driven by CD4+ T-cells. WT and miR-451−/− CD4+ T-cells had similar activation responses, but miR-451−/− CD4+ cells had significantly increased proliferation, both in vitro and in vivo. Myc is a miR-451 target with a central role in cell cycle progression and cell proliferation. CD4+ T-cells from miR-451−/− mice had increased post-activation Myc expression. RNA-Seq analysis of CD4+ cells demonstrated over 5000 differentially expressed genes in miR-451−/− mice post-infection, many of which are directly or indirectly Myc regulated. This study demonstrates that miR-451 regulates T-cell proliferative responses in part via a Myc dependent mechanism.
Introduction
MicroRNA (miRNA) are 21–22 nucleotide non-coding RNA that regulate post transcriptional gene expression by targeting the 3′ untranslated region (UTR) of mRNA transcripts. MiRNAs have an important role in diverse cell functions, including immune responses [1]. For example, miR-125b regulates CD4+ T-cell differentiation [2], miR-223 regulates neutrophil proliferation [3], and miR-29 negatively regulates T helper type 1 (Th1) responses to Mycobaterium tuberculosis [4]. However, little is known about the role of miRNA in regulating host immune responses to most infections, including blood stage malaria infection. MicroRNA-451 (miR-451) is a regulator of erythrocyte development in mice [5, 6] and directly inhibits Plasmodium falciparum growth in vitro [7]. In mouse models of cancer miR-451 has a role in regulating tumor cell proliferation [8, 4] and innate immune cell function [9]. MiR-451 has also been identified in CD4+ T-cells in both healthy and disease states [10, 11], but a role for miR-451 in adaptive immunity is not known.
More than 250 million individuals are infected with the malaria causing Plasmodium parasite each year [12]. The immune system has a critical role in limiting malaria infection and mediating parasite clearance. In the initial stages of infection innate immune cells, such as natural killer T cells, macrophages, and neutrophils, are responsible for direct parasite clearance. During blood stage infection CD4+ T helper cells are the major mediators of the adaptive immune responses that limit infection and promote parasite clearance. Several groups have used mouse models of blood-stage malaria to demonstrate that increased CD4+ T-cell responses correlate with reduced parasite burden [13, 14], however, effective means to initiate and sustain acquired immune responses to infection have not been found.
We have now found that miR-451−/− mice have improved clearance of non-lethal blood stage Plasmodium infection, that is in part mediated by a protective CD4+ response.
Material and Methods
1.1. Mouse Experiments
MiR-451−/− mice were created by the laboratory of Dr. Eric Olson (UT Southwestern) and are on a C57Bl6/J (B6) background. Control B6 mice were obtained from Jackson Laboratory. All mice were housed and experiments performed in accordance with University of Rochester Medical Center (URMC) Institutional Animal Care and Use Committee (IACUC) approved protocols and standards.
Mice were infected via intraperitoneal (i.p.) injection with ~0.5 × 107 cryopreserved and mouse passaged Plasmodium yoelii 17XNL blood stage parasites (nonlethal malaria). Parasitemia was measured in 3-day intervals post infection using Giemsa stained blood smears.
1.2. Lymphocyte staining and flow cytometry
Spleens were collected and single cell suspensions made. For T helper cell analysis, cells were surfaced stained with anti-CD4 (RM 4-5; eBioscience), permiabalized and intracellular stained with IFN-γ (XMG1.2; eBioscience), IL-4 (BVD6-24G2; eBioscience), RORγ (B2D; eBioscience), CD25 (PC61.5; eBioscience) or FoxP3 (FJK-16s; eBioscience). Representative gating in Supplemental Data Figure S1. For CD8+ T cells, cell surfaces were stained with anti- CD8a (53-6.7; eBioscience) and anti- CD3e (145-2C11; eBioscience). For B cells, cell surfaces were stained with anti- CD21/CD35 (8D9; eBioscience) and anti- CD19 (1D3; eBioscience). T cell activation markers CD25 (eBio3C7; eBioscience) and CD69 (H1.2F3, Biolegend) were measured using flow cytometry.
For CBC exams ≈ 50 μL of blood was collected into EDTA coated tubes via retro-orbital bleed. VetScan HM5 Analyzer was used.
1.3. CD4+ and CD8+ T-cell in vivo depletion
Monoclonal antibodies specific for mouse CD4+ T-cells (GK1.5, source: Rat) were injected intravenously (0.167 mg in 100 μL sterile PBS) on days 0, 7, and 14 and depletion confirmed by flow cytometry[15]. Monoclonal antibodies specific for mouse CD8+ T-cells (YTS 169.4, source: Rat) were injected intravenously (0.167 mg in 100 μL sterile PBS) on days 0, 7, and 14 and depletion confirmed by flow cytometry. The same concentration and volume of control IgG (clone LTF-2) was used in a separate infection experiment.
1.4. Bone marrow transplantation
Femurs and tibias were collected and donor cells isolated as previously described [16]. Recipients were lethally irradiated by exposure to 830 cGy irradiation. 1 × 106 bone marrow cells were injected into recipient mice intravenously. Sulfa-trimethoprim was added to the water bottles for 4 weeks post transplant.
1.5. Cell culture
Spleens were harvested and processed to generate a single cell suspension. Red blood cells were lysed using ACK lysis buffer (Life Technologies). CD4+ T cells were isolated via negative selection (EasySep; Stemcell Technologies). Purity was verified using flow cytometry. For all proliferation studies, cells were cultured in 96-well α-CD3 coated plates (1μg/mL; BioXCell), with 1μg/mL α-CD28 (PV-1; BioXCell) [17, 18]. Cell counts were measured using an automated hemocytometer (VetScan HM5). For Myc inhibition, cells were treated with 100 μM 10058-F4 at D0 (Tocris). Cell death was measured via 7-AAD staining (Life Technologies), and analysis via flow cytometry with analysis using FlowJo 7.6 software.
1.6. Quantitative Real-Time PCR
For in vitro miR-451 measurement, purified splenic CD4+ T cells were stimulated for 5 days in plates coated with 1 μg/mL α-CD3 (BioXCell) and 1 μg /mL α-CD28 (BioXCell). Small RNAs were isolated with a miRVana microRNA Isolation Kit (Ambion) and cDNA generated using a Taqman microRNA reverse transcription kit (Applied Biosystems) according to the manufacturer’s protocol. For in vitro and in vivo measurements of m RNAs, total RNA was isolated from negatively selected splenic CD4+ T cells using RNeasy Plus Micro Kit (Qiagen). A myIQ Real Time PCR Detector system based on FAM fluorescence was used for qRT-PCR. The probes used included: miR-451 (mmu-miR-451 mouse microRNA-451), cyclin B1 (Mm03053893_gH), Myc (Mm00487804_m1), Infg (Mm01168134_m1), IL4 (Mm00445259_m1), IL17f (Mm00521423_m1), IL10 (Mm01288386_m1), Sdad1 (Mm00554780_m1), and Apex1 (Mm01319526_g1). Analysis of change in threshold values (2−ΔΔCT) were assessed using an approach previously described by our group [19]. All mRNA values were normalized to at least 2 housekeeping genes (Tbp [Mm00446971_m1], GAPDH [Mm99999915_g1], 18s ribosomal RNA [Mm03928990_g1], Tubulin [Mm00446971_m1]); miRNA values were normalized to U6 small nuclear RNA (U6 snRNA 001973) and/or sno202.
1.7. T Cell Transfection
miR-451 Inhibition. Activated T helper cells were transfected with a miR-451 antagomiR locked nucleic acid (LNA, Exiqon) or an LNA scrambled control (Exiqon).
miR-451 overexpression. Unstimulated T helper cells were transfected with Pre-miR-451 (50 pM; Ambion), and scrambled Pre-miR negative control (50 pmol; Ambion).
Cells were transfected via electroporation (Lonza).
1.8. RNA sequencing
CD4+ T cells were purified from WT and miR-451−/− spleens pre and post P. yoelii infection and total RNA was isolated using RNeasy Plus Micro Kit (Qiagen). RNA samples were sequenced on an Illumina HiSeq2500 Sequencer using a TruSeq Total stranded RNAseq library preparation. Reads were mapped to the Mus musculus genome (mm10) with SHRiMP2.2.3, and differential expression was determined using cufflinks 2.0.2. Myc regulated genes identified in miR-451−/− CD4+ T-cells post infection were displayed using the Google Charts API (Sankey Diagram) [20]. RNA-Seq individual data is in Supplemental data.
Data were analyzed using QIAGEN’s Ingenuity® Pathway Analysis (IPA®, QIAGEN Redwood City, www.qiagen.com/ingenuity).
1.9. CD4+ T cell Proliferation
In vivo BrdU
Mice were infected with 5 × 105 P. yoellii XNL pRBCs. On day 8 post infection, mice were injected with 1 mg 5-Bromo-2′-Deoxyuridine (BrdU) (Life Technologies) intraperitoneally. Spleens were collected on day 9 post infection and cells were stained with anti-BrdU (BU20a; eBioscience), and BrdU incorporation was measured by flow cytometry.
In vitro BrdU
CD4+ T cells were isolated from spleens and 25 μM BrdU was added 24 hours prior to cell analysis on day 3, 5, 7 post stimulation. Cells were stained with anti-BrdU (BU20a; eBioscience), and BrdU incorporation was measured by flow cytometry.
In vitro pH3
Intracellular detection of phosphorylated histone 3 (pH3) in negatively selected splenic CD4+ T cells cultured in vitro, was performed by first staining the cell surface with anti-mouse anti-CD4 (RM 4–5; eBioscience). Cells were then permiabilized and stained with anti-human/mouse phospho-Histone H3 (HTA28; eBioscience) for 30 minutes at 4°C in the dark. Cells were washed and fluorescence was detected using flow cytometry.
1.10. Western blotting
CD4+ T cells were isolated from the spleens of mice and cell lysates were loaded and run on 4–15% Mini-PROTEAN TGX Precast gels (Bio-Rad). Proteins were then transferred onto nitrocellulose membranes using a Trans-Blot SD semi-dry electrophoresis transfer system. Membranes were immunoblotted using mouse polyclonal antibodies against GAPDH (FL-335), c-Myc (9402; Cell Signaling), or monoclonal antibody to cyclin B1 (GNS1). Horseradish peroxidase-conjugated anti-rabbit and anti-mouse (for cyclin B1 detection only) (Sigma-Aldrich) were used along with chemiluminescence to detect protein expression via X-ray film. Protein expression was quantified using Image J software.
1.11. Statistical Analysis
GraphPad Prism software and R version 3.1.0 were used for statistical analysis. For studies that involve the comparison of 2 groups, unpaired student t-tests were used to determine statistical significance unless otherwise noted. For comparison of means for groups of 3 or more, 2-way repeated measures ANOVA with a Tukey post test was used.
Results
1.12. miR-451 negatively regulates parasite clearance during malaria infection
In vitro studies have indicated that miR-451 limits Plasmodium parasite growth [7]. To determine whether miR-451 has a similar in vivo function, WT and miR-451−/− mice were infected with blood stage Plasmodium yoelii XNL (P. yoelii XNL, non-lethal malaria infection model). P. yoelii XNL is a self-limiting infection and induces host immune responses to clear the infection [21]. Parasitemia was determined by blood smear on multiple days post infection. MiR-451−/− mice cleared P. yoelii XNL significantly more rapidly than did WT mice (Fig 1A–B). To determine whether the accelerated miR-451−/− parasite clearance was hematopoietic cell dependent, WT mice were lethally irradiated, bone marrow reconstituted with either WT or miR-451−/− bone marrow, and infected with P. yoelii XNL. WT mice reconstituted with miR-451−/− bone marrow cleared the infection at a much faster rate than did mice reconstituted with WT bone marrow (Fig 1C–D). These data demonstrate that in contrast to in vitro studies, miR-451 does not limit P. yoelii in vivo, and that enhanced parasite clearance in miR-451−/− mice is driven by a hematopoietic derived cell response to infection.
Figure 1.
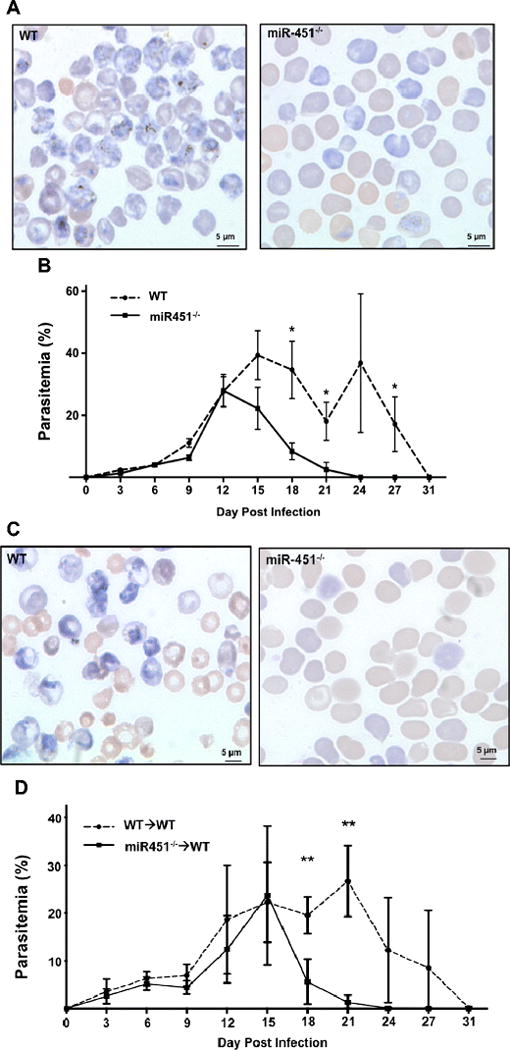
miR-451−/− mice have accelerated P. yoelii XNL clearance. a) WT and miR-451−/− mice were infected and parasitemia assessed by Giemsa stained blood smears. Representative smears (100X, day 21 post-infection). b) Parasitemia quantification (mean ± SEM, N=5, *P < 0.05, 2-way ANOVA with Tukey post-test). Representative of 2 separate experiments with similar results. c) Hematopoietic cells mediate miR-451−/− protective effects. WT mice were lethally irradiated and marrow reconstituted with WT or miR-451−/− bone marrow. Mice were infected with P yoelii XNL and parasitemia assessed (representative smear, 100X, day 21 post-infection). d) Parasitemia quantification (mean ± SEM, N=5, **P<0.01, 2-way ANOVA with Tukey post-test). Representative of 2 separate experiments with similar results.
1.13. miR-451−/− mice have increased immune responses to infection
To characterize hematopoietic responses to P. yoelii in WT and miR-451−/− mice complete blood counts (CBC) were performed over the course of infection. Total lymphocyte counts pre-infection were similar between WT and miR-451−/− mice, but total lymphocytes post-infection was increased in miR-451−/− mice, peaking at about post-infection day 9 (Supplemental Table 1). We then determined the changes in specific lymphocyte populations. Innate immune cells were similar between WT and miR-451−/− mice both pre and post infection, but post-infection neutrophils were somewhat increased in miR-451−/− mice (Supplemental Table 1). To characterize acquired immune cells, WT and miR-451−/− spleens were harvested pre-infection and on day 9 post infection, a time point with peak lymphocyte numbers and before the parasitemia diverged between the 2 groups of mice. There was no difference in B cell numbers either pre or post infection (Fig 2A). WT and miR-451−/− mice had a similar fold change in CD4+ and CD8+ T-cell numbers from pre to post-infection, but the total numbers of each T-cell type were greatly increased in miR-451−/− mice, particularly post-infection (Fig 2B–C).
Figure 2.
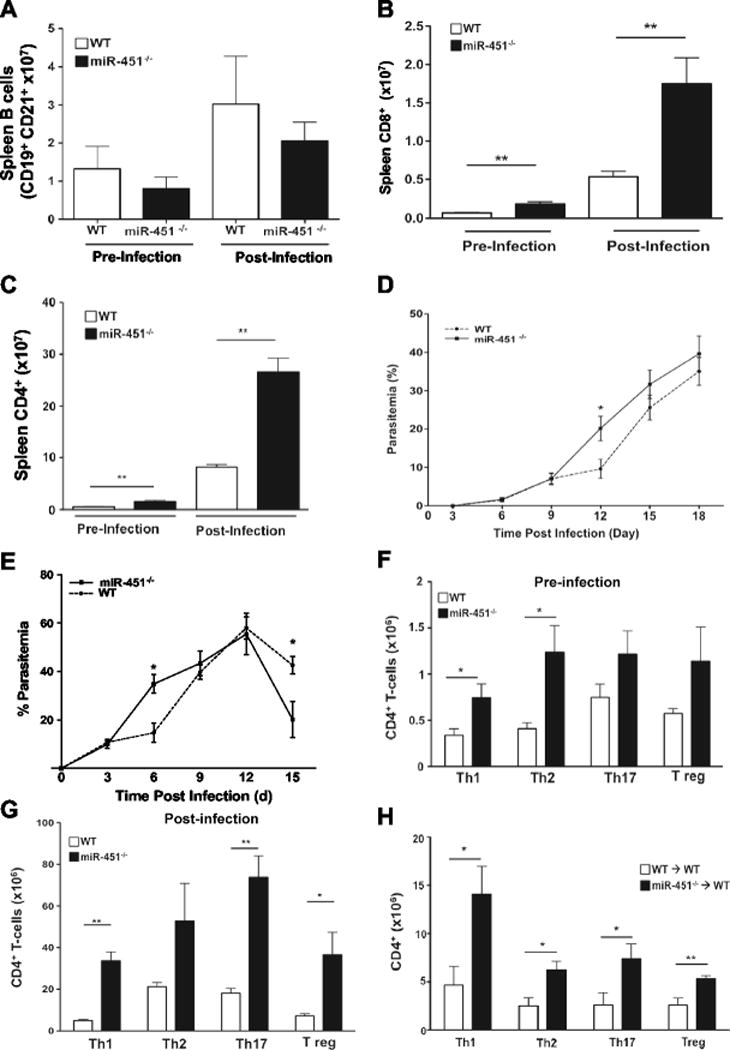
miR-451−/− mice have increased CD4+ T-cell responses to infection. Spleens were isolated from WT and miR-451−/− mice either pre or day 9 postinfection. The numbers of a) B cells, b) CD8+ and c) CD4+ T-cells were quantified by flow cytometry. MiR-451−/− mice had increased CD8+ and CD4+ T-cells compared to WT mice (mean ± S.E.M., N=4-5, * P<0.01). d–e) Accelerated parasite clearance in miR-451−/− mice is in part CD4+ T-cell dependent. Infected WT and miR-451−/− mice were treated with d) CD4+ depleting antibody or e) CD8+ depleting antibody. Parasitemia was determined over time (mean ± S.E.M., N=4–8, *P<0.05, 2-way ANOVA with Tukey post-test). f) Baseline T-helper and T reg cells are increased in miR-451−/− mice compared to WT mice (mean ± S.E.M, N=4-5, * P<0.01). g) CD4+ cells are increased in miR-451−/− mice compared to WT mice post-infection. Mice were infected with P yoelii XNL and T helper and T reg numbers determined on day 9 (mean ± S.E.M., N=4-5, * P<0.05, **P<0.01). h) Increased CD4+ cell responses are hematopoietic dependent. WT mice were lethally irradiated and bone marrow transplanted with WT or miR-451−/− bone marrow and infected with P yoelii XNL. MiR-451−/− reconstituted mice had increased T helper and T reg responses on day 9 post infection (mean ± S.E.M., N=4-5, * P<0.05, **P<0.01; representative of 2 separate experiments).
CD4+ T-cells express miR-451 (Supplemental Fig S2) [11, 10] and CD4+ T-cells are critical in driving P. yoelii XNL clearance [13]. To determine whether CD4+ cells mediate the protective miR-451−/− phenotype we treated WT and miR-451−/− mice with either CD4+ or CD8+ T-cell depleting antibody or control IgG, (Supplemental Figures S3–S5), infected the mice, and measured the parasite burden. CD4+ depleted WT and miR-451−/− mice had similar parasite burdens (Fig 2D). Because the protective phenotype was lost when CD4+ cells were depleted, it suggested that CD4+ T-cells have a central role in increased miR-451−/− blood stage parasite clearance. In contrast, when CD8+ T-cells were depleted the early parasite burden was increased in miR-451−/− mice, but by day 15 post-infection CD8+ depleted miR-451−/− mice still had significantly improved parasite clearance compared to WT mice (Fig 2E). These data indicate that the miR-451−/− protective phenotype is at least in part CD4+ T-cell dependent.
We therefore further characterized miR-451−/− and WT CD4+ T-cell subsets both pre and post-infection. MiR-451−/− mice had significantly increased Th1 cells and Th2 pre-infection (Th17 and T reg cells were also increased, but not significantly, Fig 2F). Post-infection, the total number of Th1, Th17 and T regs were significantly increased in miR-451−/− mice compared to WT mice (Fig 2G). This was further represented by the fold change in cell numbers from pre to post-infection (WT vs KO, ± SEM; Th1 17±3.6 vs 58±15, Th17 31±8.6 vs 73±14.6, T reg 12±1.8 vs 53±16.5). Similar results were found in WT mice bone marrow reconstituted with WT and miR-451−/− bone marrow (Fig 2H). These data demonstrate that miR-451 limits CD4+ proliferative responses to P yoelii XNL.
1.14. miR-451 negatively regulates CD4+ T-cell proliferation
We next determined whether increased miR-451−/− CD4+ T-cell numbers is due to increased cell activation, survival, or proliferation. To compare activation responses, WT and miR-451−/− CD4+ T-cells were isolated and stimulated with anti-CD3/CD28 antibodies for 48 hrs and CD25 and CD69 positive cells quantified by flow cytometry. There was not a significant difference in WT and miR-451−/− CD4+ T-cell activation (Fig 3A). To compare T-cell proliferation isolated CD4+ T cells were stimulated and T-cell proliferation measured by total cell count and quantification of phosphorylated histone (pH3) positive cells. There was a significant increase miR-451−/− CD4+ T-cell numbers (Fig 3B) and pH3 positive cells beginning about day 5 post stimulation (Fig 3C). Stimulated WT and miR-451−/− CD4+ T cells were also stained with 7-amino-actinomycin D (7AAD) to quantify cell death. There was no significant difference in cell death between WT and miR-451−/− CD4+ T-cells (Supplemental Figure S6). Together these studies suggest that miR-451−/− CD4+ cells have increased proliferation, but no change in activation or survival. The demonstrate a direct effect of miR-451 on CD4+ T-cell proliferation, WT CD4+ T-cells were transfected with scramble control or pre-miR-451 to increase miR-451 expression, and cells were activated to stimulate cell proliferation. Additional miR-451 decreased T-cell proliferation compared to control transfected T-cells (Fig 3D).
Figure 3.
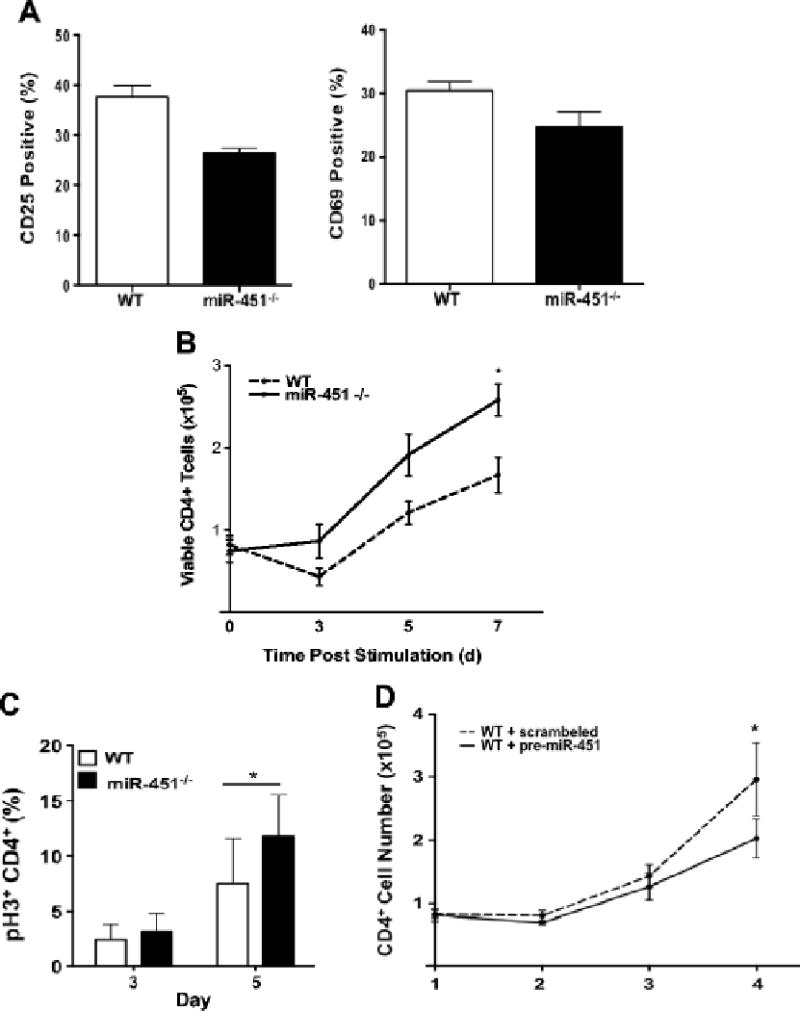
miR-451 inhibits CD4+ T-cell proliferation in vitro. a) WT and miR-451−/− CD4+ cells have similar activation. WT and miR-451−/− CD4+ cells were stimulated with anti-CD3 and CD28 antibodies and CD25 and CD69 expression determined 48 hrs later (mean ± S.E.M., N=4). b–c) miR-451−/− CD4+ T cells have increased proliferation. CD4+ cells from WT and miR-451−/− mice were stimulated and b) cell numbers (mean ± S.E.M., N=8, *P<0.05, 2-way ANOVA with Tukey post-test) and c) pH3 as a marker of active cell cycle by flow cytometry was determined on multiple days (mean ± S.E.M., N=4, Students t-test, *P<0.05). d) Increased miR-451 decreases CD4+ T-cell proliferation. CD4+ cells were treated with miR-451 pre-miRNA or a scrambled control and co-stimulated. Cell numbers were determined (mean ± S.E.M., N=4, *p<0.05, paired t-test).
A lack of miR-451 also increased CD4+ T-cell proliferation in vivo. WT and miR-451−/− mice were infected with P. yoelii XNL and proliferation determined on day 9 post infection by BRDU uptake. MiR-451−/− CD4+ T-cells had increased cell proliferation as indicated by the total number of BRDU+ CD4+ cells (Fig 4A). As a further reflection of an increased proliferative state we determined CD4+ T-cell cytokine expression by qRT-PCR on cells isolated from WT and miR-451−/− mice on day 9 post-infection. The expression of Ifng, Il4, Il10 and Il17 were all increased in miR-451−/− T-cells (Fig 4B–E, Tbp and Rna18s5 for expression normalization).
Figure 4.
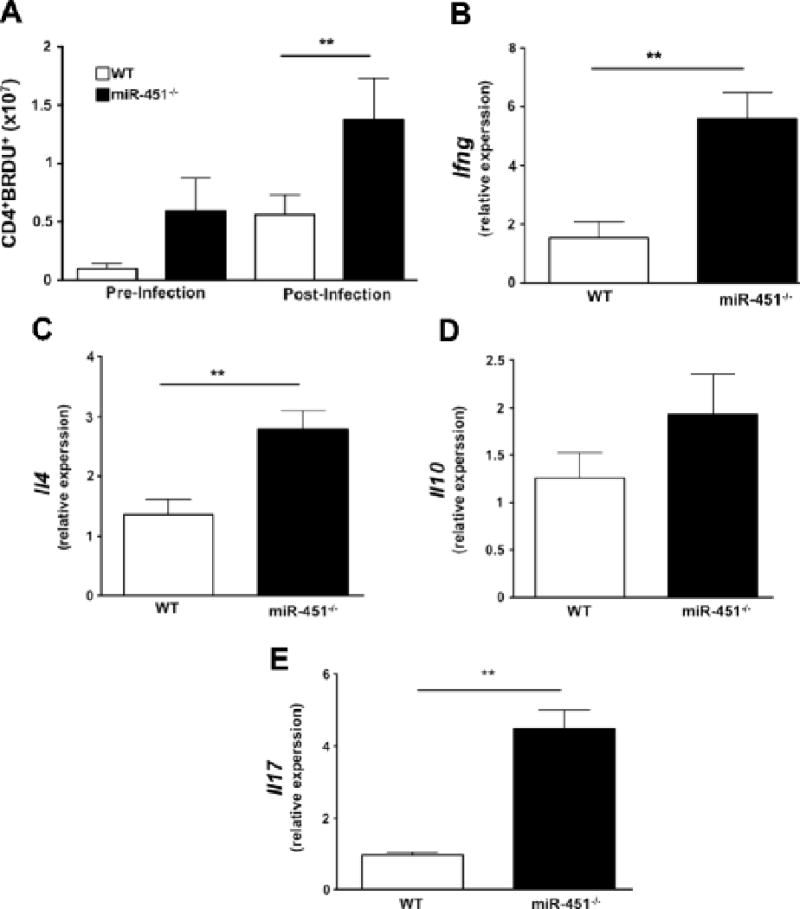
a) Mice lacking miR-451 have more in vivo CD4+ T-cell proliferation both pre- and post- infection as determined by the number of BRDU+ CD4+ cells (mean ± S.E.M., N=4-5 pre and N=8 post infection, **p<0.01, representative of 2 independent experiments). b–e) miR-451−/− T cells have increased cytokines post infection. mRNA was isolated from WT and miR-451−/− T cells on day 9 postinfection and qRT-PCR performed for Ifng, Il4, Il10 and Il17. Ifng, Il4, and Il17, but not Il10, were significantly increased in T cells from miR-451−/− mice (mean ± S.E.M., N= 3-4, **p<0.01, relative to Tbs and Rna18s5).
These in vivo and in vitro data indicate that miR-451 is a negative regulator of cell proliferation and that the protective T helper cell response in miR-451−/− mice is driven by increased CD4+ T-cell proliferation.
1.15. miR-451 targets Myc to regulate CD4+ T-cell proliferation
If miR-451 negatively regulates T cell proliferation, miR-451 expression is expected to decrease following T-cell activation when cells enter a proliferative state. To demonstrate this we measured miR-451 expression in WT CD4+ T-cells pre and on multiple days post in vitro activation. There was an approximately 50% reduction in miR-451 expression 48 hrs post stimulation and a 70% reduction after 96 hrs (Fig 5A). This suggests that miR-451 is down-regulated following T-cell stimulation.
Figure 5.
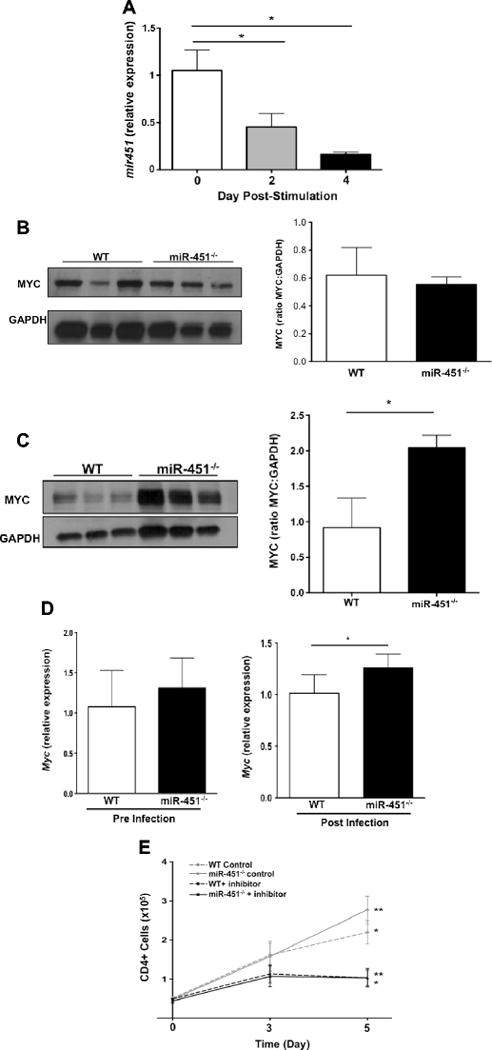
miR-451 regulates CD4+ Myc expression. a) miR-451 expression is decreased in activated T-cells. CD4+ T-cells were stimulated with anti-CD3 and CD28 antibody and miR-451 expression determined on multiple days (mean ± S.E.M., N=4, * P<0.05,). b) WT and miR-451−/− CD4+ T-cells have similar basal Myc protein expression. CD4+ cell lysates were prepared from control WT and miR-451−/− mice and immunoblotted for Myc. Myc was quantified as relative ratio to GAPDH (mean ±S.E.M., N=3). c) miR-451−/− CD4+ T-cells have increased post-infection Myc protein expression compared to WT. CD4+ T-cell lysates were prepared from WT and miR-451−/− mice on day 9 post-infection, immunoblotted for Myc and quantified relative to GAPDH (mean ±S.E.M., N=3; *P<0.05). d) qRT-PCR analysis of Myc expression in WT and miR-451−/− CD4+ T-cells pre- and post-malaria infection (N=4-5, paired T-test * p < 0.05). e) Myc mediates increased miR-451−/− cell proliferation. CD4+ T-cells from WT and miR-451−/− mice were isolated, stimulated, and treated with a Myc inhibitor. WT and miR-451−/− CD4+ T-cells had similar cell proliferation in the presence of Myc inhibitor (mean ±S.E.M., N=3; 2-way ANOVA with Tukey post-test; *P<0.05, WT control versus WT plus inhibitor; **P<0.05, miR-451−/− control versus miR-451−/− plus inhibitor).
Myc was identified as a direct miR-451 gene target in a mouse acute lymphoblastic leukemia (T-ALL) model [4]. Myc is a major regulator of cell cycle progression and proliferation of most tissues. To demonstrate that miR-451 regulated Myc expression may contribute to increased miR-451−/− CD4+ proliferation we isolated WT and miR-451−/− CD4+ T-cells and measured Myc protein expression before and 9 days after P. yoelii XNL infection. There was no difference in Myc protein expression between WT and miR-451−/− CD4+ T-cells at baseline (Fig 5B). Myc was markedly increased post-infection in miR-451−/− CD4+ T-cells (Fig 5C). These results were supported by qRT-PCR for Myc (Fig 5D).
To further demonstrate that Myc is a potential mediator of increased miR-451−/− CD4+ T-cell proliferation we stimulated WT and miR-451−/− CD4+ T-cells in the presence of a small molecule inhibitor that directly targets Myc (10058-F4) [22]. Myc inhibitor treatment resulted in WT and miR-451−/− cells having very similar T-cell proliferation (Fig 5E). These results suggest that miR-451 suppression of Myc regulated genes has a direct role in T-cell proliferation.
To identify gene expression changes in miR-451−/− CD4+ T-cells we conducted an Illumina TruSeq Total stranded RNA-seq screen on WT and miR-451−/− CD4+ T-cells isolated both before and on days 6 and 9 post P. yoelii XNL infection (data in Supplemental Excel Table). Pre-infection there were only 82 differentially expressed genes between WT and miR-451−/− T-cells (not shown). Post-infection D9 there were over 5000 differentially expressed genes, 4474 of which were up-regulated in miR-451−/− compared to WT CD4+ T-cells (Fig 6A). Pathway analysis indicated that a large number of the altered genes were regulators of cell growth and proliferation (Table 1). From these data we identified Myc regulated genes increased in miR-451−/− T-cells that are involved in cell growth and proliferation [23–26] (confirmed by the gene ontology reference, Panther Classification System) (Fig 6A red squares). Our survey identified at least 31 directly Myc regulated genes involved in cell proliferation that were increased miR-451−/− CD4+ cells (Fig 6B). Among these genes were Ccnb1, Sdad1, and Apex1 [25] that we confirmed using qRT-PCR (Fig 6C–E). To further confirm the effects of miR-451 on Myc regulated gene expression pathways we applied Ingenuity Pathway Analysis (IPA). The predicted functional pathways for Myc regulated genes that were increased in miR-451−/− CD4+ T-cells included increased cell proliferation (Fig 6F) and limiting of cell death (Fig 6G). These data support a central role for miR-451 in regulating CD4+ proliferative responses.
Figure 6.
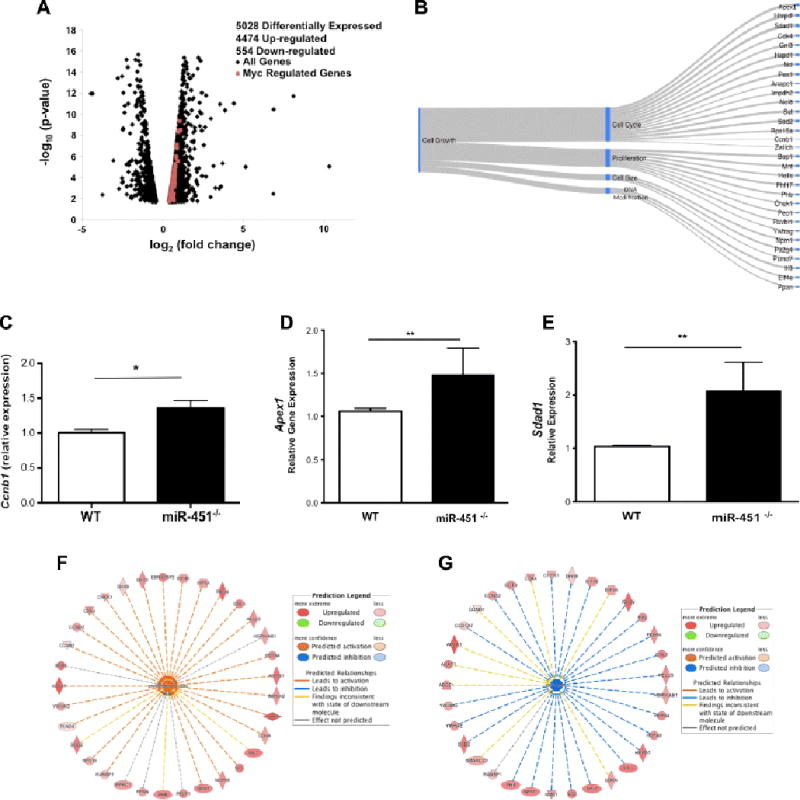
Expression of Myc target genes is increased in miR-451−/− mice postinfection. CD4+ T-cells were isolated from WT and miR-451−/− mice on day 9 postinfection and RNA-seq performed. MiR-451−/− vs WT gene expression was compared. a) Volcano plot (N=2), Myc regulated genes highlighted. b) Myc regulated cell growth related genes are upregulated in miR-451−/− CD4+ T-cells post P. yoelii XNL infection. Google Sankey Chart. c–e) Myc regulated gene expression validation by qRT-PCR. e) Ccnb1, f) Apex1, g) Sdad1 (N=6, *P<0.05, **P <0.01). F) Predicted effect of Myc regulated genes with increased expression in miR-451−/− mice. 25 of the 33 genes lead to cell proliferation. g) Effect of Myc regulated genes increased in miR-451−/− mice on cell death.
Table 1.
Functional classification of genes significantly increased in post-infection miR-451−/− CD4+ cells compared to WT CD4+ cells.
| Category Term | Gene Count | P-Value | Benjamini |
|---|---|---|---|
| phosphoprotein | 1984 | 7.90E-69 | 1.60E-66 |
| nucleus | 1231 | 2.20E-42 | 2.60E-40 |
| alternative splicing | 1157 | 1.70E-04 | 1.60E-03 |
| acetylation | 1081 | 1.60E-147 | 9.30E-145 |
| cytoplasm | 975 | 2.20E-31 | 1.60E-29 |
| ion binding | 962 | 1.30E-02 | 2.40E-01 |
| nucleotide binding | 733 | 2.00E-34 | 1.70E-31 |
| mitochondrion | 617 | 1.80E-92 | 1.10E-89 |
| regulation of transcription | 560 | 2.40E-02 | 3.60E-01 |
| organelle lumen | 544 | 1.10E-85 | 2.40E-83 |
| hydrolase | 481 | 2.40E-14 | 7.30E-13 |
| transferase | 461 | 3.30E-17 | 1.20E-15 |
| transport | 450 | 2.70E-06 | 3.80E-05 |
| protein localization | 304 | 1.80E-26 | 1.20E-23 |
| endoplasmic reticulum | 300 | 4.00E-19 | 1.40E-17 |
| RNA binding | 298 | 2.10E-35 | 3.50E-32 |
| proteolysis | 292 | 1.50E-04 | 6.90E-03 |
| cellular catabolic process | 244 | 8.10E-21 | 2.80E-18 |
| envelope | 244 | 1.30E-31 | 7.00E-30 |
| ribonucleoprotein complex | 241 | 2.20E-44 | 1.70E-42 |
| RNA processing | 237 | 7.00E-45 | 2.90E-41 |
| oxidation reduction | 225 | 1.40E-09 | 1.90E-07 |
| Golgi apparatus | 221 | 2.70E-09 | 6.10E-08 |
| kinase | 198 | 6.10E-03 | 3.80E-02 |
| nucleolus | 194 | 1.20E-51 | 1.30E-49 |
| oxidoreductase | 186 | 1.50E-06 | 2.20E-05 |
| cell cycle | 179 | 5.20E-04 | 2.00E-02 |
| death | 178 | 1.30E-08 | 1.40E-06 |
| ubl conjugation pathway | 178 | 1.20E-10 | 2.60E-09 |
| programmed cell death | 166 | 6.70E-09 | 7.80E-07 |
| homeostatic process | 163 | 7.70E-03 | 1.70E-01 |
| translation | 158 | 2.10E-24 | 9.90E-22 |
| vesicle | 146 | 2.70E-03 | 2.50E-02 |
| cellular response to stress | 144 | 2.40E-08 | 2.50E-06 |
| ligase | 122 | 8.20E-12 | 2.20E-10 |
| chromosome | 108 | 6.00E-03 | 4.80E-02 |
| ribosome | 103 | 2.20E-20 | 8.40E-19 |
| lipid biosynthetic process | 102 | 2.60E-06 | 2.00E-04 |
| protein dimerization activity | 101 | 4.10E-03 | 1.10E-01 |
| chromatin modification | 86 | 7.60E-06 | 5.20E-04 |
| immune system development | 83 | 4.50E-02 | 5.00E-01 |
| protein biosynthesis | 82 | 2.20E-18 | 1.30E-16 |
| cell division | 78 | 6.70E-02 | 6.10E-01 |
| hemopoiesis | 72 | 4.10E-02 | 4.80E-01 |
| regulation of growth | 72 | 6.10E-02 | 5.80E-01 |
DISCUSSION
Our data demonstrated that miR-451 is a regulator of the host immune responses to Plasmodium infection. In particular, miR-451 may in part limit CD4+ T cell proliferation by regulating Myc expression, although a large number of genes are altered post-infection in miR-451−/− CD4+ T cells. Mice lacking miR-451 had a more rapid P. yoelii XNL clearance. The increase in parasite clearance was diminished when CD4+ cells were antibody depleted indicating that CD4+ T cells play a significant role in the miR-451−/− protective phenotype. Other cell types were also increased post infection in the knockout mice, including neutrophils and CD8+ cells, which may be a direct result of a lack of miR-451, or an indirect result of increased T-helper cell responses. Malaria pathogenesis is driven by many parts of the immune response and miR-451 may have a potentially broad immune regulatory role in other cell types as well, either directly or indirectly. This will require more cell by cell analysis.
Our mutant mouse data are in contrast to in vitro studies indicating that increased miR-451 in human sickle cell disease RBCs may contribute to limiting Plasmodium growth [27]. The different effects for miR-451 on Plasmodium growth in vitro and in vivo may reflect differences between model systems (in vitro vs in vivo models) or the role of individual cell types in isolation vs whole organism systems. This may also indicate that miR-451 has different effects depending on whether its expression is increased or decreased. Our studies show that decreased miR-451 leads to protective immune defects. However, over-expression of miR-451 may not affect the immune response, but rather have a direct effect on parasite growth. These studies clearly indicate that caution is needed when considering the in vivo implications of in vitro miRNA studies, as a miRNA may influence the in vivo responses of many cells and organ systems.
The miR-451−/− protective phenotype is largely CD4+ T-cell dependent, although it may not be a direct CD4+ T-cell effect on infected RBCs. The primary function of T helper cells is to direct the activity of other immune cells. We have not yet elucidated the cell type or types that clear infected RBCs in miR-451−/− mice, but because all classes of T helper cells are increased in infected miR-451−/− mice, it is likely that many cell types mediate the accelerated clearance in a manner dependent on CD4+ cells. MiR-451−/− mice had more CD8+ T-cells post infection, but our cell depletion studies indicated that a role for CD8+ cells in parasite clearance was minimal compared to CD4+ T-cells. Increased CD8+ numbers may indicate that miR-451 also has a direct role in regulating CD8+ cell proliferation, or increased CD8+ cells is an indirect effect of the increased CD4+ response. For example, IFN-γ can directly increases CD8+ cells during viral infection [28]. A reduction in miR-451 expression also promotes neutrophil chemotaxis [9] and may contribute to the increased post-infection neutrophil count noted in the KO mice. Increased neutrophils may also be an indirect effect of the T helper cell responses, particularly secondary to the increase in Th17 cells.
In this non-lethal malaria model a lack of miR-451 is protective by driving increased T-cell proliferation. In other disease models, such as transplant rejection, the presence of miR-451 may be protective by limiting T-cell responses. A number of studies have demonstrated that CD4+ T-cells are critical in blood stage malaria clearance in both humans [29] and mice [13, 30]. Our study indicates that miR-451 regulation of Myc expression in CD4+ T-cells impacts host immune responses. These data are consistent with a previous study demonstrating that miR-451 regulates Myc expression in a mouse model of T-ALL. However, a functional role of miR-451 in the context of host immune responses has not been described. Our RNA-Seq data revealed over 5000 differentially expressed genes in miR-451−/− CD4+ T-cells compared to WT CD4+ T-cell gene expression post infection. Many Myc target genes that promote cell proliferation were upregualted in miR-451−/− CD4+ cells suggesting that miR-451 regulation of Myc expression is a key regulator of activated T-cell proliferation. Given the large number of genes significantly increased post-infection, Myc is unlikely to be the only gene involved in the miR-451−/− T-cell phenotype. It is likely that miR-451 directly or indirectly regulates multiple gene pathways involved in cell proliferation. Further studies are needed to elucidate these other relevant pathways.
MiRNA has important roles in regulating host immune responses. Our study identifies a unique role for miR-451 in CD4+ proliferation potentially impacting our understanding of the immune responses in multiple inflammatory and infectious diseases.
Supplementary Material
Acknowledgments
This work is funded by NIH grants to LMC (F31AI108187) and CNM (R01HL124018) and an American Heart Association (13EIA14250023) to CNM. The project was also supported by the following University of Rochester CTSA awards: TL1 RR024135, TL1 TR000096, UL1 RR024160 and UL1 TR000042 from the National Center for Research Resources and the National Center for Advancing Translational Sciences of the National Institutes of Health. Acknowledgements
The authors would like to thank Dr. Eric Olson for the miR-451−/− mice.
Footnotes
The authors have no financial conflicts.
References
- 1.O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10(2):111–22. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 2.Rossi RL, Rossetti G, Wenandy L, Curti S, Ripamonti A, Bonnal RJP, et al. Distinct microRNA signatures in human lymphocyte subsets and enforcement of the naive state in CD4+ T cells by the microRNA miR-125b. Nat Immunol. 2011;12(8):796–803. doi: 10.1038/ni.2057. http://www.nature.com/ni/journal/v12/n8/abs/ni.2057.html-supplementary-information. [DOI] [PubMed] [Google Scholar]
- 3.Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451(7182):1125–9. doi: 10.1038/nature06607. http://www.nature.com/nature/journal/v451/n7182/suppinfo/nature06607_S1.html. [DOI] [PubMed] [Google Scholar]
- 4.Li X, Sanda T, Look AT, Novina CD, von Boehmer H. Repression of tumor suppressor miR-451 is essential for NOTCH1-induced oncogenesis in T-ALL. The Journal of Experimental Medicine. 2011;208(4):663–75. doi: 10.1084/jem.20102384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patrick DM, Zhang CC, Tao Y, Yao H, Qi X, Schwartz RJ, et al. Defective erythroid differentiation in miR-451 mutant mice mediated by 14-3-3ζ. Genes & Development. 2010;24(15):1614–9. doi: 10.1101/gad.1942810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu D, dos Santos CO, Zhao G, Jiang J, Amigo JD, Khandros E, et al. miR-451 protects against erythroid oxidant stress by repressing 14-3-3ζ. Genes & Development. 2010;24(15):1620–33. doi: 10.1101/gad.1942110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.LaMonte G, Philip N, Reardon J, Lacsina Joshua R, Majoros W, Chapman L, et al. Translocation of Sickle Cell Erythrocyte MicroRNAs into Plasmodium falciparum Inhibits Parasite Translation and Contributes to Malaria Resistance. Cell Host & Microbe. 12(2):187–99. doi: 10.1016/j.chom.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H-P, Zeng X-C, Zhang B, Long J-T, Zhou B, Tan G-S, et al. miR-451 inhibits cell proliferation in human hepatocellular carcinoma through direct suppression of IKK-β. Carcinogenesis. 2013;34(11):2443–51. doi: 10.1093/carcin/bgt206. [DOI] [PubMed] [Google Scholar]
- 9.Murata K, Yoshitomi H, Furu M, Ishikawa M, Shibuya H, Ito H, et al. MicroRNA-451 Down-Regulates Neutrophil Chemotaxis via p38 MAPK. Arthritis & Rheumatology. 2014;66(3):549–59. doi: 10.1002/art.38269. [DOI] [PubMed] [Google Scholar]
- 10.Li X, Sanda T, Look AT, Novina CD, von Boehmer H. Repression of tumor suppressor miR-451 is essential for NOTCH1-induced oncogenesis in T-ALL. J Exp Med. 2011;208(4):663–75. doi: 10.1084/jem.20102384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smigielska-Czepiel K, van den Berg A, Jellema P, van der Lei RJ, Bijzet J, Kluiver J, et al. Comprehensive analysis of miRNA expression in T-cell subsets of rheumatoid arthritis patients reveals defined signatures of naive and memory Tregs. Genes and immunity. 2014;15(2):115–25. doi: 10.1038/gene.2013.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Organization WH. World Malaria Report 2009. 2014 Dec; http://www.who.int/features/factfiles/malaria/en/
- 13.Butler NS, Moebius J, Pewe LL, Traore B, Doumbo OK, Tygrett LT, et al. Therapeutic blockade of PD-L1 and LAG-3 rapidly clears established blood-stage Plasmodium infection. Nat Immunol. 2012;13(2):188–95. doi: 10.1038/ni.2180. http://www.nature.com/ni/journal/v13/n2/abs/ni.2180.html-supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vinetz JM, Kumar S, Good MF, Fowlkes BJ, Berzofsky JA, Miller LH. Adoptive transfer of CD8+ T cells from immune animals does not transfer immunity to blood stage Plasmodium yoelii malaria. The Journal of Immunology. 1990;144(3):1069–74. [PubMed] [Google Scholar]
- 15.Scanga CA, Mohan VP, Yu K, Joseph H, Tanaka K, Chan J, et al. Depletion of Cd4+ T Cells Causes Reactivation of Murine Persistent Tuberculosis despite Continued Expression of Interferon γ and Nitric Oxide Synthase 2. The Journal of Experimental Medicine. 2000;192(3):347–58. doi: 10.1084/jem.192.3.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.George TC, Ortaldo JR, Lemieux S, Kumar V, Bennett M. Tolerance and Alloreactivity of the Ly49D Subset of Murine NK Cells. The Journal of Immunology. 1999;163(4):1859–67. [PubMed] [Google Scholar]
- 17.Fecci PE, Sweeney AE, Grossi PM, Nair SK, Learn CA, Mitchell DA, et al. Systemic Anti-CD25 Monoclonal Antibody Administration Safely Enhances Immunity in Murine Glioma without Eliminating Regulatory T Cells. Clinical Cancer Research. 2006;12(14):4294–305. doi: 10.1158/1078-0432.ccr-06-0053. [DOI] [PubMed] [Google Scholar]
- 18.Kruisbeek AM, Shevach E, Thornton AM. Proliferative Assays for T Cell Function Current Protocols in Immunology. John Wiley & Sons, Inc; 2001. [DOI] [PubMed] [Google Scholar]
- 19.Shi G, Field DJ, Ko K-a, Ture S, Srivastava K, Levy S, et al. Platelet factor 4 limits Th17 differentiation and cardiac allograft rejection. The Journal of Clinical Investigation. 2014;124(2):543–52. doi: 10.1172/JCI71858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Google Charts API Homepage. https://developers.google.com/chart/. 2015. Accessed November 18, 2014.
- 21.Li C, Seixas E, Langhorne J. Rodent malarias: the mouse as a model for understanding immune responses and pathology induced by the erythrocytic stages of the parasite. Med Microbiol Immunol. 2001;189(3):115–26. doi: 10.1007/s430-001-8017-8. [DOI] [PubMed] [Google Scholar]
- 22.Wang H, Hammoudeh DI, Follis AV, Reese BE, Lazo JS, Metallo SJ, et al. Improved low molecular weight Myc-Max inhibitors. Molecular Cancer Therapeutics. 2007;6(9):2399–408. doi: 10.1158/1535-7163.mct-07-0005. [DOI] [PubMed] [Google Scholar]
- 23.Ji H, Wu G, Zhan X, Nolan A, Koh C, De Marzo A, et al. Cell-Type Independent MYC Target Genes Reveal a Primordial Signature Involved in Biomass Accumulation. PLoS ONE. 2011;6(10):e26057. doi: 10.1371/journal.pone.0026057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marinkovic D, Marinkovic T, Kokai E, Barth T, Möller P, Wirth T. Identification of novel Myc target genes with a potential role in lymphomagenesis. Nucleic Acids Research. 2004;32(18):5368–78. doi: 10.1093/nar/gkh877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menssen A, Hermeking H. Characterization of the c-MYC-regulated transcriptome by SAGE: Identification and analysis of c-MYC target genes. Proceedings of the National Academy of Sciences. 2002;99(9):6274–9. doi: 10.1073/pnas.082005599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeller K, Jegga A, Aronow B, O’Donnell K, Dang C. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biology. 2003;4(10):R69. doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LaMonte G, Philip N, Reardon J, Lacsina JR, Majoros W, Chapman L, et al. Translocation of sickle cell erythrocyte microRNAs into Plasmodium falciparum inhibits parasite translation and contributes to malaria resistance. Cell host & microbe. 2012;12(2):187–99. doi: 10.1016/j.chom.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitmire JK, Tan JT, Whitton JL. Interferon-gamma acts directly on CD8+ T cells to increase their abundance during virus infection. J Exp Med. 2005;201(7):1053–9. doi: 10.1084/jem.20041463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pombo DJ, Lawrence G, Hirunpetcharat C, Rzepczyk C, Bryden M, Cloonan N, et al. Immunity to malaria after administration of ultra-low doses of red cells infected with Plasmodium falciparum. The Lancet. 360(9333):610–7. doi: 10.1016/S0140-6736(02)09784-2. [DOI] [PubMed] [Google Scholar]
- 30.Muxel SM, Freitas do Rosário AP, Zago CA, Castillo-Méndez SI, Sardinha LR, Rodriguez-Málaga SM, et al. The Spleen CD4<sup>+</sup> T Cell Response to Blood-Stage <italic>Plasmodium chabaudi</italic> Malaria Develops in Two Phases Characterized by Different Properties. PLoS ONE. 2011;6(7):e22434. doi: 10.1371/journal.pone.0022434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


