Abstract
Background
We have recently shown in both non-human primates and in rodents that fetal and neonatal hepatic expression of the circadian transcription factor, Npas2, is modulated by a high fat maternal diet and plays a critical role in establishing life-long metabolic homeostasis. Similarly, we and others have also established the importance of the maternal and early postnatal diet on establishment of the early gut microbiome.
Objective
We hypothesized that altered circadian gene expression solely in the neonatal liver would result in gut microbiome dysbiosis, especially with diet-induced metabolic stress (i.e., restricted feeding). Using a murine model where we conditionally knock out of Npas2 in the neonatal liver, we aimed to determine the role of the circadian machinery in gut dysbiosis with restricted feeding.
Design
We collected fecal samples from liver Npas2 conditional knock out (cKO; n=11) and wild type (wt: n=13) reproductive-aged mice before (study day 0) and after the restricted feeding study (study day 17). Extracted DNA was sequenced using the MiSeq Illumina platform using primers specific for the V4 region of the 16S rDNA gene. The resulting sequences were quality filtered, aligned, and assigned taxonomy. Principal coordinate analysis was performed on unweighted and weighted UniFrac distances between samples with Permutation ANOVA to assess clustering significance between groups. Microbial taxa that significantly differ between groups of interest was determined using Linear Discriminate Analysis Effect Size and randomForrest.
Results
Principal coordinate analysis performed on weighted UniFrac distances between male cKO and wt cohorts revealed that the gut microbiome of the mice did not differ by genotype at the start of the restricted feeding study, but did differ by virtue of genotype at the end of the study (p=0.001). Moreover, these differences could be at least partially attributed to restricted feeding-associated alterations in relative abundance of the Bacteroides genus, which has been implicated as crucial to establishing a healthy gut microbiome early in development.
Conclusions
Here we have provided an initial key insight into the interplay between neonatal establishment of the peripheral circadian clock in the liver and the ability of the gut microbiome to respond to dietary and metabolic stress. As Npas2 expression in the liver is a target of maternal high fat diet induced metabolic perturbations during fetal development, we speculate that these findings have potential implications in the long term metabolic health of their offspring.
Keywords: peripheral circadian clock, Npas2, restricted feeding, gut microbiome, mouse model
Introduction
Overview
The study of how circadian biology is established, and when and how it is modified, is of importance to obstetrical and neonatal providers since intact circadian rhythms are responsible for the coordination of metabolism with key behaviors such as sleep and feeding (1). With increasing attention on the role of the human microbiome in health and disease, it is evident that many aspects of our lifelong metabolic health are influenced not by only by our genetics, but rather by the totality of our genomic and metagenomic inheritance (2,3). This includes both changes upon the genome (or “epigenetics”) such as post-translational histone modifications and DNA methylation, as well as the microbiome's metagenome (4,5). While it has previously been shown that there is bi-directional communication between the host circadian biology and the gut microbiome (3,6–8), the timing and mechanisms by which this occurs remains unknown.
Establishing Circadian Clock Function in Early Life: Fetal Disruption of Npas2 and its Long-term Health Effects
Previous work in our nonhuman primate model of maternal high-fat diet feeding led to the discovery that the circadian gene, neuronal PAS domain protein 2 (Npas2), acted as an epigenomically modified regulator of fetal hepatic metabolism (9,10). Fetuses exposed to a maternal high-fat diet (HFD) in utero demonstrated a significant increase in acetylation of histone H3 at lysine 14 (H3K14ac); these alterations persisted in juvenile animals (9–11). Using differential display chromatin immunoprecipitation (ChIP) approaches to determine where this modification was enriched throughout the fetal hepatic genome, we found differential occupancy of the Npas2 promoter region by H3K14ac H3 between control and HFD exposed offspring (9). Further analysis using ChIP followed by site-specific qPCR revealed an increase of H3K14ac occupancy in the Npas2 promoter in animals exposed to a HFD in utero (10), a finding which persisted in juvenile animals despite later consumption of a control diet (3). This alteration in promoter occupancy associated with differential Npas2 transcription between the control diet (CD) and HFD exposed offspring (10), and was accompanied by persistent metabolic dysfunction and non-alcoholic fatty liver disease through at least three years of age (12–18). These findings not only implicate a lasting role of a high fat maternal diet in regulating fetal and postnatal Npas2, but provide additional evidence of the importance of the liver circadian clock in maintaining optimal metabolic health in the offspring, starting in utero and persisting into their juvenile years. However, precisely when, where and how epigenomic reprogramming of the offspring liver exerts such broad metabolic disarray is poorly understood.
When and Where: Generation of Neonatal Liver-Specific Npas2 cKO Mice and Overall Study Design
In order to first separate the impact of the role of immediate neonatal versus circadian gene expression in the liver on the lifelong regulation of metabolic homeostasis, we generated a novel conditional knock out mouse model in which Npas2 is specifically deleted in the neonatal (but not the fetal) liver.
Previous studies have characterized the metabolic phenotype of an Npas2 KO (“whole body knock out”) mouse model and under normal metabolic conditions no phenotype was reported (19–22). However, under a restricted feeding regimen, Npas2 KO mice lose a greater body weight percentage when food is restricted over a 2 week period, suggesting a morbid maladaptation and abnormal satiety signals (23,24). In one study the weight loss was so severe that the KO mice wasted resulting in the death of 20% of mice (23). However, in these previous studies, Npas2 was knocked out in all tissues, including the brain (i.e., the light-dark responsive master clock in the superchiasmatic nucleus, or SCN) which is presumed to be the master pacemaker of the circadian clock. To determine if neonatal Npas2 peripheral expression in the liver is necessary for adaptive feeding, we exposed Npas2 cKO mice to a restricted feeding regimen, which is the only previously published condition to elucidate a unique role for Npas2 in regulating metabolic homeostasis.
How: Circadian Interactions & the Gut Microbiome
We and others have recently shown that the neonate is not born sterile, and that the primate and human gut microbiome is persistently altered with maternal high fat diet exposure (25,26). Emerging evidence has indicated that the host circadian clock is closely intertwined with the gut microbiome (8,27–33). Work in the germ-free mouse has demonstrated that the peripheral circadian clock, particularly in the liver, is potentially regulated by the gut microbiome, as mice devoid of any microbes demonstrate altered patterns of hepatic circadian gene expression (34,35). However, given the poor overall metabolic health of gnotobiotic animals, discerning cause from effect is problematic. Similarly, the composition of the gut microbiome has been shown to exhibit a circadian rhythmicity that is abolished in Clock and Bmal1 knockout mice (36,37), indicating a bi-directional dependency between circadian clock rhythmicity and gut microbiome composition.
Subsequent studies have further indicated that the impact of circadian clock disruption on the gut microbiome manifests most prominently when the animals are placed under a dietary stress, like a high-fat diet challenge (38). However, whether or not the dependency of gut microbiome on host circadian clock gene expression is due to the central clock in the brain or through the liver has not been explored. Although evidence points to the hepatic clock as the likely regulator of the gut environment, the impact of the central clock cannot be ruled out as the gut and the central nervous system are closely interconnected through the many neural projections via the gut-brain-axis.
To determine how isolated postnatal disruption of the liver circadian clock impacts the later composition of the gut microbiome, in the current study we generated a conditional knockout mouse model whereby Npas2 was specifically deleted in hepatocytes during neonatal life and henceforth. Considering ours and others past work demonstrating the impact of the dietary stress on circadian clock disruption, we hypothesized that under normal conditions, the gut microbiome would be unchanged, but would be differentially altered during a period of a restricted feeding. The aims of the current study were therefore to address the mechanistic role of neonatal Npas2 expression on later in life metabolism and gut dysbiosis.
Materials and Methods
Npas2 hepatic cKO generation
The Npas2 floxed allele was designed to delete exon 3 through Cre mediated recombination. Deletion of exon 3 eliminates the coding region for the basic helix-loop-helix DNA binding domain of Npas2 as well as the BMAL1 binding domain, encoding for a non-functional NPAS2 protein (39,40). Mice heterozygous for the Npas2 floxed allele (Npas2 fl/+) mice were crossbred to generate mice homozygous for the floxed Npas2 allele (Npas2 fl/fl). These and subsequent mice were housed at the ABBR mouse facility at Baylor College of Medicine and all animal procedures were in accordance with the guidelines of Institutional Animal Care and Use Committee (IACUC, AN-4826). Npas2 fl/fl mice were bred with B6.Cg-Tg(Alb-cre)21Mgn/J mice purchased from Jackson Laboratories (Alb-cre, Jackson Laboratories, Bar Harbor, Maine) which are homozygous for the transgenic cre gene with an albumin promotor generating mice heterozygous for the floxed Npas2 and Alb-cre transgenic alleles (Npas2 fl/+/Alb- cre). In Alb-cre mice, cre is expressed in the post-natal liver (41). Npas2 fl/+/Alb- cre females were mated to Npas2 fl/fl males to generate hepatocyte specific Npas2 conditional knock out (cKO, Npas2 fl/fl/Alb- cre) mice. Npas2 fl/fl (WT) offspring were used as controls in this study (Supplemental Figure 1).
Restricted feeding study
Mice are nocturnal animals, and preferentially feed at night during the dark. To measure the adaptation of mice to a change in timing and frequency in meals, 16 week-old Npas2 cKO and WT mice were placed in comprehensive lab animal monitoring system (CLAMS, Oxymax-CLAMS, Columbus Instruments, Columbus, OH) to measure the amount of food consumed by the mouse. PicoLab Select Rodent 50 1F/6F (LabDiet, St. Louis, MO), in powdered form, was placed in the feeding assembly. The mouse accesses food by eating from a spring loaded dish through an anti-foraging guard; this guard also prevents full body access to the dish. Spillage is accounted for by a larger dish, beneath the main food cup, that catches anything that spills over the edge. This spillage collection cup also rests on the balance, and though spilled food is not accessible to the mouse, the mass remains on the balance. The mass of food consumed is monitored continuously each minute. Mice were housed individually at 14 weeks old before being transferred to the CLAMS cages at 16 weeks old. Mice were checked daily for health and proper maintenance of the monitoring system. The mice were conditioned to the restricted feeding by gradually reducing the time allotted for food access from ad libitum to 4 hours per day over a 5-day period. Once initiated, mice were given access to food for only 4 hours per day for 17 days (Figure 1A). Mice were weighed every two days. As Npas2 is a circadian gene, these experiments were performed under normal light conditions (12hrs light and 12hrs darkness per day) with regulated temperature.
Figure 1. Restricted feeding challenge of 16 week old Npas2 cKO and WT mice.
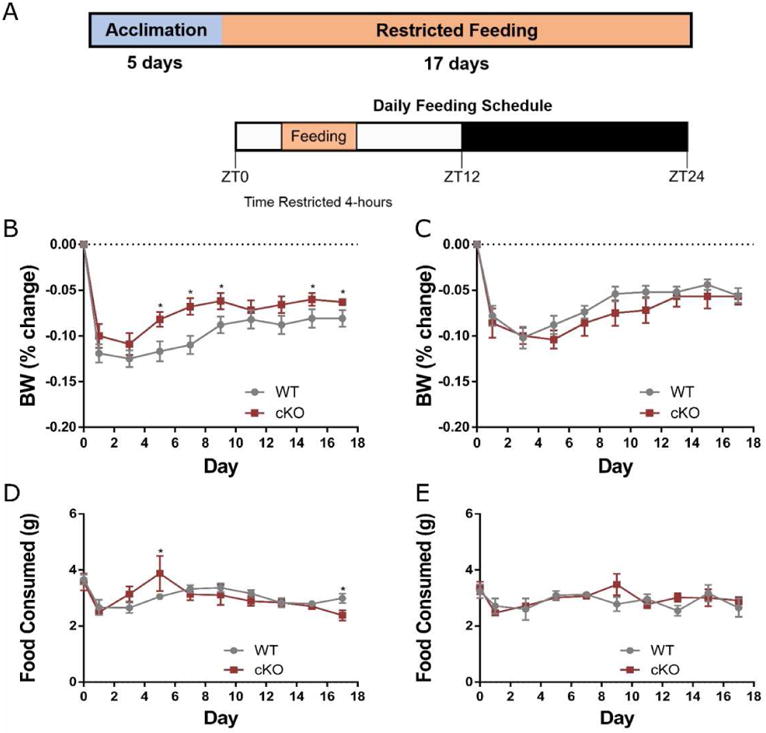
(A) Study Design. Briefly, mice were placed in CLAMS cages and acclimated to a 4-hour/day feeding period over 5 days, then the mice were allowed access to food for 4 hours/day (9:00 to 13:00) for 17 days. (B-C) Weights shown as percentage of body weight (BW) change in relation to the starting weight in males (B) and females (C) over a 17 day period. Male cKO mice lost significantly less weight than WT mice starting at day 5 and continuing through day 9, and again on days 15 and 17 of the restricted feeding study. (D-E) Daily total food intake (grams) in males (D) and females (E) over the 17 day course of the study. All data shown as mean with error bars representing 1 SE. * = P<0.05.
Microbiome 16S Sequencing and Analysis
Fecal samples were collected into PowerSoil Bead Tubes (MoBio Laboratories, Inc., Carlsbad, CA) from mice at the same time of day (1300-1330 hrs) before entering the CLAMS cages and again at the end of the restricted feeding regimen (day 17). DNA was extracted from mouse stool using the PowerSoil DNA Isolation Kit according to the manufactures suggested protocol and outlined in previous publications. Extracted DNA was sequenced using the MiSeq Illumina platform (2×250 bp) using primers specific for the V4 region of the 16S rDNA gene (515F, 806R). The resulting sequences were quality filtered, aligned, and assigned taxonomy via pipelines developed in-house (25,42–46). Briefly, read pairs were demultiplexed and merged using USEARCH v7.0.01090. Merging thresholds allowed for zero mismatches and a minimum of 50 bp overlap. Merged reads were subsequently trimmed at the first base with a Q ≤ 5 and reads with >0.5% expected errors were discarded. Operational taxonomic units (OTUs) were called at 97% similarity using the UPARSE algorithm. Chimeric sequences were detected and removed using the USEARCH and UCHIME algorithms. OTUs were assigned taxonomy by mapping to the SILVA database trimmed to the 16S V4 region. Abundances were recovered by mapping the merged reads to the UPARSE OTUs. A custom script constructed a rarefied OTU table from the output files generated in the previous two steps for downstream analyses of alpha-diversity, beta-diversity (including UniFrac), and phylogenetic trends.
Statistical Analysis
Statistical significance for weights and food intake were determined by unpaired t-test with Welch's correction. p<0.05 was considered statistically significant with data shown as mean ± SEM. Statistical analysis of 16S sequencing data was performed using in-house pipelines based on the QIIME and R platforms. Principal coordinate analysis was performed on unweighted and weighted UniFrac distances between samples. Permutation ANOVA (PERMANOVA) was used to assess clustering significance between groups. Microbial taxa that significantly differ between groups of interest was determined using Linear Discriminate Analysis Effect Size (LEfSe) and randomForrest (47,48).
Results
Male Npas2 cKO Recover More Weight on Restricted Feeding Challenge
To determine if neonatal Npas2 peripheral expression in the liver is necessary for adaptive feeding, Npas2 cKO mice were restricted food access to four hours a day (9:00 to 13:00 hours) for 17 days (Figure 1A). Body weight was significantly reduced from baseline in both cKO and WT groups regardless of gender, and no group recovered their body weight to baseline levels during the restricted feeding study (Figure 1B,C). 16-week old cKO males recovered significantly more body weight by day 5 through day 9 (p<0.05 cKO (n=6) compared to WT (n=10), Figure 1B). At the end of the 17 day restricted feeding study cKO male mice lost a significantly lower percentage of their weight compared to WT mice. Female cKO (n=9) mice had no significant difference in weight loss compared to WT (n=10) mice (Figure 1C). Daily food intake did not vary significantly between cKO and WT mice except at day 5 (p=0.033) and day 17 for the males (p=0.009, Figure 1D,E).
A Restricted Feeding Diet Results in Specific Changes to the Gut Microbiota
To determine the concomitant changes in the gut microbiota as a result of restricted feeding (RF) on a normal chow diet, fecal samples were collected immediately before the period of restricted feeding, and at the end of the 2-week dietary challenge. Extracted DNA was subjected to 16S rRNA gene sequencing, resulting in 1197502 high-quality sequences after filtering (24947 +/- 2423 per sample), and representing 318 unique OTUs.
After a 17-day period of restricted feeding, mice were found to harbor a significantly different gut microbiota profile, independent of gender or genotype (Figure 2). These differences were apparent by both unweighted (p=0.001) and weighted (p=0.001) UniFrac measures (Figure 2A,B), indicating a robust difference in the gut microbial communities related to the presence and relative abundance of the gut microbiota and its proportional relative abundance, respectively. At the end of the restricted feeding period, mice harbored significantly increased relative abundance of Bacteroides and other related members of the order Bacteroidales (p<0.05). Conversely, the relative abundances of several unclassified taxa within the Lachnospiraceae family were significantly decreased following restricted feeding (all p<0.05, Figure 2C). This trend was significant regardless of gender or genotype.
Figure 2. The gut microbiome is significantly altered after restricted feeding.
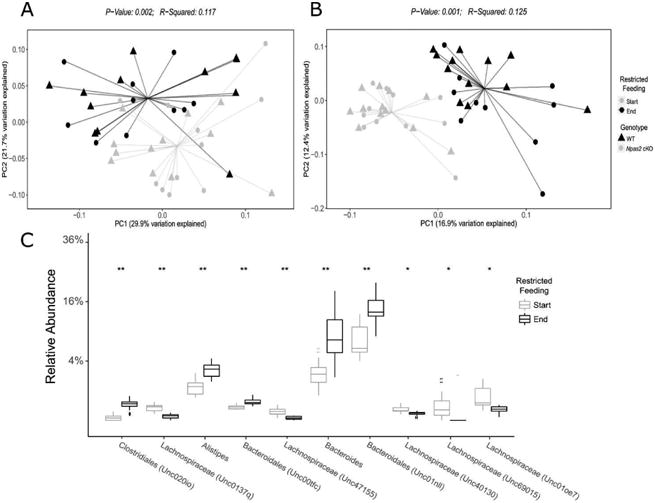
Principal coordinate analysis was performed on weighted (A) and unweighted (B) UniFrac distances between samples. PC1 (A 29.9%, B 16.9%) and PC2 (A 21.7%, B 12.4%) represent the first and second Principal Coordinate (PC) Axes determined by Principal Coordinate Analysis (PCoA), which respectively represent the percentage of the total variation within the dataset. The gut microbiome of the mice at the start of the restricted feeding study was significantly different from the gut microbiome at the end of the study (weighted p=0.001, unweighted p=0.001). (C) Significantly different relative abundance of various taxa between the start and end of the restricted feeding study (*p<0.05, **p<0.01, ***p<0.001).
Male Npas2 cKO Mice Have an Altered Gut Microbiome after Restricted Feeding
We next sought to determine if there were specific changes to the gut microbiota that were associated with reduced weight loss in the male Npas2 cKO mice compared to WT. Because the male Npas2 cKO mice, and not the females, were protected from RF diet-induced weight loss, we focused our analysis to only the male mice. First, however, we compared composition of the gut microbiome between males (n=16) and females (n=8). The alpha diversity fell during the restricted feeding for both males and females and there was no significant differences between gender or genotype in this regard (Supplemental Figure 2A). Before restricted feeding no significant difference in the gut microbiome between males and females was observed, but a non-significant trend is notable at the end of restricted feeding, with female samples clustering together and distinctly from the majority of male samples (Supplemental Figure 2B). As shown in Figure 3A, the greatest difference in the gut microbiome appeared to be driven by the restricted feeding diet, best seen along the PC1 axis. However, along the PC2 axis, while the WT and Npas2 cKO mice clustered similarly at the onset of the RF diet, they separated significantly along the PC2 axis at the conclusion of the RF challenge (Figure 3A, light blue circles vs. light red triangles). This difference in the gut microbiome between the WT and Npas2 cKO mice as a result of the diet can be best appreciated when samples from the same mouse were analyzed in a pair-wise regression, indicating a difference in the overall rate of change or trajectory of the gut microbiome over time (Figure 3B, C).
Figure 3. The gut microbiome is differentially altered in male Npas2 cKO mice after restricted feeding.
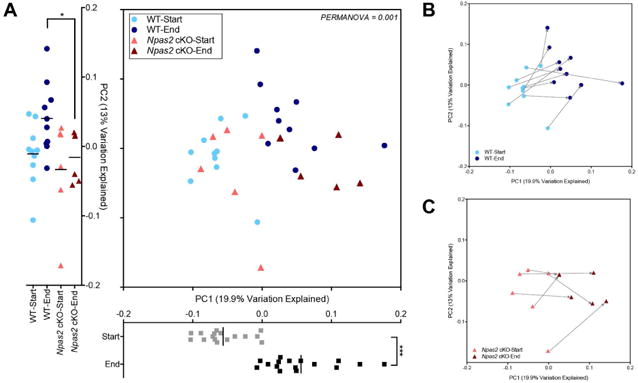
(A) Principal coordinate analysis performed on unweighted UniFrac distances between male samples only, before and after restricted feeding. PC1 and PC2 represent the first and second Principal Coordinate (PC) Axes determined by Principal Coordinate Analysis, which respectively represent 19.9% and 13.0% of the total variation within the dataset. The difference in the gut microbiome before and after the restricted feeding diet shown along the PC1 axis (grey, start; black, end), while the difference between the genotypes after a restricted feeding diet best seen along the PC2 axis. Blue colors indicate WT mice, while red colors indicate Npas2 cKO mice (Significance determined by a Mann-Whitney U test; *p<0.05, ***<0.001). (B, C) PCoA with samples obtained from same mouse connected by an arrow (arrow start, before RF diet; arrow end, after RF diet). WT (B) and Npas2 cKO (C) mice shown separately.
Specific Changes to the Gut Microbiome of Male Npas2 cKO Mice
Comparing the weight loss (%) from day 0 to day 17 in male mice only showed distinct correlations with the gut microbiota based on genotype (Figure 4). A distinct cluster consisting primarily of the Ruminiclostridium genus correlated positively with increased weight loss in the WT, but showed opposing correlations in the cKO group. Other genera from the WT group correlated with increased weight loss included Bacteroides, Roseburia, Oscillibacter, Staphylococcus, and Veillonella, with these genera also showing opposing correlations in the cKO group. Conversely, increased weight loss from the cKO group correlated with Eubacterium_coprostanoligenes_group, Corynebacterium, Parasutterella, Bacteroidales_S24_7_group, and Allobaculum, with these genera being negatively correlated with weight loss in the WT group. These findings show distinct correlations associated with the loss of hepatic Npas2 expression, with altered bacterial genera associated with the significant difference in weight loss between WT and cKO mice.
Figure 4. Alpha diversity of gut microbiome altered in response to a RF diet, but is unchanged in the male Npas2 cKO mice.
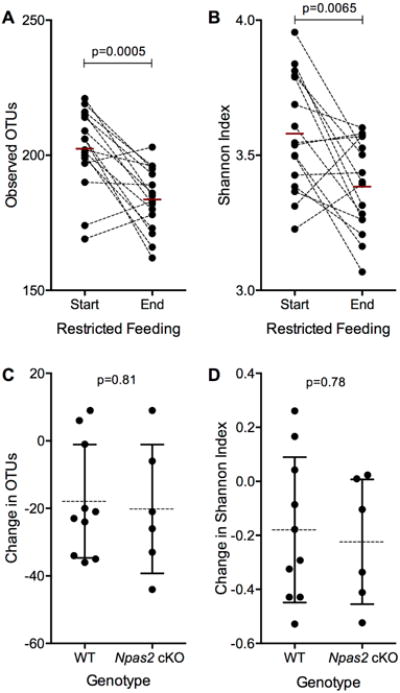
(A,B) Difference in the number of (A) observed OTUs and the (B) Shannon Index before and after a restricted feeding diet. Samples from the same mouse are connected by the dotted line, with the red bar indicating the mean for each time point. Significance determined by a paired t-test. (C, D) The change in the (C) number of OTUs and (D)Shannon Index before and after the RF diet compared between the WT and Npas2 cKO mice. The mean and standard deviation is shown for each group. Significance was determined by a Student's t-test.
Previous reports have indicated that changes in dietary composition differentially supports the growth of particular strains of Bacteroides. To interrogate how different Bacteroides species respond to a RF diet, we more closely examined all OTUs assigned to Bacteroides. Within our study, three OTUs were confidently assigned to Bacteroides at the genus level, likely representing three distinct strains (BcmNL440, Unc00nc3 and Unc41782). When summated, we observed a significant increase Bacteroides in both WT and Npas2 cKO mice as a result of the RF diet (Figure 6A), consistent with our findings across the whole cohort shown in Figure 2. However, when examining the overall change of Bacteroides between the genotypes, we found that Npas2 cKO mice tended have a greater but non-significant gain in overall Bacteroides representation (Figure 6B). When examining the individual OTUs that represented the Bacteroides genus, we found that in WT mice, the response of each OTU in the face of RF challenge was inconsistent – in some animals, the relative abundance decreased, while in others, in significantly increased (Figure 6C). In comparison, in all Npas2 cKO mice, Bacteroides was consistently elevated in response to a RF challenge (Figure 6C). Despite this observation, only one OTU (Unc41782), saw a significantly different change in relative abundance as a result of the RF diet (Figure 6D).
Figure 6.
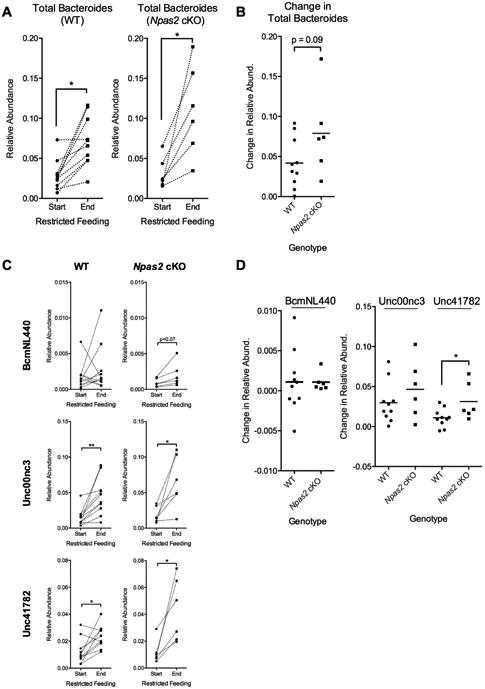
Specific Bacteroides OTUs differentially altered in male NPAS2 cKO mice in response to a RF diet. (A) The total relative abundance of all OTUs assigned to Bacteroides compared between the start and end of the RF diet. Individual mice are connected by a dotted line. Significance determined by a paired t-test (*p<0.05). (B) Comparison of the relative change of Bacteroides before and after a RF diet between male WT and NPAS2 cKO mice. (C) The relative abundance of the three individual OTUs assigned to Bacteroides at high confidence before and after a RF diet, in both WT and NPAS2 cKO mice. Significance determined by a paired t-test (*p<0.05, **<0.01). (D) Comparisons of the change in relative abundance of each Bacteroides OTU between male WT and NPAS2 cKO mice. Significance determined by a Student's t-test (*p<0.05).
Comments
Main findings
Disruption of circadian rhythms, whether by environmental factors such as inadequate sleep, dietary alterations, and shift work, or by genetic factors such as gene mutations or epigenetic remodeling, is associated with various factors of metabolic syndrome (49–55). However, how and when these alterations to the circadian clock are affecting metabolic homeostasis is unclear. During fetal life there is no light-dark signaling and the circadian rhythmicity is thought to be primarily imparted by the maternal metabolic cycle (9-11). In contrast, early in neonatal life light and dark signals presumptively begin to supersede the maternal-driven regulation of the circadian clock (1-3). However, we and others have previously demonstrated that during both fetal and infant developmental time points, a high fat maternal diet appears to have a dominant impact on the molecular regulation of the offspring circadian clock machinery, particularly epigenomic modifications to crucial gene promoter regions of Npas2 (9-11; 56–60). Prior to the studies undertaken herein, it was unknown whether it is maternal signaling, diet, or light/dark cycles that regulated the neonatal and infant circadian directed metabolism. Moreover, although several studies have examined the potential interplay between the microbiome, circadian rhythms, and metabolism, teasing apart association and causation was challenging (1-8).
Utilizing a postnatal conditional knock out mouse model whereby we deleted Npas2 in the neonatal liver, we took an innovative mechanistic approach to begin to tackle the question of what the lifelong effects of the disruption of the peripheral circadian clock are on metabolic homeostasis and microbiome dysbiosis. We observed that neonatal Npas2 peripheral expression in the liver is necessary for an adaptive response to alterations in food availability in reproductive aged male but not female mice. Specifically, we found that cKO male mice lost a significantly lower percentage of their weight compared to WT mice even though daily food intake did not vary significantly between cKO and WT mice except at day 5 (p=0.033) and day 17 for the males (p=0.009, Figure 1).
Of interest, after a 17-day period of restricted feeding, all mice were found to harbor a significantly different gut microbiota profile, independent of gender or genotype (Figure 2). Specifically, at the end of the restricted feeding period, mice harbored significantly increased relative abundance of Bacteroides and other related members of the order Bacteroidales (p<0.05). Conversely, the relative abundances of several unclassified taxa within the Lachnospiraceae family were significantly decreased following restricted feeding (all p<0.05, Figure 2C). Given our findings of reduced weight loss during restricted feeding specifically to our male Npas2 cKO mice, we next determined if there were specific changes to the gut microbiota that were associated with this significantly reduced weight loss. We found that the greatest difference in the gut microbiome appeared to be driven by the restricted feeding diet, best seen along the PC1 axis as shown in Figure 3. However, along the PC2 axis, while the WT and Npas2 cKO mice clustered similarly at the onset of the RF diet, they separated significantly along the PC2 axis at the conclusion of the RF challenge (Figure 3A, light blue circles vs. light red triangles). This difference in the gut microbiome between the WT and Npas2 cKO mice as a result of the diet can be further appreciated when samples from the same mouse underwent a pair-wise analysis, indicating a difference in the overall rate of change or trajectory of the gut microbiome over time (Figure 3B,C). Finally, by comparing the weight loss (%) from day 0 to day 17 in male mice we were able to correlate accelerated or decelerated weight loss with specific bacterial genera (Figure 4). A distinct cluster consisting primarily of the Ruminiclostridium genus correlated positively with increased weight loss in the WT, but showed opposing correlations in the cKO group. Other genera from the WT group correlated with increased weight loss included Bacteroides, Roseburia, Oscillibacter, Staphylococcus, and Veillonella, with these genera also showing opposing correlations in the cKO group. Conversely, increased weight loss from the cKO group correlated with Eubacterium_coprostanoligenes_group, Corynebacterium, Parasutterella, Bacteroidales_S24_7_group, and Allobaculum, with these genera being negatively correlated with weight loss in the WT group. With respect to changes in bacteria due to restricted feeding per se, we found that in all Npas2 cKO mice, Bacteroides was consistently elevated in response to a RF challenge (Figure 6).
Taken together, these novel findings suggest that changes in the expression of a host hepatic circadian gene, Npas2, specifically during neonatal life can have a lasting impact on the rate of weight change during restricted feeding in a male sex-specific manner. These alterations are accompanied by an overall genotype-associated change in the gut-microbiome, as well as dietary specific changes in Bacteroides species.
Comparison with existing literature and prior studies
Importance of the neonatal hepatic circadian machinery to later in life metabolic function and weight loss with restricted feeding
We did not anticipate that Npas2 cKO mice would plateau sooner and recover more percent body weight during the restricted feeding diet. Since we knocked Npas2 out in the immediate postnatal period and the liver only, we expected a more modest response than in the studies Dudley et al. and Wu et al. carried out in the Npas2 “whole body” KO mouse model (23,24), but anticipated that the Npas2 cKO mice would still lose more weight than WT mice. We have shown that male cKO mice lose significantly less percent body weight than WT mice on a restricted feeding diet and this difference in weight loss is independent of food intake. However, mice exposed to the restricted feeding diet had a significant accompanying shift to the gut microbiome, with a significant difference in the relative abundance of several bacteria taxa when comparing male cKO to WT mice.
We have shown that our findings in a novel cKO mouse model differed from the Npas2 “whole body” KO restricted feeding study. One of the major reasons for the differences between the cKO and KO mouse models could be that in the Npas2 KO mice the circadian clock in the forebrain is significantly altered. It is speculated that Npas2 is entrained through sensory stimulation, such as changes to food availability and timing of meals. Therefore, when Npas2 is not functioning the entrainment of mouse feeding behavior occurs through the SCN, whose circadian rhythm is not entrained through external feeding cues, but instead through light/dark cues (61). For this reason, Npas2 KO mice do not adapt well to restricted feeding (23). Differences in weight loss in the Npas2 cKO mice could be attributed to a change in metabolic homeostasis, as the forebrain circadian clock should be intact and able to adapt to a restricted feeding diet.
Importance of neonatal hepatic Npas2 expression on the gut microbiome, independent of restricted feeding
The interconnectedness of the importance of early in life circadian rhythms, diet and the gut microbiome is becoming increasingly evident. In the absence of microbiota, germ-free mice exhibit altered patterns of circadian gene expression in the liver and brain, while several classes of gut microbiota oscillate over the course of the day in a circadian dependent manner (30,33). Dietary patterns and composition are thought to similarly play a critical role in modulating host-microbial interactions within the circadian context as a high-fat diet challenge has been shown to alter gut microbial composition and circadian oscillations (33,38). In the current study, we report a novel finding that a restricted feeding diet, whereby the timing of feeding is altered, but not the overall caloric consumption or macromolecule composition, significantly impacts the gut microbiota in male mice with a neonatal disrupted peripheral clock. Interestingly, this change in the gut microbiota was accompanied by an altered (lesser) weight loss phenotype that only manifested in male mice. Previous reports have indicated that the microbial influence on the circadian rhythms differs between genders, which may potentially explain the sex specific phenotype seen in our model (37).
Impact of restricted feeding on the gut microbiome and speculations as to why this matters to obstetricians and neonatologists
As a result of a restricted feeding diet and irrespective of Npas2 cKO geneotype, Bacteroides appeared to be the most dramatically impacted taxa in the gut microbiome. Bacteroides species have been widely implicated as being an important early colonizer of the gut microbiome in the neonate, and some debate exists over what early environmental or host factors may regulate its presence or absence in the neonatal gut.
Of potential interest to obstetricians and neonates, mode of delivery has been reported to highly influence the presence of Bacteroides in the early neonatal gut, with Cesarean delivery typically attributed to reduced or delayed colonization in the early months (32,64). However, work by Wu et al. has demonstrated through in vivo modeling that individual Bacteroides species are highly sensitive and responsive to dietary perturbations, an observation that is consistent with published observations of longitudinally sampled infants whereby Bacteroides is associated with the introduction of formula and/or solid foods (65–67). We have additionally demonstrated that a maternal gestational high-fat diet persistently reduces the abundance of Bacteroides in the early neonatal gut microbiome, further indicating the influence on diet on Bacteroides within the gut (26). Our work thus further expands these published observations, indicating that dietary restriction, particularly in the context of an abnormal circadian rhythm, is an important environmental modifying factor that can impact the composition of the gut microbiome specifically with respect to Bacteroides spp.
Although our study did not set out to address potential correlations between weight loss following restricted feeding and the gut microbiome, our findings are of additional interest nonetheless. Our observation that at the end of the restricted feeding period, male cKO mice demonstrated significantly reduced weight loss enabled us to correlate accelerated or decelerated weight loss with specific bacterial genera (Figure 5). A distinct cluster consisting primarily of the Ruminiclostridium genus but also included Bacteroides, Roseburia, Oscillibacter, Staphylococcus, and Veillonella, was negatively associated with decreased weight loss in the cKO mice. Conversely, the Eubacterium_coprostanoligenes_group, Corynebacterium, Parasutterella, Bacteroidales_S24_7_group, and Allobaculum genera positively correlated with the decreased weight loss of the cKO group and were negatively correlated with increased weight loss of the WT group. The implications of these associations for managing with rate of weight loss with dietary restrictions in humans have yet to be examined, but may suggest the importance of circadian entrainment in neonatal life on dietary responsiveness and the microbiome. In contrast, alterations in dietary specific changes in Bacteroides species are presumptively circadian independent. We speculate that these observations may have important implications for the management of weight loss and gain later in life.
Figure 5. Correlation between gut microbiota and % weight loss between WT and cKO male mice.
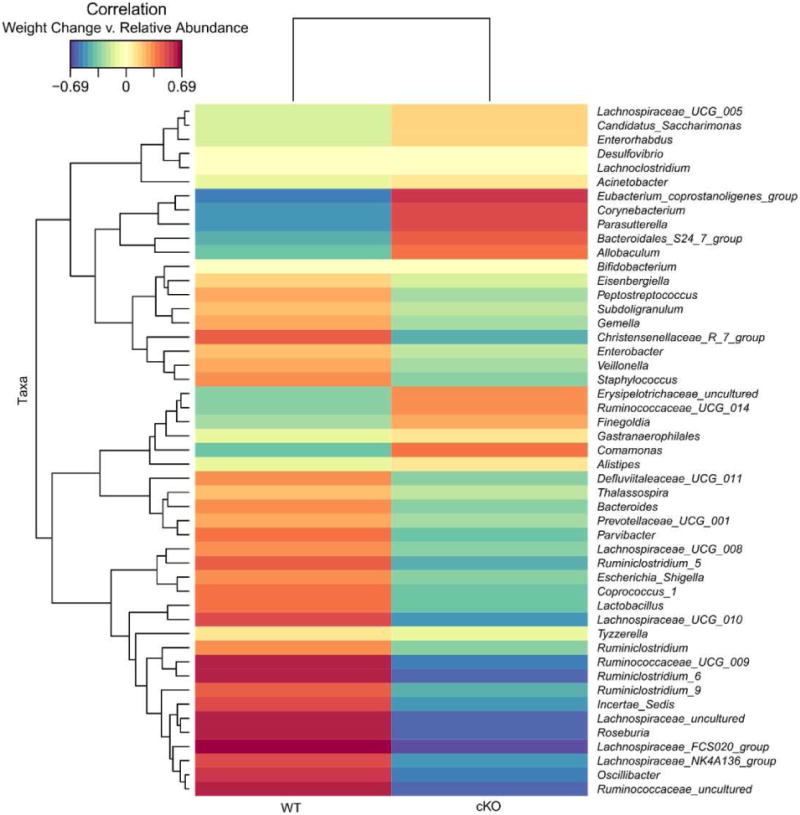
Top 50 most correlated bacterial genera are shown based on the strength of correlation between the relative abundance of bacterial genera and the % weight loss for each genotype. Red shows positive correlation and blue shows negative correlations.
Strengths and limitations of our study
The strengths of our study lie in its innovative conditional genetic modeling, and specifically with the deletion of a liver specific regulator during neonatal life. This novel approach led to several important observations, and enabled us to distinguish the impact of the host genetics from diet on the interrelationship between circadian regulation and the diet on gut dysbiosis. In parallel with these strengths of scientific approach and innovation, there are inherent limitations to our study. First, we are limited in this initial report to descriptions of weight loss and the gut microbiome. However, ongoing further studies will be aimed at determining the impact of peripheral circadian regulators on hormones governing energy homeostasis, such as leptin, corticosterone, insulin, and thyroxine, are needed (62,63). In addition, activity levels of mice on restricted feeding will be measured to ensure that the difference in weight loss is not caused by decreased energy expenditure. Second, in addition to ongoing more exhaustive metabolic studies, one shortcoming of our current study was the limited number of time points to analyze the shifting microbiome. Additional studies will need to be carried out to interrogate how the rapid weight loss and adaptation to the restricted feeding diet affect the microbiome and how Npas2 plays a role in mediating those changes. Third, in the current study we focused on conditional deletion of the neonatal circadian machinery. Ongoing studies focusing on deletion of Npas2 in the liver during fetal life are of future likely importance.
Conclusions and implications
There is no time period in mammalian development during which the circadian rhythms under greater change than during neonatal life. Due to the dependency of the offspring on the mom for lactation, both mother and baby are subjected to the temporal disruptions in circadian rhythmicity. While our previous work in both humans and primates suggested a dominant effect of a high fat maternal diet on disrupting the offspring hepatic circadian machinery, we could not tie these observations mechanistically to later in life metabolic disruption nor gut dysbiosis. However, use of an innovative murine cKO genetic model enabled us to specifically delete Npas2 in the liver from neonatal life onward.
Taken together, our collective work suggests that disruption of the Npas2 circadian rhythm transcriptional machinery early in fetal or neonatal life, whether by a maternal high fat diet or via liver specific gene mutations or epigenetic remodeling, is associated with risk for later in life metabolic syndrome (49–55). In the current study, we show a parallel effect on the gut microbiome when challenged by restricted feeding and can specifically ascribe different taxa based on host genotype, or alternately dietary restriction. These findings suggest the potential for complex interplay between the circadian clock, metabolism, and the gut microbiome. As Npas2 expression in the liver is a target of maternal high fat diet induced metabolic perturbations during fetal development, we speculate that these findings have likely implications in the long term metabolic health of their offspring. Our findings also suggest potential targets for future interventions aimed at restoring or establishing appetite satiety, as well as regulating the rate of weight loss with restricted feeding.
Supplementary Material
Supplemental Figure 1. Schematic of temporal deletion of Npas2 in cKO mouse model. Top: Npas2 cKO mice lose Npas2 expression in postnatal life proportional to increased Cre accumulation in the developing liver. However, Cre is not expressed in the brain (as well as all other tissues) preserving normal Npas2 expression. Bottom: Wild type mice have no Cre expression and have normal Npas2 expression in the liver, brain and all other tissues.
Supplemental Figure 2. The gut microbiome is not significantly altered by gender. (A) Difference in the number of observed OTUs and the Shannon Index before (start) and after (end) a restricted feeding by gender and genotype. The mean and standard deviation is shown for each group. Significance was determined by a Student's t-test. (B) Principal coordinate analysis performed on weighted UniFrac distances between male and female samples, before (start) and after (end) restricted feeding.
Acknowledgments
The authors would like to thank the Alkek Center for Metagenomics and Microbiome Research (CMMR) at Baylor College of Medicine for generating the 16S sequencing data. Measurements of food intake were performed in the Mouse Metabolic Research Unit (MMRU) at the USDA/ARS Children's Nutrition Research Center, Baylor College of Medicine, which is supported by funds from the USDA ARS (www.bcm.edu/cnrc/mmru). The authors acknowledge the expert assistance of Mr. Firoz Vohra and the MMRU Core Director, Dr. Marta Fiorotto with the latter experiments.
Funding: The effort and resources for this study was partially funded by the NIH (grant numbers 1RO1DK089201-01A (K.M.A.), R24DK090964-06 (K.M.A.), T32GM088129 (D.S.O.), and T32GM07526-37 (D.S.O.)).
Glossary of Terms
- Conditional knock-out (cKO) mouse
A genetically engineered mouse in which a specific gene is deleted in a specific tissue (in this study, the liver) using Cre-promoter technology. Depending on when the specific Cre driver used is expressed during development, there can be both developmental stage and site specific deletion. In the current study use of the Albumin Cre-driver enables conditional deletion in the liver starting on day two of postnatal life.
- Metagenome
The total genomic DNA of all organisms within a community.
- Metagenomics
The study of uncultured microbial communities, typically relying on high-throughput experimental data and bioinformatic techniques.
- Microbiome
The total microbial community and biomolecules within a defined environment.
- Microbiota
The total collection of microbial organisms within a community, typically used in reference to an animal host
- Microflora
An older term used synonymously with microbiota.
- OTU
Operational Taxonomic Unit, a cluster of organisms similar at the sequence level beyond some threshold (e.g., 95%) used in place of species, genus, etc.
- 16S rRNA
The transcript of the 16S ribosomal subunit gene, the smaller RNA component of the prokaryotic ribosome, used as the most common taxonomic marker for microbial communities.
- 16S Variable region (e.g., V4)
The variable region of the 16S rRNA, designated in sequence 5′ to 3′ as V1-V9. V4 is the 4th variable region, which detects and distinguishes many bacterial taxa in the stool
Footnotes
Disclosure statement: The authors report no conflicts of interest.
SMFM Presentation: Oral presentation (abstract #22) to be presented at 37th Annual Scientific Meeting of the Society of Maternal Fetal Medicine.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Panda S. Circadian physiology of metabolism. Science. 2016;354(6315):317–22. doi: 10.1126/science.aah4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ussar S, Griffin NW, Bezy O, Bry L, Gordon JI, Kahn CR, et al. Interactions between Gut Microbiota, Host Genetics and Diet Modulate the Predisposition to Obesity and Article Interactions between Gut Microbiota, Host Genetics and Diet Modulate the Predisposition to Obesity and Metabolic Syndrome. Cell Metab Elsevier. 2015;22(3):1–15. doi: 10.1016/j.cmet.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Govindarajan K, MacSharry J, Casey PG, Shanahan F, Joyce SA, Gahan CGM. Unconjugated bile acids influence expression of circadian genes: A potential mechanism for microbe-host crosstalk. PLoS One. 2016;11(12):1–13. doi: 10.1371/journal.pone.0167319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buffington SA, Di Prisco GV, Auchtung TA, Ajami NJ, Petrosino JF, Costa-Mattioli M. Microbial Reconstitution Reverses Maternal Diet-Induced Social and Synaptic Deficits in Offspring. Cell. Elsevier Inc. 2016;165(7):1762–75. doi: 10.1016/j.cell.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masuyama H, Mitsui T, Nobumoto E, Hiramatsu Y. The effects of high-fat diet exposure in utero on the obesogenic and diabetogenic traits through epigenetic changes in Adiponectin and Leptin gene expression for multiple generations in female mice. Endocrinology. 2015 Jul;156:2482–91. doi: 10.1210/en.2014-2020. [DOI] [PubMed] [Google Scholar]
- 6.Joyce SA, MacSharry J, Casey PG, Kinsella M, Murphy EF, Shanahan F, et al. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc Natl Acad Sci. 2014;111(20):7421–6. doi: 10.1073/pnas.1323599111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paulose JK, Wright JM, Patel AG, Cassone VM. Human gut bacteria are sensitive to melatonin and express endogenous circadian rhythmicity. PLoS One. 2016;11(1):1–13. doi: 10.1371/journal.pone.0146643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, Bayless AJ, et al. Cell Host Microbe. 5. Vol. 17. Elsevier Inc; 2015. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function; pp. 662–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aagaard-Tillery KM, Grove K, Bishop J, Ke X, Fu Q, McKnight R, et al. Developmental origins of disease and determinants of chromatin structure: maternal diet modifies the primate fetal epigenome. J Mol Endocrinol. 2008 Jun 03;41(2):91–102. doi: 10.1677/JME-08-0025. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suter M, Bocock P, Showalter L, Hu M, Shope C, McKnight R, et al. Epigenomics: maternal high-fat diet exposure in utero disrupts peripheral circadian gene expression in nonhuman primates. FASEB J. 2010 Nov 26;25(2):714–26. doi: 10.1096/fj.10-172080. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suter Ma, Takahashi D, Grove KL, Aagaard KM. Postweaning exposure to a high-fat diet is associated with alterations to the hepatic histone code in Japanese macaques. Pediatr Res. 2013;74(3):252–8. doi: 10.1038/pr.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCurdy CE, Bishop JM, Williams SM, Grayson BE, Smith MS, Friedman JE, et al. Maternal high-fat diet triggers lipotoxicity in the fetal livers of nonhuman primates. J Clin Invest. 2009;119(2):323–35. doi: 10.1172/JCI32661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grant WF, Gillingham MB, Batra AK, Fewkes NM, Comstock SM, Takahashi D, et al. Maternal high fat diet is associated with decreased plasma n-3 fatty acids and fetal hepatic apoptosis in nonhuman primates. PLoS One. 2011;6(2) doi: 10.1371/journal.pone.0017261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suter M, Bocock P, Showalter L, Hu M, Shope C, McKnight R, et al. Epigenomics: maternal high-fat diet exposure in utero disrupts peripheral circadian gene expression in nonhuman primates. FASEB J. 2011;25:714–26. doi: 10.1096/fj.10-172080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suter MA, Chen A, Burdine MS, Choudhury M, Harris RA, Lane RH, et al. A maternal high-fat diet modulates fetal SIRT1 histone and protein deacetylase activity in nonhuman primates. FASEB J. 2012;26(12):5106–14. doi: 10.1096/fj.12-212878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grant WF, Nicol LE, Thorn SR, Grove KL, Friedman JE, Marks DL. Perinatal Exposure to a High-Fat Diet Is Associated with Reduced Hepatic Sympathetic Innervation in One-Year Old Male Japanese Macaques. PLoS One. 2012;7(10) doi: 10.1371/journal.pone.0048119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thorn SR, Baquero KC, Newsom SA, El Kasmi KC, Bergman BC, Shulman GI, et al. Early life exposure to maternal insulin resistance has persistent effects on hepatic NAFLD in juvenile nonhuman primates. Diabetes. 2014;63(8):2702–13. doi: 10.2337/db14-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCurdy CE, Schenk S, Hetrick B, Houck J, Drew BG, Kaye S, et al. Maternal obesity reduces oxidative capacity in fetal skeletal muscle of Japanese macaques. JCI Insight. 2016;1(16):1–17. doi: 10.1172/jci.insight.86612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia JA, Zhang D, Estill SJ, Michnoff C, Rutter J, Reick M, et al. Impaired cued and contextual memory in NPAS2-deficient mice. Science (80-) 2000 Jun 24;288(5474):2226–30. doi: 10.1126/science.288.5474.2226. 2000. [DOI] [PubMed] [Google Scholar]
- 20.Reick M, Garcia JA, Dudley C, McKnight SL. NPAS2: An analog of clock operative in the mammalian forebrain. Science (80-) 2001;293(5529):506–9. doi: 10.1126/science.1060699. [DOI] [PubMed] [Google Scholar]
- 21.Dudley CA, Erbel-Sieler C, Estill SJ, Reick M, Franken P, Pitts S, et al. Altered patterns of sleep and behavioral adaptability in NPAS2-deficient mice. Science. 2003 Jul;301(5631):18. 379–83. doi: 10.1126/science.1082795. [DOI] [PubMed] [Google Scholar]
- 22.Franken P, Dudley CA, Estill SJ, Barakat M, Thomason R, O'Hara BF, et al. NPAS2 as a transcriptional regulator of non-rapid eye movement sleep: genotype and sex interactions. Proc Natl Acad Sci U S A. 2006 Apr 26;103(18):7118–23. doi: 10.1073/pnas.0602006103. 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dudley CA, Erbel-Sieler C, Estill SJ, Reick M, Franken P, Pitts S, et al. Altered patterns of sleep and behavioral adaptability in NPAS2-deficient mice. Science (80-) 2003;301(5631):379–83. doi: 10.1126/science.1082795. [DOI] [PubMed] [Google Scholar]
- 24.Wu X, Wiater MF, Ritter S. NPAS2 deletion impairs responses to restricted feeding but not to metabolic challenges. Physiol Behav. 2009 Dec 23;99(4):466–71. doi: 10.1016/j.physbeh.2009.12.010. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma J, Prince AL, Bader D, Hu M, Ganu R, Baquero K, et al. High-fat maternal diet during pregnancy persistently alters the offspring microbiome in a primate model. Nat Commun. 2014;5(May):3889. doi: 10.1038/ncomms4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chu DM, Antony KM, Ma J, Prince AL, Showalter L, Moller M, et al. The early infant gut microbiome varies in association with a maternal high-fat diet. Genome Med. 2016;8(1):77. doi: 10.1186/s13073-016-0330-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shirasu-Hiza MM, Dionne MS, Pham LN, Ayres JS, Schneider DS. Interactions between circadian rhythm and immunity in Drosophila melanogaster. Curr Biol. 2007;17(10):353–5. doi: 10.1016/j.cub.2007.03.049. [DOI] [PubMed] [Google Scholar]
- 28.Eckel-Mahan KL, Patel VR, De Mateo S, Orozco-Solis R, Ceglia NJ, Sahar S, et al. Reprogramming of the circadian clock by nutritional challenge. Cell. 2013;155(7):1464–78. doi: 10.1016/j.cell.2013.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukherji A, Kobiita A, Ye T, Chambon P. Cell. 4. Vol. 153. Elsevier Inc; 2013. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs; pp. 812–27. [DOI] [PubMed] [Google Scholar]
- 30.Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014;159(3):514–29. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 31.Zarrinpar A, Chaix A, Yooseph S, Panda S. Cell Metab. 6. Vol. 20. Elsevier Inc; 2014. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome; pp. 1006–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17(5):690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 33.Leone V, Gibbons SM, Martinez K, Hutchison AL, Huang EY, Cham CM, et al. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe. 2015;17(5):681–9. doi: 10.1016/j.chom.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murakami M, Tognini P, Liu Y, Eckel-Mahan KL, Baldi P, Sassone-Corsi P, et al. Gut microbiota directs PPARγ-driven reprogramming of the liver circadian clock by nutritional challenge. EMBO Rep. 2016;330(6009):1349–54. doi: 10.15252/embr.201642463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montagner A, Korecka A, Polizzi A, Lippi Y, Blum Y, Canlet C, et al. Hepatic circadian clock oscillators and nuclear receptors integrate microbiome-derived signals. Sci Rep. 2016;6:20127. doi: 10.1038/srep20127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voigt RM, Summa KC, Forsyth CB, Green SJ, Engen P, Naqib A, et al. Alcohol Clin Exp Res. 2. Vol. 40. England: 2016. Feb, The Circadian Clock Mutation Promotes Intestinal Dysbiosis; pp. 335–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang X, Bushman FD, FitzGerald Ga. Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc Natl Acad Sci. 2015;112(33):10479–84. doi: 10.1073/pnas.1501305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voigt RM, Forsyth CB, Green SJ, Mutlu E, Engen P, Vitaterna MH, et al. Circadian disorganization alters intestinal microbiota. PLoS One. 2014;9(5) doi: 10.1371/journal.pone.0097500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia JA, Zhang D, Estill SJ, Michnoff C, Rutter J, Reick M, et al. Impaired cued and contextual memory in NPAS2-deficient mice. Science (80-) 2000;288(5474):2226–30. doi: 10.1126/science.288.5474.2226. [DOI] [PubMed] [Google Scholar]
- 40.Yoshii K, Ishijima S, Sagami I. Effects of NAD(P)H and its derivatives on the DNA-binding activity of NPAS2, a mammalian circadian transcription factor. Biochem Biophys Res Commun. 2013;437(3):386–91. doi: 10.1016/j.bbrc.2013.06.086. [DOI] [PubMed] [Google Scholar]
- 41.Postic C, Magnuson MA. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis. 2000 Feb;26(2):149–50. doi: 10.1002/(sici)1526-968x(200002)26:2<149::aid-gene16>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 42.Aagaard KM, Ganu R, Ma J, Racusin D, Arndt M, Riehle K, et al. Whole metagenomic shotgun sequencing reveals a vibrant placental microbiome harboring metabolic function. Am Jouranl Obstet Gynecol. 2013;208(1):S5. [Google Scholar]
- 43.Ma J, Prince A, Aagaard KM. Use of whole genome shotgun metagenomics: A practical guide for the microbiome-minded physician scientist. Semin Reprod Med. 2014;32(1):5–13. doi: 10.1055/s-0033-1361817. [DOI] [PubMed] [Google Scholar]
- 44.Prince AL, Antony KM, Chu DM, Aagaard KM. The microbiome, parturition, and timing of birth: more questions than answers. J Reprod Immunol. 2014:104–105. 12–9. doi: 10.1016/j.jri.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Antony KM, Ma J, Mitchell KB, Racusin DA, Versalovic J, Aagaard K. The preterm placental microbiome varies in association with excess maternal gestational weight gain. Am J Obstet Gynecol. 2015;212(5):653.e1–653.e16. doi: 10.1016/j.ajog.2014.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aagaard K, Prince A, Ma J, Antony K, Chu D, Benjamin R, et al. Distinct alterations in the placental microbiome among spontaneous preterm births. Am J Obstet Gynecol. 2015;212(1):S53–4. [Google Scholar]
- 47.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liaw A, Wiener M. Classification and Regression by randomForest. R news. 2002 Dec;2:18–22. [Google Scholar]
- 49.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005 May;308(5724):13. 1043–5. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woon PY, Kaisaki PJ, Braganca J, Bihoreau MT, Levy JC, Farrall M, et al. Aryl hydrocarbon receptor nuclear translocator-like (BMAL1) is associated with susceptibility to hypertension and type 2 diabetes. Proc Natl Acad Sci U S A. 2007;104(36):14412–7. doi: 10.1073/pnas.0703247104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sookoian S, Gemma C, Gianotti TF, Burgueño A, Castaño G, Pirola CJ. Genetic variants of Clock transcription factor are associated with individual susceptibility to obesity. Am J Clin Nutr. 2008;87(6):1606–15. doi: 10.1093/ajcn/87.6.1606. [DOI] [PubMed] [Google Scholar]
- 52.Scott EM, Carter AM, Grant PJ. Association between polymorphisms in the Clock gene, obesity and the metabolic syndrome in man. Int J Obes. 2008;32(4):658–62. doi: 10.1038/sj.ijo.0803778. [DOI] [PubMed] [Google Scholar]
- 53.Scheer FAJL, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci U S A. 2009;106(11):4453–8. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Englund A, Kovanen L, Saarikoski ST, Haukka J, Reunanen A, Aromaa A, et al. NPAS2 and PER2 are linked to risk factors of the metabolic syndrome. J Circadian Rhythms. 2009;7:5. doi: 10.1186/1740-3391-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Milagro FI, Gómez-Abellán P, Campión J, Martínez JA, Ordovás JM, Garaulet M. CLOCK, PER2 and BMAL1 DNA Methylation: Association with Obesity and Metabolic Syndrome Characteristics and Monounsaturated Fat Intake. Chronobiol Int. 2012;29(9):1180–94. doi: 10.3109/07420528.2012.719967. [DOI] [PubMed] [Google Scholar]
- 56.Sekaran S, Lupi D, Jones SL, Sheely CJ, Hattar S, Yau KW, et al. Melanopsin-dependent photoreception provides earliest light detection in the mammalian retina. Curr Biol. 2005;15(12):1099–107. doi: 10.1016/j.cub.2005.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnson J, Wu V, Donovan M, Majumdar S, Renteria RC, Porco T, et al. Melanopsin-dependent light avoidance in neonatal mice. Proc Natl Acad Sci U S A. 2010;107(40):17374–8. doi: 10.1073/pnas.1008533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fernandez DC, Chang YT, Hattar S, Chen SK. Architecture of retinal projections to the central circadian pacemaker. Proc Natl Acad Sci. 2016 doi: 10.1073/pnas.1523629113. 201523629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clayton ZE, Vickers MH, Bernal A, Yap C, Sloboda DM. Early life exposure to fructose alters maternal, fetal and neonatal hepatic gene expression and leads to sex-dependent changes in lipid metabolism in rat offspring. PLoS One. 2015;10(11) doi: 10.1371/journal.pone.0141962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mouralidarane A, Soeda J, Sugden D, Bocianowska A, Carter R, Ray S, et al. Maternal obesity programs offspring non-alcoholic fatty liver disease through disruption of 24-h rhythms in mice. Int J Obes (Lond) 2015;39(9):1339–48. doi: 10.1038/ijo.2015.85. [DOI] [PubMed] [Google Scholar]
- 61.Hara R, Wan K, Wakamatsu H, Aida R, Moriya T, Akiyama M, et al. Restricted feeding entrains liver clock without participation of the suprachiasmatic nucleus. Genes Cells. 2001 Mar;6(3):269–78. doi: 10.1046/j.1365-2443.2001.00419.x. [DOI] [PubMed] [Google Scholar]
- 62.Zhang L, Rubins NE, Ahima RS, Greenbaum LE, Kaestner KH. Foxa2 integrates the transcriptional response of the hepatocyte to fasting. Cell Metab. 2005;2(2):141–8. doi: 10.1016/j.cmet.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 63.Zvonic S, Ptitsyn AA, Conrad SA, Scott LK, Floyd ZE, Kilroy G, et al. Characterization of peripheral circadian clocks in adipose tissues. Diabetes. 2006 Apr;55(4):962–70. doi: 10.2337/diabetes.55.04.06.db05-0873. [DOI] [PubMed] [Google Scholar]
- 64.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107(26):11971–5. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu M, McNulty NP, Rodionov DA, Khoroshkin MS, Griffin NW, Cheng J, et al. Genetic determinants of in vivo fitness and diet responsiveness in multiple human gut Bacteroides. Science (80-) 2015;350(6256):aac5992. doi: 10.1126/science.aac5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, et al. Factors Influencing the Composition of the Intestinal Microbiota in Early Infancy. Pediatrics. 2006;118(2):511–21. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 67.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(Suppl):4578–85. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Schematic of temporal deletion of Npas2 in cKO mouse model. Top: Npas2 cKO mice lose Npas2 expression in postnatal life proportional to increased Cre accumulation in the developing liver. However, Cre is not expressed in the brain (as well as all other tissues) preserving normal Npas2 expression. Bottom: Wild type mice have no Cre expression and have normal Npas2 expression in the liver, brain and all other tissues.
Supplemental Figure 2. The gut microbiome is not significantly altered by gender. (A) Difference in the number of observed OTUs and the Shannon Index before (start) and after (end) a restricted feeding by gender and genotype. The mean and standard deviation is shown for each group. Significance was determined by a Student's t-test. (B) Principal coordinate analysis performed on weighted UniFrac distances between male and female samples, before (start) and after (end) restricted feeding.


