Abstract
mTOR [mechanistic target of rapamycin] is a serine/threonine protein kinase that, as part of mTORC1 (mTOR complex 1), acts as an important molecular connection between nutrient signals and the metabolic processes indispensable for cell growth. While there has been pronounced interest in the upstream mechanisms regulating mTORC1, the full range of downstream molecular targets through which mTORC1 signaling stimulates cell growth is only recently emerging. It is now evident that mTORC1 promotes cell growth primarily through the activation of key anabolic processes. Through a diverse set of downstream targets, mTORC1 promotes the biosynthesis of macromolecules, including proteins, lipids, and nucleotides to build the biomass underlying cell, tissue, and organismal growth. Here, we focus on the metabolic functions of mTORC1 as they relate to the control of cell growth. As dysregulated mTORC1 underlies a variety of human diseases, including cancer, diabetes, autoimmune diseases, and neurological disorders, understanding the metabolic program downstream of mTORC1 provides insights into its role in these pathological states.
Introduction
Cellular metabolic homeostasis requires the coordinated conversion of nutrients (e.g., glucose and amino acids) and energy (ATP) into macromolecules (e.g., proteins, nucleic acids, and lipids) through anabolic processes and the recycling of these macromolecules back into their nutrient components, which can be further catabolized to produce energy (Figure 1A). With a continuous input of nutrients, this interconversion of nutrients and macromolecules can, theoretically, be self-sustaining. However, cells and organisms possess distinct systems to sense fluctuations in nutrients and energy to properly adapt their metabolic state to nutrient availability (feast) or depletion (famine) [1]. At the cellular level, these nutrient sensing mechanisms involve transcription factors, such as hypoxia-inducible factor 1 (HIF1) and sterol regulatory element-binding protein (SREBP), which can alter the metabolic program of the cell through the expression of nutrient transporters and metabolic pathway-specific enzymes. In addition, acute adaptations to changes in intracellular nutrients and energy can be achieved through nutrient sensing signaling proteins, such as the protein kinases glucose non-fermenting 2 (GCN2) and AMP-activated protein kinase (AMPK). Here, we discuss the mechanistic (or mammalian) target of rapamycin (mTOR) complex 1 (mTORC1) as a nutrient sensing protein kinase that acts as a master regulator of cellular metabolism. Through a network of upstream signaling pathways, mTORC1 integrates signals from intracellular nutrients and energy with endocrine and paracrine signals reflecting the status of the system or tissue, in the form of exogenous growth factors, hormones, and cytokines, to reciprocally regulate a variety of anabolic and catabolic processes (Figure 1B). Through its control of cellular metabolism, mTORC1 promotes the production of biomass to support growth and proliferation or the stable storage of energy in highly reduced macromolecules, such as lipids. The importance of coordinated regulation of distinct biosynthetic processes and metabolic pathways that support anabolic metabolism downstream of mTORC1 is discussed below.
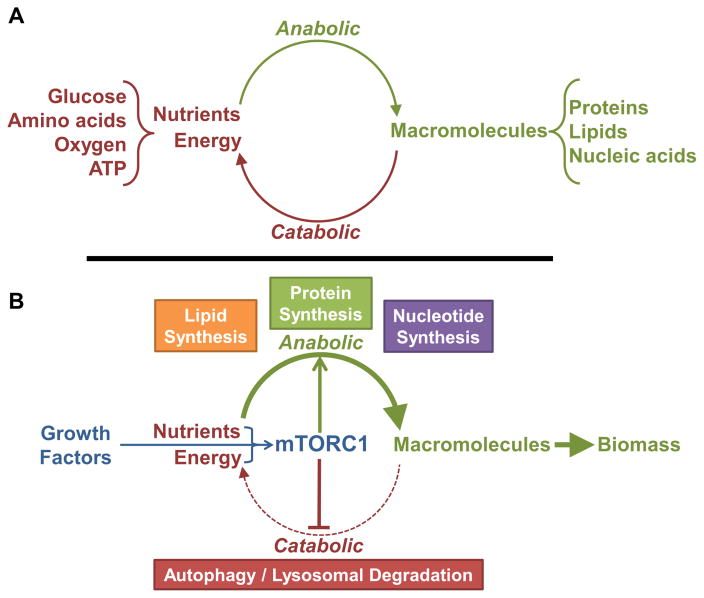
Figure 1. mTORC1 is a molecular switch between catabolic and anabolic processes.
(A) A standard metabolic cycle. Anabolic processes convert nutrients and energy into macromolecules, and catabolic processes can convert macromolecules into their nutrient components and energy. (B) Through the sensing of exogenous growth factors and endogenous nutrients and energy, mTORC1 serves to tip the metabolic balance away from catabolic processes and toward anabolic processes to produce biomass.
An upstream signaling network regulates mTORC1 through the Rag and Rheb GTPases
The activation state of mTORC1 is tightly controlled by numerous upstream signaling pathways that respond to either exogenous growth factors through cell surface receptors or changes in intracellular nutrients and energy through cytosolic sensors. These mTORC1 regulatory pathways, which have been extensively reviewed in recent years [2,3], impinge on two systems of small G proteins and their regulators that reside, at least in part, on the cytosolic face of lysosomes (Figure 2). The Rag GTPases function as a heterodimer of RagA or B bound to RagC or D and their guanine-nucleotide binding state is regulated by several multi-protein complexes, including the Ragulator and GATOR complexes [4–7]. The molecular functions of these protein complexes are still emerging but are influence by the presence or absence of specific amino acids, such as leucine and arginine [8,9], which promote the accumulation of Rag heterodimers containing GTP-bound RagA/B in complex with GDP-bound RagC/D. In this amino acid-stimulated state the Rag heterodimer, bound to the Ragulator, serves as a docking site for mTORC1 at the lysosome, thereby facilitating its subsequent activation by the Rheb GTPase [4].
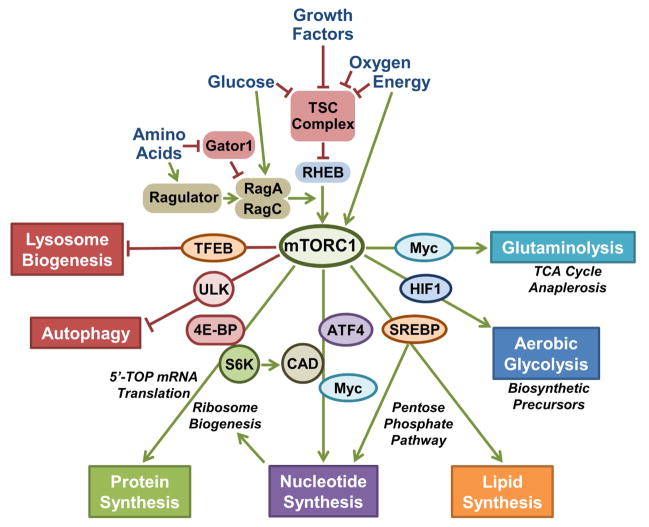
Figure 2. The mTORC1 network senses cellular growth signals and relays their status to an array of downstream metabolic processes.
The activation state of mTORC1 in response to growth factors and nutrients depends on regulatory inputs into small GTPases of the Rheb and Rag families. The TSC complex integrates a variety of growth signals and, under conditions of nutrient or growth factor withdrawal, functions as a GTPase-activating protein (GAP) to negatively regulate Rheb, which is an essential activator of mTORC1. A heterodimer of RagA or B bound to RagC or D is regulated by amino acids through the Gator1 and Ragulator complexes. In the presence of amino acids, the Rag proteins bind to mTORC1 and promote its activation by Rheb. When activated, mTORC1 regulates various cellular processes that promote macromolecular synthesis and cell growth. This occurs through parallel transcriptional, translational, and post-translational effects on downstream targets. This figure emphasized the metabolic outputs of mTORC1 signaling.
A sub-population of Rheb (Rheb1 and Rheb2/Rhebl1) resides at the lysosomal surface and, in the GTP-bound state, is essential for mTORC1 activation [10,11]. The only established regulator of Rheb is a GTPase-activating protein (GAP) complex comprised of the tuberous sclerosis complex (TSC) proteins TSC1 and TSC2 and the TBC domain protein TBC1D7, referred to as the TSC complex [2,12]. Within this complex, TSC1 scaffolds TSC2 and TBC1D7 together and stabilizes them. While the molecular function of TBC1D7 is unknown, TSC2 possesses the GAP activity toward Rheb and is regulated by multi-site phosphorylation downstream of several protein kinase signaling pathways. In this manner, the TSC complex integrates signals from growth factor pathways, such as the PI3K-Akt and Ras-Erk pathways, and stress response kinases, such AMPK, which is activated in response to cellular energy depletion. Pro-growth signals from growth factor and nutrient sensing pathways inhibit the TSC complex to promote the accumulation of Rheb-GTP and its activation of mTORC1, while growth-suppressing signals, including growth factor withdrawal, nutrient depletion or cellular stress, activate the TSC complex to decrease Rheb-GTP levels and shut down mTORC1 signaling.
Growth factor signaling through the PI3K-Akt pathway, and perhaps other signals, regulate mTORC1 by influencing co-localization of the TSC complex with Rheb at the lysosomal surface [11,13]. The Akt-mediated phosphorylation of TSC2 results in immediate release of the TSC complex from Rheb, allowing Rheb to become GTP loaded and activates mTORC1 [11]. While signals relaying the presence of exogenous growth factors and sufficient intracellular nutrients and energy are both required for full activation of mTORC1, nutrient and energy signals from within the cell are dominantly required for mTORC1 to perceive signals propagated by growth factor pathways. This hierarchy stems, at least in part, from differential spatial regulation of mTORC1 and the TSC complex [11]. For instance, in the absence of amino acids, growth factor signaling pathways that stimulate release of the TSC complex from Rheb at the lysosome will fail to activate mTORC1 because Rag heterodimers will be in a conformation unable to dock mTORC1 at the lysosomal surface where GTP-bound Rheb is present.
Importantly, the TSC proteins are tumor suppressors and some of the most commonly mutated oncogenes (e.g., PI3K, Ras) and tumor suppressors (e.g., PTEN, LKB1, NF1) lie within the signaling pathways that regulate the TSC complex [14]. As such, the TSC complex is aberrantly inhibited in the majority of human cancers, resulting in uncontrolled mTORC1 signaling in tumors. The metabolic program discussed below, which is activated downstream of mTORC1 might play a particularly important role in the growth of tumors [15].
mTORC1 activation promotes anabolic metabolism
Through a variety of transcriptional, translational, and post-translational mechanisms, mTORC1 stimulates an increase in biosynthetic processes that are otherwise maintained at basal, homeostatic states. As such, it should be emphasized that mTORC1 is not required for the anabolic processes described below, but serves as an essential link between growth signals and heightened biosynthetic activities.
Protein synthesis
Proteins constitute approximately 50% of the biomass of cells. mTORC1 plays a central role in the induction of protein synthesis in response to cellular growth signals. mTORC1 directly phosphorylates the eukaryotic translation initiation factor 4E (eIF4E)-binding proteins (4E-BP1 and 2), thereby stimulating release of 4E-BP from its inhibitory binding of eIF4E at the 5′-cap of mRNAs [16,17]. The release of eIF4E allows it to bind to eukaryotic translation initiation factors 4G (eIF4G) and 4A (eIF4A) to form the active eIF4F translation initiation complex [18]. While mTORC1 can influence global protein synthesis, it acutely controls the translation of a subset of mRNAs possessing 5′-terminal oligopyrimidine (TOP) tracts [19]. Importantly, this class of mRNAs encode for components of the translation apparatus, including ribosomal proteins and translation factors, an increase in which enhances the total protein synthesis capacity of the cell. The 5′-TOP sequence has a cytosine at the transcriptional start site, comprising the 5′-end of the mRNA, followed by a stretch of 4 to 14 pyrimidines. Recent studies using high-resolution transcriptome-scale ribosome profiling have confirmed that the translation of mRNAs containing 5′-TOP and TOP-like sequences is highly sensitive to mTOR inhibitors [20,21]. The selective regulation of 5′-TOP mRNA translation by mTORC1 depends on its regulation of the 4E-BPs. However, the molecular mechanism by which 4E-BP regulation selectively influences ribosome occupancy for this subset of mRNAs is unknown. The La-related protein 1 (LARP1) has been found to directly interact with mRNAs containing a 5′TOP motif to influence their stability and translation [22–24]. Evidence suggests that mTORC1 regulates LARP1, but conflicting data currently exist regarding whether LARP1 is a positive or negative regulator of 5′-TOP mRNA translation.
The ribosomal S6 protein kinases, S6K1 (comprised of the p70 and p85 isoforms) and S6K2, are also direct targets of mTORC1 that, through a variety of downstream substrates, regulate aspects of mRNA translation [25]. As their name indicates, these proteins were identified based on their ability to phosphorylate the 40S ribosomal protein S6. While the molecular function of the highly conserved phosphorylation sites on S6 remains elusive, the S6Ks have been found to engage other proteins in the translation machinery. S6K activates the eIF4A RNA helicase activity within the eIF4F complex by phosphorylating and recruiting its binding partner eIF4B, while also phosphorylating the eIF4A inhibitory protein programmed cell death 4 (PDCD4), which targets it for degradation [26–28]. In this manner, S6K is believed to promote the eIF4A-mediated unwinding of mRNAs with highly structured 5′-untranslated regions to facilitate their translation. S6K also phosphorylates and inhibits the eukaryotic elongation factor 2 (eEF2) kinase (eEF2K), thereby relieving the inhibition of eEF2 by eEF2K and enhancing translation elongation [29,30]. Finally, S6K1 has been found to influence mRNA maturation by phosphorylating and regulating components of the splicing machinery [31,32].
Enhancing ribosome biogenesis is a major function of mTORC1 [33]. In addition to stimulating the synthesis of ribosomal proteins through the induction of 5′-TOP mRNA translation, mTORC1 also positively regulates the Pol I- and Pol III-dependent transcription of the different classes of ribosomal RNAs through mechanisms that are still emerging [34]. One such mechanism involves the direct phosphorylation and inhibition of MAF1, a key repressor of Pol III-mediated transcription [35–38]
Nucleotide synthesis
In addition to protein, enhanced synthesis of the nucleic acids RNA and DNA, comprised of purine and pyrimidine nucleotides is required for cell growth. As described above, mTORC1 signaling promotes an increase in ribosome biogenesis, a hallmark of growing cells. The majority of intracellular nucleotides reside in the ribosome, as rRNA molecules comprise approximately 55% of the mass of ribosomes. As such, an increase in ribosome biogenesis is accompanied by a great increase in the demand for more nucleotides, which can be acquired through either de novo synthesis or exogenous uptake (nucleotide salvage). Proliferating cells, including activated lymphocytes and cancer cells, strongly induce the de novo nucleotide synthesis pathways, likely due to limiting concentrations of nucleosides and nucleobases available in our blood [39]. Recent studies have demonstrated that mTORC1 signaling promotes de novo pyrimidine and purine synthesis through a variety of transcriptional and post-translational mechanisms. Activation of mTORC1 leads to the acute stimulation of de novo pyrimidine synthesis through the S6K1-dependent phosphorylation of a multifunctional enzyme called CAD (carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydro-orotase), which catalyses the first three steps in pyrimidine synthesis [15,40,41] (Figure 3). Importantly, the S6K1-mediated phosphorylation of CAD is not required for the basal activity of this metabolic pathway but serves to increase pathway activity in response to growth signals that stimulate mTORC1 signaling.
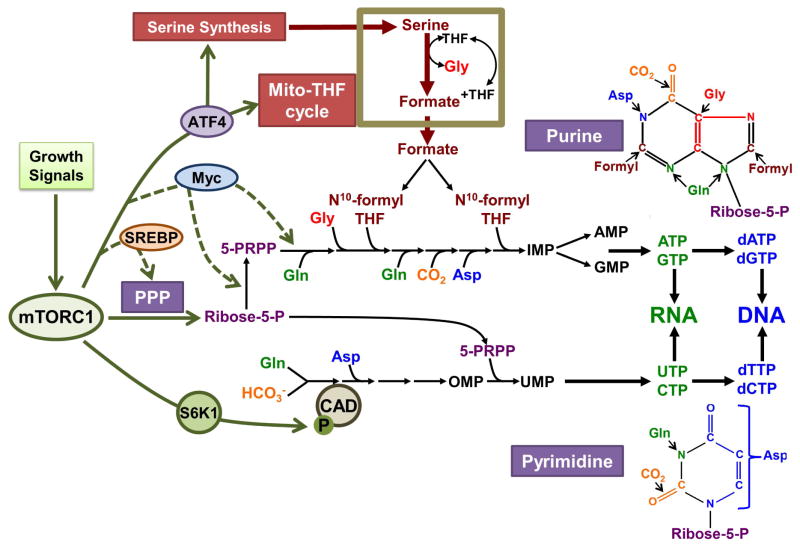
Figure 3. mTORC1 signaling stimulates de novo nucleotide synthesis through multiple downstream mechanisms.
In response to growth signals, mTORC1 activation leads to S6K1 mediated-phosphorylation of CAD, the rate-limiting enzyme of the de novo pyrimidine synthesis pathway, thereby acutely stimulating an increase in pyrimidine synthesis. Through downstream effects on transcription factors that induce expression of key metabolic enzymes, mTORC1 activation also increases the de novo purine synthesis pathway. One such mechanism involves activation of ATF4 and its induction of serine synthesis and mTHF enzymes, which contribute one-carbon formyl units to the purine ring. Purine and pyrimidine nucleotides are depicted and color coded by the source of their carbon, nitrogen, and oxygen moieties assembled through the de novo synthesis pathways.
mTORC1-activating stimuli also enhance de novo purine synthesis through transcriptional effects on multiple enzymes that feed into the purine synthesis pathway, including those of the pentose phosphate pathway (PPP), serine and glycine synthesis, and the mitochondrial tetrahydrofolate (mTHF) pathway [42]. The enzyme MTHFD2 (Methylene Tetrahydrofolate Dehydrogenase 2) is particularly sensitive to the activation state of mTORC1 and, as part of the mTHF cycle, provides cytosolic one-carbon formyl units required for purine synthesis (Figure 3). mTORC1 signaling elevates MTHFD2 expression by increasing translation of the mRNA encoding the ATF4 transcription factor, the increased level of which directly stimulates MTHFD2 transcription [42]. Interestingly, an accompanying study suggested that mTOR promotes assembly of a complex of purine synthesis enzymes, referred to as the purinosome, proximal to the outer surface of mitochondria [43]. This observation could provide a mechanism that facilitates the use of mitochondrial-derived one-carbon units in two essential enzymatic steps of the cytosolic purine synthesis pathway. Two additional transcription factors known to function downstream of mTORC1, Myc and SREBP, were also found to contribute to enhanced purine synthesis in response to mTORC1 activation [42] (Figure 3). Myc has been established to induce the expression of multiple enzymes in both the pyrimidine and purine synthesis pathways [44]. Thus, in parallel to its induction of ribosome biogenesis, mTORC1 promotes the de novo production of nucleotides to sustain rRNA synthesis and, in proliferating cells, DNA synthesis.
A major metabolic pathway required for the synthesis of both pyrimidine and purine nucleotides is the PPP, which is the sole source of glucose-derived ribose. Ribose 5-phosphate is the product of two distinct branches of the PPP: the irreversible oxidative branch, which through the oxidation of glucose 6-phosphate also generates reducing equivalents in the form of NADPH, and the non-oxidative branch, a series of reversible reactions generating ribose from glycolytic intermediates. In order to be used for nucleotide synthesis, ribose 5-phosphate is converted to phosphoribosyl pyrophosphate (PRPP) through the action of 5-phosphoriboylsynthetase (PRPS) (Figure 3). A recent study found that overexpressed Myc enhances the eIF4E-driven translation of PPRS2 to promote nucleotide synthesis [45], suggesting another level of regulation by downstream components of the mTORC1 pathway. Another study measuring the effects of mTORC1 activation and inhibition on relative flux through the two branches of the PPP found that mTORC1 preferentially stimulates flux through the NADPH-producing oxidative branch [46]. This occurs predominantly through the transcriptional induction of genes encoding oxidative PPP enzymes, which is driven, at least in part, by SREBP1. Through this regulation, mTORC1 promotes not only the production of ribose 5-phosphate, but also NADPH, providing essential reducing power to drive the anabolic processes of nucleotide and lipid synthesis.
Lipid synthesis
The stimulation of cell growth, as well as the uncontrolled growth of cancer cells, is often accompanied by an increase in de novo lipid synthesis to meet the demand for increased lipids required for organellar and plasma membrane biogenesis [47]. mTORC1 appears to play a key role in relaying growth factor and oncogenic signals to downstream regulators of lipid synthesis. The SREBPs (SREBP1a, 1c and 2) are a family of transcription factors that directly induce the expression of genes encoding the major lipogenic enzymes involved in the synthesis of both fatty acids and sterols [48]. It is now well established in different settings that mTORC1 signaling promotes SREBP processing events that lead to its activation of lipogenic gene expression and enhanced lipid synthesis [46,49–54]. Several downstream targets of mTORC1 have been found to influence SREBP processing and activation, including S6K1 [46,53,55], lipin1 [52], and CREB regulated transcription coactivator 2 (CRTC2) [24]. While the precise molecular mechanism(s) remains to be characterized, it seems likely that, by analogy to protein and nucleotide synthesis, mTORC1 controls SREBP and lipid synthesis through multiple inputs. An important question requiring further investigation is how mTORC1 activation influences the lipid content of cells. It is possible that in actively growing cells, mTORC1 signaling channels de novo synthesized lipids toward membrane phospholipids and away from storage lipids (e.g., triglycerides) through additional downstream targets, such as the phosphatidic acid phosphatase lipin1 [56]. The importance of coupling the mTORC1-driven increase in protein synthesis with its stimulated increase in lipid synthesis and membrane expansion has been well illustrated in studies demonstrating that perturbing lipid synthesis in settings of mTORC1 activation can cause endoplasmic reticulum (ER) stress [57,58]. Studies such as these provide a further rationale for the biosynthesis of the three major classes of macromolecules (proteins, nucleotides, and lipids) being tightly coordinated through parallel mechanisms downstream of mTORC1.
Anabolic support pathways
In addition to the production of reducing power in the form of NADPH through the mTORC1-mediated control of the oxidative PPP [46], mTORC1 also induces the processes of glycolysis and glutaminolysis to further support its anabolic functions. The phenomenon known as the Warburg Effect (aerobic glycolysis), where cells take up more glucose and convert a majority of this additional glucose to lactate, even under normal oxygen concentrations (normoxia), is now well recognized to support anabolic cell growth under both physiological (immune cell activation) and pathological (cancer) states [59]. Aerobic glycolysis is believed to provide ample glycolytic intermediates to promote metabolic flux into the side branches of glycolysis required for the synthesis of ribose, glycerol, non-essential amino acids, and one-carbon units, thereby supporting the synthesis of macromolecules. mTORC1 signaling can promote aerobic glycolysis through upregulation of the hypoxia inducible factor (HIF1α) transcription factor [46], which induces expression of glucose transporters and nearly all enzymes of glycolysis [60]. As its name implies, HIF1α is predominantly activated under conditions of hypoxia, where its levels can increase some 50-fold due to increased protein stability. However, under normoxic conditions mTORC1, through its phosphorylation and inhibition of 4EBP1, can enhance the cap-dependent translation of the HIF1α mRNA leading to an approximately 5-fold increase in protein levels, without changes in stability, which is sufficient to activate the glycolytic program [46,61–63]. One implication of this regulation is that in a growing tumor, the capacity of mTORC1 to promote HIF1α activity would enhance glycolysis even in oxygenated areas of the tumor with high mTORC1 signaling. The mTORC1-mediated regulation of aerobic glycolysis through HIF1α is also important in expanding immune cell populations [64,65]. Like HIF1α, Myc can also induce the expression of genes involved in glucose uptake and glycolysis and can be translated in an mTORC1-dependent manner in some settings [66,67]. The mTORC1-mediated control of Myc has also been found to contribute to glutaminolysis and its role in anaplerosis. Glutamine is catabolized to the TCA cycle intermediate α-ketoglutarate (or oxoglutarate), through two deamination reactions involving glutaminase (GLS) and glutamate dehydrogenase (GDH). Activation of the mTORC1 pathway has been shown to promote the anaplerotic entry of glutamine-derived carbon into the TCA cycle via regulation of both of these enzymes, at least in part, through the S6K1-dependent regulation of c-Myc [68,69]. This regulation serves to replenish TCA cycle intermediates that are consumed as essential precursors for the synthesis of proteins, nucleotides and lipids.
Inhibition of catabolic processes
Autophagy and Lysosomal Degradation
Autophagy is a highly conserved catabolic pathway that is activated in response to nutrient and energy deprivation, at least in part, through the inhibition of mTORC1 signaling. The process of autophagy serves to recycle cytosolic components, such as proteins and organelles, into their nutrient components through engulfment in a membranous structure called the autophagosome, which subsequently fuses with the lysosome for degradation of its components. The autophagy pathway is made up of several distinct regulatory complexes, among which the ULK1 (Unc51-like kinase 1) complex functions as important gatekeeper for the induction of autophagy [70–73]. ULK1 is a Ser/Thr protein kinase that functions in a complex with ATG13, FIP200, and ATG101 to initiate the process of autophagy and is reciprocally regulated by mTORC1 and AMPK [74–76]. The reversible phosphorylation of ULK1 by mTORC1 under nutrient and energy rich conditions and AMPK under states of energy depletion serve to, respectively, inhibit and activate the initial steps of autophagy through effects on subsequent ULK1-regulated processes [75,77,78]. Through less well-defined mechanisms, mTORC1 also targets the function of other proteins involved in the early stages of autophagy, such as Atg13 [70,79] Atg14L [80], Ambra1 (Autophagy/beclin-1 regulator 1) [81]. mTORC1 has recently been shown to phosphorylate the autophagy pathway component UVRAG (UV radiation resistance-associated gene product) to mediate regulation of autophagy at later stages, including autophagosome maturation [82]. Through these, and likely other downstream targets, the regulation of mTORC1 is a key link between the sensing of intracellular nutrients and the control of autophagy as a key adaptive mechanism to recycle macromolecules into nutrients in times of need.
It is now clear that the crosstalk between mTORC1 activity and lysosomal function extends beyond the regulation of autophagy. As mentioned above, mTORC1 is activated by Rheb at the lysosomal surface in response to signals from amino acids acting on the Ragulator-Rag complex. The Ragulator has been found to associate with the large vacuolar H+-adenosine triphosphatase (v-ATPase) complex, which functions to acidify lysosomes, and mTORC1 signaling requires a functional v-ATPase [83]. In this manner, mTORC1 activity can monitor lysosomal function. It is now evident that mTORC1 inhibition in response to nutrient deprivation, lysosomal dysfunction, or pharmacological inhibition, induces a transcriptional program to promote lysosome biogenesis and enhance the activity of lysosomes [84–88]. TFEB and its related transcription factors, TFE3 and MITF, induce the expression of genes encoding components of the v-ATPase, lysosomal enzymes, and autophagy proteins [89,90]. When mTORC1 is active, it phosphorylates TFEB and sequesters it in the cytosol, thereby attenuating the production of lysosome and autophagy components, whereas mTORC1 inhibition promotes TFEB nuclear translocation and induction of its transcriptional program. TFEB regulation by mTORC1 thus acts as another mode of adaptive regulation to fluctuations in intracellular nutrients. Through its enhancement of lysosome-mediated degradation, mTORC1 inhibition has also been found to facilitate the consumption and degradation of extracellular materials, from exogenous proteins or dead cells, as an adaptive response to nutrient depletion, or the perception there of [91,92].
Lipolysis and fatty acid oxidation
In addition to promoting lipid synthesis, there is evidence that mTORC1 signaling blocks the release and catabolism of lipids for energy [93]. Cells can maintain energy-rich lipid stores in the form of lipid droplets [94]. While lipid droplets are not unique to adipocytes, adipose tissue plays a particularly important role in the dynamic and systemic storage and release of fatty acids during feeding and fasting cycles. Lipid droplets store fatty acids as triacylglycerols (TAG), and the mobilization of fatty acids from these stores requires lipolysis, a process that mTORC1 signaling is believe to suppress, at least in adipocytes. An increase in circulating lipids is a feature of systemic rapamycin treatment in humans [95,96], and this is also observed in mice with loss of function mutations in the mTOR pathway [97]. Rapamycin can stimulate lipolysis in cultured adipocytes, and mTORC1 signaling appears to suppress the function of hormone sensitive lipase (HSL) and adipose triacylglycerol lipase (ATGL), the major lipases responsible for fatty acid release from lipid droplets [98–101]. Free fatty acids can be subsequently oxidized in the mitochondria to produce energy, and mTORC1 signaling appears to suppress the expression of key mitochondrial transporters and enzymes required for fatty acid oxidation, at least in part, through attenuation of PPARα transcriptional activity [102]. Further studies are needed to understand the mechanistic details of mTORC1-mediated inhibition of lipolysis and fatty acid oxidation and how it influences metabolic homeostasis at the cellular, tissue-specific, and systemic levels.
Conclusions and impact on human diseases
It is now evident that mTORC1 serves as a key molecular link between signals that control cell growth and the metabolic processes that underlie growth (Figure 2). It should be emphasized that, while mTORC1 is essential for the survival of organisms from yeast to mammals [103,104], it is not generally required for the basal activity of the metabolic pathways it controls. As such, inhibition of mTORC1 is not equal to direct inhibition of a metabolic enzyme in the given pathway. However, the signal-integrating capacity of mTORC1 acts as a rheostat for cellular metabolism, reinforcing shifts between anabolic and catabolic processes as the nutrient state of the local and systemic environment changes.
Given the diversity of metabolic processes controlled by mTORC1, it is not surprising that dysregulated mTORC1 signaling has been implicated in a variety of human diseases. These diseases include those characterized by uncontrolled growth, including somatic overgrowth disorders (e.g. macrocephaly, proteus syndrome), tumor syndromes (e.g., Cowden syndrome, TSC, lymphangioleiomyomatosis), and the majority of sporadic cancers [105,106]. These settings are likely to offer the clearest insights into the biological consequences of aberrant uncoupling of normal control mechanisms upstream of mTORC1 from the pro-growth metabolic program that mTORC1 induces. While rapamycin (sirolimus) is being used alone and in combination with other drugs for the treatment of these growth and neoplastic diseases, it is most commonly used as an immunosuppressant following organ transplantation or in a variety of autoimmune disorders [107,108]. Importantly, mTORC1 activation is closely associated with dynamic changes in T cell metabolism that allow differentiation and expansion of T-cell populations [109–112]. One might speculate that mTORC1 activates T cells by driving a similar metabolic program to that of tumor cells. It is interesting to note that, like rapamycin, inhibitors of nucleotide synthesis are commonly used immunosuppressants [113–115], as activated lymphocytes exhibit increased de novo nucleotide synthesis on which they are highly dependent [116,117]. As discussed above, rapamycin also attenuates de novo nucleotide synthesis [40–42,102], suggesting an important underlying mechanism that might contribute to its immunosuppressant effects.
While some progress has been made in understanding the role of mTORC1-mediated metabolic changes in cancer and immunity, it has been more difficult to determine the contributions from alterations in cellular metabolism in other disease settings where mTORC1 has been implicated [118]. This is most evident with a diverse array of neurological disorders with distinct etiologies, including epilepsy, autism, and neurodegenerative diseases, for which there is intense interest in the mTORC1 network. The metabolic program associated with mTORC1 signaling will vary, at least in part, between different cell types and tissues. Understanding this complexity is key to defining the role of mTORC1 in the control of systemic metabolic homeostasis and metabolic diseases, including obesity, type-2 diabetes, and cardiovascular disease. Finally, dietary restriction and rapamcyin treatment, both of which inhibit mTORC1, are among the most evolutionarily conserved perturbations that prolong organismal lifespan, from yeast to mice. However, it is currently unknown how changes to the metabolic processes downstream of mTORC1 contribute to the beneficial effects of its inhibition for the promotion of longevity and prevention of age-related diseases, including cancer, neurodegeneration, and type-2 diabetes.
Acknowledgments
We thank Gerta Hoxhaj and Alexander Valvezan for discussions and critical comments. Studies in the Manning laboratory on this topic were supported by grants from the LAM Foundation (I.B.-S.), Tuberous Sclerosis Alliance (B.D.M.), Ellison Medical Foundation (B.D.M.), and NIH grants K99/R00-CA194192 (I.B.-S.) and P01-CA120964 and R35-CA197459 (B.D.M.).
Abbreviations
- ATF4
Activating transcription factor 4
- CAD
carbamoyl-phosphate synthetase 2, aspartate transcarbamoylase, dihydroorotase
- eIF4E
eukaryotic translation initiation factor 4E
- 4EBP1
eukaryotic translation initiation factor 4E binding protein 1
- G6PD
glucose-6-phosphate dehydrogenase
- HIF1-α
hypoxia-inducible factor 1α
- NADPH
Nicotinamide adenine dinucleotide phosphate reduced
- PPP
pentose phosphate pathway
- PRPP
5-phosphoribosylpyrophosphate
- Rheb
Ras homology enriched in brain
- SREBP1
sterol regulatory element-binding protein 1
- and S6K1/2
ribosomal protein S6 kinase 1 and 2
- TSC
tuberous sclerosis complex
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Efeyan A, Zoncu R, Sabatini DM. Amino acids and mTORC1: from lysosomes to disease. Trends Mol Med. 2012;18:524–533. doi: 10.1016/j.molmed.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dibble CC, Manning BD. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat Cell Biol. 2013;15:555–564. doi: 10.1038/ncb2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bar-Peled L, Sabatini DM. Regulation of mTORC1 by amino acids. Trends Cell Biol. 2014;24:400–406. doi: 10.1016/j.tcb.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340:1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••8.Wolfson RL, Chantranupong L, Saxton RA, Shen K, Scaria SM, Cantor JR, Sabatini DM. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science. 2016;351:43–48. doi: 10.1126/science.aab2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••9.Chantranupong L, Scaria SM, Saxton RA, Gygi MP, Shen K, Wyant GA, Wang T, Harper JW, Gygi SP, Sabatini DM. The CASTOR Proteins Are Arginine Sensors for the mTORC1 Pathway. Cell. 2016;165:153–164. doi: 10.1016/j.cell.2016.02.035. The activation state of mTORC1 is particularly sensitive to the amino acids leucine and arginine. These studies identify long sought sensors of these amino acids that link to mTORC1 regulation by the Rag GTPases. Sestrin2 is identified and characterized as a sensor of leucine and CASTOR1 as an arginine sensor, both impinging on the GATOR2 complex that regulates mTORC1 signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••11.Menon S, Dibble CC, Talbott G, Hoxhaj G, Valvezan AJ, Takahashi H, Cantley LC, Manning BD. Spatial Control of the TSC Complex Integrates Insulin and Nutrient Regulation of mTORC1 at the Lysosome. Cell. 2014;156:771–785. doi: 10.1016/j.cell.2013.11.049. This study demonstrated that the TSC complex localizes to Rheb at the lysosome in the absence of growth factors and acutely dissociates from this location in response to PI3K-Akt signaling. The direct phosphorylation of TSC2 by Akt is responsible for this spatial regulation, which is required for the ability of insulin to activate Rheb and mTORC1. The study in Ref. 13 found a correlation between lysosomal localization of the TSC complex and mTORC1 inhibition in response to various cellular stresses, suggesting that effects on TSC complex localization might be a more broadly applicable mechanism of mTORC1 regulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dibble CC, Elis W, Menon S, Qin W, Klekota J, Asara JM, Finan PM, Kwiatkowski DJ, Murphy LO, Manning BD. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol Cell. 2012;47:535–546. doi: 10.1016/j.molcel.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demetriades C, Plescher M, Teleman AA. Lysosomal recruitment of TSC2 is a universal response to cellular stress. Nat Commun. 2016;7:10662. doi: 10.1038/ncomms10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menon S, Manning BD. Common corruption of the mTOR signaling network in human tumors. Oncogene. 2008;27(Suppl 2):S43–51. doi: 10.1038/onc.2009.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howell JJ, Ricoult SJ, Ben-Sahra I, Manning BD. A growing role for mTOR in promoting anabolic metabolism. Biochem Soc Trans. 2013;41:906–912. doi: 10.1042/BST20130041. [DOI] [PubMed] [Google Scholar]
- 16.Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 1999;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci U S A. 1998;95:1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jefferies HB, Reinhard C, Kozma SC, Thomas G. Rapamycin selectively represses translation of the “polypyrimidine tract” mRNA family. Proc Natl Acad Sci U S A. 1994;91:4441–4445. doi: 10.1073/pnas.91.10.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aoki K, Adachi S, Homoto M, Kusano H, Koike K, Natsume T. LARP1 specifically recognizes the 3′ terminus of poly(A) mRNA. FEBS Lett. 2013;587:2173–2178. doi: 10.1016/j.febslet.2013.05.035. [DOI] [PubMed] [Google Scholar]
- •23.Tcherkezian J, Cargnello M, Romeo Y, Huttlin EL, Lavoie G, Gygi SP, Roux PP. Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5′TOP mRNA translation. Genes Dev. 2014;28:357–371. doi: 10.1101/gad.231407.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •24.Fonseca BD, Zakaria C, Jia JJ, Graber TE, Svitkin Y, Tahmasebi S, Healy D, Hoang HD, Jensen JM, Diao IT, et al. La-related Protein 1 (LARP1) Represses Terminal Oligopyrimidine (TOP) mRNA Translation Downstream of mTOR Complex 1 (mTORC1) J Biol Chem. 2015;290:15996–16020. doi: 10.1074/jbc.M114.621730. These two studies demonstrate that LARP1 associates with mTORC1 and regulates 5′TOP mRNA translation. Ref 23 concludes that LARP1 promotes 5′TOP mRNA translation, whereas Ref 24 concludes that it is a repressor of translation for this class of mRNAs. Further studies are required to remedy this discrepancy and to define the molecular function of this regulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magnuson B, Ekim B, Fingar DC. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem J. 2012;441:1–21. doi: 10.1042/BJ20110892. [DOI] [PubMed] [Google Scholar]
- 26.Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467–471. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- 27.Shahbazian D, Roux PP, Mieulet V, Cohen MS, Raught B, Taunton J, Hershey JW, Blenis J, Pende M, Sonenberg N. The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. Embo J. 2006;25:2781–2791. doi: 10.1038/sj.emboj.7601166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dennis MD, Jefferson LS, Kimball SR. Role of p70S6K1-mediated phosphorylation of eIF4B and PDCD4 proteins in the regulation of protein synthesis. J Biol Chem. 2012;287:42890–42899. doi: 10.1074/jbc.M112.404822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Li W, Williams M, Terada N, Alessi DR, Proud CG. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J. 2001;20:4370–4379. doi: 10.1093/emboj/20.16.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •30.Faller WJ, Jackson TJ, Knight JR, Ridgway RA, Jamieson T, Karim SA, Jones C, Radulescu S, Huels DJ, Myant KB, et al. mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature. 2015;517:497–500. doi: 10.1038/nature13896. As previously shown in Ref. 28 and 29, S6K1 phosphorylates and inactivates eEF2K to promote eEF2-dependent mRNA translation elongation. Ref. 30 demonstrated that intestinal proliferation associated with Wnt-signaling requires mTORC1 signaling to S6K and eEF2K, highlighting a functional role for eEF2K and translation elongation in GI cancers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richardson CJ, Broenstrup M, Fingar DC, Julich K, Ballif BA, Gygi S, Blenis J. SKAR is a specific target of S6 kinase 1 in cell growth control. Curr Biol. 2004;14:1540–1549. doi: 10.1016/j.cub.2004.08.061. [DOI] [PubMed] [Google Scholar]
- 32.Ma XM, Yoon SO, Richardson CJ, Julich K, Blenis J. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell. 2008;133:303–313. doi: 10.1016/j.cell.2008.02.031. [DOI] [PubMed] [Google Scholar]
- 33.Iadevaia V, Liu R, Proud CG. mTORC1 signaling controls multiple steps in ribosome biogenesis. Semin Cell Dev Biol. 2014;36:113–120. doi: 10.1016/j.semcdb.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 34.Zaragoza D, Ghavidel A, Heitman J, Schultz MC. Rapamycin induces the G0 program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol Cell Biol. 1998;18:4463–4470. doi: 10.1128/mcb.18.8.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Upadhya R, Lee J, Willis IM. Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Mol Cell. 2002;10:1489–1494. doi: 10.1016/s1097-2765(02)00787-6. [DOI] [PubMed] [Google Scholar]
- 36.Oficjalska-Pham D, Harismendy O, Smagowicz WJ, Gonzalez de Peredo A, Boguta M, Sentenac A, Lefebvre O. General repression of RNA polymerase III transcription is triggered by protein phosphatase type 2A-mediated dephosphorylation of Maf1. Mol Cell. 2006;22:623–632. doi: 10.1016/j.molcel.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 37.Roberts DN, Wilson B, Huff JT, Stewart AJ, Cairns BR. Dephosphorylation and genome-wide association of Maf1 with Pol III-transcribed genes during repression. Mol Cell. 2006;22:633–644. doi: 10.1016/j.molcel.2006.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michels AA, Robitaille AM, Buczynski-Ruchonnet D, Hodroj W, Reina JH, Hall MN, Hernandez N. mTORC1 directly phosphorylates and regulates human MAF1. Mol Cell Biol. 2010;30:3749–3757. doi: 10.1128/MCB.00319-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 40.Ben-Sahra I, Howell JJ, Asara JM, Manning BD. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 2013;339:1323–1328. doi: 10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robitaille AM, Christen S, Shimobayashi M, Cornu M, Fava LL, Moes S, Prescianotto-Baschong C, Sauer U, Jenoe P, Hall MN. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science. 2013;339:1320–1323. doi: 10.1126/science.1228771. [DOI] [PubMed] [Google Scholar]
- ••42.Ben-Sahra I, Hoxhaj G, Ricoult SJ, Asara JM, Manning BD. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science. 2016;351:728–733. doi: 10.1126/science.aad0489. This study demonstrated a new role for mTORC1 signaling in the regulation of de novo purine synthesis. mTORC1 stimulates de novo purine synthesis through multiple transcriptional effects, including an ATF4-dependent mechanism involving increased production of one carbon units through the mitochondrial tetrahydrofolate cycle. Taken together with previous studies (Ref. 40 and Ref. 41), it is now recognized that mTORC1 can stimulate the de novo synthesis of both purine and pyrimidine nucleotides. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.French JB, Jones SA, Deng H, Pedley AM, Kim D, Chan CY, Hu H, Pugh RJ, Zhao H, Zhang Y, et al. Spatial colocalization and functional link of purinosomes with mitochondria. Science. 2016;351:733–737. doi: 10.1126/science.aac6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu YC, Li F, Handler J, Huang CR, Xiang Y, Neretti N, Sedivy JM, Zeller KI, Dang CV. Global regulation of nucleotide biosynthetic genes by c-Myc. PLoS One. 2008;3:e2722. doi: 10.1371/journal.pone.0002722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •45.Cunningham JT, Moreno MV, Lodi A, Ronen SM, Ruggero D. Protein and nucleotide biosynthesis are coupled by a single rate-limiting enzyme, PRPS2, to drive cancer. Cell. 2014;157:1088–1103. doi: 10.1016/j.cell.2014.03.052. This study concluded that oncogenic Myc promotes the expression of eIF4E leading to translational upregulation of PRPS2, a key rate-limiting enzyme linking pentose phosphate pathway-derived ribose to nucleotide synthesis pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 48.Horton JD, Goldstein JL, Brown MS. SREBPs: transcriptional mediators of lipid homeostasis. Cold Spring Harb Symp Quant Biol. 2002;67:491–498. doi: 10.1101/sqb.2002.67.491. [DOI] [PubMed] [Google Scholar]
- 49.Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung YL, Schulze A. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li S, Brown MS, Goldstein JL. Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc Natl Acad Sci U S A. 2010;107:3441–3446. doi: 10.1073/pnas.0914798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yecies JL, Zhang HH, Menon S, Liu S, Yecies D, Lipovsky AI, Gorgun C, Kwiatkowski DJ, Hotamisligil GS, Lee CH, et al. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab. 2011;14:21–32. doi: 10.1016/j.cmet.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Owen JL, Zhang Y, Bae SH, Farooqi MS, Liang G, Hammer RE, Goldstein JL, Brown MS. Insulin stimulation of SREBP-1c processing in transgenic rat hepatocytes requires p70 S6-kinase. Proc Natl Acad Sci U S A. 2012;109:16184–16189. doi: 10.1073/pnas.1213343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ricoult SJ, Yecies JL, Ben-Sahra I, Manning BD. Oncogenic PI3K and K-Ras stimulate de novo lipid synthesis through mTORC1 and SREBP. Oncogene. 2016;35:1250–1260. doi: 10.1038/onc.2015.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li S, Ogawa W, Emi A, Hayashi K, Senga Y, Nomura K, Hara K, Yu D, Kasuga M. Role of S6K1 in regulation of SREBP1c expression in the liver. Biochem Biophys Res Commun. 2011;412:197–202. doi: 10.1016/j.bbrc.2011.07.038. [DOI] [PubMed] [Google Scholar]
- 56.Harris TE, Finck BN. Dual function lipin proteins and glycerolipid metabolism. Trends Endocrinol Metab. 2011;22:226–233. doi: 10.1016/j.tem.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Griffiths B, Lewis CA, Bensaad K, Ros S, Zhang Q, Ferber EC, Konisti S, Peck B, Miess H, East P, et al. Sterol regulatory element binding protein-dependent regulation of lipid synthesis supports cell survival and tumor growth. Cancer Metab. 2013;1:3. doi: 10.1186/2049-3002-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Young RM, Ackerman D, Quinn ZL, Mancuso A, Gruber M, Liu L, Giannoukos DN, Bobrovnikova-Marjon E, Diehl JA, Keith B, et al. Dysregulated mTORC1 renders cells critically dependent on desaturated lipids for survival under tumor-like stress. Genes Dev. 2013;27:1115–1131. doi: 10.1101/gad.198630.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 60.Keith B, Johnson RS, Simon MC. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2011;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhong H, Chiles K, Feldser D, Laughner E, Hanrahan C, Georgescu MM, Simons JW, Semenza GL. Modulation of hypoxia-inducible factor 1alpha expression by the epidermal growth factor/phosphatidylinositol 3-kinase/PTEN/AKT/FRAP pathway in human prostate cancer cells: implications for tumor angiogenesis and therapeutics. Cancer Res. 2000;60:1541–1545. [PubMed] [Google Scholar]
- 62.Laughner E, Taghavi P, Chiles K, Mahon PC, Semenza GL. HER2 (neu) signaling increases the rate of hypoxia-inducible factor 1alpha (HIF-1alpha) synthesis: novel mechanism for HIF-1-mediated vascular endothelial growth factor expression. Mol Cell Biol. 2001;21:3995–4004. doi: 10.1128/MCB.21.12.3995-4004.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hudson CC, Liu M, Chiang GG, Otterness DM, Loomis DC, Kaper F, Giaccia AJ, Abraham RT. Regulation of hypoxia-inducible factor 1alpha expression and function by the mammalian target of rapamycin. Mol Cell Biol. 2002;22:7004–7014. doi: 10.1128/MCB.22.20.7004-7014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakamura H, Makino Y, Okamoto K, Poellinger L, Ohnuma K, Morimoto C, Tanaka H. TCR engagement increases hypoxia-inducible factor-1 alpha protein synthesis via rapamycin-sensitive pathway under hypoxic conditions in human peripheral T cells. J Immunol. 2005;174:7592–7599. doi: 10.4049/jimmunol.174.12.7592. [DOI] [PubMed] [Google Scholar]
- 65.Finlay DK, Rosenzweig E, Sinclair LV, Feijoo-Carnero C, Hukelmann JL, Rolf J, Panteleyev AA, Okkenhaug K, Cantrell DA. PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8+ T cells. J Exp Med. 2012;209:2441–2453. doi: 10.1084/jem.20112607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007;12:108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, Metabolism, and Cancer. Cancer Discov. 2015;5:1024–1039. doi: 10.1158/2159-8290.CD-15-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Csibi A, Fendt SM, Li C, Poulogiannis G, Choo AY, Chapski DJ, Jeong SM, Dempsey JM, Parkhitko A, Morrison T, et al. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell. 2013;153:840–854. doi: 10.1016/j.cell.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••69.Csibi A, Lee G, Yoon SO, Tong H, Ilter D, Elia I, Fendt SM, Roberts TM, Blenis J. The mTORC1/S6K1 pathway regulates glutamine metabolism through the eIF4B-dependent control of c-Myc translation. Curr Biol. 2014;24:2274–2280. doi: 10.1016/j.cub.2014.08.007. This study, together with Ref. 68, showed that mTORC1 promotes glutaminolysis, at least in part, through translational upregulation of the transcription factor Myc. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 70.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hosokawa N, Sasaki T, Iemura S, Natsume T, Hara T, Mizushima N. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy. 2009;5:973–979. doi: 10.4161/auto.5.7.9296. [DOI] [PubMed] [Google Scholar]
- 72.Chan EY. mTORC1 phosphorylates the ULK1-mAtg13-FIP200 autophagy regulatory complex. Sci Signal. 2009;2:pe51. doi: 10.1126/scisignal.284pe51. [DOI] [PubMed] [Google Scholar]
- 73.Wirth M, Joachim J, Tooze SA. Autophagosome formation--the role of ULK1 and Beclin1-PI3KC3 complexes in setting the stage. Semin Cancer Biol. 2013;23:301–309. doi: 10.1016/j.semcancer.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 74.Lee JW, Park S, Takahashi Y, Wang HG. The association of AMPK with ULK1 regulates autophagy. PLoS One. 2010;5:e15394. doi: 10.1371/journal.pone.0015394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Egan D, Kim J, Shaw RJ, Guan KL. The autophagy initiating kinase ULK1 is regulated via opposing phosphorylation by AMPK and mTOR. Autophagy. 2011;7:643–644. doi: 10.4161/auto.7.6.15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Loffler AS, Alers S, Dieterle AM, Keppeler H, Franz-Wachtel M, Kundu M, Campbell DG, Wesselborg S, Alessi DR, Stork B. Ulk1-mediated phosphorylation of AMPK constitutes a negative regulatory feedback loop. Autophagy. 2011;7:696–706. doi: 10.4161/auto.7.7.15451. [DOI] [PubMed] [Google Scholar]
- 77.Shang L, Chen S, Du F, Li S, Zhao L, Wang X. Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proc Natl A. doi: 10.1073/pnas.1100844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Puente C, Hendrickson RC, Jiang X. Nutrient-regulated Phosphorylation of ATG13 Inhibits Starvation-induced Autophagy. J Biol Chem. 2016;291:6026–6035. doi: 10.1074/jbc.M115.689646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yuan HX, Russell RC, Guan KL. Regulation of PIK3C3/VPS34 complexes by MTOR in nutrient stress-induced autophagy. Autophagy. 2013;9:1983–1995. doi: 10.4161/auto.26058. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cad Sci U S A. 2011;108:4788–4793. [Google Scholar]
- 81.Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J, Piacentini M, Fimia GM, et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol. 2013;15:406–416. doi: 10.1038/ncb2708. [DOI] [PubMed] [Google Scholar]
- •82.Kim YM, Jung CH, Seo M, Kim EK, Park JM, Bae SS, Kim DH. mTORC1 phosphorylates UVRAG to negatively regulate autophagosome and endosome maturation. Mol Cell. 2015;57:207–218. doi: 10.1016/j.molcel.2014.11.013. This study showed that mTORC1 inhibits autophagy by phosphorylating UVRAG and inducing its interaction with Rubicon, precluding autophagosome and endosome maturation. The findings indicate that, in addition to mTORC1 phosphorylating and inhibiting the autophagy-initiating kinase ULK1 (Ref. 78), mTORC1 also regulates this later step in the autophagy pathway. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pena-Llopis S, Vega-Rubin-de-Celis S, Schwartz JC, Wolff NC, Tran TA, Zou L, Xie XJ, Corey DR, Brugarolas J. Regulation of TFEB and V-ATPases by mTORC1. EMBO J. 2011;30:3242–3258. doi: 10.1038/emboj.2011.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, Facchinetti V, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5:ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Perera RM, Zoncu R. The Lysosome as a Regulatory Hub. Annu Rev Cell Dev Biol. 2016;32:223–253. doi: 10.1146/annurev-cellbio-111315-125125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 90.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••91.Palm W, Park Y, Wright K, Pavlova NN, Tuveson DA, Thompson CB. The Utilization of Extracellular Proteins as Nutrients Is Suppressed by mTORC1. Cell. 2015;162:259–270. doi: 10.1016/j.cell.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••92.Krishna S, Palm W, Lee Y, Yang W, Bandyopadhyay U, Xu H, Florey O, Thompson CB, Overholtzer M. PIKfyve Regulates Vacuole Maturation and Nutrient Recovery following Engulfment. Dev Cell. 2016;38:536–547. doi: 10.1016/j.devcel.2016.08.001. These two studies demonstrate that under conditions of complete leucine starvation but availability of exogenous protein, cellular material, or dead cells, the inhibition of mTORC1 signaling promotes lysosome-dependent degradation of these materials for use as intracellular nutrients. Thus, the findings further indicate that, beyond the inhibition of autophagy, mTORC1 plays a functional role in attenuating lysosomal functions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ricoult SJ, Manning BD. The multifaceted role of mTORC1 in the control of lipid metabolism. EMBO Rep. 2013;14:242–251. doi: 10.1038/embor.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wilfling F, Haas JT, Walther TC, Farese RV., Jr Lipid droplet biogenesis. Curr Opin Cell Biol. 2014;29:39–45. doi: 10.1016/j.ceb.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Morrisett JD, Abdel-Fattah G, Hoogeveen R, Mitchell E, Ballantyne CM, Pownall HJ, Opekun AR, Jaffe JS, Oppermann S, Kahan BD. Effects of sirolimus on plasma lipids, lipoprotein levels, and fatty acid metabolism in renal transplant patients. J Lipid Res. 2002;43:1170–1180. [PubMed] [Google Scholar]
- 96.Kasiske BL, de Mattos A, Flechner SM, Gallon L, Meier-Kriesche HU, Weir MR, Wilkinson A. Mammalian target of rapamycin inhibitor dyslipidemia in kidney transplant recipients. Am J Transplant. 2008;8:1384–1392. doi: 10.1111/j.1600-6143.2008.02272.x. [DOI] [PubMed] [Google Scholar]
- 97.Um SH, Frigerio F, Watanabe M, Picard F, Joaquin M, Sticker M, Fumagalli S, Allegrini PR, Kozma SC, Auwerx J, et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431:200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- 98.Zhang C, Yoon MS, Chen J. Amino acid-sensing mTOR signaling is involved in modulation of lipolysis by chronic insulin treatment in adipocytes. Am J Physiol Endocrinol Metab. 2009;296:E862–868. doi: 10.1152/ajpendo.90651.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chakrabarti P, English T, Shi J, Smas CM, Kandror KV. Mammalian target of rapamycin complex 1 suppresses lipolysis, stimulates lipogenesis, and promotes fat storage. Diabetes. 2010;59:775–781. doi: 10.2337/db09-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Soliman GA, Acosta-Jaquez HA, Fingar DC. mTORC1 inhibition via rapamycin promotes triacylglycerol lipolysis and release of free fatty acids in 3T3-L1 adipocytes. Lipids. 2010;45:1089–1100. doi: 10.1007/s11745-010-3488-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chakrabarti P, Kim JY, Singh M, Shin YK, Kim J, Kumbrink J, Wu Y, Lee MJ, Kirsch KH, Fried SK, et al. Insulin inhibits lipolysis in adipocytes via the evolutionarily conserved mTORC1-Egr1-ATGL-mediated pathway. Mol Cell Biol. 2013;33:3659–3666. doi: 10.1128/MCB.01584-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sengupta S, Peterson TR, Laplante M, Oh S, Sabatini DM. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature. 2010;468:1100–1104. doi: 10.1038/nature09584. [DOI] [PubMed] [Google Scholar]
- 103.Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva NR, Hall MN. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73:585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- 104.Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 105.Keppler-Noreuil KM, Parker VE, Darling TN, Martinez-Agosto JA. Somatic overgrowth disorders of the PI3K/AKT/mTOR pathway & therapeutic strategies. Am J Med Genet C Semin Med Genet. 2016 doi: 10.1002/ajmg.c.31531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ilagan E, Manning BD. Emerging role of mTOR in the response to cancer therapeutics. Trends Cancer. 2016;2:241–251. doi: 10.1016/j.trecan.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol. 2012;12:325–338. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pollizzi KN, Powell JD. Regulation of T cells by mTOR: the known knowns and the known unknowns. Trends Immunol. 2015;36:13–20. doi: 10.1016/j.it.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Waickman AT, Powell JD. mTOR, metabolism, and the regulation of T-cell differentiation and function. Immunol Rev. 2012;249:43–58. doi: 10.1111/j.1600-065X.2012.01152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.MacIver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annu Rev Immunol. 2013;31:259–283. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Verbist KC, Guy CS, Milasta S, Liedmann S, Kaminski MM, Wang R, Green DR. Metabolic maintenance of cell asymmetry following division in activated T lymphocytes. Nature. 2016;532:389–393. doi: 10.1038/nature17442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pollizzi KN, Patel CH, Sun IH, Oh MH, Waickman AT, Wen J, Delgoffe GM, Powell JD. mTORC1 and mTORC2 selectively regulate CD8(+) T cell differentiation. J Clin Invest. 2015;125:2090–2108. doi: 10.1172/JCI77746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kay AB. Immunosuppressive agents in chronic severe asthma. Allergy Proc. 1994;15:147–150. doi: 10.2500/108854194778702838. [DOI] [PubMed] [Google Scholar]
- 114.Antin JH, Kim HT, Cutler C, Ho VT, Lee SJ, Miklos DB, Hochberg EP, Wu CJ, Alyea EP, Soiffer RJ. Sirolimus, tacrolimus, and low-dose methotrexate for graft-versus-host disease prophylaxis in mismatched related donor or unrelated donor transplantation. Blood. 2003;102:1601–1605. doi: 10.1182/blood-2003-02-0489. [DOI] [PubMed] [Google Scholar]
- 115.Martel RR, Klicius J, Galet S. Inhibition of the immune response by rapamycin, a new antifungal antibiotic. Can J Physiol Pharmacol. 1977;55:48–51. doi: 10.1139/y77-007. [DOI] [PubMed] [Google Scholar]
- 116.Fairbanks LD, Bofill M, Ruckemann K, Simmonds HA. Importance of ribonucleotide availability to proliferating T-lymphocytes from healthy humans. Disproportionate expansion of pyrimidine pools and contrasting effects of de novo synthesis inhibitors. J Biol Chem. 1995;270:29682–29689. [PubMed] [Google Scholar]
- 117.Quemeneur L, Gerland LM, Flacher M, Ffrench M, Revillard JP, Genestier L. Differential control of cell cycle, proliferation, and survival of primary T lymphocytes by purine and pyrimidine nucleotides. J Immunol. 2003;170:4986–4995. doi: 10.4049/jimmunol.170.10.4986. [DOI] [PubMed] [Google Scholar]
- 118.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]