Abstract
African trypanosomes evade the immune response of their mammalian hosts by sequentially expressing genes for different variant surface glycoproteins (VSGs) from telomere-linked VSG expression sites. In the Trypanosoma brucei clone whose genome is being sequenced (GUTat 10.1), we show that the expressed VSG (VSG 10.1) is duplicated from a silent donor VSG located at another telomere-linked site. We have determined two 130 kb sequences representing the VSG 10.1 donor and expression sites. The telomere-linked donor VSG 10.1 resembles metacyclic VSG expression sites, and is preceded by a cluster of 35 or more tandem housekeeping genes, all of which are transcribed away from the telomere. The 45 kb telomere-linked VSG 10.1 expression site contains a promoter followed by seven expression site-associated genes (ESAGs), three pseudo ESAGs, two pseudo VSGs and VSG 10.1. The 80 kb preceding the expression site has few, if any, functional ORFs, but contains 50 bp repeats, INGI retrotransposon-like elements, and novel 4–12 kb repeats found near other telomeres. This analysis provides the first look over a 130 kb range of a telomere-linked donor VSG and its corresponding telomere-linked VSG expression site and forms the basis for studies on antigenic variation in the context of a completely sequenced genome.
INTRODUCTION
The African trypanosome Trypanosoma brucei is transmitted by tsetse flies to its mammalian host where it causes a fatal disease commonly called sleeping sickness in humans and nagana in domestic livestock. The World Health Organization estimates that 300 000 new cases of human sleeping sickness occur annually, primarily in Sudan, Congo, Uganda and Angola, but the actual number is unknown because most infected persons live in areas with little or no medical care (1,2). Trypanosoma brucei replicates in the bloodstream of its mammalian host, where it is in constant contact with the immune system. To survive in this hostile environment, African trypanosomes have evolved molecular mechanisms to evade the immune responses. The best characterized of these mechanisms is antigenic variation, a phenomenon whereby bloodstream trypanosomes switch from one variant surface glycoprotein (VSG) on their surface to another at a rate of 10–2–10–7 switches/doubling time of 5–10 h (3–5).
Although hundreds of VSG genes (VSGs) are present in the genome, under normal circumstances one, and only one, VSG is expressed at a time in a given bloodstream parasite (6). The unexpressed VSGs are scattered throughout the genome, but all expressed VSGs studied to date are located near telomeres (for recent reviews, see 7–10). These telomere-linked VSG expression sites (ESs) have been defined as the sequences that extend from the VSG promoters to the telomeric repeats of (TTAGGG)n located downstream of the VSGs (7,11). Two types of ESs have been identified. Metacyclic ESs (M-ESs) express VSGs in metacyclic trypanosomes, the final developmental stage of the parasite in the tsetse fly (12). After the parasite infects a mammalian host, VSG expression generally switches to a bloodstream ES (B-ES). There are ∼10–15 telomere-linked B-ESs and a similar number of M-ESs in the genome, only one of which is active at a time (13–16).
The conventional model of a B-ES is based primarily on a detailed characterization of the B-ES for the AnTat 1.3 VSG (reviewed in 7). This B-ES contains a promoter and a polycistronic transcription unit of 45–60 kb and is preceded by 20–40 kb of a 50 bp repeat element. The polycistronic transcription unit includes several ES-associated genes (ESAGs), 5–20 kb of a 70 bp repeat and the VSG. The AnTat 1.3 B-ES differs significantly from the few M-ESs that have been characterized (11,17,18). These M-ESs have short (3–5 kb) monocistronic transcription units and generally lack all or most of the associated 50 bp repeats, 70 bp repeats and ESAGs of B-ESs.
To determine if other bloodstream VSG expression sites have a similar organization to the AnTat 1.3 B-ES, we examined the VSG expressed by T.brucei TREU 927/4 clone GUTat 10.1 (Glasgow University Trypanozoon antigen type 10.1). The complete sequence determination of the GUTat 10.1 genome is currently underway (19), but no information is available on its expressed VSG or, for that matter, any of the VSGs in the genome of this trypanosome stock. We identified the VSG expressed by GUTat 10.1 and show here that VSG 10.1 is duplicated into a B-ES from a silent telomere-linked donor gene. We sequenced two telomere-linked genomic regions of ∼130 kb each, one that contains the GUTat 10.1 VSG donor copy and one that bears its B-ES. We find that the VSG 10.1 donor copy is located in an M-ES-like site and was duplicated into a B-ES. The VSG 10.1 ES is similar, but not identical, to the AnTat 1.3 B-ES. This work provides the first complete sequence analysis of both a telomere-linked donor VSG and its expressed copy, and generates the foundation for future studies on antigenic variation within the context of a fully sequenced African trypanosome genome.
MATERIALS AND METHODS
Trypanosome clones
Trypanosoma brucei TREU (Trypanosomiasis Research Edinburgh University) 927/4 (GPAL/KE/70/EATRO 1534) was isolated from a tsetse fly in Kiboko, Kenya in 1970 (20), passaged in mice ∼12 times and cloned. This isolate is pleomorphic, tsetse transmissible, genetically competent and gives rise to chronic infections in mice (21,22). To generate a more virulent line with better stability of expression of variant antigen types (VAT) of this genotype, TREU 927/4 parasites were passaged in adult female CFLP mice 27 times at 2–3 day intervals (23). A single cell was then isolated optically and a sub-clone grown in a BALB/c mouse. The predominant VAT expressed by this clone has been designated GUTat 10.1. Mouse and rat VAT-specific antisera were generated using an infection/cure procedure as previously described (24). To obtain a second clone expressing a different VAT, a patent infection of GUTat 10.1 was sub-curatively treated with 1.0 mg/kg cymelarsan (a kind gift from Rhone Merieux) on day four of infection and a trypanosome line sub-cloned from the subsequent relapse population on day 15. This line has been designated GUTat 10.3. Antibody analysis of the GUTat 10.1 and 10.3 trypanosomes indicated that these clones are ≥95% pure for expression of their respective VATs. Genomic DNA from the GUTat 10.1 clone is being used for sequence determination of the African trypanosome genome.
cDNA library construction
Total RNA was isolated from GUTat 10.1 bloodstream trypanosomes with Trizol reagent (Gibco BRL). Poly(A)+ RNA was selected using an Oligotex mRNA kit (Qiagen) and used to generate a unidirectional λ phage cDNA library via the ZAP-cDNA Synthesis Kit (Stratagene). Aliquots of 1 ml (>1010 p.f.u.) of the amplified library were prepared and are available upon request.
Inverse PCR
Inverse PCR was performed as described by Willis et al. (25). GUTat 10.1 and 10.3 DNA (5 µg) was digested to completion with HindIII or NgoMIV and purified with the QIAEX II Gel Extraction Kit (Qiagen) following the manufacturer’s protocol for desalting DNA solutions. One-tenth of the recovered DNA (∼0.5 µg) was incubated for 12 h at 15°C with 5 U of T4 DNA ligase (Boehringer Mannheim) in 500 µl total volume. DNA was isolated with QIAEX II as described above and eluted in 100 µl. Sequences from the VSG 10.1 B-ES were PCR-amplified from 2.5 µl of circularized DNA with a 70 bp repeat-specific primer (5′-TATTCGTATATACACACTCACAACACTCTCCTAT-3′) and a primer specific for the sequence located between the 70 bp repeats and VSG 10.1 (5′-AAAATACGACAGCAACCTATGACGAC-3′).
BAC clone sequence determination
A BAC library made from T.brucei GUTat 10.1 was used to select clones. Sheared BAC DNA (1.6–2 kb) was ligated to a modified pUC19 vector and transformed into Escherichia coli. Sequencing reactions were performed using BigDye primers or terminators (PE Biosystems), and were run on ABI377 and ABI3700 sequencers (PE Biosystems). Shotgun clones were sequenced to generate 7–8-fold coverage of each BAC. BACs were assembled from the shotgun sequences using the TIGR assembler program (26) and closed using a combination of BAC walking, directed PCR and transposition of individual shotgun clones. Annotation of the BAC sequences involved both DNA and protein database searches and gene prediction programs. Gene predictions were made using a modified version of GLIMMER (27). Predicted protein sequences were searched against a non-redundant amino acid database and against hidden Markov models (HMMs) of the protein domains from pFAM3 and TIGR-built HMMs.
RESULTS AND DISCUSSION
Identification of the cDNAs for the VSGs expressed by T.brucei clones 10.1 and 10.3
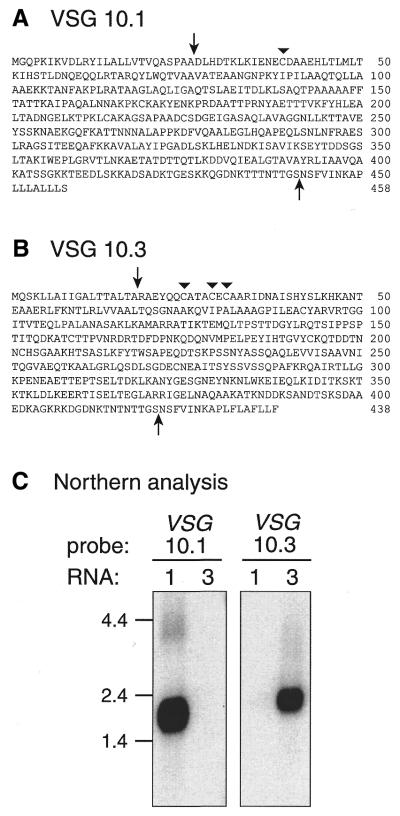
Poly(A)+ RNA was isolated from bloodstream forms of GUTat 10.1 and 10.3 and used as template for reverse transcriptase-PCR (RT–PCR) (28). The expressed VSGs were RT–PCR-amplified by using a 3′ primer complementary to the conserved 14mer found in the 3′ UTRs of all VSGs and a 5′ primer derived from the 39 nt spliced leader sequence at the 5′ ends of all trypanosomatid mRNAs (29,30). A single 1.7 kb product was obtained from GUTat 10.1 and 10.3 RNA, cloned into pBluescript plasmid and sequenced. The deduced amino acid sequences (Fig. 1A and B) have significant homology to those of other VSGs in the GenBank database and possess the conserved cysteine residue near the mature VSG N-termini but, curiously, lack the several cysteines that usually occur near the mature VSG C-termini (31).
Figure 1.
Deduced amino acid sequences of (A) VSG 10.1 and (B) VSG 10.3. The VSG genes expressed by T. brucei clones GUTat 10.1 and 10.3 were amplified by RT–PCR, cloned and sequenced. Arrows indicate predicted N- and C-terminal cleavage sites. Triangles mark conserved cysteine residues typically found ∼13 residues from the N-termini of mature VSGs. (C) Northern blot analysis of VSG 10.1 and 10.3 expression. GUTat 10.1 (lane 1) and 10.3 (lane 3) total RNA (5 µg) was subjected to gel electrophoresis, transferred to positively charged nylon, and hybridized to radiolabeled VSG 10.1 and 10.3 cDNAs. RNA size markers, in kb, are indicated.
Northern blots confirmed that these two cDNAs were derived from the VSG mRNAs expressed in GUTat clones 10.1 and 10.3 (Fig. 1C). VSG 10.1 cDNA hybridizes to mRNA from GUTat 10.1, but not to mRNA from GUTat 10.3, whereas the VSG 10.3 cDNA had the opposite hybridization pattern. In addition, 14% of the plaques in a GUTat 10.1 bloodstream cDNA library hybridized to VSG 10.1 cDNA under high stringency wash conditions. These results, along with the sequence data, demonstrate that the cDNAs for VSGs 10.1 and 10.3 have been correctly identified.
VSG 10.1 is expressed from a duplicated gene copy
VSGs can be introduced into an active B-ES by several molecular mechanisms (reviewed in 7–10). The best-studied switching mechanism is gene conversion, or ‘duplicative transposition’, in which the VSG in an active B-ES is replaced with a duplicated copy of an unexpressed VSG. Other switching mechanisms include the duplicative conversion of an entire telomere plus its adjacent VSG to another chromosomal end (telomere conversion), and reciprocal exchange of two telomeres and their associated VSGs (telomere exchange). In addition, transcription of one B-ES can switch to another B-ES in situ without associated DNA rearrangements (in situ activation). In contrast, M-ESs appear to be only activated by an in situ mechanism; there is no evidence that DNA rearrangements are associated with their expression.
To determine if a DNA rearrangement was associated with expression of VSG 10.1, Southern blots were conducted (Fig. 2). VSG 10.1 cDNA (probe A) hybridized strongly to two restriction fragments in GUTat 10.1 DNA, and to only one fragment in GUTat 10.3 DNA. This result indicates that VSG 10.1 is a single-copy gene in GUTat 10.3 trypanosomes and is expressed as a duplicated, expression-linked copy (ELC) in GUTat 10.1 trypanosomes. The ELC band is slightly weaker than the donor VSG 10.1 band, a phenomenon observed previously with other ELCs (11,32) and thought to be due to heterogeneity in the number of telomere repeats in the fragment containing the expressed telomere-linked VSG ES (33,34). In addition, as is most apparent in the XhoI digest, the fragment containing the donor VSG 10.1 is a different size in the GUTat 10.1 and 10.3 DNAs. This result suggests the single-copy donor VSG 10.1 is also located near a telomere, and that the size difference is due to differing numbers of telomere repeats between the VSG 10.1 and the telomere in the two genomes. We confirmed that VSG 10.1 is telomere-linked in both GUTat 10.1 and 10.3 by PCR amplification with primers specific for VSG 10.1 and the telomere repeats. Sequence analysis of the PCR product revealed telomere repeats and 10.1 VSG sequences on opposite ends of the fragment (not shown).
Figure 2.
VSG 10.1 is duplicated in trypanosome clone GUTat 10.1. VSG 10.1 rearrangements were analyzed by Southern blot. The locations of the probes and restriction sites are indicated on the schematic diagram of the VSG 10.1 donor region (gray bar, 70 bp repeats; black bar, VSG 10.1; B, BamHI; E, EcoRI; Bg, BglI; X, XhoI; solid lines, probes A, B and C, as labeled; black circle, telomere repeats of several kb). GUTat 10.1 (lane 1) and 10.3 (lane 3) genomic DNA (5 µg) was digested with the indicated restriction enzymes, subjected to gel electrophoresis, and transferred to positively charged nylon. Blots were hybridized to radiolabeled probes derived from the 5′-end of the VSG 10.1 cDNA (probe A), or the regions 3′ (probe B) or 5′ (probe C) of the 70 bp repeats in BAC clone 45I2. Probes B and C were sequentially hybridized to the same blot (probe B was stripped prior to reprobing with C). DNA size markers in kb are indicated.
Organization of the VSG 10.1 donor region
The 18 000 clones in the BAC library of GUTat 10.1 genomic DNA (19) were screened with the VSG 10.1 cDNA to identify clones that contain the VSG 10.1 donor region and B-ES. This genomic library statistically contains 84-fold coverage of the genome, but is under-represented in sequences near the ends of chromosomes. Nevertheless, we expected to identify two groups of positive BAC clones—one group containing the donor (telomere-linked) VSG 10.1 and another group possessing its ELC (also telomere-linked). Surprisingly, only one clone (BAC 45I2) in the BAC library hybridized to the VSG 10.1 cDNA. In contrast, a probe for VSG 10.3, which in preliminary Southern blots appears to be located at an internal site in a GUTat 10.1 megachromosome (D.J.LaCount, unpublished data), hybridized to 55 BAC clones.
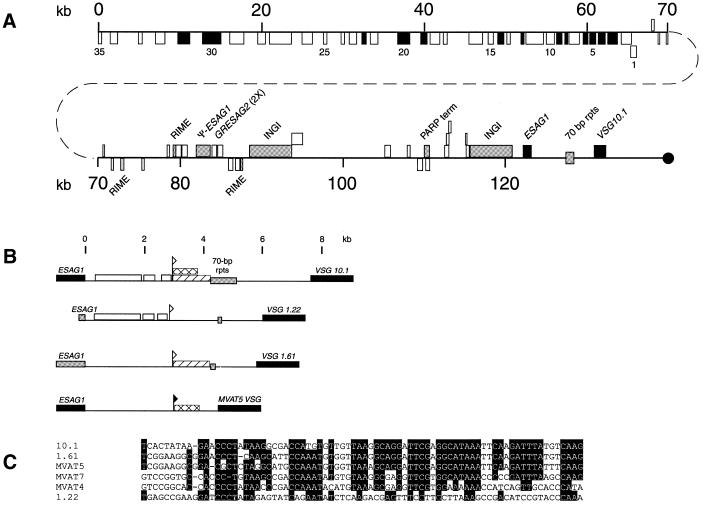
Sequencing across the ends of BAC 45I2 revealed that the last 186 nt at one end are identical to the first 186 nt of the VSG 10.1 cDNA. Thus, we anticipated that this BAC insert extended from a site far upstream of either the telomere-linked donor VSG 10.1 or its ELC to the coding region of gene itself. To determine if BAC 45I2 included the donor or the expressed VSG 10.1, we sequenced the 132 kb insert (Fig. 3). When the locations of the restriction sites near the VSG 10.1 end of this sequence were compared with the sizes of the different restriction fragments on Southern blots (Fig. 2 and data not shown), we discovered that this BAC clone is derived from the region immediately upstream of the telomere-linked donor VSG 10.1, and does not contain the corresponding ELC. DNA probes from the region immediately downstream from the 70 bp repeats hybridized to both the donor copy and the ELC, but probes from the region immediately upstream from the 70 bp repeats hybridized only to the donor copy (Fig. 2B and C), indicating that the duplicated region in the ELC must include the region from the 70 bp repeats through at least the VSG gene. The large-sized fragment observed in all lanes with probe C is due to cross-hybridization of this probe to another region(s) of the genome.
Figure 3.
Gene organization of the VSG 10.1 donor copy. (A) The location and coding strand of the genes and repeat elements in BAC clone 45I2 are indicated by boxes. Genes or repeat elements shown above the line are located on the ‘top’ strand and are oriented towards the telomere (black circle), whereas those shown below the line are located on the ‘bottom’ strand and oriented away from the telomere. Black boxes indicate known genes and genes with significant homology to known genes (see Table 1). White boxes designate hypothetical genes with no significant homology to known genes. Numbers 1–35 refer to the 35 adjacent ORFs oriented away from the telomere. Gray boxes specify repeat elements (RIME, INGI and 70 bp repeats), pseudo genes, and a PARP transcription terminator. The region beginning in the 70 bp repeats and extending through VSG 10.1 has been duplicated in the GUTat 10.1 B-ES. (B) Comparison of the putative VSG 10.1 M-ES to the M-ESs of VSG 1.22, VSG 1.61 and MVAT5 VSG. Regions in the putative VSG 10.1 M-ES with >80% sequence identity to other M-ESs are indicated (VSG 1.22, white box; VSG 1.61, diagonal lined box; MVAT5 VSG, crosshatched box). The MVAT5 promoter is indicated by a closed flag. Sequences within the VSG 10.1, VSG 1.22 and VSG 1.61 M-ESs that have homology to the MVAT5 promoter are marked by an open flag. Black boxes indicate ESAG 1 and VSG genes. Gray boxes indicate pseudo-ESAG 1 genes and 70 bp repeats. (C) Nucleotide sequence comparison of the MVAT4, 5 and 7 M-ES promoters to sequences from the VSG 10.1, VSG 1.22 and VSG 1.61 M-ESs. Shaded residues are conserved in at least four of the six sequences shown.
The sequence of BAC clone 45I2 has several striking features. The first half of this ‘donor VSG 10.1’ region, i.e. ∼65 kb, is comprised of 35 closely spaced, tandemly arrayed open reading frames (ORFs), all of which are on the ‘bottom’ strand and are predicted to be transcribed away from the telomere (Fig. 3A). This 35 gene cluster extends to the extreme end of the sequence, and may contain additional genes outside the region sequenced. Thus, the gene organization in T.brucei appears to be similar to the polycistronic clusters of genes in the published 270 kb sequence of Leishmania major chromosome 1 (35). Also as observed in Leishmania, there is no apparent relationship or metabolic connection among these trypanosome genes. None of these genes encode VSGs or appear to be involved with antigenic variation. Rather, the proteins encoded by these genes are involved in a variety of cellular processes including cell division, DNA and protein metabolism, signal transduction, and translation (Table 1). Two-thirds of the predicted genes code for proteins with no substantial homology to known proteins, and are therefore designated hypothetical proteins. None of these 35 genes are tandemly repeated, as is the case for some other T.brucei genes.
Table 1. ORFs in BAC clone 45I2 with homology to known proteins.
| ORF number |
Closest match |
| 3 | Dyskerin |
| 4 | ATP synthase γ subunit |
| 5 | Ribosomal protein S6 |
| 6 | Vacuolar-type H(+) ATPase |
| 8 | Ribosomal protein L37A |
| 9 | Proteosome subunit |
| 12 | Ribosomal protein L32 |
| 14 | Proteosome subunit |
| 19 | Kinetoplast structure-specific endonuclease 1 |
| 20 | Protein kinase |
| 22 | Calcineurin b subunit |
| 30 | Serine/threonine kinase |
| 31 | Choline dehydrogenase |
The remaining 67 kb of this donor VSG 10.1 region, extending down to the donor VSG 10.1 and telomere repeats, is gene-poor and much less organized. Among the notable features in this region are three ribosomal mobile element (RIME) sequences (36,37), a pseudo-ESAG 1 (containing internal termination codons), two tandemly arranged GRESAG 2.1 genes, and two copies of the 5 kb INGI retrotransposon-like element (38,39). The two INGIs are separated by ∼22 kb that contains a procyclic acidic repetitive protein gene (PARP) transcription terminator sequence (40) and 10 hypothetical ORFs, none of which have substantive matches in the databases. The sequence downstream of the second INGI displays substantial similarity to the sequences of M-ESs, including the M-ESs for MVAT5 VSG, VSG 1.61 and VSG 1.22 (17,41–44). This region includes an ESAG 1 gene followed by 13 copies of 70 bp repeats and the donor VSG 10.1. Based on this organization, the VSG 10.1 donor region appears either to be, or to have been, an M-ES. Within this 132 kb sequence, there are no 50 bp repeats that have been found to precede B-ESs.
In other known M-ESs, the region located downstream from ESAG 1 contains conserved metacyclic VSG promoter elements (32,42,44–47). This region within the 132 kb has a high degree of sequence identity with the corresponding regions from MVAT5 VSG, VSG 1.22, and VSG 1.61 M-ESs (Fig. 3B and C). Over the first 2900 bp following ESAG 1, three blocks of sequence covering 2257 bp have >80% sequence identity with the VSG 1.22 M-ES. In addition, the last 1300 bp before the 70 bp repeats are ∼84% identical to sequences from the VSG 1.61 and MVAT5 VSG M-ESs. The sequence immediately prior to the MVAT5 homology region contains 28 of 32 residues previously found to be conserved among the MVAT4, 5 and 7 promoters (Fig. 3C) and 15 of 16 residues conserved among the MVAT promoters and the bloodstream VSG promoters (data not shown) (32). The conservation of these promoter sequences suggests that this region is an M-ES, although this must still be verified experimentally.
Organization of the B-ES for VSG 10.1
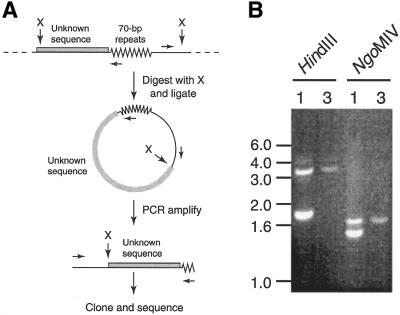
Since BAC clone 45I2 is derived from the region containing the donor VSG 10.1 and no other BAC clones hybridize to the VSG 10.1 cDNA, we cloned the DNA sequence upstream from the 70 bp repeats in the VSG 10.1 ELC by inverse PCR (Fig. 4). As depicted in Figure 4A, GUTat 10.1 and 10.3 genomic DNA was digested with restriction enzymes that cleave on both sides of the 70 bp repeats and ligated to form predominately circular DNA. These circles were then used as templates for PCR amplification using two divergent primers—one within the 70 bp repeats and one upstream of the restriction site between the 70 bp repeats and VSG 10.1. The subsequent PCR product(s) were then cloned and sequenced.
Figure 4.
Inverse PCR identification of VSG 10.1 ELC sequence upstream from the 70 bp repeats. (A) Schematic representation of the inverse PCR strategy. (B) Inverse PCR on GUTat 10.1 (lane 1) and 10.3 (lane 3) genomic DNA digested with HindIII and NgoMIV. DNA size markers, in kb, are indicated.
When GUTat 10.1 genomic DNA was digested with either HindIII or NgoMIV and the fragments circularized, two PCR products were obtained from GUTat 10.1 DNA (one each from the donor copy and the ELC) and one product was obtained from GUTat 10.3 DNA (from the donor copy only) (Fig. 4B). The sequence of the 3.2 kb product from the HindIII digest and the 1.6 kb product from the NgoMIV digest exactly matched the sequence from BAC 45I2, confirming that these products were derived from the donor VSG. In contrast, one end of the 1.8 kb product from the HindIII digest and the 1.6 kb product from the NgoMIV digest (the PCR products derived for the ELC) contained new sequence. This sequence has significant homology to ESAG9 from Trypanosoma equiperdum (48) and exactly matched the end sequences of two DpnII-BAC clones in the TIGR T.brucei Genome Project database (http://www.tigr.org/tdb/mdb/tbdb/). The sequence of BAC clone 26P8, which has the larger insert, was determined. A summary of the annotation of this 135 kb sequence is shown in Figure 5A.
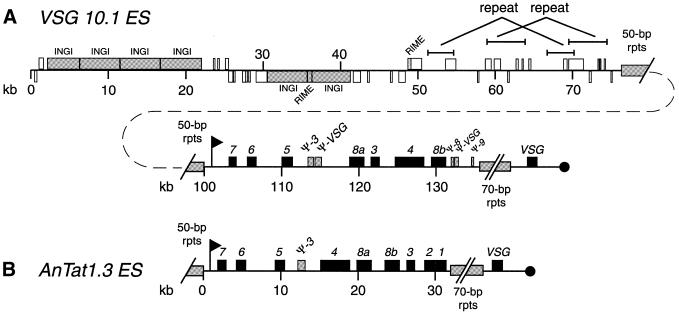
Figure 5.
Comparison of the B-ESs of VSG 10.1 (A, top two lines) and AnTat 1.3 VSG (B) (7). Boxes, flags and the black circle indicate the same features as described in Figure 3 legend. The ESAGs are numbered 1–8.
The first 80 kb of this sequence is highly complex, but surprisingly gene-poor. This region contains two clusters of four and two tandemly repeated INGIs, respectively, separated by ∼8 kb of sequence with nine short hypothetical ORFs. Twenty hypothetical ORFs are found between the last INGI pair and 50 bp repeats that begin ∼80 kb into the BAC clone. These ORFs are nearly equally distributed on both strands and do not appear to form a polycistronic transcription unit. Seven ORFs have limited homology to RNA polymerase subunits, but are likely to be pseudogenes. This entire region appears to have been generated by multiple duplication and/or recombination events. Sequences from 51.3 to 54.7 kb and from 59.0 to 64.0 kb are imperfectly repeated at 66.7–70.3 kb and at 69.5–74.5 kb, respectively. The 26 kb from 48.5–74.5 kb are repeated multiple times in an extremely complex pattern in BAC 3B10 and BAC 25N24 from T.brucei chromosome II and near one telomere in chromosome I (http://www.sanger.ac.uk/Projects/T_brucei and http://www.tigr.org/tdb/mdb/tbdb). Closely related sequences are also found in a long-distance anchored PCR product described as originating from the region upstream from the 50 bp repeats in the AnTat 1.3 ES (accession number AF193542, direct submission by G.Rudenko, A.Dirks-Mulder, T.van Welsem and P.Borst). Much of the sequence between the two INGI clusters is highly homologous to sequences from chromosomes I, II and VI. Thus, this repetitive region preceding the VSG 10.1 ES is highly conserved in other chromosomes, suggesting that it is functionally important.
The entire sequence following the 50 bp repeats resembles a B-ES (Fig. 5A). A potential bloodstream VSG promoter sequence with only four nucleotide differences from the 75 bp AnTat 1.3 promoter region occurs immediately after the 50 kb repeats. This promoter is followed by seven ESAGs (ESAGs 7, 6, 5, 8a, 3, 4 and 8b) and five interspersed pseudogenes (pseudo-ESAG 3 and -VSG after ESAG 5 and pseudo-ESAG 8, -VSG, and -ESAG 9 after ESAG 8b) (Fig. 5A). The pseudogenes have internal stop codons or truncations and are unlikely to encode functional proteins, but may represent the vestiges of past ESs. Approximately 15 kb of 70 bp repeats are found after pseudo-ESAG 9, the exact sequence of which was not determined because the 70 bp repeats are downstream of the sequence present in BAC 26P8. The only ORF between these 70 bp repeats and the telomere repeats is VSG 10.1. The sequence of the expressed VSG 10.1 exactly matches the sequence of the VSG 10.1 donor copy, indicating that no point mutations occurred in the formation of the ELC, in contrast to the MVAT5 VSG ELCs derived from the MVAT5 VSG M-ES in another serodeme that we have examined previously (41,45).
The overall organization of the GUTat 10.1 B-ES (Fig. 5A, second line) is similar to the AnTat 1.3 B-ES (15) (Fig. 5B). Both B-ESs are ∼45 kb long, have nearly identical promoters located shortly after the 50 bp repeats, and include several ESAGs (Fig. 5C). Consistent with the designations of GUTat 10.1 and AnTat 1.3 as T.brucei brucei, both lack the serum resistance-associated gene that confers resistance to normal human serum and which is found in some B-ESs (49). The similarity between the ESs extends beyond the level of overall organization down to the nucleotide sequences. The 14 kb after the 50 bp repeats through pseudo-ESAG3 are 95% identical in the GUTat 10.1 and AnTat 1.3 B-ESs. The high level of sequence identity suggests that this region of the B-ESs was derived from the same ancestral B-ES. However, the remaining sequences differ substantially between the two ESs. After pseudo-ESAG 3, the GUTat 10.1 B-ES has fewer ESAG genes than the AnTat 1.3 B-ES, and the ESAGs present are in a different order. The GUTat 10.1 B-ES lacks ESAGs 1 and 2, but contains several pseudogenes not found in AnTat 1.3. Thus, if the 5′-end of the GUTat 10.1 and AnTat 1.3 B-ESs had a similar origin, the 3′-ends must have been extensively modified or derived from different ancestral B-ESs.
The GUTat 10.1 B-ES bears little similarity to the VSG 10.1 donor region, other than the presence of INGI and RIME sequences. The putative VSG 10.1 M-ES (Fig. 3) is smaller and less complex than the B-ES, having only a promoter, 70 bp repeats and the VSG. The sequences upstream of the VSG 10.1 B-ES and M-ES differ extensively. The distance between the putative M-ES and the first upstream polycistronic transcription unit in the interior of the chromosome is ∼58 kb, whereas the corresponding distance upstream of the B-ES is at least 100 kb, and possibly much more. In addition, the region adjacent to the M-ES lacks the repetitive sequences described above and the 50 bp repeats that occur immediately upstream of the B-ES promoter. It is not clear why these sequences, which are conserved in several B-ESs, are unnecessary in the VSG 10.1 M-ES.
In conclusion, this study provides the first look over a 130 kb range of a telomere-linked donor VSG and its corresponding telomere-linked B-ES. In addition, this report lays the foundation for future studies on antigenic variation in GUTat 10.1, the T.brucei clone whose genome is being sequenced. We have shown that VSG 10.1 is expressed from a B-ES that is remarkably similar to the AnTat 1.3 B-ES, which may indicate that this is the general organization for B-ESs in T.brucei. Furthermore, our study has uncovered additional complexity in B-ESs. The region upstream of the 50 bp repeats contains sequences that are repeated both internally and in the regions near other T.brucei telomeres. These sequences are not found in the donor VSG 10.1 M-ES, which also lacks 50 bp repeats. The significance of these repetitive sequences is unclear, but the fact that these sequences are present near telomeres in multiple chromosomes suggest that they are important. What, if any, role do they play in recombination, chromatin remodeling, or ES silencing? The answers to these questions will provide further insights into the molecular biology of gene expression and antigenic variation in African trypanosomes.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by NIH grants AI10512 (D.J.L.), AI40591 (J.E.D.) and AI43062 (N.M.E.). C.M.R.T. is grateful to The Wellcome Trust for financial support.
DDBJ/EMBL/GenBank accession nos+ To whom correspondence should be addressed. Tel: +1 319 335 7932; Fax: +1 319 335 9570; Email: AF335471, AF335472, AC087700–AC087702
References
- 1. Thirteenth Programme Report of the UDNP World Bank/WHO Special Programme for Research and Training in Tropical Diseases (1996). World Health Organization, pp. 124–133.
- 2.Smith D.H., Pepin,J. and Stich,A.H. (1998) Human African trypanosomiasis: an emerging public health crisis. Br. Med. Bull., 54, 341–355. [DOI] [PubMed] [Google Scholar]
- 3.Turner C.M. and Barry,J.D. (1989) High frequency of antigenic variation in Trypanosoma brucei rhodesiense infections. Parasitology, 99, 67–75. [DOI] [PubMed] [Google Scholar]
- 4.Turner C.M. (1997) The rate of antigenic variation in fly-transmitted and syringe-passaged infections of Trypanosoma brucei. FEMS Microbiol. Lett., 153, 227–231. [DOI] [PubMed] [Google Scholar]
- 5.Davies K.P., Carruthers,V.B. and Cross,G.A. (1997) Manipulation of the vsg co-transposed region increases expression-site switching in Trypanosoma brucei. Mol. Biochem. Parasitol., 86, 163–177. [DOI] [PubMed] [Google Scholar]
- 6.Van der Ploeg L.H., Valerio,D., De Lange,T., Bernards,A., Borst,P. and Grosveld,F.G. (1982) An analysis of cosmid clones of nuclear DNA from Trypanosoma brucei shows that the genes for variant surface glycoproteins are clustered in the genome. Nucleic Acids Res., 10, 5905–5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pays E. and Nolan,D.P. (1998) Expression and function of surface proteins in Trypanosoma brucei. Mol. Biochem. Parasitol., 91, 3–36. [DOI] [PubMed] [Google Scholar]
- 8.Borst P., Bitter,W., Blundell,P.A., Chaves,I., Cross,M., Gerrits,H., van Leeuwen,F., McCulloch,R., Taylor,M. and Rudenko,G. (1998) Control of VSG gene expression sites in Trypanosoma brucei. Mol. Biochem. Parasitol., 91, 67–76. [DOI] [PubMed] [Google Scholar]
- 9.Cross G.A., Wirtz,L.E. and Navarro,M. (1998) Regulation of vsg expression site transcription and switching in Trypanosoma brucei. Mol. Biochem. Parasitol., 91, 77–91. [DOI] [PubMed] [Google Scholar]
- 10.Donelson J.E. (1996) Genome research and evolution in trypanosomes. Curr. Opin. Genet. Dev., 6, 699–703. [DOI] [PubMed] [Google Scholar]
- 11.Pedram M. and Donelson,J.E. (1999) The anatomy and transcription of a monocistronic expression site for a metacyclic variant surface glycoprotein gene in Trypanosoma brucei. J. Biol. Chem., 274, 16876–16883. [DOI] [PubMed] [Google Scholar]
- 12.Lenardo M.J., Esser,K.M., Moon,A.M., Van der Ploeg,L.H. and Donelson,J.E. (1986) Metacyclic variant surface glycoprotein genes of Trypanosoma brucei subsp. rhodesiense are activated in situ, and their expression is transcriptionally regulated. Mol. Cell. Biol., 6, 1991–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zomerdijk J.C., Ouellette,M., ten Asbroek,A.L., Kieft,R., Bommer,A.M., Clayton,C.E. and Borst,P. (1990) The promoter for a variant surface glycoprotein gene expression site in Trypanosoma brucei. EMBO J., 9, 2791–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Navarro M. and Cross,G.A. (1996) DNA rearrangements associated with multiple consecutive directed antigenic switches in Trypanosoma brucei. Mol. Cell. Biol., 16, 3615–3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pays E., Tebabi,P., Pays,A., Coquelet,H., Revelard,P., Salmon,D. and Steinert,M. (1989) The genes and transcripts of an antigen gene expression site from T.brucei. Cell, 57, 835–845. [DOI] [PubMed] [Google Scholar]
- 16.Turner C.M., Barry,J.D., Maudlin,I. and Vickerman,K. (1988) An estimate of the size of the metacyclic variable antigen repertoire of Trypanosoma brucei rhodesiense. Parasitology, 97, 269–276. [DOI] [PubMed] [Google Scholar]
- 17.Barry J.D., Graham,S.V., Fotheringham,M., Graham,V.S., Kobryn,K. and Wymer,B. (1998) VSG gene control and infectivity strategy of metacyclic stage Trypanosoma brucei. Mol. Biochem. Parasitol., 91, 93–105. [DOI] [PubMed] [Google Scholar]
- 18.Alarcon C.M., Pedram,M. and Donelson,J.E. (1999) Leaky transcription of variant surface glycoprotein gene expression sites in bloodstream African trypanosomes. J. Biol. Chem., 274, 16884–16893. [DOI] [PubMed] [Google Scholar]
- 19.El-Sayed N.M., Hegde,P., Quackenbush,J., Melville,S.E. and Donelson,J.E. (2000) The African trypanosome genome. Int. J. Parasitol., 30, 329–345. [DOI] [PubMed] [Google Scholar]
- 20.Goedbloed E., Ligthart,G.S., Minter,D.M., Wilson,A.J., Dar,F.K. and Paris,J. (1973) Serological studies of trypanosomiasis in East Africa. II. Comparisons of antigenic types of Trypanosoma brucei subgroup organisms isolated from wild tsetse flies. Ann. Trop. Med. Parasitol., 67, 31–43. [PubMed] [Google Scholar]
- 21.Turner C.M., Sternberg,J., Buchanan,N., Smith,E., Hide,G. and Tait,A. (1990) Evidence that the mechanism of gene exchange in Trypanosoma brucei involves meiosis and syngamy. Parasitology, 101, 377–386. [DOI] [PubMed] [Google Scholar]
- 22.Turner C.M., Aslam,N., Smith,E., Buchanan,N. and Tait,A. (1991) The effects of genetic exchange on variable antigen expression in Trypanosoma brucei. Parasitology, 103, 379–386. [DOI] [PubMed] [Google Scholar]
- 23. Herbert,W.J. and Parratt,D. (1979) Virulence of trypanosomes in the vertebrate host. In Lumsden,W.H.R. and Evans,D.A. (eds), Biology of Kinetoplastida. Academic Press London,UK, Vol. 2, pp. 481–521.
- 24.Turner C.M., Aslam,N. and Dye,C. (1995) Replication, differentiation, growth and the virulence of Trypanosoma brucei infections. Parasitology, 111, 289–300. [DOI] [PubMed] [Google Scholar]
- 25.Willis T.G., Jadayel,D.M., Coignet,L.J., Abdul-Rauf,M., Treleaven,J.G., Catovsky,D. and Dyer,M.J. (1997) Rapid molecular cloning of rearrangements of the IGHJ locus using long-distance inverse polymerase chain reaction. Blood, 90, 2456–2464. [PubMed] [Google Scholar]
- 26.Sutton G.G., White,O., Adams,M.D. and Kerlavage,A.R. (1995) TIGR Assembler: a new tool for assembling large shotgun sequencing projects. Genome, 1, 9–19. [Google Scholar]
- 27.Salzberg S.L., Delcher,A.L., Kasif,S. and White,O. (1998) Microbial gene identification using interpolated Markov models. Nucleic Acids Res., 26, 544–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rudenko G., Chaves,I., Dirks-Mulder,A. and Borst,P. (1998) Selection for activation of a new variant surface glycoprotein gene expression site in Trypanosoma brucei can result in deletion of the old one. Mol. Biochem. Parasitol., 95, 97–109. [DOI] [PubMed] [Google Scholar]
- 29.Majumder H.K., Boothroyd,J.C. and Weber,H. (1981) Homologous 3′-terminal regions of mRNAs for surface antigens of different antigenic variants of Trypanosoma brucei. Nucleic Acids Res., 9, 4745–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walder J.A., Eder,P.S., Engman,D.M., Brentano,S.T., Walder,R.Y., Knutzon,D.S., Dorfman,D.M. and Donelson,J.E. (1986) The 35-nucleotide spliced leader sequence is common to all trypanosome messenger RNAs. Science, 233, 569–571. [DOI] [PubMed] [Google Scholar]
- 31.Carrington M., Miller,N., Blum,M., Roditi,I., Wiley,D. and Turner,M. (1991) Variant specific glycoprotein of Trypanosoma brucei consists of two domains each having an independently conserved pattern of cysteine residues. J. Mol. Biol., 221, 823–835. [DOI] [PubMed] [Google Scholar]
- 32.Kim K.S. and Donelson,J.E. (1997) Co-duplication of a variant surface glycoprotein gene and its promoter to an expression site in African trypanosomes. J. Biol. Chem., 272, 24637–24645. [DOI] [PubMed] [Google Scholar]
- 33.Myler P.J., Aline,R.F.,Jr, Scholler,J.K. and Stuart,K.D. (1988) Changes in telomere length associated with antigenic variation in Trypanosoma brucei. Mol. Biochem. Parasitol., 29, 243–250. [DOI] [PubMed] [Google Scholar]
- 34.Horn D. and Cross,G.A. (1997) Analysis of Trypanosoma brucei vsg expression site switching in vitro. Mol. Biochem. Parasitol., 84, 189–201. [DOI] [PubMed] [Google Scholar]
- 35.Myler P.J., Audleman,L., DeVos,T., Hixson,G., Kiser,P., Lemley,C., Magness,C., Rickel,E., Sisk,E., Sunkin,S., Swartzell,S., Westlake,T., Bastien,P., Fu,G.L., Ivens,A. and Stuart,K. (1999) Leishmania major Friedlin chromosome 1 has an unusual distribution of protein-coding genes. Proc. Natl Acad. Sci. USA, 96, 2902–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hasan G., Turner,M.J. and Cordingley,J.S. (1984) Complete nucleotide sequence of an unusual mobile element from Trypanosoma brucei. Cell, 37, 333–341. [DOI] [PubMed] [Google Scholar]
- 37.Hasan G., Turner,M.J. and Cordingley,J.S. (1982) Ribosomal RNA genes of Trypanosoma brucei. Cloning of an rRNA gene containing a mobile element. Nucleic Acids Res., 10, 6747–6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy N.B., Pays,A., Tebabi,P., Coquelet,H., Guyaux,M., Steinert,M. and Pays,E. (1987) Trypanosoma brucei repeated element with unusual structural and transcriptional properties. J. Mol. Biol., 195, 855–871. [DOI] [PubMed] [Google Scholar]
- 39.Kimmel B.E., ole-MoiYoi,O.K. and Young,J.R. (1987) Ingi, a 5.2-kb dispersed sequence element from Trypanosoma brucei that carries half of a smaller mobile element at either end and has homology with mammalian LINEs. Mol. Cell. Biol., 7, 1465–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berberof M., Pays,A., Lips,S., Tebabi,P. and Pays,E. (1996) Characterization of a transcription terminator of the procyclin PARP A unit of Trypanosoma brucei. Mol. Cell. Biol., 16, 914–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu Y., Hall,T., Gay,L.S. and Donelson,J.E. (1993) Point mutations are associated with a gene duplication leading to the bloodstream reexpression of a trypanosome metacyclic VSG. Cell, 72, 397–406. [DOI] [PubMed] [Google Scholar]
- 42.Nagoshi Y.L., Alarcon,C.M. and Donelson,J.E. (1995) The putative promoter for a metacyclic VSG gene in African trypanosomes. Mol. Biochem. Parasitol., 72, 33–45. [DOI] [PubMed] [Google Scholar]
- 43.Matthews K.R., Shiels,P.G., Graham,S.V., Cowan,C. and Barry,J.D. (1990) Duplicative activation mechanisms of two trypanosome telomeric VSG genes with structurally simple 5′ flanks. Nucleic Acids Res., 18, 7219–7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Graham S.V., Terry,S. and Barry,J.D. (1999) A structural and transcription pattern for variant surface glycoprotein gene expression sites used in metacyclic stage Trypanosoma brucei. Mol. Biochem. Parasitol., 103, 141–154. [DOI] [PubMed] [Google Scholar]
- 45.Lu Y., Alarcon,C.M., Hall,T., Reddy,L.V. and Donelson,J.E. (1994) A strand bias occurs in point mutations associated with variant surface glycoprotein gene conversion in Trypanosoma rhodesiense. Mol. Cell. Biol., 14, 3971–3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alarcon C.M., Son,H.J., Hall,T. and Donelson,J.E. (1994) A monocistronic transcript for a trypanosome variant surface glycoprotein. Mol. Cell. Biol., 14, 5579–5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Graham S.V. and Barry,J.D. (1995) Transcriptional regulation of metacyclic variant surface glycoprotein gene expression during the life cycle of Trypanosoma brucei. Mol. Cell. Biol., 15, 5945–5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Florent I.C., Raibaud,A. and Eisen,H. (1991) A family of genes related to a new expression site-associated gene in Trypanosoma equiperdum. Mol. Cell. Biol., 11, 2180–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xong H.V., Vanhamme,L., Chamekh,M., Chimfwembe,C.E., Van Den Abbeele,J., Pays,A., Van Meirvenne,N., Hamers,R., De Baetselier,P. and Pays,E. (1998) A VSG expression site-associated gene confers resistance to human serum in Trypanosoma rhodesiense. Cell, 95, 839–846. [DOI] [PubMed] [Google Scholar]